Abstract
High hyperdiploid acute lymphoblastic leukemia (ALL) is one of the most common malignancies in children. It is characterized by gain of chromosomes, typically +X, +4, +6, +10, +14, +17, +18, and +21,+21; little is known about additional genetic aberrations. Approximately 20% of the patients relapse; therefore it is clinically important to identify risk-stratifying markers. We used SNP array analysis to investigate a consecutive series of 74 cases of high hyperdiploid ALL. We show that the characteristic chromosomal gains are even more frequent than previously believed, indicating that karyotyping mistakes are common, and that almost 80% of the cases display additional abnormalities detectable by SNP array analysis. Subclonality analysis strongly implied that the numerical aberrations were primary and arose before structural events, suggesting that step-wise evolution of the leukemic clone is common. An association between duplication of 1q and +5 was seen (P = 0.003). Other frequent abnormalities included whole-chromosome uniparental isodisomies (wUPIDs) 9 and 11, gain of 17q not associated with isochromosome formation, extra gain of part of 21q, deletions of ETS variant 6 (ETV6), cyclin-dependent kinase inhibitor 2A (CKDN2A) and paired box 5 (PAX5), and PAN3 poly(A) specific ribonuclease subunit homolog (PAN3) microdeletions. Comparison of whole-chromosome and partial UPID9 suggested different pathogenetic outcomes, with the former not involving CDKN2A. Finally, two cases had partial deletions of AT rich interactive domain 5B (ARID5B), indicating that acquired as well as constitutional variants in this locus may be associated with pediatric ALL. Here we provide a comprehensive characterization of the genetic landscape of high hyperdiploid childhood ALL, including the heterogeneous pattern of secondary genetic events.
Acute lymphoblastic leukemia (ALL) is the most common form of childhood malignancy, with a yearly incidence of ~4/100,000 person-years in children 1–14 y old (1). Twenty to twenty-five percent display high hyperdiploidy (51–67 chromosomes), making high hyperdiploid cases one of the largest subgroups of pediatric cancer (2). Clinically, high hyperdiploid ALL is associated with age of 3–5 y, a relatively low WBC count, and a B-cell precursor immunophenotype (3–7). The prognosis is favorable, with 5-y overall survival rates (OS) close to 90% (5, 7). Several lines of evidence suggest that the initiating transforming event may occur in utero (8–10), but the etiology remains unknown. Recently, two independent, large, genome-wide association studies reported linkage to a locus in the gene AT rich interactive domain 5B (ARID5B) at 10q21.2, but it is unclear how this region affects the risk of developing high hyperdiploid childhood ALL (11, 12).
Genetically, high hyperdiploid ALL is characterized by massive aneuploidy, manifesting as a nonrandom gain of chromosomes, typically including some or all of +X, +4, +6, +10, +14, +17, +18, and +21,+21, but other trisomies and tetrasomies are seen also. Monosomies, on the other hand, are exceedingly rare (2, 13). The pathogenetic consequences of the chromosomal gains remain poorly understood, but it generally is believed that gene dosage effects are of importance (2, 14, 15). No other characteristic genetic abnormality, such as a fusion gene, has been found in the vast majority of high hyperdiploid ALL cases; however, it is possible that as yet unidentified primary aberrations are present but are not detected because of the low resolution of most genetic screening techniques. Such hidden primary events have been reported previously in aneuploid tumors, e.g., the recent identification of structural rearrangements resulting in deregulation of cytokine receptor-like factor 2 (CRLF2) in a large proportion of ALL in patients with Down syndrome and microdeletions leading to the transmembrane protease, serin 2 (TMPRSS2)/v-ets erythroblastosis virus E26 oncogene homolog (ERG) hybrid gene in prostate cancer (16, 17). Identification of a fusion gene in high hyperdiploid ALL would be of major clinical importance, possibly simplifying diagnostic procedures and providing novel treatment options.
Although high hyperdiploidy generally confers a favorable prognosis in childhood ALL, ~20% of the patients suffer a relapse, and 10% succumb to the disease (5, 7). Identification of additional recurrent changes could aid in the identification of the high-risk cases and would be of great clinical importance. The only specific genetic markers that now are used for risk stratification in some centers are the “triple trisomies” (i.e., +4, +10, and +17), which denote a lower-risk group in the Children's Oncology Group (COG) protocol (18, 19). None of the other genetic abnormalities that have been shown to be recurrent in addition to the chromosomal gains—including point mutations of fms-related tyrosine kinase 3 (FLT3), neuroblastoma RAS viral oncogene homolog (NRAS), v-Ki ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), and protein tyrosine phosphatase, non receptor type 11 (PTPN11), microdeletions of, e.g., cyclin-dependent kinase inhibitor 2A (CDKN2A) (9p21.3), ETS variant 6 (ETV6) (12p13.2), and paired box 5 (PAX5) (9p13.2), and structural aberrations such as dup(1q) and i(17q)—has proved to affect outcome, and hence there is a need for novel prognostic markers in high hyperdiploid childhood ALL (2).
In the present study, we used SNP array analyses to investigate the genomes of a consecutive series of high hyperdiploid childhood ALL. We show that the pattern of chromosomal gains is even more specific than indicated by standard cytogenetic analysis and that there is a genetic heterogeneity in this subgroup regarding the additional genetic events.
Results
Chromosomal Gains in High Hyperdiploid ALL.
