Abstract
Microinvasive carcinomas and high-grade intraepithelial neoplasms are commonly discovered within the fallopian tube of BRCA1 mutation carriers at the time of risk-reducing salpingo-oophorectomy, suggesting that many BRCA1-mutated ovarian carcinomas originate in tubal epithelium. We hypothesized that changes in gene expression profiles within the histologically normal fallopian tube epithelium of BRCA1 mutation carriers would overlap with the expression profiles in BRCA1-mutated ovarian carcinomas and represent a BRCA1 preneoplastic signature. Laser capture microdissection of frozen sections was used to isolate neoplastic cells or histologically normal fallopian tube epithelium, and expression profiles were generated on Affymetrix U133 Plus 2.0 gene expression arrays. Normal-risk controls were 11 women wild type for BRCA1 and BRCA2 (WT-FT). WT-FT were compared with histologically normal fallopian tube epithelium from seven women with deleterious BRCA1 mutations who had foci of at least intraepithelial neoplasm within their fallopian tube (B1-FTocc). WT-FT samples were also compared with 12 BRCA1 ovarian carcinomas (B1-CA). The comparison of WT-FT versus B1-FTocc resulted in 152 differentially expressed probe sets, and the comparison of WT-FT versus B1-CA resulted in 4079 differentially expressed probe sets. The BRCA1 preneoplastic signature was composed of the overlap between these two lists, which included 41 concordant probe sets. Genes in the BRCA1 preneoplastic signature included several known tumor suppressor genes such as CDKN1C and EFEMP1 and several thought to be important in invasion and metastasis such as E2F3. The expression of a subset of genes was validated with quantitative reverse transcription-polymerase chain reaction and immunohistochemistry.
Introduction
Ovarian carcinoma is the leading cause of death from gynecologic malignant neoplasms in the developed world. Identification of the early molecular events leading to ovarian carcinoma has been hindered by the lack of an identifiable preneoplastic lesion and the limited occurrence of early-stage neoplasms. Although it has been proposed that ovarian carcinoma originates from the surface epithelium of the ovary and/or the epithelial lining of ovarian inclusion cysts, there have been few reports of intraepithelial neoplasms at these sites [1,2].
Alternatively, there has been increasing evidence that many ovarian carcinomas originate within the fallopian tube [3]. Fallopian tube epithelium can exhibit areas of increased proliferation and cytologic atypia, called intraepithelial neoplasia (IEN). Most ovarian carcinomas are of serous histology and frequently exhibit mutations in the critical cell cycle regulator p53 [4]. Severe IEN in fallopian tubes has been found in conjunction with mullerian malignant neoplasms, particularly serous carcinomas of ovarian, uterine, or peritoneal origin [5,6]. Identical p53 mutations have been identified in tubal IEN and coexisting sporadic serous carcinoma [7], suggesting that genetic disruption within the fallopian tube may progress to ovarian carcinoma.
Further evidence for a tubal origin is suggested by the high prevalence of occult fallopian tube carcinomas identified among BRCA1 and BRCA2 mutation carriers undergoing risk-reducing salpingo-oophorectomy (RRSO). Although the lifetime risk of ovarian carcinoma in the general population is only 1% to 2%, women who inherit mutations in the BRCA1 and BRCA2 genes have up to a 50% lifetime risk of ovarian carcinoma [8]. These high-risk women are frequently discovered to have occult neoplasms at the time of RRSO, and 57% to 100% of these lesions arise in the fallopian tube [9–11]. Fallopian tube epithelium frequently contains areas that have been termed p53 foci (also referred to as p53 signatures), which overexpress p53 and have increased expression of the proliferation marker Ki-67 [12]. These tubal p53 foci are more frequent in tubes from BRCA1 and BRCA2 mutation carriers compared with normal-risk women, and they have also been shown to exhibit decreased expression of the tumor suppressor protein p27 [13]. These observations have resulted in the proposal of a new paradigm for ovarian carcinoma, in which the fallopian tube epithelium acquires a sequence of molecular abnormalities leading to an in situ or invasive neoplasm, which exfoliates and spreads to the ovary and peritoneum [3]. Validating the role of the fallopian tube in ovarian carcinoma carcinogenesis will require additional studies, such as comparative analysis of gene expression between wild-type and high-risk fallopian tubes.
We obtained frozen fallopian tube tissue from seven women with BRCA1 mutations found to have occult invasive carcinomas or severe IEN in the fallopian tube on final pathologic examination. We hypothesized that the histologically normal tubal epithelium from these women would possess a gene expression profile that would reflect early alterations in gene expression contributing to the development of carcinoma. By comparing the gene expression profiles between these high-risk fallopian tubes and histologically normal fallopian tubes from women with wild-type BRCA1 and BRCA2, we identified a set of genes potentially important in the development of BRCA1-associated carcinomas. We hypothesized that genes important in BRCA1 ovarian carcinogenesis would have similarly altered expression patterns in BRCA1 carcinomas. Therefore, we used the expression patterns in BRCA1 ovarian carcinomas to further define the genes of interest in BRCA1 tubal epithelium.
Materials and Methods
Study Design and Sample Selection
All tissues and clinical information were obtained from the University of Washington Gynecologic Oncology Tissue Bank according to an institutional review board-approved protocol. To maximize the likelihood of identifying biologically important gene differentially expressed between histologically normal BRCA wild-type fallopian tubes and high-risk fallopian tubes from BRCA1 mutation carriers, we specifically selected BRCA1 mutation carriers possessing occult microinvasive or high-grade intraepithelial fallopian tube neoplasm to create the gene profile (B1-FTocc). In addition, to minimize the false discovery rate, we also identified genes differentially expressed between the BRCA wild-type fallopian tube epithelium (WT-FT) and invasive BRCA1 carcinomas (B1-CA). We limited our BRCA1 preneoplastic profile to genes showing concordant up-regulation or down-regulation in both B1-FTocc and B1-CA. Thirty patients were analyzed to create the BRCA1 preneoplastic gene signature: 11 histologically normal fallopian tube epithelium from women with wild-type BRCA1 and BRCA2 (WT-FT), 7 histologically normal fallopian tube epithelium from women with deleterious BRCA1 mutations and documented occult microinvasive or high-grade intraepithelial fallopian tube carcinoma (B1-FTocc), and 12 high-grade serous ovarian carcinomas from women with deleterious BRCA1 mutations (B1-CA). The characteristics of these patients are shown in Table 1. We chose WT-FT samples to match the age and menopausal distribution of the B1-FTocc cases. Some women in the WT-FT group had a personal history of breast cancer or a family history of breast cancer; however, women were excluded from the WT-FT group if they had a family history of ovarian cancer. All WT-FT control women had had full gene sequencing by Myriad genetics, and those who did not have comprehensive rearrangement testing performed by Myriad were screened with Multiplex Ligation-dependent Probe Amplification (MRC-Holland BV, Amsterdam, Holland) according to the manufacturer's instructions in our laboratory using normal DNA extracted from lymphocytes. For the B1-FTocc samples, the histologically normal epithelium was obtained from the same fallopian tube discovered to contain the occult fallopian tube neoplasm. Three of the seven B1-FTocc women were premenopausal at the time of oophorectomy.
Table 1.
