Abstract
The messenger RNAs containing the thirteen protein coding sequences of the human mitochondrial genome have frequently been regarded as a single functional category, alike in arrangement and hence in mode of expression. The “generic” mitochondrial mRNA is perceived as having (i) an arrangement within the polycistronic unit that permits its liberation following mt-tRNA processing, (ii) no 5′ cap structure or introns, (iii) essentially no untranslated regions, and (iv) a poly(A) tail of approximately fifty nucleotides that is required in part to complete the termination codon. Closer inspection reveals that only two molecules fit this pattern. This article examines the extent to which human mitochondrial mRNA species differ from one another.
Abbreviations: mt-mRNA, mitochondrial messenger RNA; mtDNA, mitochondrial genome; ORF, open reading frame; UTR, untranslated region; nt, nucleotide
Keywords: Mitochondria, RNA, Poly(A), Processing, Transcript, Translation, Termination
1. Introduction
Evolution from being an intracellular parasite to an organelle has resulted in a reduced human mitochondrial genome that now encodes a mere 37 genes; 13 of these encode proteins, these are in addition to the complete set of 22 mitochondrial (mt-)tRNAs and the 2 mitochondrial (mt-)rRNAs that are required for their expression [1]. All 13 of these mitochondrially expressed proteins are indispensable polypeptides of the inner membrane multi-subunit complexes I, III, IV and V that couple oxidative phosphorylation. The genes that encode the remainder (greater than 99%) of the mitochondrial proteome are encoded by the nucleus.
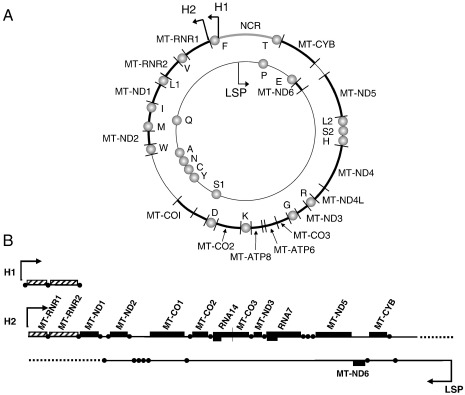
Great progress has been made over the past 15°years in characterising the molecular mechanisms and key players underlying mammalian mitochondrial transcription (detailed in [2]). This article, however, is more concerned with the mitochondrial (mt)-mRNAs themselves and exploring cis-acting elements in these RNA units. All mt-mRNAs are produced from large polycistronic RNA. To generate these polycistronic molecules, the genome (mtDNA, Fig. 1A) is transcribed from both strands in almost its entirety. Historically, the two strands of mtDNA have been referred to as the heavy (H-) and light (L-) strands dependent on their buoyancy in CsCl density gradients. Thus, transcription from the H-strand results in an RNA unit that is processed into 2 mt-rRNAs, 14 mt-tRNAs and mt-mRNAs that encode 12 proteins. Conversely, transcription of the L-strand produces RNA that is cleaved to produce 8 mt-tRNAs and only 1 mt-mRNA that encodes a single protein. Promoters that are found in the mtDNA non-coding region recruit mitochondrial RNA polymerase to drive this transcription, leading to the production of three polycistronic transcription units; two originating from the heavy strand promoter that initiate at independent sites (H1 and H2) and one from the light strand promoter (LSP) (Fig. 1B). Polycistronic RNA initiating from H1 starts upstream of MT-TF, the gene encoding mt-tRNAPhe and has a defined termination site within MT-TL1 (mt-tRNALeu(UUR)). The longer transcription unit initiates at H2, close to the start of MT-RNR1 (gene encoding the 12 S mt-tRNA) and extends throughout the entire downstream coding region of the genome. In exponentially dividing HeLa cells, initiation at H1 is favoured fifty to hundred times over initiation at H2 [3,4]. Termination of the shorter transcript is believed to be mediated by binding of mTERF, the first identified member of the mTERF protein family [5–7]. Transcription driven from LSP generates the second long polycistronic unit [8]. The termination site for neither of the long polycistronic RNA units has been precisely mapped ([2], Fig. 1B). Further, these two long polycistronic transcripts are generally not observed as full length units under experimental conditions, indicating that transcript cleavage occurs concurrently or rapidly after synthesis [9]. Cleavage is not 100% efficient at all sites and unprocessed forms can be found in wild type cells (see below). The efficiency of cleavage may be affected by tissue type or by mutations that cause sequence alterations [10]. Additional RNA species that are rarely observed at steady state by northern analysis are the anti-sense transcripts [11] and RNA corresponding to the entire noncoding region [12].