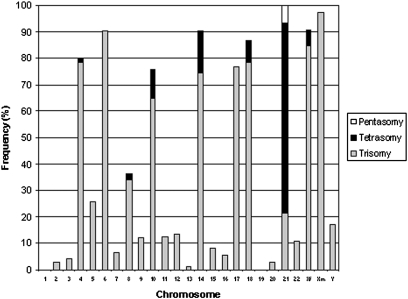
The SNP array analysis showed that the median modal number was 55 (range 51–63) among the 74 cases. Chromosome 21 was gained in 100% of the cases, followed by chromosome X in 95%, chromosomes 6 and 14 in 91%, chromosome 18 in 86%, chromosome 4 in 80%, chromosome 17 in 77%, chromosome 10 in 76%, chromosome 8 in 36%, and chromosome 5 in 26% (Figs. 1 and 2 and Table S1; for examples of SNP array results, see Fig. 3). The remaining chromosomes were all gained in less than 20% of the cases (range 0–17%); extra chromosomes 1 and 19 were not present in any of the cases (Fig. 1). The majority of the gains were trisomies, but tetrasomies were detected for chromosomes 21 (72% of the 74 cases), 14 (16%), 10 (11%), 18 (8.1%), X (6.1% of girls), 8 (2.7%), and 4 (1.4%), and pentasomy was seen for chromosome 21 (6.8%) (Fig. 1). All but one tetrasomy were “2:2”; i.e., both homologs had been duplicated. Thirty-nine cases (53%) had concurrent gains of chromosomes 4, 10, and 17 [i.e., were positive for the “triple trisomies” (18, 19)]. There were no significant differences in event-free survival (EFS) or OS between cases with and without the triple trisomies or between cases positive and negative for gain of chromosome 18, as previously has been debated (5) (Fig. S1). Sixteen cases (22%) displayed only numerical aberrations (i.e., had no large structural changes or microdeletions) (Table S1). Gains of only parts of the characteristically trisomic and tetrasomic chromosomes of high hyperdiploid ALL (chromosomes X, 4, 6, 10, 14, 17, 18, and 21) were rare, with the exception of chromosome 17, for which there was a clear preference for partial gains favoring 17q. The only chromosomal loss seen was for the Y chromosome; two cases had subclonal nullisomy Y. Twenty (27%) of the 74 cases harbored one to four acquired whole-chromosome uniparental isodisomies (wUPIDs). These wUPIDs comprised chromosomes 9 (11%), 11 (8.1%), 12 (5.4%), X and 1 (4.1% each), 7 (2.7%), and 2, 3, 5, 8, 13, 15, and 19 (1.4% each) (Tables S1 and S2). Quantitative fluorescent PCR (QF-PCR) of leukemic and parental samples showed that wUPID9 was of paternal origin in two cases and maternal origin in one case; wUPID11 was of paternal origin in one case and maternal origin in one case.
Fig. 1.
Chromosome gains detected by SNP array analysis in 74 cases of high hyperdiploid childhood ALL. Xf denotes gain of the X chromosome in females; Xm in males. “Trisomy” and “Tetrasomy” of Xm and Y correspond to one and two extra copies of these chromosomes, respectively. Chromosomes X, 4, 6, 10, 14, 17, 18, and 21 were all gained in more than 70% of the cases; chromosome 21 was gained in 100% of the cases. Extra copies of chromosomes 8 and 5 were present in 36% and 26% of the cases, respectively, whereas all other chromosomes were gained in <20% of the 74 cases. Tetrasomies were seen for chromosomes 4, 8, 10, 14, 18, 21, and X, and pentasomy was seen for chromosome 21 only.
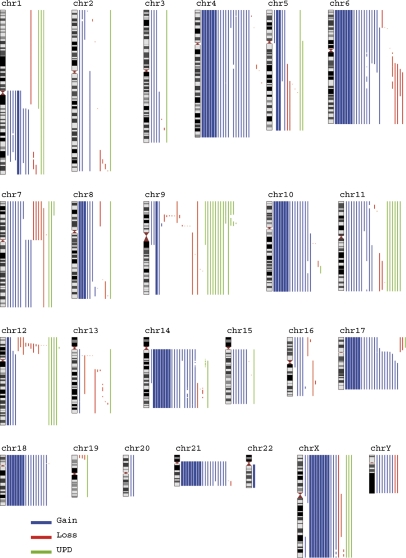
Fig. 2.
Overview of acquired genetic imbalances and UPIDs detected by SNP array analysis in 74 cases of high hyperdiploid childhood ALL. Each line corresponds to an aberration in the chromosome on the left. Thin lines correspond to aberrations detected in one case; thicker lines indicate aberrations detected in eight cases. Blue lines denote gains, red lines denote losses, and green lines denote UPIDs. This figure was made using the Genome-Wide Viewer software, freely available at http://www.well.ox.ac.uk/~jcazier/GWA_View.html.
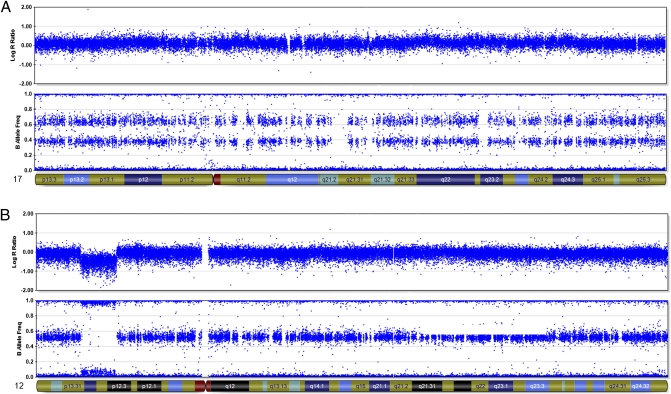
Fig. 3.