Cases Used to Generate the BRCA1 Preneoplastic Gene Signature.
| Case Identifier | Age (years) | Menopausal Status | BRCA1/2 Status* | Other Characteristics |
| WT-FT no. 1 | 46 | Pre | Negative | Personal history of breast cancer |
| WT-FT no. 2 | 47 | Post | Negative | Personal history of breast cancer |
| WT-FT no. 3 | 48 | Post | Negative | Personal history of breast cancer |
| WT-FT no. 4 | 48 | Pre | Negative | No personal history of cancer |
| WT-FT no. 5 | 49 | Post | Negative | Personal history of breast cancer |
| WT-FT no. 6 | 50 | Pre | Negative | Personal history of breast cancer |
| WT-FT no. 7 | 52 | Pre | Negative | Personal history of breast cancer |
| WT-FT no. 8 | 54 | Post | Negative | Personal history of breast cancer |
| WT-FT no. 9 | 55 | Post | Negative | Personal history of breast DCIS |
| WT-FT no. 10 | 61 | Post | Negative | No personal history of cancer |
| WT-FT no. 11 | 61 | Post | Negative | Personal history of breast cancer |
| B1-FTocc no. 1 | 39 | Pre | B1.3109insAA | Microinvasion left fallopian tube |
| B1-FTocc no. 2 | 40 | Post | B1.M1V (120A>G) | Microinvasion left fallopian tube |
| B1-FTocc no. 3 | 47 | Pre | B1.2800delAA | High-grade intraepithelial |
| B1-FTocc no. 4 | 49 | Pre | B1.3795del4 | Microinvasion right fallopian tube† |
| B1-FTocc no. 5 | 53 | Post | B1.del ex14–20 | High-grade intraepithelial |
| B1-FTocc no. 6 | 62 | Post | B1.C61G | High-grade intraepithelial |
| B1-FTocc no. 7 | 63 | Post | B1.2800delAA | Microinvasion left fallopian tube |
| B1-CA no. 1 | 40 | Pre | B1.2576.delC | Stage IIIC, grade 3, serous carcinoma |
| B1-CA no. 2 | 41 | Pre | B1.185delAG | Stage IIIC, grade 3, serous carcinoma |
| B1-CA no. 3 | 44 | Pre | B1.2798del4 | Stage IIIC, grade 3, serous carcinoma |
| B1-CA no. 4 | 49 | Pre | B1.3795del4 | Stage IIIC, grade 3, serous carcinoma† |
| B1-CA no. 5 | 50 | Post | B1.5382insC | Stage IA, grade 3, undifferentiated |
| B1-CA no. 6 | 51 | Post | B1.3171ins5 | Stage IIIC, grade 3, serous carcinoma |
| B1-CA no. 7 | 54 | Post | B1.2594delC | Stage IV, grade 3, serous carcinoma |
| B1-CA no. 8 | 54 | Post | B1.del_exon14 | Stage IIIC, grade 3, serous carcinoma |
| B1-CA no. 9 | 55 | Post | B1.M1V (120A>G) | Stage IIC, grade 3, serous carcinoma |
| B1-CA no. 10 | 57 | Post | B1.5382insC | Stage IIIC, grade 3, mixed serous/endo |
| B1-CA no. 11 | 57 | Post | B1.5382insC | Stage IIIC, grade 3, serous carcinoma |
| B1-CA no. 12 | 65 | Post | B1.5382insC | Stage IIIC, grade 3, serous carcinoma |
Negative cases were wild-type by full sequencing as well as by comprehensive testing for gene rearrangements. WT-FT indicates histologically normal fallopian tube epithelium from BRCA1 wild-type patients; B1-FTocc, histologically normal fallopian tube epithelium from patients with deleterious BRCA1 mutations and at least high-grade intraepithelial fallopian tube neoplasm; B1-CA, tumor tissue from patients with deleterious BRCA1 mutations.
B1-FTocc no. 4 and B1-CA no. 4 are from the same individual who had both microscopic invasive neoplasm within the fallopian tube and peritoneal metastasis. DCIS indicates ductal carcinoma in situ. Menstrual phase of WT-FT cases: WT-FT no. 1 and WT-FT no. 7 had proliferative endometrium and WT-FT no. 4 and WT-FT no. 6 did not have hysterectomy performed.
Laser Capture Microdissection, RNA Amplification, and Gene Expression Chips
Tissues samples had been collected at the time of RRSO or ovarian carcinoma cytoreductive surgery and were immediately frozen in the operating room in liquid nitrogen in Tissue-Tek OCT (Alphen aan den Rijn, the Netherlands). For RRSO specimens, a small piece of tubal fimbriae was collected for the tissue bank. A frozen section of that tissue was stained with hematoxylin and eosin to confirm normal histologic diagnosis and rule out neoplasia in the research specimen. The remaining fallopian tube tissues from these cases were then subjected to serial sectioning by the pathologist to look for intraepithelial carcinoma or invasive carcinoma. All stored samples were subjected to the identical protocol of laser capture microdissection (LCM), linear RNA amplification, and microarray production. Hematoxylin and eosin slides from the frozen tissue OCT blocks were reviewed to select blocks with adequate distal fimbriated fallopian tube epithelium. Before LCM, 7-µm frozen sections were cut, adhered onto glass membrane slides (Arcturus, Mountain View, CA), and immediately stored on dry ice. Before LCM, the slides were dehydrated and stained with hematoxylin with the Histogene LCM Frozen Section Staining Kit (Arcturus). Slides were immediately transferred to the Veritas Laser Capture Microdissection system (Arcturus). Fallopian tube epithelium from the distal fimbriated fallopian tube was selectively captured for the fallopian tube samples, and ovarian carcinoma cells were selectively captured for neoplastic samples (Figure W1). Total RNA was isolated, and contaminating DNA was removed using the PicoPure RNA Isolation Kit (Arcturus) as per the company's protocol. The MessageAmp II aRNA amplification Kit (Ambion, Austin, TX) was used to amplify the total RNA once. The quality of each amplified RNA sample was confirmed using Agilent 2000 Bioanalyzer RNA 6000 Pico LabChip Kit (Agilent Technologies, Inc, Santa Clara, CA), and quantity was measured using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). All labeling, hybridization, and scanning were performed at the University of Washington Centre for Array Technology core facility. Amplified complementary DNA (cDNA) was purified, enzymatically fragmented, and labeled with biotin. Quality and quantity of the purified labeled cDNA product were confirmed before hybridizing to Affymetrix GeneChip U133A Plus 2.0 Arrays (Affymetrix, Inc, Santa Clara, CA). To minimize batch effect, several samples from each study group were included in each batch of array runs.
Development of the Gene Expression Profile
GeneSifter (Seattle, WA) software was used for pairwise gene expression analysis and clustering analysis (Manhattan, Complete Linkage). For pairwise gene expression analysis, a Welch t test was used when generating P values. To develop a potential BRCA1 preneoplastic gene expression profile, two independent comparisons were made. First, the 11 WT-FT were compared with the seven B1-FTocc samples, and probe sets were selected, which showed a 1.8-fold differential expression at P < .01. To minimize the false discovery rate, we also performed a comparison between the 11 WT-FT and 12 B1-CA and selected probe sets, which showed a 1.8-fold differential expression at P < .01. Probe sets were only selected for the BRCA1 preneoplastic gene expression profile if they demonstrated concordant up-regulation or down-regulation in both the B1-FTocc and the B1-CA. The 1.8-fold was the cutoff at which we had most overlapping genes while still optimizing the ratio of overlapping genes to nonoverlapping genes.
