Abstract
Epigenetic alterations in the pattern of DNA and histone modifications play a crucial role in cancer development. Analysis of patient samples, however, is hampered by technical limitations in the study of chromatin structure from pathology archives that usually consist of heavily fixed, paraffin-embedded material. Here, we present a methodology [pathology tissue–ChIP (PAT-ChIP)] to extract and immunoprecipitate chromatin from paraffin-embedded patient samples up to several years old. In a pairwise comparison with canonical ChIP, PAT-ChIP showed a high reproducibility of results for several histone marks and an identical ability to detect dynamic changes in chromatin structure upon pharmacological treatment. Finally, we showed that PAT-ChIP can be coupled with high-throughput sequencing (PAT-ChIP-Seq) for the genome-wide analysis of distinct chromatin modifications. PAT-ChIP therefore represents a versatile procedure and diagnostic tool for the analysis of epigenetic alterations in cancer and potentially other diseases.
Global epigenetic alterations have been described in most cancer patients and have been linked to the clinical outcome of the disease (1–3). Indeed, pathological changes in DNA methylation and in the combinatorial patterns of histone posttranslational modifications (PTMs) have a profound impact on gene expression and other nuclear events, driving inappropriate proliferation of cancer cells. New technologies are beginning to allow global analyses of the epigenetic profile of the genome (the so-called “epigenome”) (4, 5), which have revealed at an unprecedented resolution the position of both DNA and histone modifications and dramatically extended our understanding of the structural determinants of the epigenome. These technologies, based on massively parallel sequencing of DNA fragments, are being applied systematically to the study of the epigenome in model cell lines (6–8). In these studies, chromatin-associated DNA is immunoprecipitated using antibodies against specific histone PTMs and then sequenced to obtain a genome-wide map of each specific PTM (ChIP-Seq) (9). Application of these technologies to clinically derived patient samples represents the logical next step of these approaches. It would allow a better definition of the role of epigenetic alterations in human diseases and provide useful biomarkers for their management (10). Unfortunately, the study of patient samples presents a number of technical hurdles. The small number of cells that can be obtained in most cases (such as from bioptic material) is limiting for several methodologies, although recent approaches have been described to partially circumvent this problem (11–13). Paraffin-embedded tissues, linked to patient clinical information, represent an invaluable source of clinical samples, because their preparation is indispensable for the diagnosis and clinical management of the vast majority of diseases. Large archives of paraffin-embedded tissues are available in most hospitals and have been extensively used for DNA and RNA analyses. To preserve tissue architecture and allow sectioning for histology-based analyses, patient-derived materials are first fixed and subsequently embedded in paraffin, a procedure that is generally perceived as too harsh to preserve complex cellular structures such as chromatin. Interestingly, however, fixation of tissue samples for tissue inclusion involves the usage of formaldehyde (FA), the same reagent that is currently used to fix fresh cells for standard chromatin preparations. Here we report the setup of a protocol (PAT-ChIP, for pathology tissue–chromatin immunoprecipitation) that allows high-quality chromatin purification from paraffin-embedded samples for genome-wide epigenetic studies (PAT-ChIP-Seq).
Results
Setup of a Protocol for ChIP from Paraffin-Embedded Pathology Tissues.
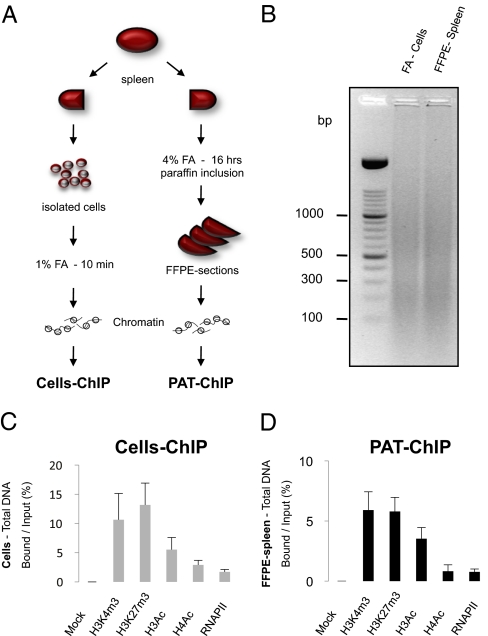
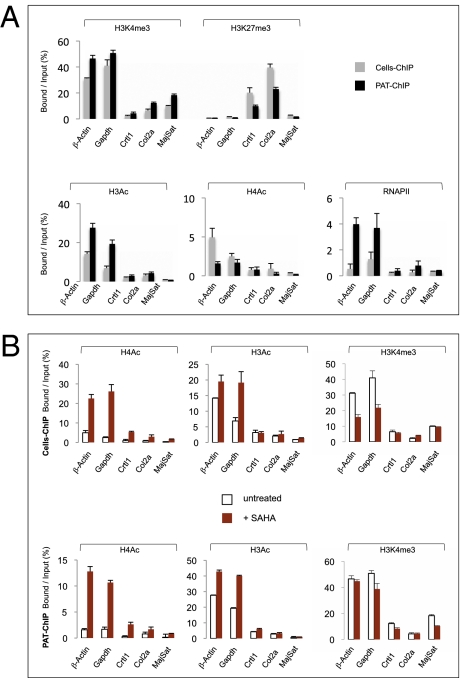