Examples of SNP array data from case 56. Top panels show log2 ratios along the chromosomes. Each dot represents the log2 ratio of one marker. A log2 ratio of zero corresponds to a normal, diploid copy number. Increased and decreased log2 ratios correspond to gained and deleted regions, respectively. Lower panels show B allele frequencies (BAF), which are calculated as (signal intensity for allele B)/(signal intensities for allele A + allele B). Homozygous SNPs have a value of 0 or 1, and heterozygous SNPs a value of 0.5 in a diploid chromosome segment. (A) Chromosome 17. Trisomy for this chromosome is apparent as an increased average log2 ratio and BAF values of 0, ~0.33, ~0.67, and 1.0, where the middle values correspond to the heterozygous SNPs. (B) Chromosome 12. Disomy for this chromosome is apparent as an average log2 ratio of ~0 and BAF values of 0, ~0.5, and 1.0. A hemizygous deletion is seen in 12p12.3–13.3 including ETV6, visible as a log2 ratio of less than −0.5 and complete loss of heterozygosity (i.e., BAF values of 0 and 1.0 only). The panels were extracted from the BeadStudio 3.1.2.0 software with Illumina Genome Viewer 3.2.9.
Structural Changes and Partial UPIDs.
Thirty-nine (53%) of the 74 cases harbored gain or loss of chromosomal regions >10 Mb, potentially cytogenetically visible (Table S3). The most common abnormality was partial gain of 1q which was found in 16 cases (22%), with two minimally gained regions between 149,615,798–185,104,377 and 188,096,756–213,315,040 bp (1q21.2–q25.3 and 1q31.1–q32.3, respectively) (Table S1). The 1q duplications were significantly associated with trisomy 5; 9/16 (56%) dup(1q)-positive cases also had +5, as compared with 10/58 (17%) dup(1q)-negative cases (Fisher's exact test, P = 0.003). Duplication of parts of chromosome 17 was seen in seven cases (9.4%), with a minimally gained region of 38,779,627–78,774,742 bp (17q21.2-qter). Only one of these cases had concurrent loss of 17p material consistent with an isochromosome 17q (Table S1). Five cases (6.8%) harbored deletions involving 6q; two of these had retention of one copy and three had retention of two copies (i.e., had trisomy 6 with deletion on one of the homologs). Two minimally deleted regions were identified at 93,537,222–99,926,277 bp and 107,467,536–117,963,320 bp (6q16.1–q16.2 and 6q21–q22.2, respectively) (Table S1). Partial gains of 21q were seen in five cases (6.8%), including two trisomic and three tetrasomic cases harboring additional 21q material, with a minimally gained region of 32,170,575–39,165,107 bp (21q22.11–q22.13) (Table S1). Four cases (5.4%) had loss of one copy of 7p and concurrent gain of 7q consistent with an isochromosome 7q. Chromosome 14 displayed complex patterns of losses and gains in three cases (4.1%) but without any commonly deleted or gained region. Partial UPID (pUPID) for 9p was seen in five cases (6.8%); four of these also had homozygous deletions of CDKN2A (Table S2 and Fig. 2).
Subclonal Genetic Changes.
To ascertain the minimum percentage of a clone that was detectable by the Illumina system, dilution series of a mixture of a trisomic sample and the corresponding remission sample and of a mixture of a sample with a wUPID and the corresponding remission sample were analyzed by SNP array analysis. The experiment showed that trisomies present in 20% or more of the cell population and wUPIDs present in 10% or more of the cell population were easily detectable (Fig. S2). SNP array analysis of leukemic samples showed that 10 (14%) of the 74 cases harbored subclones with one or two extra chromosomes. These cases comprised eight instances of subclonal disomy/trisomy, two of trisomy/tetrasomy 2:2, one of trisomy/tetrasomy 3:1, and one of tetrasomy 2:2/pentasomy. Six cases harbored a subclonal trisomy/wUPID, involving chromosome 9 in three cases and chromosomes 7, 11, and 12 in one case each (Table S1). Interphase FISH confirmed the subclonality for extra chromosomes in the seven cases for which material was available. In total, of the 647 aberrations comprising whole chromosomes (i.e., numerical aberrations and wUPIDs), 18 (2.8%) were subclonal. As regards the aberrations involving parts of chromosomes (i.e., unbalanced structural aberrations and microdeletions) 50/245 (20%) were subclonal. Thus, subclonality was more common for subchromosomal abnormalities (P < 0.0001, Yate's χ2).
Targets of Deletion.
