Abstract
Human colorectal cancer (CRC) arises from activating mutations in the Wnt/β-catenin pathway that converge with additional molecular changes to shape tumor development and patient prognosis. We report here that Na+/H+ exchanger 3 regulating factor 1 (NHERF1)/EBP50, an adaptor molecule that interacts with β-catenin, undergoes successive alterations during the colorectal adenoma-to-carcinoma transition, ranging from loss of normal apical membrane distribution to ectopic cytoplasmic overexpression. NHERF1 depletion in human intestinal epithelial polarized cells induced epithelial-mesenchymal transition, β-catenin nuclear translocation with elevation of Wnt/β-catenin transcriptional targets, and increased cell migration and invasion. Ectopic cytoplasmic NHERF1 expression additionally intensified the transformed phenotype by increasing cell proliferation. The epithelial morphology and reduced cell motility could only be restored by re-expression of NHERF1 specifically at the apical plasma membrane. We conclude that alterations in the apical membrane localization of NHERF1 contribute to CRC through the disruption of epithelial morphology. This study identifies NHERF1 as a new player in CRC progression and supports the notion that the expression or subcellular distribution of NHERF1 may be used as diagnostic marker for CRC.
Introduction
Human colorectal cancer (CRC) is the third most common type of cancer and the third cause of mortality due to cancer in the United States [1]. It typically evolves from a benign polyp, or adenoma, to malignant carcinoma that starts in the mucosa (carcinoma in situ) and spreads through the other layers of the colon. In advanced stages, malignant cells metastasize to the lymph nodes or to distant organs. The ability of CRC cells to invade and metastasize renders the tumors unresectable and resistant to chemotherapy and diminishes abruptly the overall survival rate for patients with metastatic disease to approximately 10%at 5 years.
The acquired invasiveness of malignant cells results from the disorganization of epithelial morphology leading to an epithelial-mesenchymal-like transition (EMT) [2]. This process occurs naturally during the embryonic development of some structures and is reactivated in malignant cells of epithelial origin. In neoplastic cells, EMT is reflected by various degrees of epithelial change, from mild loss of cell polarity to the acquisition of frank mesenchymal/fibroblastic characteristics with increased cell motility [3]. The main event in EMT is the loss of E-cadherin from adherens junctions, and several pathways have been involved in triggering it [4]. The Wnt/β-catenin pathway can promote EMT through activation of the Slug and Snail transcription factors, which are known to repress the E-cadherin promoter [5,6]. Both Slug and Snail have been shown to be upregulated in advanced CRC tumors [7,8]. Wnt/β-catenin signaling also activates promoters of genes usually expressed in fibroblasts that enhance invasiveness, such as fibronectin and matrix metalloproteinase 7 (MMP7) [4].
The Wnt/β-catenin pathway is activated in more than 90% of CRCs by mutations in either the Apc gene (80%–85%) or the gene encoding β-catenin (5%–10%) [9]. These mutations interfere with the degradation of cytoplasmic β-catenin, allowing its translocation to the nucleus and the subsequent activation of genes involved in proliferation and EMT. However, the pathology of CRC is complex and cannot be fully explained by this central pathway alone [10]. Modifying influences are present within and outside the Wnt/β-catenin central pathway, and we investigate here whether NHERF1/EBP50 (Na+/H+ exchanger 3 regulating factor 1; ezrin-radixin-moesin (ERM) binding phosphoprotein 50), an adaptor protein that interacts directly with β-catenin [11], plays a role in CRC.
NHERF1 is a 50-kDa adaptor protein composed of two tandem PSD-95/Disc-large/ZO-1 (PDZ) domains and a carboxyl (C)-terminal ERM-binding region [12,13]. It associates with β-catenin through its PDZ2 domain [11] and with ezrin through its ERM-binding region [12]. NHERF1 is localized mainly at the apical plasma membrane (PM) in human epithelial tissues [14] and its inactivation in mice induces ultrastructural abnormalities of the intestinal brush border membrane [15,16]. Our previous studies in mouse embryonic fibroblasts showed that NHERF1 behaves as a tumor suppressor through its effects on β-catenin and PTEN [17,18]. We have now examined the involvement of NHERF1 in human CRC, first by examining patient tumor samples for changes in the expression and intracellular distribution of NHERF1 and second by modeling these changes in polarized intestinal epithelial cells. The NHERF1 alterations observed in CRC induced a malignant cell phenotype, implicating NHERF1 as a new and important modifier of CRC progression.
Materials and Methods
CRC Resection Specimens
A total of 18 tissue resection specimens from patients with CRC (6 men and 12 women, 63.3 ± 2.3 years) were obtained from the CRC Bank at MD Anderson Cancer Center. Surgeries for all cases had been performed in a 15-year period between 1992 and 2007, and no patient received previous therapy. Of the 18 specimens, 11 were paraffin-embedded, with areas of adenoma, carcinoma, and adjacent normal mucosa on the same slide. The remaining seven cases were matched sets of frozen specimens with the pathologist's annotations as normal, deep tumor (carcinoma), and tumor edge (adenoma).