In standard pathology analyses, tissues derived from surgery or biopsies are cross-linked with FA at high concentrations (4%) for an extended period (usually overnight). Subsequently, fixed tissues are dehydrated and included in paraffin (Materials and Methods). Extraction of chromatin from tissue sections after the harsh conditions of FA fixation and paraffin embedding (FFPE) are considered a limiting step for efficient ChIP protocols, in which single-cell suspensions are cross-linked for short periods (10–15 min) using lower concentrations of FA (1%). In the setup of a ChIP protocol for paraffin-embedded pathology tissues (PAT-ChIP), we used an in-house murine model of human acute promyelocytic leukemia (APL) to compare the PAT-ChIP procedure with the canonical ChIP assay performed on isolated cells (14). In this model, leukemic blasts invade the spleen and completely subvert its normal architecture, resulting in massive splenomegaly. Importantly, splenic tissue under these conditions is easily minced to obtain a single-cell suspension, which can then be treated according to standard ChIP protocols. We therefore divided the spleen of each leukemic mouse into two halves that were subjected either to conventional ChIP (“Cells-ChIP” in Fig. 1A and below), or to overnight fixation with high concentrations of FA followed by paraffin embedding (FFPE-spleen). Chromatin preparation from FFPE-spleen started with the rehydratation and deparaffination of 10-μm FFPE-spleen sections (Fig. S1 and Materials and Methods for more details) followed by chromatin fragmentation and extraction (Fig. 1A and Materials and Methods). The sonication step was adjusted to achieve comparable size of DNA fragments from chromatin of both cells and FFPE-spleen samples (Fig. 1B and Fig. S1). Chromatin from both sources was then subjected to parallel ChIP assays using a set of ChIP-grade antibodies specific for well-studied, histone PTMs: trimethylated lysine-4 and -27 of histone H3 (H3K4me3 and H3K27me3, which are preferentially associated to active and silent promoters, respectively), and hyperacetylated histone H3 or H4 [H3Ac and H4Ac, both associated with active regulatory regions (15–17)]. Additionally, we performed one ChIP assay for RNA polymerase II (RNAPII), chosen as an example of nonhistone chromatin-associated proteins. The amount of immunoprecipitated DNA obtained by Cells-ChIP or PAT-ChIP was similar for each of the used antibodies (Fig. 1 C and D; average of three independent experiments). To compare the resolution of the two approaches, the immunoprecipitated DNAs were analyzed by real-time quantitative PCR (qPCR) of five genomic DNA sequences, including the promoter regions of genes expressed (βActin and Gapdh) or silent (Crtl1 and Col2a) in APL blasts (18). As a control region, we also included the major satellite repetitive sequence, which is poorly enriched in the PTMs under study (19–21). Results showed that the regions associated to active genes were enriched in H3K4me3, H3Ac, H4Ac, and RNAPII and poorly represented in H3K27me3 (Fig. 2A) in both samples. Likewise, DNA sequences associated with silent genes were enriched in H3K27me3 and poorly represented in all of the other histone PTMs (Fig. 2A). The major satellite repeat was not enriched in any modification under study (Fig. 2A). Altogether, these data revealed a robust overlap between results obtained with the two ChIP strategies [correlation coefficient (r) = 0.876, 0.947, and 0.916 in three independent experiments; Table S1A].
Fig. 1.
Overview of the paraffin-embedded ChIP method (PAT-ChIP). (A) Schematic representation of canonical ChIP and PAT-ChIP procedures starting from the spleen of a leukemic mouse. (B) Agarose gel electrophoresis of DNA samples representative of chromatin used as starting material (input) to perform Cells-ChIP and PAT-ChIP. (C and D) Analysis of the amounts of DNA recovered by Cells-ChIP (C) or PAT-ChIP (D), using the indicated antibodies. The amount of DNA is expressed as the ratio between the amount of total immunoprecipitated DNA (bound) and the amount of input DNA (mock: ChIP performed in absence of antibody). Data are expressed as means ± SD.
Fig. 2.