There were a total of 97 acquired microdeletions [here defined as <10 Mb and either not seen in remission or not reported in the Database of Genomic Variants; http://projects.tcag.ca/variation/ (20)] in 44 (59%) of the 74 cases (Table S1). The median number of microdeletions was 1 (range 0–6). All but 12 microdeletions were hemizygous. The most common target of deletion was ETV6, which was hemizygously deleted in 11 (15%) of the cases (Fig. 3B); one further case had reduction from trisomy to disomy for this locus. Furthermore, one additional case harbored a hemizygous deletion including exon 2 of ETV6 (Tables S1 and S2) both at diagnosis and at remission. SNP array analysis of DNA extracted from peripheral blood from the patient's parents revealed the same deletion in the mother, confirming that this constitutional deletion had been inherited. Of the other well-known targets of deletion in childhood ALL, seven cases had homozygous deletions and one case a hemizygous deletion of CKDN2A (8/74; 11%); five cases had PAX5 deletions (6.8%), of which four (5.4%) were focal; and five cases (6.8%) had deletion of IKAROS family zinc finger 1 (IKZF1) (7p12.1), of which one was focal (1.4%) (Tables S1 and S2). In addition, three cases (4.1%) had deletion of 19p13.3-pter, including transcription factor 3 (TCF3) at 19p13.3 (Tables S1 and S2), and two cases had microdeletions of parts of ARID5B in 10q21.2 (see below). Another recurrent site of deletion was situated in 13q12.2, which was targeted by microdeletions in three cases and was included in a larger deletion in one additional case (4/74; 5.4%) (Tables S1 and S2). The only gene in this region is PAN3 poly(A) specific ribonuclease subunit homolog (PAN3) which is immediately 5′ of FLT3. Real-time quantitative RT-PCR (qRT-PCR) analyses of five cases, including cases 23 and 27 which harbored focal PAN3 deletions, showed that PAN3 was expressed at a lower level in cases 23 and 27 than in the three samples without del(13q); FLT3 was highly expressed in case 23 but not in case 27 (Table S4).
Risk Allele G of rs7089424 in ARID5B is Common in High Hyperdiploid Patients.
An association between constitutional SNPs in the ARID5B locus and risk of high hyperdiploid childhood ALL was reported recently (11, 12). To address this issue, we extracted the genotype of the risk-associated SNP rs7089424 in 10q21.2 from the array data set. The analysis showed that the constitutional frequency of the high-risk allele G was 0.47 in 71 cases where data were available for this SNP. As regards the leukemic genotype, data for this SNP were available from 69 cases, showing that the T allele was duplicated in eight (32%) of the 25 heterozygous cases with trisomy 10, and the G allele was duplicated in 17 cases (68%) (P = 0.054, binomial distribution test) (Table 1). In addition, two cases harbored nonoverlapping, acquired microdeletions of part of ARID5B involving exons 1–4 and 5–8, respectively (Table S1). Both cases had disomy 10 with the genotype TT for rs7089424 (Table S2).
Table 1.
Genotype of rs7089424 in ARID5B on chromosome 10 in 69 samples of high hyperdiploid leukemia
| Disomic cases |
Trisomic cases |
Tetrasomic cases |
|||
| Genotype | No. of cases | Genotype | No. of cases | Genotype | No. of cases |
| TT | 6 | TTT | 11 | TTTT | 2 |
| TG | 7 | TTG | 8 | TTGG | 3 |
| GG | 4 | TGG | 17 | GGGG | 1 |
| GGG | 10 | ||||
Discussion
Despite the high frequency of high hyperdiploidy in childhood ALL, the underlying genetic features of this subtype remain relatively poorly characterized, primarily because the chromosome morphology often is substandard, rendering cytogenetic analyses difficult and prone to classification mistakes. SNP array analysis therefore is expected to provide a more accurate picture of the unbalanced genetic aberrations associated with this leukemia subtype. As expected, the present analysis showed that a high proportion of the 74 investigated cases harbored gains of chromosomes X, 4, 6, 10, 14, 17, 18, and 21. However, the frequency of these aneusomies was even higher than reported in the literature (2). For example, +14 was present in 91% of the cases in this study (vs. 73% in the literature) (2), +6 in 91% (vs. 75%), and +18 in 86% (vs. 65%) (Fig. 1). Moreover, gains of chromosomes other than the characteristic trisomies and tetrasomies were less frequent than previously reported, and were all seen in <20% of the cases, with the exception of +5 and +8. These results support the notion that karyotyping errors are common when high hyperdiploid childhood ALL is investigated with standard cytogenetic techniques. In light of this unreliability, it is notable that studies performed with G-banding have been the basis for associating specific trisomies with a prognostic impact (e.g., the triple trisomies—concurrent +4, +10, and +17—in the COG protocol) (18, 19). We did not detect any difference in EFS or OS between cases positive and negative for these trisomies or between cases with trisomy 18 and disomy 18, as has been reported previously (Fig. S1) (21). Our series, admittedly, is small; nevertheless, our results could imply that investigations using more specific methods, such as SNP array or FISH analyses, instead of standard cytogenetic techniques may yield different results regarding the clinical impact of specific trisomies.
Our data thus indicate that there is an even stronger association between gains of chromosomes X, 4, 6, 10, 14, 17, 18, and 21 and the high hyperdiploid subgroup than previously surmised. The pathogenetic basis for this association is unknown. It has been reported that there is a general up-regulation of genes located at the gained chromosomes in high hyperdiploid ALL [previously also shown in a high proportion of the cases included in the present study (14)], and thus a gene-dosage effect is a possibility (14, 15). However, one then could envision that gain of additional chromosome copies (i.e., tetrasomies and pentasomies) might be even more beneficial; a pattern of tetrasomies similar to that of trisomies then would be expected. In contrast, tetrasomies seem to occur almost exclusively for chromosomes 21, 14, 10, and 18, and although chromosome 6 is gained in more than 90% of the cases, it never was tetrasomic in our series (Fig. 1). Furthermore, with the exception of 17q and possibly 21q, partial gains of the characteristically aneusomic chromosomes are rare (Fig. 2), also arguing against general dosage effects. Unfortunately, alternative models for the pathogenetic outcome are lacking. In fact, it is possible that only some of the common chromosomal gains, in particular gain of chromosome 21, are pathogenetically important “driver mutations” and that the remaining trisomies and tetrasomies are “passenger mutations” (22), i.e., epiphenomena that are tolerated by the cell but that do not give any selective advantage.