Immunofluorescence (IF)
The CRC specimens were deparaffinized and hydrated as previously described [17], followed by antigen retrieval in a steamer for 20 minutes. The sections were cooled for 10 minutes, rinsed with ddH2O for 5 minutes three times, and blocked from nonspecific binding for 30 minutes with 20% to 50% donkey serum in PBS. The NHERF1 antibody (Affinity BioReagents/Thermo, Rockford, IL) was applied at 1:200 in PBS overnight at 4°C, and unbound antibody was removed by washing three times with PBS for 5 minutes. The secondary antibody, Alexa Fluor 488 donkey antirabbit IgG (Molecular Probes/Invitrogen, Carlsbad, CA), was applied at 1:500 in PBS for 45 minutes followed by three PBS washes for 5 minutes. The sections were incubated in ToPro-3 iodide (Molecular Probes/Invitrogen) at 1:2000 in PBS for 20 minutes for nuclear staining and were then washed twice with PBS for 5 minutes. The Slowfade Gold antifade reagent (Molecular Probes/Invitrogen) was used for the mounting of the slides. Images were acquired with the Zeiss 510 confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY), using 40x/1.30 objective with oil immersion. The densitometric analysis of the IF intensity was performed with the ImageJ software (National Institutes of Health, Bethesda, MD).
Cells, Plasmids, and Retroviral Infections
The Caco-2, HT29, and RKO human CRC cell lines were grown in Dulbecco modified Eagle medium/Ham's F-12 nutrients (DMEM/F-12) supplemented with 10% FBS. The 293T cells were grown in DMEM supplemented with 10% FBS. The retroviral construct encoding wild-type NHERF1 (358 residues) in the pCXb vector (blasticidin selection) was described elsewhere [17]. The short hairpin RNA (shRNA) 1 (ACCCCATCCTAGACTTCAA) and 4 (GGGAAACTGACGAGTTCTT) used for NHERF1 knockdown were cloned in pSIREN-RetroQ vector (puromycin selection) [19]. The pSIREN-RetroQ control vector carries an irrelevant shRNA for enhanced green fluorescent protein. The myristoylation signal of v-Src, MGSSKSKPKDPSQR, was added N-terminally to NHERF1 in the pCXb vector, generating Myr-NHERF1. The Myr signal was also added to the NHERF1 PDZ1-2 construct that lacks the terminal ERM-binding region. Transfections and retroviral infections were performed as previously described [20]. All functional assays were performed on cell pools resulting after selection of infected cells with the appropriate selection marker.
Cell Morphology, Migration, and Invasion Assays
For assessment of cell morphology, 5 x 105 cells in 2% FBS DMEM were plated in duplicate in six-well dishes and cultured for 10 days at 37°C. On day 10, cells were washed with PBS twice, fixed in 10% acetic acid for 20 minutes, washed with PBS once, and stained with 0.1% crystal violet for 20 minutes. Cell migration was assessed by scratching a confluent cell monolayer with a 2- to 200-µl tip in serum-free DMEM. The width of the scratch was measured at different time points with ImageJ software. For cell invasion, 1 x 106 cells were prepared in duplicate in 250 µl of serum-free DMEM. The cells were placed in transwells with 8-µm pore size polycarbonate filters (Corning Incorporated, Corning, NY), precoated with 100 µl of 0.7 mg/ml Matrigel (BD Biosciences, San Jose, CA). To obtain a chemogradient across the transwells, the lower wells were filled with 750 µl of 10% FBS DMEM. The cells were incubated at 37°C for 48 hours, fixed with methanol, and stained with hematoxylin and eosin (H&E). Nonmigratory cells on the upper surface of the transwells were removed with a cotton swab, and the number of cells remaining on the lower surface of the transwell was counted.
Gelatin Zymography
To analyze the activity of secreted MMPs, 2.5 x 105 cells in 10% FBS DMEM were plated in 12-well dishes, incubated overnight at 37°C, and changed in serum-free DMEM for 24 hours. This was collected and spun at 10,000g for 5 minutes. The cell-free supernatant was normalized to the cell number, and equivalent amounts were loaded without boiling on a 12% polyacrylamide gel copolymerized with 1 mg/ml gelatin and separated in nonreducing conditions. The gel was washed twice for 30 minutes in 2.5% Triton X buffer with 50 mM Tris (pH 7.5), 5 mM CaCl2, 1 µM ZnCl2, and 0.02% NaN3. It was then equilibrated for 30 minutes at room temperature in the same buffer devoid of 1% Triton X, thereafter incubated in this buffer for 48 hours at 37°C. Coomassie blue was used to stain the gel for 30 minutes, and the white bands of gelatin-cleared areas were sought in the dark blue background.
Protein and IF Analysis of Cells
Cell lysis, fractionation, immunoprecipitation, Western blot, and IF of formaldehyde-fixed cells were performed as previously described [18,21]. The CRC frozen specimens were homogenized with a pestle and lysed similarly in a buffer containing 1% Triton X-100, 50 mM HEPES, pH 7.5, 100 mM NaCl, 10 mM EDTA, and 10% glycerol. Antibodies for Western blot were obtained as follows: NHERF1, E-cadherin, fibronectin, and β-catenin from BD Biosciences and GAPDH (0411) and Erk2 (C-14) were from Santa Cruz Biotechnology (Santa Cruz, CA). For IF, the primary antibodies and corresponding dilutions were NHERF1 (Affinity BioReagents/Thermo) at 1:500, E-cadherin (BD Biosciences) at 1:250, and β-catenin (Sigma-Aldrich, St Louis, MO) at 1:500. The secondary antibodies were Alexa Fluor 568 goat antirabbit IgG and Alexa Fluor 488 goat antimouse IgG (Molecular Probes/Invitrogen) at 1:500, with ToPro-3 iodide at 1:2000.
Statistics
All numerical data were examined for normality of distribution. Parametric or nonparametric methods of one- or two-way analysis of variance and t-test were used, as appropriate, to analyze the differences between groups or groups by day. A 3 x 3 contingency table of categorical data from CRC case study was analyzed by Fisher exact test. Statistical significance was considered for P < .05. Unless otherwise indicated, values are presented as means ± SEM.