PAT-ChIP validation and its application to monitor dynamic changes of the epigenome induced by in vivo treatment of APL mice with SAHA. (A) Comparison of Cells-ChIP and PAT-ChIP results obtained by qPCR analysis of selected genomic regions. Enrichment of the promoter sequences associated with the indicated genes for several histone marks (H3K4me3, H3K27me3, H3Ac, H4Ac, and RNAPII) is expressed as the ratio between bound and input (percentage). Gray bars: canonical Cells-ChIP. Black bars: PAT-ChIP. (B) Enrichments of the promoter sequences associated with the indicated genes are reported as bound/input ratio for untreated (white bars) and SAHA-treated (red bars) mice. Upper: Cells-ChIP results. Lower: PAT-ChIP results. Data are expressed as means ± SD (for statistical analysis see Table S1 A and B).
PAT-ChIP Efficiently Monitors Dynamic Changes in Histone Modifications, After in Vivo Pharmacological Treatment with Epigenetic Drugs.
In our model system, treatment with histone deacetylase (HDAC) inhibitors induces apoptosis of APL leukemic cells and extends mouse survival (22). We treated APL mice for 4 d with the known HDAC inhibitor suberoylanilide hydroxamic acid (SAHA; at 150 mg/kg per die). SAHA treatment increased levels of histone-H4 acetylation (Fig. S2A) and prevented splenomegaly (Fig. S2B), as expected. Spleen tissues were subjected to the procedures outlined in Fig. 1A and ChIP assays performed by either canonical ChIP or PAT-ChIP. The immunoselected DNA (bound) was analyzed by qPCR of βActin, Gapdh, Crtl1, and Col2a promoter sequences and major satellite repeats. Results showed a 5- to 10-fold increase in histone H4 acetylation of all promoter regions tested after SAHA treatment, a more modest increase of histone H3 acetylation, and no variations of histone H3K4 methylation (Fig. 2B). Notably, data obtained by PAT-ChIP and canonical ChIP were very similar (r = 0.844; P < 0.003; Table S1B). Taken together, these data indicate that PAT-ChIP can be used to follow dynamic changes in histone PTMs.
Application of PAT-ChIP to Human FFPE Tissue Samples.
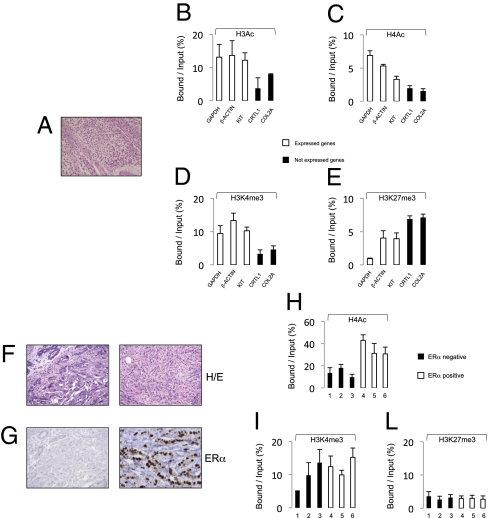
We prepared chromatin from FFPE sections of one seminoma sample [testicular germ cell tumor (TGCT); year, 1999; Fig. 3A] and six mammary tumors [either positive or negative for the expression of estrogen receptor α (ERα); years, 1999 to 2002; Fig. 3 F and G]. The TGCT chromatin was immunoprecipitated using antibodies against the H3Ac, H4Ac, H3K4me3, and H3K27me3 histone modifications and analyzed by qPCR of promoter regions of GAPDH and βACTIN (two housekeeping genes), KIT (a gene highly expressed in TGCT) (23), and CRTL1 and COL2A (not expressed in TCGT). Results showed a consistently higher enrichment of chromatin marks associated with active chromatin in the transcribed genes and an enrichment of the repressive H3K27Me3 histone mark in the silent genes (Fig. 3 B–E). Mammary tumor chromatin was immunoprecipitated against H4Ac, H3K4me3, and H3K27me3 and then analyzed by qPCR of the ERα regulatory region (24) (Fig. 3 F and G). Notably, higher levels of H4Ac were detected in ERα-positive samples, whereas we did not observe consistent changes in histone H3 methylation (H3K4me3 and H3K27me3; Fig. 3 H–L). Taken together, these results demonstrate that PAT-ChIP can be used to explore the epigenome of patient-derived, FA-embedded tissue samples stored for several years in pathology archives.
Fig. 3.
Application of PAT-ChIP to human FFPE samples. (A–E) PAT-ChIP analysis of a TGCT FFPE sample tissue. (A) H&E staining of human seminoma (TGCT). PAT-ChIP assays conducted against (B) H3Ac, (C) H4Ac, (D) H3K4me3, and (E) H3K27me3 are shown. (F–L) PAT-ChIP analysis of the ERα promoter in human breast carcinoma. (F) H&E staining of breast cancer cases representative of ERα negative and positive samples and (G) immunohystochemical analysis of relative ERα expression. PAT-ChIP assays conducted against (H) H4Ac, (I) H3K4me3, and (L) H3K27me3 are shown. Enrichment of the promoter sequences associated with the indicated genes is reported as bound/input ratio for expressed genes (white bars), not-expressed genes (black bars, B–E), and for ERα negative (black bars) and ERα positive (white bars, H–L) breast cancer cases. Data are expressed as means ± SD.