Equally enigmatic is the frequent occurrence of wUPIDs in high hyperdiploid childhood ALL. Several studies have shown that wUPIDs are present in 25–30% of cases from this subgroup (23–25), in line with our finding of 27%. We have proposed previously that this overrepresentation may reflect the underlying mechanism by which the hyperdiploidy originates (2), in particular because the previous studies have not shown wUPID for specific chromosomes (23–25). However, our present data indicate that there also is a nonrandom pattern of wUPIDs in high hyperdiploid ALL (Fig. 2). In particular, chromosomes 9 and 11 seem to be commonly involved (11% and 8.1% of cases, respectively). pUPIDs or wUPIDs frequently result in homozygosity for gene mutations or deletions in hematological malignancies (26); in ALL, the most common association is between pUPID for 9p and homozygous CDKN2A deletions (24, 27). The latter was seen in four of the five cases with this aberration in the present analysis (Table S2). In contrast, however, none of the eight cases with wUPID for chromosome 9 harbored CDKN2A deletion, strongly suggesting that the pathogenetic impact of wUPID9 may differ from that of pUPD9p and may be independent of CDKN2A deletions. Although one cannot exclude the possibility that wUPID9 may result in homozygosity for a gene mutation, it also is possible that wUPIDs may mainly affect imprinted loci. Such genes are differentially expressed, depending on the parental origin, and appear to be present in the majority of human chromosomes, including chromosome 9 (28). Furthermore, chromosome 11, which displayed wUPID in six cases, contains one of the best-known imprinting clusters in the human genome (28). However, neither wUPID9 nor wUPID11 had a consistent parental origin; in our investigations, in the three cases with wUPIDs9, the origin of wUPIDs9 was paternal in two cases and maternal in one case, whereas UPID11 was of paternal origin in one case and of maternal origin in one case.
Approximately half of the high hyperdiploid cases harbored subchromosomal imbalances expected to be cytogenetically visible (Table S3). The most common of these subchromosomal imbalances was dup(1q), which was seen in 22% of the cases (Fig. 3 and Table S2). Notably, dup(1q) had been detected in only 9.6% of our cases with G-banding (Table S3), in line with previously published reports of dup(1q) in 10–15% of high hyperdiploid ALL (4–6). Thus the present data suggest that the frequency of this aberration may be underestimated with conventional chromosome banding techniques. Interestingly, a significant association was seen between the presence of dup(1q) and trisomy 5 (P = 0.003). The pathogenetic basis for this association is unknown. It is noteworthy that both +5 and the presence of structural abnormalities have been associated with a worse prognosis in some G-banding studies, although data have been conflicting (5, 6, 29, 30). Considering that dup(1q) appears to be missed frequently with standard cytogenetic techniques, it is tempting to speculate that these studies would have yielded more consistent results had SNP array data been included. A negative association between the presence of dup(1q) and deletions of the long arm of chromosome 6 [del(6q)] has been reported previously in high hyperdiploid childhood ALL (5). In contrast, we found that as many as three of five cases with del(6q) also carried a dup(1q). Duplication of parts of chromosome 17 was seen in seven cases; only one of these cases had concurrent loss indicative of an isochromosome 17q (Fig. 2 and Table S2). This indicates that unbalanced aberrations other than i(17q) may lead to gain of 17q material in high hyperdiploid ALL. The presence of such abnormalities suggests that dosage effects resulting from 17q gain may be the underlying pathogenetic effect of trisomy 17 in high hyperdiploid ALL. Similarly, extra gain of part of 21q in addition to trisomy or tetrasomy for the entire chromosome was seen in five cases, with a minimally gained region encompassing ~7 Mb in 21q22. This region overlaps with the common region of amplification detected in childhood ALL with intrachromosomal amplification of chromosome 21 (iAMP21) (31). Although iAMP21-positive pediatric ALL is associated with a poor outcome (32), all the high hyperdiploid cases identified herein as having extra gain of the same chromosome 21 region remain alive and well in first remission; hence, there is no indication that extra gain of this part of chromosome 21 confers a more dismal prognosis in high hyperdiploid childhood ALL.
We recently reported that the chromosomal gains appear to arise before deletions and gene mutations in high hyperdiploid childhood ALL, based on comparisons of diagnostic and relapse samples (33). The SNP array results in the present study show that subclonality is much more common for structural aberrations/microdeletions than for numerical changes (20% vs. 3% of abnormalities, respectively; P < 0.0001). Furthermore, when subclonality for extra chromosomes occurs, it always involves a minority of the chromosomal gains; the high hyperdiploid pattern as such is present in all leukemic cells. These data support the notion that the massive aneuploidy is the primary genetic event in high hyperdiploid pediatric ALL.