Real-time Quantitative Reverse Transcription-Polymerase Chain Reaction Analysis
Real-time quantitative reverse transcription-polymerase chain reaction (RT-PCR) was used to validate the Affymetrix array results for four genes from the BRCA1 preneoplastic signature. For each group (WT-FT, B1-FTocc, and B1-CA), five representative cases were selected and analyzed for expression of the four genes (EFEMP1, p57, CYP3A5, and CSPG5). TaqMan Gene Expression Assays (Applied Biosystems, Carlsbad, CA) were used for EFEMP1 (Hs01013942_m1), p57 (Hs00908986_g1), CYP3A5 (Hs02511768_s1), and CSPG5 (Hs00962721_m1), and GAPDH was used as the reference gene. All samples were run in triplicate, and the comparative Ct method was used for relative quantitation using ABI PRISM Sequence Detection Software (Applied Biosystems). Target gene Ct values were normalized to GAPDH.
Interrogation of the BRCA1 Preneoplastic Gene Signature Using Independent Samples
Additional fallopian tube samples that were not used to create the gene expression signature were selected to interrogate the BRCA1 preneoplastic gene signature. To test the intrasample reproducibility of the tubal expression profiles, three duplicates of the B1-FTocc samples were analyzed (Table W1). For two of these B1-FTocc - DUP cases (B1-FTocc no. 2 - DUP and BT-FTocc no. 6 - DUP), expression arrays were created using tissue from the fallopian tube contralateral to the microinvasive or high-grade intraepithelial lesion. For the duplicate of B1-FTocc no. 1, LCM was performed a second time using separate sections obtained approximately 100 µm further into the frozen tissue block. In addition, 12 BRCA1-mutated fallopian tubes from postmenopausal women, which did not contain occult lesions (B1-FT), were also analyzed (Table W2). The expression profiles were generated by comparing to the same set of WT-FT cases. These additional expression profiles were then analyzed by combining them one at a time with the original 30 samples that had been used to create the premaligant gene signature. Unsupervised hierarchical clustering (Manhattan, Complete Linkage) using the probe sets from the BRCA1 preneoplastic gene signature was performed for each combination to determine whether the additional cases contained the BRCA1 preneoplastic expression profile and would therefore cluster with the B1-FTocc cases. In addition, for 10 of the B1-CA carcinomas with adequate DNA available, DNA was sequenced for p53 exons 4 to 10 as previously described [14].
Ki-67 Immunohistochemistry
To validate the array gene expression data for MKI67 (antigen identified by monoclonal antibody Ki-67), we performed immunohistochemistry on a larger set of fallopian tube samples, which had been stored as paraffin blocks. Most of these cases had right and left distal fallopian tube tissues available, and an average Ki-67 staining score was obtained from both tubes. Fallopian tube tissues from 26 BRCA1 wildtype cases were compared with fallopian tube tissues from 52 BRCA1 mutation carriers without carcinoma obtained at RRSO. Paraffin sections were deparaffinized and sequentially rehydrated, and endogenous peroxidases were blocked. Heat-mediated antigen retrieval was performed in a citrate buffer (Antigen Unmasking Solution; Vector Laboratories, Burlingame, CA). Slides were incubated with the Ki-67 mouse monoclonal antibody MIB-1, diluted 1:100 (Dako, Copenhagen, Denmark). Sections were washed with phosphate-buffered saline and incubated with secondary antibody (horseradish peroxidase-antimouse; Vector Laboratories). DAB was used to visualize antibody complexes, and sections were counterstained with hematoxylin. Negative and positive controls were assessed for each run. Slides were scored by two independent observers blinded to case designation. The percentages of positive epithelial cells were scored (0 = none, 1 = 1%, 2 = 2%–4%, 3 = 5%–15%, 4 ≥ 15%). A Mann-Whitney test was used to compare the staining results.
Results
Affymetrix Gene Expression
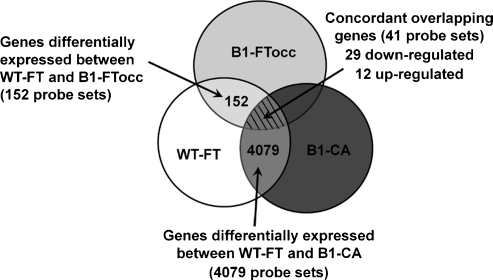
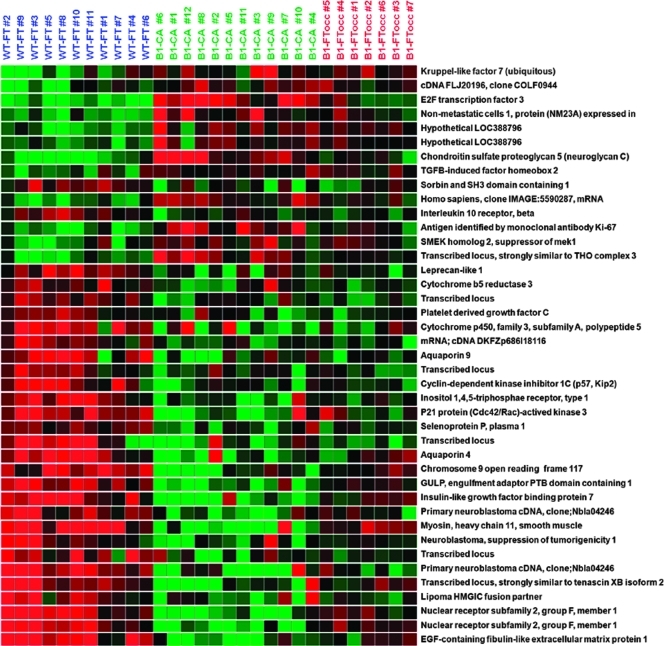
There were 18,600 probe sets expressed on the Affymetrix chips, which showed quality more than 0.7 in all samples. There were 152 probe sets with significant differential expression (>1.8-fold, P < .01) between the WT-FT and B1-FTocc. There were 4079 probe sets with significant differential expression (>1.8-fold, P < .01) between the WT-FT and B1-CA. The 152 probe sets differentially expressed from the BI-FTocc were compared with the 4079 differentially expressed probe sets in the B1-CA (Figure 1). The overlap between the two differentially expressed probe sets consisted of 29 probe sets downregulated in both groups, 12 probe sets upregulated in both groups, and 7 probe sets showing contradictory expression (up-regulation in one comparison and downregulated in the other). The 41 probe sets demonstrating concordant up-regulation or down-regulation in both comparisons comprised the BRCA1 preneoplastic gene signature and are shown in Table 2 and Figure 2.
Figure 1.
Diagram illustrating the protocol used to define the BRCA1 preneoplastic gene signature: Pairwise comparison between the WT-FT group and the B1-FTocc group (fold change ≥ 1.8, P < .01) identified 152 differentially expressed probe sets. Pairwise comparison between the WT-FT group and the B1-CA group (fold change ≥ 1.8, P < .01) identified 4079 differentially expressed probe sets. To minimize the false discovery rate, probe sets were only included in the gene signature with concordant down-regulation or up-regulation in both pairwise comparisons (hatched region).
Table 2.