Results
NHERF1 Intracellular Localization and Levels Are Progressively Changed in CRC Samples
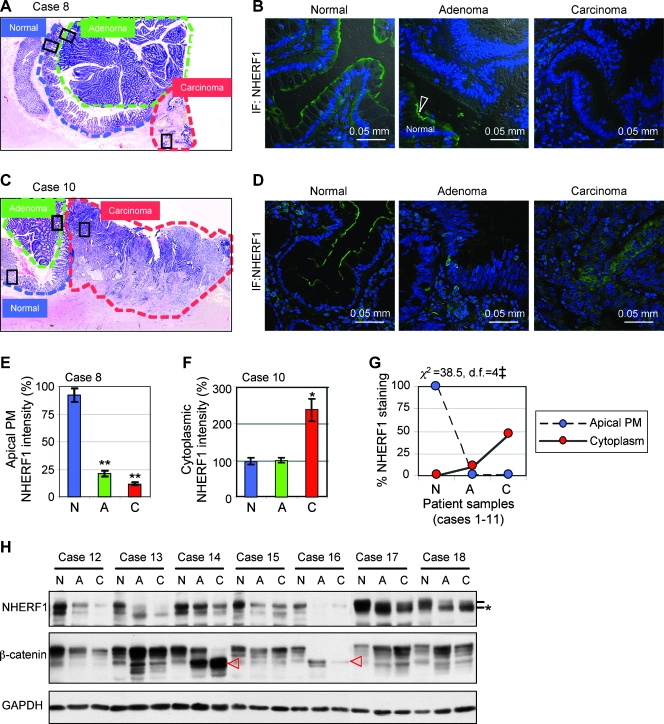
To investigate the role of NHERF1 in human CRC, we analyzed a total of 18 CRC specimens for which matched sets of normal, adenoma, and carcinoma tissues were available from the surgically removed tumors. Of these, 11 paraffin-embedded resection specimens contained normal, adenoma, and carcinoma tissue on the same slide (Figure 1, A and C). The regions were delineated by H&E histopathologic analysis, and the expression levels and subcellular localization of NHERF1 were determined by confocal IF microscopy (Figure 1, A–D). NHERF1 was expressed at the apical PM in normal colonic epithelium, and its expression was lost in adenoma (Figure 1E). In carcinoma, two expression patterns were observed: either absent/low expression (Figure 1, B and E) or ectopic overexpression in the cytoplasm (Figure 1, D and F). Overall, 10 of 11 adenomas and 6 of 11 carcinomas showed absent/low NHERF1 staining, and 5 of 11 carcinomas had tumor areas with high NHERF1 cytoplasmic staining (Figure 1G).
Figure 1.
Progressive changes of NHERF1 levels and intracellular localization in CRC. (A–G) Analysis of 11 resection specimens containing normal (blue outlining), adenoma (green outlining) and carcinoma (red outlining) tissues in the same sample. CRC case 8 (A–B) and 10 (C–D) are exemplified by H&E (A, C) and confocal IF with NHERF1 antibody and ToPro-3 (B, D). Boxes indicate the selected analyzed regions of normal colonic mucosa and adjacent adenoma and carcinoma. Note apical PM staining of normal mucosa—arrowhead in the middle panel in panel B—but not of adenoma or carcinoma. Note also NHERF1 cytoplasmic overexpression in carcinoma in panel D. (E and F) Area comparisons of membrane (E) and cytoplasmic (F) NHERF1 intensity by densitometry. N indicates normal; A, adenoma; C, carcinoma. Data are means ± SEM of the triplicate measurements for cases 8 and 10. *P < .05, **P < .001 versus N. (G) Area comparisons of NHERF1 intracellular localization by IF either at the apical PM or in the cytoplasm show significant differences (‡P < .001) between N (normal), A (adenoma), and C (carcinoma samples). n = 11 (100%), df = degrees of freedom. (H) Western blot of whole tissue lysates from seven additional frozen specimens showing NHERF1 expression in matched sets of normal (N), adenoma (A), and carcinoma (C) samples. Proteins were loaded at 20 µg per lane. Lines mark the phosphorylation species of NHERF1 and star marks the less phosphorylated form, most likely ectopically localized to the cytoplasm. Red arrowheads mark N-terminally truncated β-catenin oncogenic proteins.
To confirm these results, seven CRC cases with matched normal, adenoma. and carcinoma frozen samples were processed for Western blot analysis with NHERF1 antibody (Figure 1H). As internal control, β-catenin showed N-terminal in-frame truncation in adenoma and carcinoma but not in normal tissue in two cases (Figure 1H, arrowheads). In five cases, NHERF1 showed significant depletion in both adenoma and carcinoma samples compared with the corresponding normal tissue (Figure 1H, cases 12–16). In the remaining two cases, NHERF1 was present in both adenoma and carcinoma samples but as a lower-molecular weight form in comparison to the normal tissue (Figure 1H, cases 17–18, star). The low-molecular weight NHERF1 form most likely represents the less phosphorylated protein that has been associated with NHERF1 ectopic cytoplasmic expression in the intestine of ezrin knockout mice [22].