One or more microdeletions (<10 Mb) were present in ~60% of the investigated cases, with a median number of one deletion per case. This relatively low number of microdeletions as compared with other genetic subtypes is in line with previous reports of childhood ALL (24, 25). ETV6 was the most common target, being hemizygously deleted in 15% of the cases, makings it one of the most frequent additional genetic aberrations known to occur in high hyperdiploid pediatric ALL. Interestingly, one patient harbored a constitutional hemizygous deletion of exon 2 of ETV6 (Table S2), leading to disruption of the ORF, that had been inherited from the mother. As far as we could determine, there were no indications of a familial leukemic syndrome; neither the mother nor the patient's two siblings had any history of hematologic malignancy. Whether this constitutional deletion of part of a gene known to be a common acquired deletion target in ALL contributed to the development of leukemia in this patient therefore remains an open question. Nevertheless, it is important to note that ETV6 deletions detected in leukemia samples are not necessarily acquired; this caveat is particularly pertinent if no remission sample is available for comparison. Deletions of CDKN2A and PAX5 were seen in 11% and 6.8% of the cases, respectively, which is lower than the overall frequency of such losses in childhood leukemia (34% and 30%, respectively) (25), suggesting that alternative leukemogenic pathways may be involved in high hyperdiploid ALL. An interesting region of deletion was detected in 13q12 (Table S1). This region was targeted by focal hemizygous deletions in three cases and was included in a larger deletion in one additional case. The minimally deleted region is ~33 kb 5′ of the transcription start site of FLT3 and includes the first five exons of the PAN3 gene. PAN3 encodes a protein involved in the degradation of poly(A) tails and has not as yet been associated with leukemia or any other neoplastic disorders (34). qRT-PCR analysis of FLT3 and PAN3 expression did not show consistent expression changes in two cases with the deletion as compared with cases without the deletion (Table S4). Further studies thus are needed to determine the leukemogenic effect of the del(13q).
Recently, two independent studies reported that SNPs in the ARID5B gene in 10q21.2 were associated with an increased risk of childhood ALL, in particular high hyperdiploid ALL (11, 12). ARID5B encodes a transcription factor involved in embryogenesis and growth regulation, and Arid5b-deficient mice display abnormalities in B-lymphocyte development (35). The reason for the increased risk is as yet unknown but may involve differences in the transcription level of this gene (12). We found that the frequency of one of the risk alleles in our series of high hyperdiploid pediatric ALL was similar to that reported by Papaemmanuil et al. (11) (0.47 and 0.55, respectively), a rate that is higher than the frequency in their controls (0.34). In heterozygous leukemic samples with trisomy 10, the high-risk allele G was duplicated in 17 cases, and the T allele was duplicated in eight cases (P = 0.054). Although these data are not statistically significant, the possibility that constitutional variants of ARID5B are associated with trisomy 10 in high hyperdiploid ALL cannot be excluded. Interestingly, two cases harbored acquired, nonoverlapping hemizygous microdeletions involving exons 1–4 and exons 5–8 of ARID5B, respectively. Both cases had disomy for chromosome 10; neither had the high-risk variant. This observation indicates that, in addition to constitutional variants, somatic abnormalities of ARID5B may be important in the leukemogenesis of high hyperdiploid childhood ALL.
Taken together, our results indicate that gain of chromosomes X, 4, 6, 10, 14, 17, 18, and 21 is even more characteristic for high hyperdiploid childhood ALL than previously believed, further supporting the notion that these alterations comprise the primary genetic event in high hyperdiploid ALL. The additional abnormalities, on the other hand, are heterogeneous and acquired in a stepwise fashion. Our data add to the emerging picture of a leukemic subgroup that is characterized genetically by the specific gain of some chromosomes but with a wide range of other genetic anomalies in the majority of patients, including nonclassic trisomies; dup(1q) and other structural abnormalities; wUPIDs 9 and 11; microdeletions of ETV6, CDKN2A, PAX5, and possibly PAN3; point mutations of FLT3, NRAS, KRAS, and PTPN11; and constitutional and acquired changes of ARID5B (Table S2). Whether this heterogeneity in the additional genetic aberrations has clinical ramifications remains to be addressed.
Materials and Methods
Patients.
A total of 116 cases of high hyperdiploid (51–67 chromosomes) B-cell precursor ALL were diagnosed in children and adolescents (0–18 y old) at the Departments of Pediatrics at Lund University Hospital and Linköping University Hospital, Sweden, between 1977 and 2009. One patient (case 59) had been diagnosed previously with leukemia of unknown type in another country and hence may have been either a relapse or a secondary leukemia. Material for DNA extraction was available from 78 cases (67%). During the investigation, four cases were excluded: three (cases 9, 33, and 76) because no chromosomal gains were visible with SNP array analysis, presumably because of normal cell contamination, and one (case 28) because the SNP array analysis revealed that it had a modal number of 50 chromosomes; hence, it was, per definition, not a high hyperdiploid ALL (2). Thus, the total number of informative cases in this investigation was 74 (64%). These cases comprised 33 females and 41 males, with a median age of 4 y at diagnosis (range 1–16 y) (Table S5). Of the 116 cases, 62 (53%) had been treated uniformly according to the Nordic Society of Pediatric Hematology (NOPHO) ALL-1992/2000 protocols (36). Forty-seven (76%) of the 62 cases were included in the SNP array analysis. There were no statistically significant differences regarding sex (P = 0.37), age (P = 0.32), WBC count (P = 0.37), EFS (P = 0.23), or OS (P = 0.70) between cases included in the SNP array analysis and those excluded because of lack of DNA. Informed consent was obtained according to the Declaration of Helsinki, and the study was approved by the Ethics Committee of Lund University.