The 41 Probe Sets Demonstrating Concordant Up-regulation or Down-regulation in Both Comparisons between WT-FT and B1-FTocc or B1-CA.
| Affymetrix Probe Set | Gene Name | Gene Symbol | WT-FT vs B1-FTocc | WT-FT vs B1-CA | ||
| Fold | P | Fold | P | |||
| Downregulated | ||||||
| 230130_at | Transcribed locus | Unknown | 3.7 | .0020 | 3.4 | .0054 |
| 214078_at | Primary neuroblastoma cDNA | Unknown | 3.6 | .0003 | 7.4 | .0001 |
| 201843_s_at | EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 3.4 | .0015 | 12.4 | .0001 |
| 205568_at | Aquaporin 9 | AQP9 | 2.7 | .0021 | 3.4 | .0020 |
| 214235_at | Cytochrome P450, family 3, subfamily A | CYP3A5 | 2.7 | .0048 | 5.2 | .0023 |
| 226228_at | Aquaporin 4 | AQP4 | 2.6 | .0081 | 7.7 | .0003 |
| 213182_x_at | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | CDKN1C | 2.5 | .0008 | 3.1 | .0014 |
| 203710_at | Inositol 1,4,5-triphosphate receptor, type 1 | ITPR1 | 2.4 | .0004 | 3.7 | .0002 |
| 214607_at | p21 (CDKN1A)-activated kinase 3 | PAK3 | 2.4 | .0070 | 5.8 | .0002 |
| 231183_s_at | Jagged 1 (Alagille syndrome) | JAG1 | 2.3 | .0002 | 1.9 | .0011 |
| 218656_s_at | Lipoma HMGIC fusion partner | LHFP | 2.3 | .0032 | 2.8 | .0083 |
| 218717_s_at | Leprecan-like 1 | LEPREL1 | 2.3 | .0034 | 2.9 | .0034 |
| 229480_at | MRNA; cDNA DKFZp686I18116 | Unknown | 2.1 | .0018 | 2.2 | .0002 |
| 209506_s_at | Nuclear receptor subfamily 2, group F, member 1 | NR2F1 | 2.1 | .0036 | 7.5 | .0000 |
| 231262_at | Transcribed locus | Unknown | 2.1 | .0035 | 3.0 | .0088 |
| 201497_x_at | Myosin, heavy chain 11, smooth muscle | MYH11 | 2.1 | .0068 | 15.5 | .0000 |
| 236277_at | Primary neuroblastoma cDNA | Unknown | 2.0 | .0094 | 3.5 | .0001 |
| 201885_s_at | Cytochrome b5 reductase 3 | CYB5R3 | 2.0 | .0004 | 1.8 | .0066 |
| 230233_at | Transcribed locus | Unknown | 2.0 | .0047 | 1.9 | .0073 |
| 1557866_at | Chromosome 9 open reading frame 117 | C9orf117 | 2.0 | .0013 | 6.0 | .0001 |
| 201162_at | Insulin-like growth factor binding protein 7 | IGFBP7 | 2.0 | .0088 | 6.1 | .0000 |
| 201427_s_at | Selenoprotein P, plasma, 1 | SEPP1 | 2.0 | .0004 | 2.3 | .0010 |
| 213451_x_at | Transcribed locus, sim to tenascin XB isoform 1 | TNXB | 1.9 | .0072 | 5.3 | .0002 |
| 218087_s_at | Sorbin and SH3 domain containing 1 | SORBS1 | 1.9 | .0082 | 2.1 | .0038 |
| 204235_s_at | GULP, engulfment adaptor PTB domain containing 1 | GULP1 | 1.9 | .0006 | 4.8 | .0000 |
| 218718_at | Platelet-derived growth factor C | PDGFC | 1.9 | .0007 | 1.8 | .0059 |
| 209575_at | Interleukin 10 receptor, beta | IL10RB | 1.9 | .0010 | 1.8 | .0016 |
| 209505_at | Nuclear receptor subfamily 2, group F, member 1 | NR2F1 | 1.8 | .0096 | 7.0 | .0000 |
| 37005_at | Neuroblastoma, suppression of tumorigenicity 1 | NBL1 | 1.8 | .0066 | 2.5 | .0017 |
| Upregulated | ||||||
| 225857_s_at | Hypothetical LOC388796 | LOC388796 | 2.4 | .0000 | 2.6 | .0000 |
| 238482_at | Kruppel-like factor 7 (ubiquitous) | KLF7 | 2.4 | .0020 | 2.2 | .0050 |
| 39966_at | Chondroitin sulfate proteoglycan 5 | CSPG5 | 2.1 | .0030 | 6.2 | .0000 |
| 203693_s_at | E2F transcription factor 3 | E2F3 | 2.1 | .0054 | 6.4 | .0000 |
| 1560622_at | CDNA FLJ20196 fis, clone COLF0944 | Unknown | 2.0 | .0038 | 2.2 | .0005 |
| 201577_at | Nonmetastatic cells 1, protein (NM23A) | NME1 | 1.9 | .0066 | 3.0 | .0002 |
| 65588_at | Hypothetical LOC388796 | LOC388796 | 1.9 | .0000 | 2.3 | .0001 |
| 224474_x_at | SMEK homolog 2, suppressor of mek1 | SMEK2 | 1.9 | .0087 | 2.0 | .0037 |
| 212020_s_at | Antigen identified by monoclonal antibody Ki-67 | MKI67 | 1.9 | .0056 | 3.5 | .0000 |
| 224623_at | Transcribed locus, similar THO complex 3 | THOC3 | 1.8 | .0003 | 3.2 | .0000 |
| 1560258_a_at | Homo sapiens, clone IMAGE:5590287, mRNA | Unknown | 1.8 | .0044 | 3.0 | .0000 |
| 216262_s_at | TGFB-induced factor homeobox 2 | TGIF2 | 1.8 | .0094 | 1.8 | .0033 |
Figure 2.
The 41 probe sets in the BRCA1 preneoplastic profile include several known tumor suppressors and oncogenes.
To test the significance of the overlap in differentially expressed genes, we created a simulation in which there were 4079 randomly selected expressed clones in one group and 152 in a second group from 18,600 expressed clones and compared overlap. We repeated this simulation 10,000 times. A total of 21 overlapping clones were only observed in 1 (0.01%) of 10,000 simulations, and 22 or more overlapping genes were never observed. These data suggest that the overlap of 41 genes between our BRCA1 tubal epithelium and BRCA1 carcinomas is highly significant and that it did not occur by chance. The concordance of direction of the expression differences in 41 (85%) of 48 overlapping probes also suggests that overlap in differentially expressed genes is nonrandom.
Real-time Quantitative RT-PCR Analysis
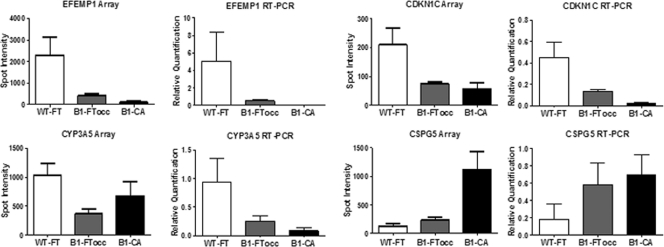
Figure 3 shows the Affymetrix expression array results imposed beside the real-time quantitative RT-PCR results for each of the four selected genes (EFEMP1, p57, CYP3A5, and CSPG5). For each gene, the real-time quantitative RT-PCR shows a similar expression pattern to the corresponding Affymetrix array.
Figure 3.
Correlation between expression array and real-time quantitative RT-PCR results: Four genes from the gene signature were selected for validation by RT-PCR with TaqMan assays. Five cases were used from each group (WT-FT, B1-FTocc, and B1-CA). The four genes included EFEMP1 (EGF-containing fibulin-like extracellular matrix protein 1), CDKN1C (cyclin-dependent kinase inhibitor 1C or p57), CYP3A5 (cytochrome P450, family 3, subfamily A), and CSPG5 (chondroitin sulfate proteoglycan 5 or neuroglycan C). For each gene, the array expression data are shown beside the corresponding RT-PCR results.