NHERF1 Depletion Induces EMT and β-Catenin Nuclear Translocation in Polarized Intestinal Cells
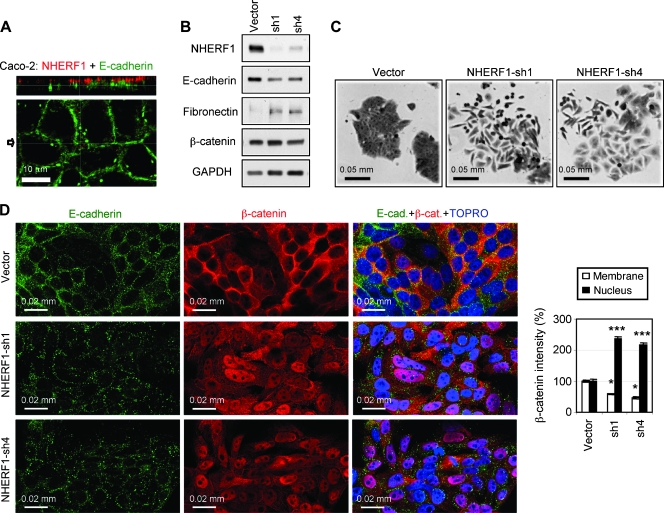
To study the effect of NHERF1 in human intestinal cells, we analyzed the subcellular distribution of NHERF1 in seven CRC cell lines (not shown) and found that only Caco-2 cells exhibit NHERF1 apical PM localization, similarly to normal colon epithelial cells (Figure 2A). Caco-2 cells have been used as a model of normal intestinal cells because they form polarized cell monolayers with well-developed apical microvilli in confluent culture [23]. We further used these cells to model the alterations of NHERF1 observed in human CRC specimens. In adenoma and carcinoma, we observed NHERF1 depletion from the apical PM. We similarly depleted NHERF1 in Caco-2 cells by two different shRNAs (Figure 2B) and observed scattering of the cells with elongated or flattened shape and process extension, consistent with a change toward mesenchymal morphology (Figure 2C). The phenotypic changes in cell shape were accompanied by a loss of the epithelial marker E-cadherin and an increase in the mesenchymal marker fibronectin (Figure 2B), indicating that NHERF1 loss from the PM induces EMT in epithelial polarized cells.
Figure 2.
NHERF1 depletion induces EMT-like changes and nuclear translocation of β-catenin in polarized Caco-2 cells. (A) Confocal IF analysis of a Caco-2 monolayer with NHERF1 (red) and E-cadherin (green) antibodies. XZ image (top) shows NHERF1 in the microvilli at the apical PM, and the XY middle-plane section (bottom) shows E-cadherin at the lateral cell-cell junctions. The arrow indicates the plane of XZ section. (B) Western blot of whole cell lysates showing NHERF1 depletion by two shRNAs (sh1 and sh4) and opposite variations of epithelial (E-cadherin) and mesenchymal (fibronectin) markers in NHERF1-depleted cells. Proteins were loaded at 30 µg per lane. (C) Crystal violet staining showing EMT-like morphologic changes in NHERF1-depleted cells compared with control (vector) cells. (D) Confocal IF with indicated antibodies shows nuclear accumulation and lateral PM effacement of β-catenin in NHERF1-depleted cells compared with vector control cells. The densitometric analysis of β-catenin intensity for control and NHERF1-depleted cells is shown. n = 25. *P < .05, ***P < .001 versus vector.
Although Caco-2 cells resemble normal intestinal cells, they are derived from CRC and have truncated Apc [24] that should interfere with the degradation of β-catenin and promote its accumulation in the nucleus. Nevertheless, β-catenin colocalized predominantly with E-cadherin at the lateral PM in Caco-2 cells and was not observed in the nucleus (Figure 2D, upper panels). This finding, by itself, suggests that other events are necessary to cooperate with Apc mutations to promote β-catenin nuclear localization. In NHERF1-depleted cells, a dramatic increase in β-catenin nuclear localization accompanied by a decrease from the lateral PM was apparent (Figure 2D). A parallel E-cadherin reduction from cell-cell junctions was also observed (Figure 2D).
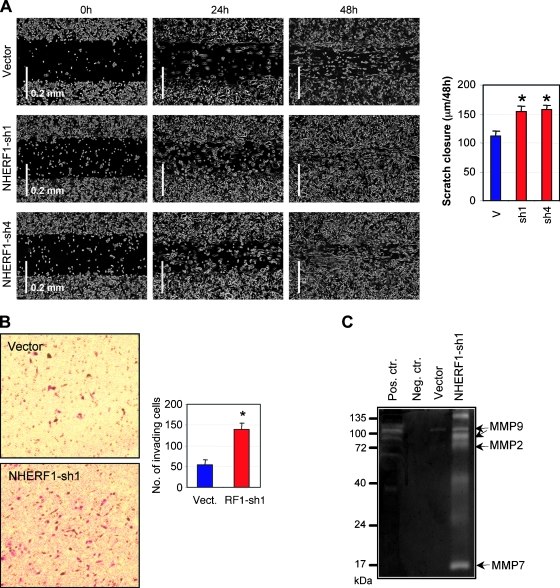
Consistent with the mesenchymal-like morphologic changes, NHERF1-depleted cells migrated significantly faster than control cells in a monolayer scratch assay (Figure 3A). We also tested the ability of these cells to invade through a Matrigel substrate. Control Caco-2 cells have limited ability to invade through Matrigel. A significant increase of invasiveness was apparent after NHERF1 depletion (Figure 3B). The ability to invade the Matrigel requires, in addition to enhanced cell motility, an increased activity of secreted MMPs. Using gelatin zymography, we found a general increased activity of MMPs in supernatant from NHERF1-depleted cells, with digestion bands suggestive for MMP2 and MMP9 activity (Figure 3C). The low-molecular band could correspond to MMP7 activity, a Wnt/β-catenin transcriptional target, or to activation products of MMP2 (Figure 3C). Altogether, these results suggested that loss of membrane NHERF1 in polarized cells determines EMT and increased cell motility, most likely as a consequence of β-catenin nuclear translocation.
Figure 3.