SNP Array Analyses.
DNA was extracted from frozen bone marrow (n = 65) or peripheral blood (n = 9) obtained at diagnosis (Table S5) using phenol-chloroform or the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions. For 35 of the cases (1–8, 10–27, 29–32, 55, 66, 71, 73, and 77), DNA also could be extracted from remission samples. For all cases, SNP array analysis was performed using the Illumina 1M-duo bead Infinium BD BeadChip platform, containing 1.2 million markers with a median physical distance between markers of 1.5 kb. All analyses were done according to the manufacturer's instructions (Illumina), and data analysis was performed using the BeadStudio 3.1.3.0 software (Illumina) with Illumina Genome Viewer 3.2.9, extracting probe positions from the NCBI36.1 genome build. In 10 cases SNP array analysis also was performed with the Affymetrix GeneChip Human Mapping 250K Nsp, 250K Sty, and 10K 2.0 arrays (Affymetrix). These analyses were performed according to the manufacturer's instructions (Affymetrix), except for post-PCR washes, which were done with Ultrafree-MC columns (Millipore). GTYPE (Affymetrix) was used for analysis of signal intensity and for genotype calling. For copy number estimates and identification of regions with UPID, the in-house Genome Orientated Laboratory File (GOLF) software package was used (freely available online at http://bioinformatics.cancerresearchuk.org/cazier01/). For both the Illumina and Affymetrix array results, constitutional copy number polymorphisms were excluded based on comparison with the corresponding remission sample or with the Database of Genomic Variants (http://projects.tcag.ca/variation/) (20). Deletions most likely corresponding to somatic rearrangements of the T-cell receptor and Ig loci were excluded also.
To determine the extent to which subclonal gains and UPIDs were detectable by the Illumina system, a dilution series was made of a sample known to have such aberrations in 100% of the leukemic cells mixed with the corresponding remission sample in concentrations of 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, and 90% and was subjected to SNP array analysis as described above.
FISH.
Interphase FISH was performed according to standard methods using centromere- or locus-specific probes (Abbott Molecular). A control probe to identify high hyperdiploid cells was included in each hybridization, and a minimum of 200 nuclei were counted in each case.
QF-PCR.
To ascertain the parental origin of wUPIDs 9 and 11, QF-PCR was performed as previously described (23) using primers for the microsatellite markers D9S768 in 9q21 and D11S1304 in 11q25 on leukemic and remission samples and on samples from both of the patients’ parents. [Samples from parents had been obtained as part of a previous investigation (23).] The analysis could be performed in three of the cases with wUPID9 (cases 3, 8, and 50) and in two of the cases with wUPID11 (cases 1 and 50).
qRT-PCR Analysis.
Expression of the PAN3 and FLT3 genes was analyzed in five cases, including two with 13q14 deletions (cases 23 and 27), one with a FLT3 point mutation (case 21), one with a wUPID13 (case 25), and one with no known chromosome 13 aberration (case 22) (Table S2), using qRT-PCR with TaqMan Gene Expression assays (Applied Biosystems) according to the manufacturer's instructions. RNA was extracted according to standard methods from frozen bone marrow samples obtained at diagnosis.
Statistical Analyses.
Statistical analyses regarding genetic aberrations were performed using the χ2 test with Yate's correction and the exact two-tailed binomial distribution test, as specified in Results (VassarStats: Website for statistical computation, http://faculty.vassar.edu/lowry/VassarStats.html). Clinical data on sex, age, WBC count, EFS, and OS were compared using the two-sided Fisher's exact test, the two-sided Mann–Whitney u test, and the Kaplan–Meier test as applicable [PASW Statistics 18, SPSS) in the 62 cases that had been treated uniformly according to the NOPHO ALL-1992/2000 protocols (36)]. P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Jean-Baptiste Cazier for help with Fig. 2. This work was supported by grants from the Swedish Childhood Cancer Foundation, the Swedish Cancer Fund, the Swedish Research Council, Åke Wibergs Stiftelse, and Kungliga Fysiografiska Sällskapet.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006981107/-/DCSupplemental.