Clustering Analysis
The 30 samples used to create the BRCA1 preneoplastic gene signature were subjected to unsupervised hierarchical clustering analysis using all 18,600 probe sets expressed with quality greater than 0.7 (Figure W2). The B1-CA formed a distinct group, but the clustering of the B1-FTocc and WT-FT did not generate any distinct pattern using all expressed probe sets. Interestingly, using only the 41 overlapping probes, the wild-type samples separated into distinct premenopausal and postmenopausal groups, which they did not do when clustering was based on the entire expressed probe set. Of 10 carcinomas evaluated, 6 contained a somatic p53 mutation determined by sequencing p53 exons 4 to 10 (data not shown). Carcinomas 2, 5, 6, 7, 10, and 11 had p53 mutations, whereas carcinomas 1, 3, 9, and 12 were wild-type. The p53 mutation status was not associated with how the carcinomas clustered when considering the 41 overlapping probe sets or all of the 18,600 expressed probes.
The duplicated samples from the B1-FTocc group were then added to the clustering analysis and subjected to unsupervised hierarchical clustering using the BRCA1 preneoplastic gene signature. As shown in Figure W3, each of the duplicated samples clustered closely with their paired sample even when obtained from the contralateral FT, demonstrating the reproducibility of the expression profile in independent experiments as well as the consistency between paired bilateral fallopian tubes.
To further interrogate the BRCA1 preneoplastic gene signature, expression profiles from 12 additional B1-FT that had not been used in developing the expression profile were each individually combined with the 30 samples used to create the signature and subjected to unsupervised hierarchical clustering using the BRCA1 preneoplastic gene signature. Five of the new samples clustered with the B1-FTocc/B1-CA group, whereas seven of the new samples clustered with the WT-FT (Table W2). A representative example of each clustering pattern is shown in Figure W4. This suggested that 5 (42%) of the 12 B1-FT had experienced sufficient molecular disruptions to resemble B1-FTocc or B1-CA samples. Although the remaining seven B1-FT test samples clustered with wild-type fallopian tubes, they always clustered with the group from premenopausal women.
Ki-67 Immunohistochemistry
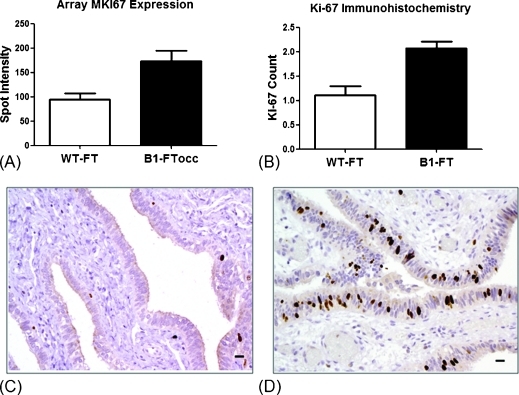
The Affymetrix array analysis showed a significantly increased expression of MKI67 (gene for the antigen identified by monoclonal antibody Ki-67) in the B1-FTocc compared with WT-FT (P = .01; Figure 4A). To confirm the generalizability of the preneoplastic expression pattern in a larger set of wild-type and BRCA1 histologically normal FT, we evaluated Ki-67 protein expression by immunohistochemistry in a larger set of paraffin-embedded FT specimens. Significantly higher Ki-67 protein expression was identified in fallopian tubes from BRCA1 mutation carriers than from BRCA1 wild-type women (P = .0002) who did not have cancer (Figure 4B). Representative images of low Ki-67 staining in WT-FT (Figure 4C) and high Ki-67 staining in B1-FT (Figure 4D) are shown.
Figure 4.
Validation of MIK67 expression data with Ki-67 immunohistochemistry in 26 WT-FT and 52 B1-FT. (A) MIK67 gene expression in the 11 WT-FT samples compared with the 7 B1-FTocc samples (P = .01). (B) Ki-67 protein expression (brown) in the fallopian tubes from 26 wild-type women compared with fallopian tubes from 52 women with deleterious BRCA1 mutations (P = .0002). Representative images of low Ki-67 expression in a wild-type case (C) and high Ki-67 staining in a BRCA1-mutated case (D).
Discussion
For many years, it was believed that ovarian carcinoma arises from the ovarian surface epithelium or in cortical inclusion cysts in the ovary. In accordance with this belief, most studies assessing disruption of gene expression in ovarian carcinomas have focused on the ovarian surface epithelium and carcinomas within the ovarian tissue [15]. However, the relevance of the ovarian surface epithelium has come under increasing scrutiny, and a comprehensive review of the literature regarding the origin of ovarian carcinoma concluded that there is insufficient evidence to support ovarian surface epithelium or inclusion glands as the origin of ovarian carcinomas [16]. In contrast, there has been increasing evidence that many ovarian and primary peritoneal carcinomas arise from neoplastic alterations within the fallopian tube epithelium. This view has been supported by the frequent discovery of occult fallopian tube neoplasms in fallopian tubes removed prophylactically from women at high risk due to hereditary BRCA1 or BRCA2 mutations [9–11]. The tubal epithelium in women with BRCA1 mutations who have up to a 50% lifetime risk of ovarian carcinoma could represent a unique opportunity to study at-risk tissues just before neoplastic transformation. We hypothesized that the epithelium in these high-risk fallopian tubes would express some of the earliest gene disruptions leading to ovarian carcinoma.
Half of all BRCA1 mutation carriers never develop ovarian carcinoma. This fact could make it difficult to identify a BRCA1 preneoplastic gene expression profile in normal BRCA1 tubal epithelium in cancer-free BRCA1 mutation carriers. Our current study is unique because we used histologically normal fallopian tube epithelium from the same fallopian tubes that contained at least a high-grade intraepithelial neoplasm. By using tissues already proven susceptible to neoplastic transformation, we improved our ability to identify a BRCA1 preneoplastic expression profile in BRCA1 mutation carriers. We predicted that gene expression differences that precede morphologically identifiable neoplastic transformation should also be present in BRCA1-associated ovarian carcinomas. Indeed, 41 of 152 differentially expressed probe sets in the normal tubal epithelium from BRCA1 mutation carriers with tubal neoplasia were also similarly differentially expressed in the BRCA1 carcinomas when compared with tubal epithelium from normal-risk women. Our computer model confirmed that the identified overlap in expression profiles between BRCA1 tubal epithelium and BRCA1 carcinoma is highly significant, suggesting that the expression profile that we termed the BRCA1 preneoplastic signature represents a true biological phenomenon. The 41 overlapping probe sets represent unique genes altered in progression from normal fallopian tube epithelium to carcinoma. Furthermore, many of these 41 probe sets represent genes that have been shown to play an important role in cancer biology, such as EFEMP1, CYP3A5, CDKN1C, NR2F1, E2F3, MKI67, NME1, and CSPG5.
One gene in the BRCA1 preneoplastic signature overexpressed in BRCA1 FT is the gene encoding the Ki-67 antigen, expressed in the nuclei of proliferating cells. To generalize our findings to other cases from women with known BRCA1 mutations, we performed immunohistochemistry in a larger series of normal FTs. Consistent with the array data, our pathologists (who were blinded to case designation) identified significantly higher Ki-67 protein expression in FT epithelium of women with BRCA1 mutations compared to women with negative genetic testing (Figure 4). These data suggest that at least some elements of the BRCA1 preneoplastic signature are generalizable to BRCA1-mutated FTs without neoplasia. These data suggest that before neoplastic transformation, there exists a higher rate of proliferation in BRCA1 tubal epithelium, which could increase the opportunity for somatic clonal genetic changes (such as loss of the wild-type allele) and subsequent neoplastic development.