NHERF1 depletion enhances motility and promotes invasion of Caco-2 cells. (A) Scratch assay showing faster migration of NHERF1-depleted (sh1 and sh2) cells relative to control cells. The graph shows the distance of cell migration from both ends in 48 hours. *P < .05 versus vector (v). (B) Matrigel invasion assay at 48 hours of NHERF1-depleted (RF1-sh1) cells. *P < .05 versus vector (Vect.). (C) Gelatin zymography showing higher activity of the secreted MMPs in supernatant from NHERF1-depleted (sh1) cells relative to vector control cells. The supernatant was collected from samples containing an equal number of cells. Positive and negative controls are DMEM with and without FBS, respectively. The putative molecular weights of MMP9, MMP2, and MMP7 are indicated.
Re-expression of Cytoplasmic NHERF1 Increases Cell Proliferation
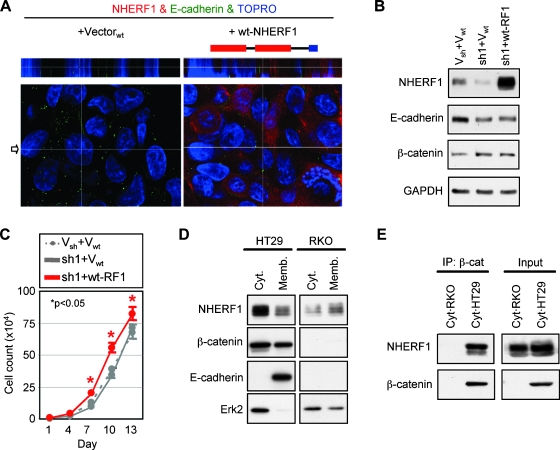
To confirm that the observed EMT is specifically caused by NHERF1 depletion, we reconstituted the NHERF1-depleted Caco-2 cells with wild-type wt-NHERF1 (Figure 4, A and B). Surprisingly, ectopically expressed untagged wt-NHERF1 did not distribute to the apical PM but to the cytoplasm and, in some instances, to the nuclei of reconstituted cells (Figure 4A). In addition, wt-NHERF1 neither rescued the epithelial morphology of the cells (not shown) nor reestablished the expression levels and lateral membrane localization of E-cadherin (Figure 4, A and B), clearly indicating that cytoplasmic NHERF1 does not establish epithelial morphology.
Figure 4.
Cytoplasmic NHERF1 increases cell proliferation and associates with β-catenin. (A) Confocal IF of NHERF1-depleted cells reconstituted with control vector or wild-type NHERF1 (wt-NHERF1). The schematic structure of NHERF1 is shown on top of the panels. (B) Western blot of whole cell lysates showing expression of wt-NHERF1 in NHERF1-depleted cells. Proteins were loaded at 30 µg per lane. (C) Growth curve of wt-NHERF1-reconstituted cells, with corresponding vector controls. *P < .05 versus control (sh1 + Vwt). (D) Subcellular fractionation of HT29 and RKO β-catenin-negative control cells. Note cofractionation of NHERF1 and β-catenin in the cytoplasm (cyt.) and membrane (memb.) fractions of HT29 cells. E-cadherin and Erk2 were used as membrane and cytoplasmic markers, respectively. Proteins were loaded at 20 µg per lane. (E) Coimmunoprecipitation of cytoplasmic β-catenin with NHERF1 in HT29 cells. RKO cells were used as negative control of β-catenin expression.
Whereas NHERF1 knockdown did not alter cell proliferation, re-expression of wt-NHERF1 that adopted cytoplasmic localization increased the proliferation of reconstituted cells compared with vector control cells (Figure 4C). A possible explanation for the increased proliferation in cells with NHERF1 expressed in the cytoplasm or nucleus could rely on the direct interaction between NHERF1 and β-catenin with further activation of the Wnt/β-catenin pathway [11]. We investigated this possibility by immunoprecipitation of β-catenin from cytoplasmic fractions of HT29 CRC cells that express endogenous cytoplasmic NHERF1 (Figure 4D). RKO CRC cells that lack β-catenin were used as a negative control. Endogenous NHERF1 could be co-immunoprecipitated with β-catenin (Figure 4E), indicating complex formation between these proteins in the cytoplasm of CRC cells. Thus, it seems that cytoplasmic NHERF1, as detected in the inner mass of carcinomas, maintains the mesenchymal phenotype and confers additional growth advantage to cells, possibly by associating with β-catenin outside the PM compartment.
Apical PM-Targeted NHERF1 Re-establishes the Epithelial Morphology
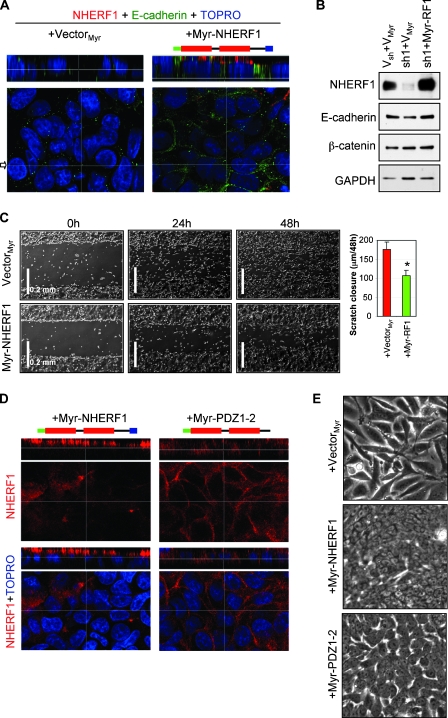
The observation that wt-NHERF1 localized to the cytoplasm of reconstituted cells but not to the apical PM prompted us to express a membrane-targeted Myr-NHERF1 in NHERF1-depleted cells (Figure 5, A and B). The Myr-NHERF1-reconstituted cells reexpressed high levels of E-cadherin, specifically at the cell-cell junctions (Figure 6, A and B), and redistributed β-catenin from the nucleus to the lateral PM (Figure W1A), similarly to parental Caco-2 cells. The epithelial morphology was also restored in Myr-NHERF1-reconstituted cells (Figure W1B), and the motility was correspondingly decreased to the values of the parental Caco-2 cells (Figure 5C). This indicated that membrane-targeted NHERF1 fully reverts both the molecular and the cellular mesenchymal changes induced by NHERF1 knockdown.