PAT-ChIP-Seq Analysis of FFPE Tissues.
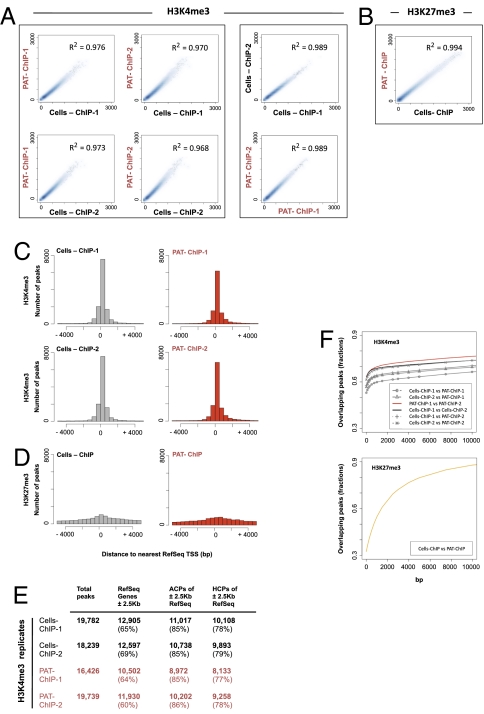
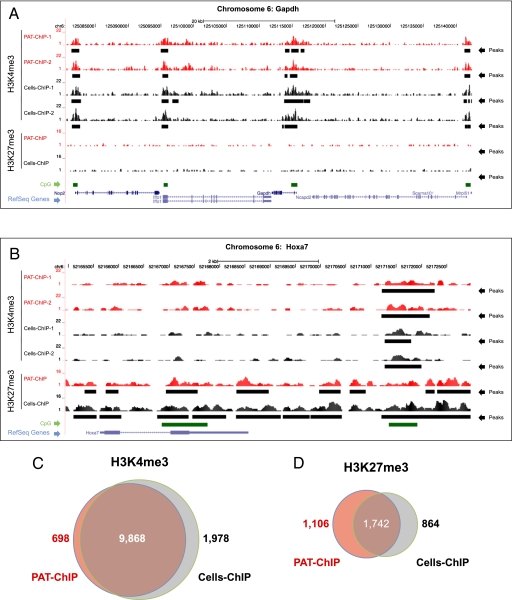
We therefore investigated the compatibility of PAT-ChIP with massive parallel sequencing methodologies (25). To compare the performance of PAT-ChIP-Seq and standard ChIP-Seq protocols (Cells-ChIP-Seq), we used spleens from the APL murine model. Chromatin was extracted from cell suspensions or FFPE tissues and immunoprecipitated with antibodies against H3K4me3. To evaluate the reproducibility of the technique, we performed this experiment in duplicate. H3K4me3 was chosen among various histone modifications, because it has been previously shown by ChIP-Seq to be preferentially associated with the promoters of active genes (6, 7, 18, 26–28). Purified DNA was then analyzed by ultra-sequencing using the Illumina Genome Analyzer IIx. For each experiment (Cells-ChIP-Seq or PAT-ChIP-Seq), we generated a dataset of H3K4me3-enriched genomic regions that were analyzed using model-based analysis of ChIP-Seq (MACS) (29) after normalization against input DNA (that was also subjected to deep sequencing). Using a high stringency threshold (P = 1e-5; Dataset S1), a total of 19,782 and 18,239 genomic peaks were identified in the two Cells-ChIP-Seq–derived samples, whereas 16,426 and 19,739 genomic peaks were identified by PAT-ChIP-Seq (Fig. 4E and Dataset S1 for summary tables and statistics; Fig. 5 A and B and Figs. S3 and S4 for snapshots from representative genomic regions). As expected, we measured a very high degree of correlation among duplicate samples, derived either from Cells-ChIP or from PAT-ChIP-Seq (Cells-ChIP-1 vs. Cells-ChIP-2 and PAT-ChIP-1 vs. PAT-ChIP-2; Fig. 4 A and F). Strikingly, we also observed a very high correlation between Cells-ChIP and PAT-ChIP datasets, showing a substantial overlapping of the two techniques (R2 ≥ 0.968, P < < 0.001; Fig. 4A; see also Fig. 4F for percentage of overlap observed among peaks derived from different samples). To further strengthen this finding, the correlation Cells-ChIP vs. PAT-ChIP was at levels comparable to those measured by comparison of the biological replicates derived from the same methodology, and therefore the different sample derivation (Cells-ChIP vs. PAT-ChIP) apparently had only a minor impact on the results (Fig. 4 A and F). Consistently, ≈60% of the peaks obtained by PAT-ChIP were found in the exact same genomic location as those obtained by conventional Cells-ChIP-Seq (Fig. 5 A, B, and F and Figs. S3 and S4 for representative genomic regions; Dataset S1), a percentage that increased to ≈90% when the peak overlap was considered within a ±2.5-Kb window surrounding the transcription start sites (TSS) of the RefSeq annotated genes (Fig. 5C and Dataset S2). As expected, H3K4me3 was mainly distributed in the proximity of TSS (Fig. 4C and Dataset S1), with the majority of peaks located in a window of ±2.5 Kb from the RefSeq annotated TSSs (Cells-ChIP replicates, 65% and 69%; PAT-ChIP replicates, 64% and 60%; Fig. 4E). As reported, the vast majority of RefSeq genes with promoter-associated H3K4Me3 peaks also contain a CpG island (26). Consistently, a high percentage of RefSeq genes (≈85%) associated with H3K4me3 peaks were found to contain a CpG island in both Cells-ChIP and PAT-ChIP datasets [all CpG containing promoters (ACPs); Fig. 4E]. Furthermore, we obtained a comparable and consistent enrichment (≈80%) of “high CpG containing promoters” (HCPs) [HCPs contain a 500-bp interval within −0.5 kb to +2 kb around the annotated TSS with a (G+C) fraction ≥0.55 and a CpG observed/expected ratio ≥0.6] in both Cells-ChIP and PAT-ChIP replicates (Fig. 4E; Fig. 5 A and B show representative snapshots) (26). Finally, we analyzed whether the results obtained by immunoprecipitation of the H3K4Me3 histone mark can be extended to other epigenetic modifications. We therefore performed additional ChIP against the H3K27me3 epigenetic mark, known to be generally associated with silenced regions (6, 7). A total of 46,552 and 48,235 genomic peaks were identified by Cells-ChIP-Seq and PAT-ChIP-Seq, respectively, using a high stringency threshold (P = 1e-5; Dataset S3). As shown previously, and differently from H3K4me3, H3K27Me3 showed a broader distribution of peaks and did not show a bias for TSSs [Figs. 4D and 5B and Fig. S3 and S4 for additional genomic loci (30)]. As for H3K4Me3, the distribution of the reads revealed a very high correlation between Cells-ChIP and PAT-ChIP datasets (Figs. 4B and 5D), and up to ≈80% overlap can be observed among the two procedures (Fig. 4F, Lower). Taken together, these results show that the PAT-ChIP approach can also be used in genome-wide studies using deep-sequencing analyses.
Fig. 4.
PAT-ChIP-Seq analysis of H3K4me3 and H3K27 me3 in APL mice. (A and B) Reads from the ChIP-Seq experiments were counted within each 100-kbp window, used to scan the mouse genome. Scatterplot analysis of the read numbers were drawn for (A) H3K4me3 and (B) H3K27me3 immunoprecipitations, identified within Cells-ChIP-Seq and PAT-ChIP-Seq experiments. Linear regression was performed for each of the analyzed comparison. Coefficients of determination are indicated. (C and D) Peaks density in proximity of the transcription start site (±5 Kb from TSS) referred to (C) H3K4me3 and (D) H3K27me3 immunoprecipitation. (E) Distribution of peaks with respect to functional genomic regions: ±2.5 Kb across TSS peak RefSeq annotated genes, percentage of ACPs, and percentage of HCPs, referred to the ±2.5 Kb surrounding RefSeq genes TSS. (F) Analysis of the overlaps between Cells-ChIP and PAT-ChIP peaks. Two peaks were considered as overlapping if they were found within a fixed number of base pairs from each other (x-axis values). Percentages of peaks in Cells-ChIP datasets that were found in the PAT-ChIP (y-axis values) were then plotted in function of the distance to consider two peaks as overlapping. Results for H3K4me3 (Upper) and H3K27me3 (Lower) are shown.
Fig. 5.
PAT-ChIP-Seq analyses of H3K4me3 and H3K27me3 in APL mice. (A and B) Profiles of H3K4me3 and H3K27me3 signals along the mouse Gapdh locus (representative of an H3K4me3 enriched region) and the Hoxa7 locus (representative of an H3K27me3 enriched region) are reported for Cells-ChIP (black) and PAT-ChIP (red). Identified peaks (black bars) are marked below the corresponding profile. HCP regions are reported as green bars, and Ref-Seq genes are indicated in blue (screenshots obtained using the University of California, Santa Cruz Genome Browser). (C and D) Venn diagrams showing common and unique peak-containing promoters identified by Cells-ChIP-Seq (gray) and PAT-ChIP-Seq (red).