Examples of downregulated probe sets in the BRCA1 preneoplastic signature include those representing EFEMP1, CDKN1C, and NR2F1. Decreased expression of each of these genes has been implicated in carcinogenesis in a variety of neoplasms. EFEMP1 (FLBN3) is a member of the fibulin family, a family of secreted glycoproteins with repeated epidermal growth factor domains and a unique C-terminal fibulin-type module. Fibulins mediate cell-to-cell and cell-to-matrix communication within the extracellular matrix [17]. Mutations in EFEMP1 cause an autosomal-dominant disorder associated with early onset macular degeneration (Doyne honeycomb retinal dystrophy), which has been associated with excessive angiogenesis [18]. EFEMP1 has antiangiogenic properties and has been shown to inhibit tumor growth in mice. The expression of EFEMP1 is reduced in many human neoplasms, including ovarian carcinoma [19], and EFEMP1 is inactivated by promoter methylation in 38% of primary lung carcinomas but not in paired normal lung tissue [20]. The cell cycle regulatory gene CDKN1C (p57/Kip2) is an imprinted maternally expressed gene on chromosome 11p15.4. Disruption of CDKN1C expression causes the cancer predisposing Beckwith-Wiedemann syndrome [21]. CDKN1C has also been implicated as a tumor suppressor gene in a number of human malignant neoplasms including breast, lung, pancreatic, bladder, esophageal, and a variety of hematological and myeloid neoplasms [22,23]. Prostate explants from a CDKN1C knockout mouse develop IEN and prostate adenocarcinoma in nude mice, providing the first mouse model that is pathologically identical to human prostate carcinoma [24]. CDKN1C dysregulation has not been extensively studied in ovarian carcinoma, but the majority (75%) of sporadic ovarian carcinomas demonstrate reduced CDKN1C protein expression (< 10% of tumor cells) using immunohistochemistry [25]. NR2F1 encodes for the protein chicken ovalbumin upstream promoter transcription factor I (COUP-TF1). COUP-TF1 is a nuclear receptor that has been shown to repress transcription, influence the tumor necrosis factor a signaling pathway [26], and modulate the retinoic acid receptor [27]. In breast carcinoma, decreased expression of COUP-TF1 is associated with the up-regulation of aromatase expression [28]. Decreased expression of COUP-TF1 has also been observed in ovarian and bladder carcinomas [29,30].
Upregulated probe sets in our BRCA1 preneoplastic signature included E2F3, NME1, CSPG5, and MKI67. The E2F3 gene is a transcription factor that has been implicated in malignant transformation of human lung [31], prostate [32], and bladder carcinomas [33]. Upregulation of E2F transcription factors has been shown to influence disruptions of the cell cycle in high-grade serous ovarian carcinomas [34], and E2F3 has been used as a biomarker for ovarian carcinoma [35]. The E2F3-Aurora-A axis has been implicated in colorectal cancer [36] and ovarian cancer [37], and recently, E2F3 has been implicated in the proliferation of ovarian cancer cells through interaction with epidermal growth factor receptor [38]. NME1 (NM23) overexpression has been associated with decreased overall survival in patients with serous ovarian carcinoma [39]. CSPG5 (neuroglycan C, neuregulin-6) is a growth factor that transactivates the ErbB2 (HER2/neu) oncogene. CSPG5 is a membrane-anchored chondroitin sulfate proteoglycan that stimulates cell proliferation in a dose-dependent fashion, acts as a specific ligand for ErbB3, and is capable of transactivation of ErbB2 (HER2) [40]. ErbB2 (HER2/neu) is a well-recognized oncogene capable of inducing cellular proliferation and disrupting epithelial cellular polarity. Although CSPG5 has not been well studied in human malignant neoplasms, CSPG5 is secreted by neural stem cells, and it promotes its own proliferation in the fetal brain [41].
The traditional clonal model of carcinogenesis states that clonal expansion and neoplastic proliferation stem from genetic disruptions within an individual cell. However, a more contemporary hypothesis called the epigenetic progenitor model proposes that before this clonal event, there are global epigenetic alterations in nonneoplastic cell lines that allow the proliferation of cell line-specific stem or progenitor cells. This results in a large population of epigenetically disrupted progenitor cells that could then be affected by an initiating mutation of a key gatekeeper gene in a single cell [42]. An epigenetic progenitor model could explain our ability to identify global alterations of gene expression of key tumor progenitor genes in at-risk epithelium in areas that do not have histologically identifiable neoplastic proliferation. Further epigenetic studies will be necessary to assess this hypothesis in BRCA1 tubal epithelium.
We assessed whether the tubal expression profile was consistent between various areas of the distal FT by performing unsupervised hierarchical clustering using independent samples from three of the B1-FTocc cases (two from the contralateral tube). Regardless of whether the duplicated samples were created from the ipsilateral or contralateral fallopian tube, all three duplicates clustered immediately adjacent to their corresponding sample when considering the preneoplastic gene signature (Figure W3). This suggests that the gene disruptions we observed in high-risk fallopian tubes represent a global field effect that affects bilateral fallopian tubes in patients with BRCA1 mutations.
p53-immunopositive foci have been frequently observed in tubal epithelium of both high-risk and normal-risk women [3,13]. We made no effort to select p53-positive cells to derive the BRCA1 preneoplastic expression profile. The resulting expression profile did not seem to be driven by p53 because the expression profiles from the p53 wild-type carcinomas were not distinct from the p53 mutant carcinomas when just considering these genes. The probe sets on the Affymetrix array representing p53 showed minimal signal regardless of BRCA1 status. This is not surprising because p53 foci are generally small, occurring in as few as 10 cells and, consequently, would only be present in a small fraction of the cells that we used for expression array analysis. p53 foci likely represent a clonal event (somatic mutation) in a small subset of tubal epithelial cells. The fact that we can detect differences in expression profiles of BRCA1 tubal epithelium despite not selecting for p53 foci implies that global alterations in gene expression including MKI67 (Ki-67 protein) occur even in cells that do not have p53 alterations or mutation. These data support an alternate model in which global alterations (including increased Ki-67) precede somatic clonal events such as p53 mutation in p53 foci [13].
Three of the seven B1-FTocc samples available from our tissue bank were collected from premenopausal women. To equalize the menopausal status in our three groups, we specifically included cases in the WT-FT and B1-CA groups that were premenopausal at the time of surgery. When unsupervised hierarchical clustering was performed using the preneoplastic gene signature, the four premenopausal WT-FT cases formed a distinct group from the seven postmenopausal WT-FT cases. Interestingly, when the 12 additional FT-B1 samples (which were all postmenopausal) were subjected to clustering analysis, the 7 samples that clustered with the WT-FT group always clustered with the premenopausal WT-FT. It seems that BRCA1-mutated fallopian tubes maintain a gene expression profile that is more similar to premenopausal tissue, even without the stimulation of the premenopausal hormonal milieu. Our group has recently demonstrated that proliferation in WT-FT as measured by Ki-67 protein expression decreases with age, but Ki-67 expression is maintained at a higher level with a less marked decrease with age in women with BRCA1 mutation [13]. Therefore, both protein expression and expression profiling suggest that BRCA1 fallopian tube epithelium maintains a premenopausal proliferative phenotype. Overall, 5 (42%) of the 12 additional B1-FT samples clustered with the B1-FTocc/B1-CA group based on the BRCA1 preneoplastic signature. This closely reflects the percentage of women with BRCA1 mutations who will go on to develop ovarian carcinoma [8]. Interestingly, the samples that clustered with the B1-FTocc/B1-CA group had higher Ki-67 staining.