Figure 5.
Membrane-targeting and the ERM-binding region of NHERF1 are required for epithelial reconversion of NHERF1-depleted Caco-2 cells. (A) Confocal IF analysis of NHERF1-depleted cells reconstituted with control vector or membrane-targeted NHERF1 (Myr-NHERF1). The added N-terminal myristoylation signal is shown in green in the NHERF1 structure cartoon. Note expression of Myr-NHERF1 at the apical PM and of E-cadherin (green) at the lateral PM. (B) Western blot of whole cell lysates showing restoration of E-cadherin expression by Myr-NHERF1 expression in NHERF1-depleted cells. Proteins were loaded at 30 µg per lane. (C) Scratch assay showing slower migration of Myr-NHERF1-reconstituted cells compared with vector control cells. The graph shows the distance of cell migration from both ends in 48 hours. *P < .05 versus + vector. (D) Confocal IF of Myr-NHERF1 and Myr-PDZ1-2-reconstituted cells showing apical and lateral PM localization of Myr-NHERF1 and Myr-PDZ1-2, respectively. (E) Phase-contrast images showing partial restoration of epithelial cell morphology by Myr-PDZ1-2 compared with full restoration by Myr-NHERF1.
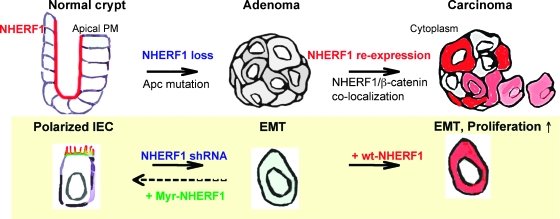
Figure 6.
Model of the NHERF1 alterations in the progression of CRC and of the molecular interventions in Caco-2 intestinal epithelial cells (IECs) designed to mimic these alterations.
Surprisingly, reconstituted Myr-NHERF1 was found exclusively at the apical PM (Figure 5, A and D, left panels), although myristoylation is a general membrane-targeting signal. We have previously shown that the ERM-binding region of NHERF1 is required for NHERF1 localization at the microvilli in polarized kidney epithelial cells [25]. To examine if this region is driving the expression of Myr-NHERF1 to the apical PM, we reconstituted the cells with a Myr-PDZ1-2 construct containing the N-terminal PDZ domains but lacking the ERM-binding region of NHERF1 (Figure 5D, cartoon). This form localized at the lateral cell-cell junctions (Figure 5D, right panels), suggesting that the ERM-binding region is the signal that targets NHERF1 to the apical PM. Interestingly, the morphology of reconstituted Myr-PDZ1-2 cells was intermediate between the elongated mesenchymal appearance of control cells and the polygonal-epithelial shape of Myr-NHERF1 cells, indicating that Myr-PDZ1-2 restored only partially the epithelial phenotype of reconstituted cells (Figure 5E). These findings underscore the importance of NHERF1 apical PM localization in maintaining epithelial morphology.
Discussion
Studies of human CRC have suggested that the accumulation of specific alterations in cell growth-regulating genes triggers the stage-wise progression to malignancy [26]. However, the complexity of the mutational signatures that accompany CRC poses the problem of which ones are relevant for progression. In this respect, a recent study landscaping the genetic changes in CRC demonstrated that individual CRC tumors accumulate approximately 90 mutant genes, most of which are not known to contribute to tumorigenesis [27]. We describe here NHERF1 as a new marker of CRC progression that undergoes progressive alterations of expression and subcellular localization during the CRC adenoma-carcinoma sequence. The normal apical PM expression of NHERF1 was lost in adenomas and replaced by cytoplasmic overexpresssion in approximately half of carcinomas, and these changes modeled in polarized epithelial cell resulted in a transformed cell phenotype (Figure 6).
We used intestinal Caco-2 cells to examine the function of NHERF1 in CRC. Caco-2 are unusual for CRC-derived cells in that, despite the presence of Apc and other genetic mutations [24], they differentiate to quasi-normal polarized monolayers on reaching confluency in culture [23]. Importantly, endogenous NHERF1 is expressed in the microvilli at the apical membrane similar to normal intestinal cells, thus providing an in vitro cell system for the study of the stage-wise progression of CRC. Loss of NHERF1, as observed in adenoma, induced prominent epithelial morphology alterations manifested as typical EMT-like changes. Previous studies using NHERF1 knockdown in undifferentiated mouse embryonal carcinoma or syncytiotrophoblast cell lines have shown a requirement for NHERF1 only in microvilli morphogenesis [28,29]. It seems that NHERF1 depletion triggers much more profound morphologic changes in intestinal epithelial cells, thereby implicating NHERF1 more broadly in epithelial morphogenesis.