References
- 1.Hjalgrim LL, et al. Age- and sex-specific incidence of childhood leukemia by immunophenotype in the Nordic countries. J Natl Cancer Inst. 2003;95:1539–1544. doi: 10.1093/jnci/djg064. [DOI] [PubMed] [Google Scholar]
- 2.Paulsson K, Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:637–660. doi: 10.1002/gcc.20671. [DOI] [PubMed] [Google Scholar]
- 3.Third International Workshop on Chromosomes in Leukemia Chromosomal abnormalities in acute lymphoblastic leukemia: Structural and numerical changes in 234 cases. Cancer Genet Cytogenet. 1981;4:101–110. doi: 10.1016/0165-4608(81)90076-5. [DOI] [PubMed] [Google Scholar]
- 4.Groupe Francais de Cytogénétique Hématologique Collaborative study of karyotypes in childhood acute lymphoblastic leukemias. Leukemia. 1993;7:10–19. [PubMed] [Google Scholar]
- 5.Moorman AV, et al. United Kingdom Medical Research Council's Childhood Leukemia Working Party Outcome heterogeneity in childhood high-hyperdiploid acute lymphoblastic leukemia. Blood. 2003;102:2756–2762. doi: 10.1182/blood-2003-04-1128. [DOI] [PubMed] [Google Scholar]
- 6.Raimondi SC, et al. Heterogeneity of hyperdiploid (51-67) childhood acute lymphoblastic leukemia. Leukemia. 1996;10:213–224. [PubMed] [Google Scholar]
- 7.Forestier E, et al. Nordic Society of Paediatric Haematology, Oncology (NOPHO); Swedish Cytogenetic Leukaemia Study Group (SCLSG); NOPHO Leukaemia Cytogenetic Study Group (NLCSG) Outcome of ETV6/RUNX1-positive childhood acute lymphoblastic leukaemia in the NOPHO-ALL-1992 protocol: Frequent late relapses but good overall survival. Br J Haematol. 2008;140:665–672. doi: 10.1111/j.1365-2141.2008.06980.x. [DOI] [PubMed] [Google Scholar]
- 8.Maia AT, et al. Prenatal origin of hyperdiploid acute lymphoblastic leukemia in identical twins. Leukemia. 2003;17:2202–2206. doi: 10.1038/sj.leu.2403101. [DOI] [PubMed] [Google Scholar]
- 9.Maia AT, et al. Identification of preleukemic precursors of hyperdiploid acute lymphoblastic leukemia in cord blood. Genes Chromosomes Cancer. 2004;40:38–43. doi: 10.1002/gcc.20010. [DOI] [PubMed] [Google Scholar]
- 10.Panzer-Grümayer ER, et al. Nondisjunction of chromosomes leading to hyperdiploid childhood B-cell precursor acute lymphoblastic leukemia is an early event during leukemogenesis. Blood. 2002;100:347–349. doi: 10.1182/blood-2002-01-0144. [DOI] [PubMed] [Google Scholar]
- 11.Papaemmanuil E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Treviño LR, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heerema NA, et al. Specific extra chromosomes occur in a modal number dependent pattern in pediatric acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2007;46:684–693. doi: 10.1002/gcc.20451. [DOI] [PubMed] [Google Scholar]
- 14.Andersson A, et al. Molecular signatures in childhood acute leukemia and their correlations to expression patterns in normal hematopoietic subpopulations. Proc Natl Acad Sci USA. 2005;102:19069–19074. doi: 10.1073/pnas.0506637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruszka-Westwood AM, et al. Comparative expressed sequence hybridization studies of high-hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2004;41:191–202. doi: 10.1002/gcc.20085. [DOI] [PubMed] [Google Scholar]
- 16.Mullighan CG, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 18.Schultz KR, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: A combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children's Cancer Group (CCG) Blood. 2007;109:926–935. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutcliffe MJ, et al. High concordance from independent studies by the Children's Cancer Group (CCG) and Pediatric Oncology Group (POG) associating favorable prognosis with combined trisomies 4, 10, and 17 in children with NCI Standard-Risk B-precursor Acute Lymphoblastic Leukemia: A Children's Oncology Group (COG) initiative. Leukemia. 2005;19:734–740. doi: 10.1038/sj.leu.2403673. [DOI] [PubMed] [Google Scholar]
- 20.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 21.Moorman AV, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 22.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsson K, et al. Evidence for a single-step mechanism in the origin of hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2005;44:113–122. doi: 10.1002/gcc.20222. [DOI] [PubMed] [Google Scholar]
- 24.Kawamata N, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 26.Fitzgibbon J, et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–9154. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 27.Paulsson K, et al. Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: Unexpected similarities with pediatric disease. Proc Natl Acad Sci USA. 2008;105:6708–6713. doi: 10.1073/pnas.0800408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luedi PP, et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pui C-H, et al. Prognostic importance of structural chromosomal abnormalities in children with hyperdiploid (>50 chromosomes) acute lymphoblastic leukemia. Blood. 1989;73:1963–1967. [PubMed] [Google Scholar]
- 30.Heerema NA, et al. Prognostic impact of trisomies of chromosomes 10, 17, and 5 among children with acute lymphoblastic leukemia and high hyperdiploidy (> 50 chromosomes) J Clin Oncol. 2000;18:1876–1887. doi: 10.1200/JCO.2000.18.9.1876. [DOI] [PubMed] [Google Scholar]
- 31.Strefford JC, et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci USA. 2006;103:8167–8172. doi: 10.1073/pnas.0602360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moorman AV, et al. UK Medical Research Council (MRC)/National Cancer Research Institute (NCRI) Childhood Leukaemia Working Party (CLWP) Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21) Blood. 2007;109:2327–2330. doi: 10.1182/blood-2006-08-040436. [DOI] [PubMed] [Google Scholar]
- 33.Davidsson J, et al. Relapsed childhood high hyperdiploid acute lymphoblastic leukemia: Presence of preleukemic ancestral clones and the secondary nature of microdeletions and RTK-RAS mutations. Leukemia. 2010;24:924–931. doi: 10.1038/leu.2010.39. [DOI] [PubMed] [Google Scholar]
- 34.Brown CE, Tarun SZ, Jr., Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lahoud MH, et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 2001;11:1327–1334. doi: 10.1101/gr.168801. [DOI] [PubMed] [Google Scholar]
- 36.Schmiegelow K, et al. Nordic Society of Paediatric Haematology and Oncology Long-term results of NOPHO ALL-92 and ALL-2000 studies of childhood acute lymphoblastic leukemia. Leukemia. 2010;24:345–354. doi: 10.1038/leu.2009.251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.