There has only been one published study by Tone et al. [43] looking at differential gene expression profiles from BRCA1-mutated fallopian tube epithelium and fallopian tube/ovarian carcinomas, which was designed differently from our study. These investigators analyzed fallopian tube epithelium only from premenopausal women, included both BRCA1 and BRCA2 mutation carriers, and focused on fallopian tubes without associated carcinoma, as opposed to our strategy of microdissecting epithelium from fallopian tubes containing at least high-grade intraepithelial neoplasm. Furthermore, their carcinoma group included sporadic fallopian tube and ovarian carcinomas, whereas we compared expression profiles specifically to BRCA1-mutated carcinomas. We felt it was important to separate BRCA1 from BRCA2 fallopian tube epithelium given that ovarian carcinomas have distinct different expression profiles according to whether they have a BRCA1 or BRCA2 mutation [44]. They observed that BRCA1-mutated fallopian tubes collected during the luteal phase of the menstrual cycle were more likely to cluster with the carcinoma samples. They hypothesized that the hormonal environment of the luteal phase causes distinct changes in high-risk fallopian tubes resulting in similar gene expression to carcinoma tissue. Because of the different approaches between this study and our current study, it is difficult to compare the specific genes identified. However, both studies suggest that fallopian tube epithelium from BRCA1 mutation carriers is susceptible to disruption in gene expression, which causes histologically normal fallopian tube tissue to exhibit gene expression resembling carcinoma.
By analyzing gene expression from histologically normal fallopian tube epithelium isolated from BRCA1-mutated fallopian tubes containing early neoplasms, we have identified a potential BRCA1 preneoplastic gene expression signature for BRCA1 serous carcinoma. This gene signature may include some of the earliest disruptions in gene expression leading to the development of serous ovarian carcinoma. Further validation will be necessary to determine which of the genes from this signature are critical in this process and to identify the mechanisms of gene expression alterations. The fact that these genes are disrupted in the fallopian tube tissue before the development of invasive carcinoma could make them useful targets for chemoprevention or early detection of ovarian carcinoma.
Supplementary Material
Acknowledgments
The authors thank Mary-Claire King for her support and guidance in this project, the staff at the Centre for Array Technology for their assistance with processing gene expression arrays, Danbin Xu for guidance with real-time PCR analysis, Robert Vessella and Colm Morrissey for assistance with LCM, Lawrence True for guidance with tissue preparation, and Peter Nelson and Ilsa Coleman for LCM and expression array protocols.
Footnotes
This study was supported by the Yvonne Betson Trust, DOD OC073389 (to E.M.S.) and 1R01CA131965 (E.M.S.). No authors have conflicts of interest to declare.
This study was presented in abstract form at the 40th Annual Society of Gynecologic Oncologists meeting in San Antonio, TX, 2009.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Cai KQ, Klein-Szanto A, Karthik D, Edelson M, Daly MB, Ozols RF, Lynch HT, Godwin AK, Xu XX. Age-dependent morphological alterations of human ovaries from populations with and without BRCA mutations. Gynecol Oncol. 2006;103:719–728. doi: 10.1016/j.ygyno.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 2.Schlosshauer PW, Cohen CJ, Penault-Llorca F, Miranda CR, Bignon YJ, Dauplat J, Deligdisch L. Prophylactic oophorectomy: a morphologic and immunohistochemical study. Cancer. 2003;98:2599–2606. doi: 10.1002/cncr.11848. [DOI] [PubMed] [Google Scholar]
- 3.Crum CP, Drapkin R, Miron A, Ince TA, Muto M, Kindelberger DW, Lee Y. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr Opin Obstet Gynecol. 2007;19:3–9. doi: 10.1097/GCO.0b013e328011a21f. [DOI] [PubMed] [Google Scholar]
- 4.Kmet LM, Cook LS, Magliocco A. A review of p53 expression and mutation in human benign, low malignant potential, and invasive epithelial ovarian tumors. Cancer. 2003;97:389–404. doi: 10.1002/cncr.11064. [DOI] [PubMed] [Google Scholar]
- 5.Stern J, Buscema J, Parmley T, Woodruff JD, Rosenshein NB. Atypical epithelial proliferations in the fallopian tube. Am J Obstet Gynecol. 1981;140:309–312. doi: 10.1016/0002-9378(81)90279-9. [DOI] [PubMed] [Google Scholar]
- 6.Yanai-Inbar I, Silverberg SG. Mucosal epithelial proliferation of the fallopian tube: prevalence, clinical associations, and optimal strategy for histopathologic assessment. Int J Gynecol Pathol. 2000;19:139–144. doi: 10.1097/00004347-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Kindelberger DW, Lee Y, Miron A, Hirsch MS, Feltmate C, Medeiros F, Callahan MJ, Garner EO, Gordon RW, Birch C, et al. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: evidence for a causal relationship. Am J Surg Pathol. 2007;31:161–169. doi: 10.1097/01.pas.0000213335.40358.47. [DOI] [PubMed] [Google Scholar]
- 8.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risk due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 9.Finch A, Shaw P, Rosen B, Murphy J, Narod SA, Colgan TJ. Clinical and pathologic findings of prophylactic salpingo-oophorectomies in 159 BRCA1 and BRCA2 carriers. Gynecol Oncol. 2006;100:58–64. doi: 10.1016/j.ygyno.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 10.Lamb JD, Garcia RL, Goff BA, Paley PJ, Swisher EM. Predictors of occult neoplasia in women undergoing risk-reducing salpingo-oophorectomy. Am J Obstet Gynecol. 2006;194:1702–1709. doi: 10.1016/j.ajog.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Leeper K, Garcia R, Swisher E, Goff B, Greer B, Paley P. Pathologic findings in prophylactic oophorectomy specimens in high-risk women. Gynecol Oncol. 2002;87:52–56. doi: 10.1006/gyno.2002.6779. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Miron A, Drapkin R, Nucci MR, Medeiros F, Saleemuddin A, Garber J, Birch C, Mou H, Gordon RW, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211:26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- 13.Norquist BM, Garcia RL, Allison KH, Jokinen CH, Kernochan LE, Pizzi CC, Barrow BJ, Goff BA, Swisher EM. The molecular pathogenesis of hereditary ovarian carcinoma: alterations in the tubal epithelium of women with BRCA1 and BRCA2 mutations. Cancer. doi: 10.1002/cncr.25439. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galic V, Willner J, Wollan M, Garg R, Garcia R, Goff BA, Gray HJ, Swisher EM. Common polymorphisms in TP53 and MDM2 and the relationship to TP53 mutations and clinical outcomes in women with ovarian and peritoneal carcinomas. Genes Chromosomes Cancer. 2007;46:239–247. doi: 10.1002/gcc.20407. [DOI] [PubMed] [Google Scholar]
- 15.Fehrmann RS, Li XY, van der Zee AG, de Jong S, Te Meerman GJ, de Vries EG, Crijns AP. Profiling studies in ovarian cancer: a review. Oncologist. 2007;12:960–966. doi: 10.1634/theoncologist.12-8-960. [DOI] [PubMed] [Google Scholar]