Surprisingly, the morphologic changes induced by NHERF1 depletion in polarized epithelial cells were not reversed by expression of wild-type NHERF1. In this instance, the expression of wild-type NHERF1 after previous depletion of endogenous NHERF1 resulted in cytoplasmic and nuclear localization of the protein and a more aggressive phenotype than the loss of NHERF1 alone. NHERF1 has been reported to interact with β-catenin and to increase β-catenin transcriptional activation in reporter assays [11]. In NHERF1-depleted cells, we could already observe a shift of β-catenin to the nucleus that most likely contributed to EMT and the increased invasiveness of cells. However, an extra layer of Wnt/β-catenin activation may be added after expression of aberrantly localized NHERF1, perhaps by direct interactions with oncogenic β-catenin, and this could explain the increased proliferation and lack of EMT rescue (Figure 6). In addition, disruption of the PM localization of NHERF1 could destabilize PM complexes with other NHERF1 ligands and lead to structural abnormalities and increased cell invasiveness and proliferation [30]. For example, NHERF1 interacts with the ERM proteins [12], and we have shown that the presence of NHERF1 at the apical PM is required for the proper brush border organization and the localization of the ERM proteins in the same compartment in intestinal epithelial cells in vivo [16]. Another member of the ERM family, the neurofibromatosis type 2 gene product merlin, interacts with NHERF1 in actin-rich membrane structures, such as ruffles, microvilli, and filopodia [31], and merlin displacement from PM has been linked to inactivation of its tumor suppressor function [21,32]. Of note is that NHERF1 also interacts with platelet-derived growth factor receptor and epidermal growth factor receptor [33,34], and its removal from the PM enhances the platelet-derived growth factor receptor-dependent signaling and cell motility [18].
Interestingly, the expression of membrane-targeted Myr-NHERF1 did rescue the EMT induced after depletion of endogenous NHERF1. Moreover, Myr-NHERF1 localized specifically to the apical PM dependent on the presence of the ERM-binding region in NHERF1. We and others have found that in polarized opossum kidney cells exogenous NHERF1 localizes at the apical microvilli, and disruption of the ERM-binding region either by truncation or mutation redirects it to the cytoplasm [25,35]. Along with our findings in this study, these observations suggest that apically localized NHERF1 is required to maintain epithelial morphology but that it is the interaction with ERM proteins that properly localizes NHERF1 at the apical PM.
In conclusion, this study shows that NHERF1 alterations correlate with the progression and enhanced invasiveness of human CRC and implicates NHERF1 as an important regulator of epithelial morphology. Further studies are necessary to elucidate how the NHERF1 loss from the PM and the cytoplasmic re-expression in CRC occur because these seem to be sequential events correlated with the CRC pathological progression. Whereas we are the first to observe alterations of NHERF1 subcellular expression in CRC and also to model the disorganization of the epithelial phenotype by which these alterations lead to progressive transformation, several similar observations have been reported in breast cancer [36–38] and hepatocellular carcinoma [11]. Thus, it seems that the NHERF1 loss or cytoplasmic overexpression is a more general oncogenic event in carcinomas. In this light, efforts to restore NHERF1 to its correct PM location deserve thorough consideration as alternative strategies for cancer treatment.
Supplementary Material
Acknowledgments
The authors thank Alan Hall for insightful discussions, Henry Adams for help with immunofluorescence analysis, and Kerry Siegler and Toni Castaneda for processing the resection specimens.
Abbreviations
- CRC
colorectal cancer
- EMT
epithelial-mesenchymal transition
- ERM
ezrin-radixin-moesin
- IF
immunofluorescence
- MMP
matrix metalloproteinase
- NHERF1
Na+/H+ exchanger 3 regulating factor 1
- PDZ
PSD-95/Disc-large/ZO-1
- PM
plasma membrane
Footnotes
This work was supported by NCI-CA107201 and G.S. Hogan Gastrointestinal Research Fund at MD Anderson Cancer Center to M.-M.G. The sequencing was partly supported by NCI-CA16672.
This article refers to supplementary material, which is designated by Figure W1 and is available online at www.neoplasia.com.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 3.Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am J Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, β-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 5.Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze'ev A. Autoregulation of E-cadherin expression by cadherincadherin interactions: the roles of β-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem. 2005;280:11740–11748. doi: 10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- 7.Roy HK, Smyrk TC, Koetsier J, Victor TA, Wali RK. The transcriptional repressor SNAIL is overexpressed in human colon cancer. Dig Dis Sci. 2005;50:42–46. doi: 10.1007/s10620-005-1275-z. [DOI] [PubMed] [Google Scholar]
- 8.Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 10.Huang D, Du X. Crosstalk between tumor cells and microenvironment via Wnt pathway in colorectal cancer dissemination. World J Gastroenterol. 2008;14:1823–1827. doi: 10.3748/wjg.14.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata T, Chuma M, Kokubu A, Sakamoto M, Hirohashi S. EBP50, a β-catenin-associating protein, enhances Wnt signaling and is overexpressed in hepatocellular carcinoma. Hepatology. 2003;38:178–186. doi: 10.1053/jhep.2003.50270. [DOI] [PubMed] [Google Scholar]