Discussion
We report here experimental evidence that histone PTMs can also be investigated in FFPE samples. We expect that the application of this technology (PAT-ChIP) to clinical and diagnostic research will accelerate the identification of epigenetic biomarkers in different pathological conditions. Comparison of Cells-ChIP and PAT-ChIP data from the same sample was critical for the setup of optimal experimental conditions and particularly for the isolation of high-quality chromatin from the paraffin-embedded tissues. Specifically, the strong FA fixation, dehydration, and paraffin embedding, routinely performed on the histological samples, imposed a substantial modification of the chromatin extraction protocol, which led to comparable results from analyses of either individual genetic loci or, more importantly, the whole genome. In this regard, PAT-ChIP-Seq was shown (i) to be highly reproducible; (ii) to generate results highly consistent with Cells-ChIP-Seq; and (iii) to be applicable to several histone marks with distinctive features (TSS-centered, narrow peaks in the case of H3K4Me3, and broader regions more distributed in the genome in the case of H3K27Me3). Analysis of peak amplitude showed that common peaks are constituted by more reads than unique peaks in the datasets either for H3K4me3 or H3K27me3 immunoselections, suggesting that in some cases overlapping between the two procedures was not detected due to incomplete saturation of the sequencing procedure (Fig. S5). We showed the feasibility of PAT-ChIP in human FFPE tissue samples as old as 10 y. Although further studies are required to investigate the maximum age of FFPE samples that are compatible with PAT-ChIP studies, the possibility of analyzing at least 10-y-old samples opens the opportunity of accessing large pathology archives with extended clinical information (long follow-up). We have also shown, using the murine APL model, that PAT-ChIP is efficient for measurement of dynamic changes in the epigenome after pharmacological treatment. Thus, PAT-ChIP can also be applied to monitor the effects of treatment with specific drugs, either conventional or epigenetic drugs, using bioptic material. As an example, the observation that ERα can be transcriptionally silenced in breast cancer through different epigenetic mechanisms, also involving the lysine 9 hypoacetylation of histone H3 (31), opens the possibility of modulating its expression using HDAC inhibitor-based therapy and to monitor the efficacy of treatment at the molecular level. In conclusion, PAT-ChIP can be considered a relevant tool for the study of the epigenome in the patient setting, for both diagnostic and therapeutic purposes.
Materials and Methods
Preparation of Cell Suspension and FFPE Tissues from APL Mice.
Leukemic blasts were isolated from APL transgenic mice and i.v. injected (1 × 106 cells) in syngeneic recipient mice to induce secondary leukemias (14). When a massive splenomegaly was established (usually, ≥9 d after injection), mice were killed, and spleens were divided into two portions. For FFPE tissues, half the spleen was rapidly washed in PBS and incubated 16 h at room temperature in 4% FA solution. FA-fixed spleen was then routinely dehydrated by increasing concentrations of ethanol, starting from 70% through to 80%, 90%, and 100% (absolute ethanol), and subsequently included in paraffin, with use of a tissue processor (Leica ASP300). The other half of the spleen was homogenized in 5 mL of ice-cold PBS using a dounce homogenizer and cross-linked for 15 min at 37 °C by adding 1/10 of cross-linking solution [50 mM Hepes (pH 7.5), 11% FA, 1 mM Na2EDTA, and 0.1 M NaCl]. The cross-linking reaction was stopped by addition of glycine at a final concentration of 0.125 M. Cells were then washed twice with ice-cold PBS and stored at +4 °C (“Cells” in the main text).
Immunohistochemistry.
Immunohistochemistry was performed on 2-mm spleen sections of untreated and SAHA-treated mice by use of the monoclonal T25 antibody (specific for the hyperacetylated form of histone H4) (32) according to a previously described procedure (33). Testicular germ cell (TGCT, 1999) and breast cancer tumor samples (from 1999 to 2002) were provided by the Pathology Department of Presidio Ospedaliero (Vimercate) and the European Institute of Oncology (EIO, Milan). Samples were stained with H&E, and ERα expression in breast cancer samples was evaluated by using an anti-ERα antibody (33). All procedures were approved by the EIO institutional ethics board.
Chromatin Preparation.