- 16.Bell DA. Origins and molecular pathology of ovarian cancer. Mod Pathol. 2005;18(suppl 2):S19–S32. doi: 10.1038/modpathol.3800306. [DOI] [PubMed] [Google Scholar]
- 17.Gallagher WM, Currid CA, Whelan LC. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11:336–340. doi: 10.1016/j.molmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Stone EM, Lotery AJ, Munier FL, Héon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, et al. A single EFEMP1 mutation associated with bothMalattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 19.Albig AR, Neil JR, Schiemann WP. Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res. 2006;66:2621–2629. doi: 10.1158/0008-5472.CAN-04-4096. [DOI] [PubMed] [Google Scholar]
- 20.Yue W, Dacic S, Sun Q, Landreneau R, Guo M, Zhou W, Siegfried JM, Yu J, Zhang L. Frequent inactivation of RAMP2, EFEMP1 and Dutt1 in lung cancer by promoter hypermethylation. Clin Cancer Res. 2007;13:4336–4344. doi: 10.1158/1078-0432.CCR-07-0015. [DOI] [PubMed] [Google Scholar]
- 21.Hatada I, Ohashi H, Fukushima Y, Kaneko Y, Inoue M, Komoto Y, Okada A, Ohishi S, Nabetani A, Morisaki H, et al. An imprinted gene p57KIP2 is mutated in Beckwith-Wiedemann syndrome. Nat Genet. 1996;14:171–173. doi: 10.1038/ng1096-171. [DOI] [PubMed] [Google Scholar]
- 22.Larson PS, Schlechter BL, King CL, Yang Q, Glass CN, Mack C, Pistey R, de Las Morenas A, Rosenberg CL. CDKN1C/p57kip2 is a candidate tumor suppressor gene in human breast cancer. BMC Cancer. 2008;8:68. doi: 10.1186/1471-2407-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi T, Toyota M, Itoh F, Suzuki H, Obata T, Yamamoto H, Kakiuchi H, Kusano M, Issa JP, Tokino T, et al. Inactivation of p57KIP2 by regional promoter hypermethylation and histone deacetylation in human tumors. Oncogene. 2002;21:2741–2749. doi: 10.1038/sj.onc.1205376. [DOI] [PubMed] [Google Scholar]
- 24.Jin RJ, Lho Y, Wang Y, Ao M, Revelo MP, Hayward SW, Wills ML, Logan SK, Zhang P, Matusik RJ. Down-regulation of p57Kip2 induces prostate cancer in the mouse. Cancer Res. 2008;68:3601–3608. doi: 10.1158/0008-5472.CAN-08-0073. [DOI] [PubMed] [Google Scholar]
- 25.Khouja MH, Baekelandt M, Nesland JM, Holm R. The clinical importance of Ki-67, p16, p14, and p57 expression in patients with advanced ovarian carcinoma. Int J Gynecol Pathol. 2007;26:418–425. doi: 10.1097/pgp.0b013e31804216a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang LJ, Liu X, Gafken PR, Kioussi C, Leid M. A chicken ovalbumin upstream transcription factor 1 (COUP-TF1) complex represses expression of the gene encoding tumor necrosis factor α-induced protein 8 (TNFAIP8) J Biol Chem. 2008;284:6156–6168. doi: 10.1074/jbc.M807713200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin B, Chen GQ, Xiao D, Kolluri SK, Cao X, Su H, Zhang XK. Orphan receptor COUP-TF is required for induction of retinoic acid receptor beta, growth inhibition, and apoptosis by retinoic acid in cancer cells. Mol Cell Biol. 2000;20:957–970. doi: 10.1128/mcb.20.3.957-970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S, Jingjing Y, Ikuko K, Yoshiyuki K, Zhou D. Positive and negative transcriptional regulation of aromatase expression in human breast cancer tissue. J Steroid Biochem Mole Biol. 2005;95:17–23. doi: 10.1016/j.jsbmb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Ham WS, Lee JH, Yu HS, Choi YD. Expression of chicken ovalbumin upstream promoter-transcription factor 1 (COUP-TF1) in bladder transitional cell carcinoma. Urology. 2008;72:921–926. doi: 10.1016/j.urology.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 30.Damiao RDS, Oshima CTF, Stavale JN, Goncalves WJ. Analysis of the expression of estrogen receptor, progesterone receptor and chicken ovalbumin upstream promoter-transcription factor 1 in ovarian epithelial cancers and normal ovaries. Oncol Rep. 2007;18:25–32. [PubMed] [Google Scholar]
- 31.Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, Roe T, Clark J, Joshi A, Norman A, et al. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer. 2006;54:155–162. doi: 10.1016/j.lungcan.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23:5871–5879. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 33.Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, et al. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004;23:1627–1630. doi: 10.1038/sj.onc.1207274. [DOI] [PubMed] [Google Scholar]
- 34.Meyer TD, Bijsmans IT, Van de Vijver KK, Bekaert S, Oosting J, Van Criekinge W, van Engeland M, Sieben NL. E2Fs mediate a fundamental cell-cycle deregulation in high-grade serous ovarian carcinomas. J Pathol. 2009;217:14–20. doi: 10.1002/path.2452. [DOI] [PubMed] [Google Scholar]
- 35.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 36.Baba Y, Nosho K, Shima K, Irahara N, Kure S, Toyoda S, Kirkner GJ, Goel A, Fuchs CS, Ogino A. Aurora-A expression is independently associated with chromosomal instability in colorectal cancer. Neoplasia. 2009;11:418–425. doi: 10.1593/neo.09154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He L, Yang H, Ma Y, Pledger WJ, Cress WD, Cheng JQ. Identification of Aurora-A as a direct target of E2F3 during G2/M cell cycle progression. J Biol Chem. 2008;283:31012–31020. doi: 10.1074/jbc.M803547200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Reimer D, Hubalek M, Riedle S, Skvortsov S, Erdel M, Conci N, Fiegl H, Muller-Holzner E, Marth C, Illmensee K, et al. E2F3a is critically involved in epidermal growth factor receptor-directed proliferation in ovarian cancer. Cancer Res. 2010;70:4613–4623. doi: 10.1158/0008-5472.CAN-09-3551. [DOI] [PubMed] [Google Scholar]
- 39.Youn BS, Kim DS, Kim JW, Kim YT, Kang S, Cho NH. NM23 as a prognostic biomarker in ovarian serous carcinoma. Mod Pathol. 2008;21:885–892. doi: 10.1038/modpathol.2008.64. [DOI] [PubMed] [Google Scholar]
- 40.Kinugasa Y, Ishiguro H, Tokita Y, Oohira A, Ohmoto H, Higashiyama S. Neuroglycan C, a novel member of the neuregulin family. Biochem Biophys Res Commun. 2004;321:1045–1049. doi: 10.1016/j.bbrc.2004.07.066. [DOI] [PubMed] [Google Scholar]
- 41.Ida M, Shuo T, Hirano K, Tokita Y, Nakanishi K, Matsui F, Aono S, Fujita H, Fujiwara Y, Kaji T, et al. Identification and functions of chondroitin sulfate in the milieu of neural stem cells. J Biol Chem. 2006;281:5982–5991. doi: 10.1074/jbc.M507130200. [DOI] [PubMed] [Google Scholar]
- 42.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 43.Tone AA, Begley H, Sharma M, Murphy J, Rosen B, Brown TJ, Shaw PA. Gene expression profiles of luteal phase fallopian tube epithelium from BRCA mutation carriers resemble high-grade serous carcinomas. Clin Cancer Res. 2008;14:4067–4078. doi: 10.1158/1078-0432.CCR-07-4959. [DOI] [PubMed] [Google Scholar]
- 44.Jazaeri AA, Yee CJ, Sotiriou C, Brantley KR, Boyd J, Liu ET. Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers. J Natl Cancer Inst. 2002;94:990–1000. doi: 10.1093/jnci/94.13.990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.