- 12.Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrinradixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinman EJ, Steplock D, Tate K, Hall RA, Spurney RF, Shenolikar S. Structure-function of recombinant Na/H exchanger regulatory factor (NHE-RF) J Clin Invest. 1998;101:2199–2206. doi: 10.1172/JCI204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stemmer-Rachamimov AO, Wiederhold T, Nielsen GP, James M, Pinney-Michalowski D, Roy JE, Cohen WA, Ramesh V, Louis DN. NHE-RF, a merlin-interacting protein, is primarily expressed in luminal epithelia, proliferative endometrium, and estrogen receptor-positive breast carcinomas. Am J Pathol. 2001;158:57–62. doi: 10.1016/S0002-9440(10)63944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broere N, Chen M, Cinar A, Singh AK, Hillesheim J, Riederer B, Lunnemann M, Rottinghaus I, Krabbenhoft A, Engelhardt R, et al. Defective jejunal and colonic salt absorption and altered Na(+)/H (+) exchanger 3 (NHE3) activity in NHE regulatory factor 1 (NHERF1) adaptor protein-deficient mice. Pflugers Arch. 2009;457:1079–1091. doi: 10.1007/s00424-008-0579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morales FC, Takahashi Y, Kreimann EL, Georgescu MM. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc Natl Acad Sci USA. 2004;101:17705–17710. doi: 10.1073/pnas.0407974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreimann EL, Morales FC, de Orbeta-Cruz J, Takahashi Y, Adams H, Liu TJ, McCrea PD, Georgescu MM. Cortical stabilization of β-catenin contributes to NHERF1/EBP50 tumor suppressor function. Oncogene. 2007;26:5290–5299. doi: 10.1038/sj.onc.1210336. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi Y, Morales FC, Kreimann EL, Georgescu MM. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 2006;25:910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molina JR, Morales FC, Hayashi Y, Aldape K, Georgescu MM. Loss of PTEN binding adapter protein NHERF1 from plasma membrane in glioblastoma contributes to PTEN inactivation. Cancer Res. 2010;70:6697–6703. doi: 10.1158/0008-5472.CAN-10-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Natl Acad Sci USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morales FC, Molina JR, Hayashi Y, Georgescu MM. Overexpresssion of ezrin inactivates NF2 tumor suppressor in glioblastoma. Neuro Oncol. 2010;12:528–539. doi: 10.1093/neuonc/nop060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saotome I, Curto M, McClatchey AI. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev Cell. 2004;6:855–864. doi: 10.1016/j.devcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Simon-Assmann P, Turck N, Sidhoum-Jenny M, Gradwohl G, Kedinger M. In vitro models of intestinal epithelial cell differentiation. Cell Biol Toxicol. 2007;23:241–256. doi: 10.1007/s10565-006-0175-0. [DOI] [PubMed] [Google Scholar]
- 24.Gayet J, Zhou XP, Duval A, Rolland S, Hoang JM, Cottu P, Hamelin R. Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines. Oncogene. 2001;20:5025–5032. doi: 10.1038/sj.onc.1204611. [DOI] [PubMed] [Google Scholar]
- 25.Morales FC, Takahashi Y, Momin S, Adams H, Chen X, Georgescu MM. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol Cell Biol. 2007;27:2527–2537. doi: 10.1128/MCB.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [see comment] [DOI] [PubMed] [Google Scholar]
- 27.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [see comment] [DOI] [PubMed] [Google Scholar]
- 28.Chiba H, Sakai N, Murata M, Osanai M, Ninomiya T, Kojima T, Sawada N. The nuclear receptor hepatocyte nuclear factor 4alpha acts as a morphogen to induce the formation of microvilli. J Cell Biol. 2006;175:971–980. doi: 10.1083/jcb.200608012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A. EPI64 regulates microvillar subdomains and structure. J Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgescu MM, Morales FC, Molina JR, Hayashi Y. Roles of NHERF1/EBP50 in cancer. Curr Mol Med. 2008;8:459–468. doi: 10.2174/156652408785748031. [DOI] [PubMed] [Google Scholar]
- 31.Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, Ramesh V. NHE-RF, a regulatory cofactor for Na(+)-H+ exchange, is a common interactor for merlin and ERM (MERM) proteins. J Biol Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- 32.Curto M, McClatchey AI. Nf2/Merlin: a coordinator of receptor signalling and intercellular contact. Br J Cancer. 2008;98:256–262. doi: 10.1038/sj.bjc.6604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazar CS, Cresson CM, Lauffenburger DA, Gill GN. The Na+/H+ exchanger regulatory factor stabilizes epidermal growth factor receptors at the cell surface. Mol Biol Cell. 2004;15:5470–5480. doi: 10.1091/mbc.E04-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maudsley S, Zamah AM, Rahman N, Blitzer JT, Luttrell LM, Lefkowitz RJ, Hall RA. Platelet-derived growth factor receptor association with Na(+)/H(+) exchanger regulatory factor potentiates receptor activity. Mol Cell Biol. 2000;20:8352–8363. doi: 10.1128/mcb.20.22.8352-8363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernando N, Deliot N, Gisler SM, Lederer E, Weinman EJ, Biber J, Murer H. PDZ-domain interactions and apical expression of type IIa Na/P(i) cotransporters. Proc Natl Acad Sci USA. 2002;99:11957–11962. doi: 10.1073/pnas.182412699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardone RA, Bellizzi A, Busco G, Weinman EJ, Dell'Aquila ME, Casavola V, Azzariti A, Mangia A, Paradiso A, Reshkin SJ. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol Biol Cell. 2007;18:1768–1780. doi: 10.1091/mbc.E06-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangia A, Chiriatti A, Bellizzi A, Malfettone A, Stea B, Zito FA, Reshkin SJ, Simone G, Paradiso A. Biological role of NHERF1 protein expression in breast cancer. Histopathology. 2009;55:600–608. doi: 10.1111/j.1365-2559.2009.03424.x. [DOI] [PubMed] [Google Scholar]
- 38.Song J, Bai J, Yang W, Gabrielson EW, Chan DW, Zhang Z. Expression and clinicopathological significance of oestrogen-responsive ezrin-radixin-moesin-binding phosphoprotein 50 in breast cancer. Histopathology. 2007;51:40–53. doi: 10.1111/j.1365-2559.2007.02730.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.