For FFPE tissues, deparaffination of tissue sample sections (four sections of 10 μm each) was carried out through sequential incubations (10 min each) in 1 mL of hystolemon solution (six to eight times) at room temperature. Then samples were rehydrated by decreasing concentrations of ethanol starting from 100% (absolute ethanol) through to 95%, 70%, 50%, and 20%, with water as the final step (5 min at room temperature for each step of rehydration). After the rehydration step, most of the subsequent procedures were carried out similarly for either FFPE-derived samples or from suspension of APL leukemic cells isolated from spleens and cross-linked ex vivo as described in Results (Fig. 1A; for a technical summary see Fig. S1A). Rehydrated FFPE sections and isolated leukemic blasts (Cells) were incubated in 0.5 mL permeabilization buffer [1× Tris-buffered saline (TBS), 0.5% Tween20, 1 mM PMSF, and 10 μg/mL RNase A] for 30 min at room temperature in a rotating platform. After centrifugation at 18,000 × g for 5 min at +4 °C, samples were resuspended in 200 μL digestion buffer [50 mM Tris-HCl (pH 7.4), 0.32 M sucrose, 4 mM MgCl2, 1 mM CaCl2, and 0.1 mM PMSF]. FFPE-derived samples were partially fragmented through mild sonication, using a Labsonic L sonicator (B. Braun, Biotech International) and then digested for 1 min at 37 °C with micrococcal nuclease (N.70196Y; USB) at the final concentration of 1 U/10 μg of chromatin. After centrifugation at 18,000 × g for 5 min at +4 °C, samples were resuspended in 200 μL sonication buffer [1× TBS, 0.1% SDS, and 1 mM Na2EDTA (pH 8.0)] and further fragmented. After centrifugation at 8,000 × g for 5 min at room temperature, the first supernatant was collected (volume of ≈170 μL). The pellets were washed once with 50 μL sonication buffer, vortexed for 5 s, and centrifuged again to obtain the second supernatant (to reach a final volume of ≈220 μL).
ChIP Assay (PAT-ChIP and Cells-ChIP).
Chromatin was quantitated fluorimetrically by Qubit (Invitrogen). Immunoselection of chromatin was carried out in ChIP buffer [30 mM Tris-HCl (pH 7.4), 50 mM NaCl, 5 mM Na2EDTA, and 0.1 mM PMSF] using 260–600 ng of chromatin for each assay (dependent on either the amount of chromatin extracted from FFPE samples in each experiment or the number of ChIP assays to perform) and incubated 16 h at +4 °C in a rotating platform with the desired antibody at a previously determined optimal amount: 5 μg of anti-H3K4me3 (ab8580, Lot 533389; Abcam); 5 μg of anti-H3K27me3 (07-449, Lot DAM1529471; Upstate); 10 μg anti-H3Ac (06-599, Lot DAM1422332; Upstate); 20 μg anti-H4Ac (32); and 5 μg anti-PolII (sc-899, Lot I2208; Santa Cruz Biotechnology). Forty microliters of 50% vol/vol slurry rec-Protein G-Sepharose 4B Conjugate (preincubated 16 h at +4 °C with 1 mg/mL of BSA in ChIP buffer; Zymed) were added to each ChIP assay and incubated for 3 h at +4 °C. After centrifugation at 2,000 × g for 5 min at +4 °C, pellets were washed sequentially with 10 mL of washing buffer A [50 mM Tris-HCl (pH 7.4), 1% TritonX-100, 50 mM NaCl, 5 mM Na2EDTA, and 0.1 mM PMSF], 10 mL of washing buffer B [50 mM Tris-HCl (pH 7.4), 1% TritonX-100, 100 mM NaCl, 5 mM Na2EDTA, and 0.1 mM PMSF], and 10 mL of washing buffer C [50 mM Tris-HCl (pH 7.4), 1% TritonX-100, 150 mM NaCl, 5 mM Na2EDTA, and 0.1 mM PMSF]. Elution was carried out by adding 200 μL of elution buffer [1× Tris-EDTA (TE)/1% SDS] and incubating for 30 min at room temperature in a rotating platform. After centrifugation at 1,200 × g for 5 min at room temperature, the supernatant was saved and the elution repeated to obtain a final volume of 400 μL (bound fraction).
DNA Isolation.
De-cross-linking was performed through an overnight incubation at 65 °C in elution buffer/0.2 M NaCl, followed by digestion with 80 μg/mL proteinase K (3 h at +45 °C). DNA was isolated by sequential extractions with one-third volume of phenol:chloroform (1:1), one volume of phenol:chloroform (1:1) and one volume of chloroform. DNA was precipitated overnight at −20 °C. After centrifugation, DNA pellets were resuspended in 50 μL of TE buffer (stored at −20 °C).
Whole-Genome ChIP-Seq Analysis.
Either genomic DNA (input) or immunoprecipitated DNA obtained by using the anti-H3K4me3 or anti-H3K27me3 antibodies (bound) were sequenced on an Illumina Genome Analyzer IIx. Bowtie (34) has been used to map the short reads obtained onto the Mm9 release of the mouse reference genome. We performed the alignments with a maximum of two mismatches and keeping only the reads that align to unique positions in the genome (see Datasets S1 and S2 for statistical information on the ChIP-Seq runs). To define the ChIP-enriched genomic regions (peaks), MACS was applied (29). As parameters for MACS, we used a bandwidth of 300 bp and an mfold of 16 (see Datasets S1 and S2 for detailed lists of peaks). Reads obtained from inputs were used as reference control for the corresponding DNA fractions immunoprecipitated with both techniques. Final regions resulting from these analyses were annotated to the nearest RefSeq gene using GIN (35). Datasets are available for download from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession no. GSE21449).
Real-Time qRT-PCR.
Real-time RT-PCR assays were performed with the Fast Start Sybr Green Master Mix (Roche). For a list of primers see Table S1C] (36).
Supplementary Material
Acknowledgments
This study was supported by grants from Associazione Italiana Ricerca sul Cancro, European Economic Community (Epitron), Ministero dell'Istruzione, Università e Ricerca, and Ministero della Salute (to M.F., P.G.P., and S.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE21449).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007647107/-/DCSupplemental.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;4:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: Time is now. Crit Rev Oncol Hematol. 2008;1:1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Seligson DB, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;5:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gargiulo G, Minucci S. Epigenomic profiling of cancer cells. Int J Biochem Cell Biol. 2009;1:127–135. doi: 10.1016/j.biocel.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;8:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 6.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhao XD, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Gargiulo G, et al. NA-Seq: A discovery tool for the analysis of chromatin structure and dynamics during differentiation. Dev Cell. 2009;3:466–481. doi: 10.1016/j.devcel.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altucci L, Minucci S. Epigenetic therapies in haematological malignancies: searching for true targets. Eur J Cancer. 2009;7:1137–1145. doi: 10.1016/j.ejca.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 11.O'Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 12.Dahl JA, Reiner AH, Collas P. Fast genomic μChIP-chip from 1,000 cells. Genome Biol. 2009;10:R13.1–R13.11. doi: 10.1186/gb-2009-10-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goren A, et al. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Methods. 2010;1:47–49. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minucci S, et al. PML-RAR induces promyelocytic leukemias with high efficiency following retroviral gene transfer into purified murine hematopoietic progenitors. Blood. 2002;100:2989–2995. doi: 10.1182/blood-2001-11-0089. [DOI] [PubMed] [Google Scholar]
- 15.Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 16.Kubicek S, et al. The role of histone modifications in epigenetic transitions during normal and perturbed development. Ernst Schering Res Found Workshop. 2006;57:1–27. doi: 10.1007/3-540-37633-x_1. [DOI] [PubMed] [Google Scholar]
- 17.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, et al. The functional landscape of mouse gene expression. J Biol. 2004;3:21.1–21.22. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32:5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 22.HE LZ, et al. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. J Clin Invest. 2001;9:1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goddard NC, et al. KIT and RAS signalling pathways in testicular germ cell tumors: New data and a review of the literature. Int J Androl. 2007;30:337–349. doi: 10.1111/j.1365-2605.2007.00769.x. [DOI] [PubMed] [Google Scholar]
- 24.Kos M, Reid G, Denger S, Gannon F. Minireview: Genomic organization of the human ERα gene promoter region. Mol Endocrinol. 2001;12:2057–2063. doi: 10.1210/mend.15.12.0731. [DOI] [PubMed] [Google Scholar]
- 25.Schweiger MR, et al. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS ONE. 2009;5:1–7. doi: 10.1371/journal.pone.0005548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137.1–R137.9. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 31.Woodfield GW, Hitchler MJ, Chen Yizhen C, Domann FE, Weigel RJ. Interaction of TFAP2C with the estrogen receptor-a promoter is controlled by chromatin structure. Clin Cancer Res. 2009;11:3672–3679. doi: 10.1158/1078-0432.CCR-08-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronzoni S, Faretta M, Ballarini M, Pelicci PG, Minucci S. New method to detect histone acetylation levels by flow cytometry. Cytometry A. 2005;66:52–61. doi: 10.1002/cyto.a.20151. [DOI] [PubMed] [Google Scholar]
- 33.Nuciforo PG, Luise C, Capra M, Pelosi G, d'Adda di Fagagna F. Complex engagement of DNA damage response pathways in human cancer and in lung tumor progression. Carcinogenesis. 2007;28:2082–2088. doi: 10.1093/carcin/bgm108. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;3:R25.1–R25.10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cesaroni M, Cittaro D, Brozzi A, Pelicci PG, Luzi L. CARPET: A web-based package for the analysis of ChIP-chip and expression tiling data. Bioinformatics. 2008;24:2918–2920. doi: 10.1093/bioinformatics/btn542. [DOI] [PubMed] [Google Scholar]
- 36.Amatori S, et al. Malten, a new synthetic molecule showing in vitro antiproliferative activity against tumor cells and induction of complex DNA structural alterations. Br J Cancer. 2010;2:239–248. doi: 10.1038/sj.bjc.6605745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.