Fig. 1.
Map of human mitochondrial DNA and the RNA transcription units that are generated. (A) The human mitochondrial genome encodes 37 genes. Circles represent the 22 genes encoding mt-tRNAs depicted by their single letter code; the 13 genes encoding mt-mRNA and 2 mt-rRNA indicated using the nomenclature given on Mitomap (http://www.mitomap.org). The initiating/promoter positions of the polycistronic transcription units H1, H2 and LSP within the non-coding region (NCR) are indicated. (B) Transcription map of the human mitochondrial genome. The three polycistronic RNA units generated by transcription initiating at H1, H2 or produced from the Light Strand Promoter (LSP), are represented here in a linear fashion. The shortest of these commences from H1, with well-defined start and stop sites initiating upstream of MT-TF and terminating part way through MT-TL1[2]. The long transcription unit from H2 incorporates MTRNR1 and extends through the genome, terminating downstream of MT-TT. Transcription from LSP generates the second of the long unit that also extends throughout the genome, terminating downstream of MT-TP, however there is no consensus on the final termination site for either of the long polycistronic units [2], which are represented by dashed lines. RNA sequences are represented as follows; mt-rRNAs as hashed boxes, mt-mRNAs as black boxes and mt-tRNAs as circles. Nomenclature of each mt-mRNA or mt-rRNA corresponds to the gene nomenclature as described above, except for the two bicistronic RNA units RNA7 and RNA14 that encodes the proteins ND4L/ND4 and ATPase8/6, respectively.
As mentioned above, the general view of a typical human mt-mRNA is that of a molecule that is liberated by the processing of upstream and downstream mt-tRNAs, lacks both 3′ and 5′ untranslated regions (UTRs), contains no introns, lacks any cap or modification at the 5′ end and is polyadenylated to approximately fifty residues at the 3′ terminus to allow completion of the stop codon. In fact only two of the human mitochondrial transcripts resemble this description, MT-ND2 and MT-ND3. Although mt-mRNAs as a group do share particular features that mark them as different from cytosolic mRNAs, they also exhibit significant variability as a class. This article outlines the principal characteristics of human mitochondrial mRNAs; their similarities and how they differ.
2. Not all mt-mRNAs are produced by processing of mt-tRNAs from a polycistronic RNA unit
Traditionally, mt-tRNA folding and cleavage is described as the nucleation event that releases the mt-mRNAs from the long polycistronic transcripts [9]. A minimum of forty-three cleavage events would be required to liberate all of the mt-mRNAs, mt-tRNAs and mt-rRNAs, however, not all of these cleavages can be accounted for by mt-tRNA processing. No sequence similarity is evident in these cut sites that might direct recognition by processing or maturation machinery [1]. Various observations of transcription units, including those incorporating the open reading frame (ORF) encoding ND6 [13–16], imply that a number of cleavage events routinely take place that would not be accounted for by the mt-tRNA punctuation model, originally proposed by Ojala et al. [9]. Irrespective, this model does still satisfy the majority of processing events. The trans-acting factor that is responsible for the 5′ processing had long been a source of debate. After many years of controversy, however, Rossmanith and colleagues have now fully characterised mitochondrial RNase P, its protein-only components and its role in mt-tRNA processing [17], although the possibility of a second, RNA containing, mitochondrial RNase P with differing substrate specificity has not been ruled out. An RNase Z, presumably ELAC2 (RNZ2), is proposed to resolve the 3′ end of the mt-tRNA [18], although this activity has yet to be formally attributed to ELAC2 in human mitochondria.
Mitochondrial mRNA transcription units were first identified by the affinity of their poly(A) tails to oligo dT columns and were numbered 1 to 18 by size [9,19]. Since then the nomenclature has become somewhat varied so this report will use the current system favoured by MitoMap (http://www.mitomap.org). The exception to this is the terminology for the 2 transcription units that remain as bicistronic elements containing overlapping open reading frames encoding ATPase8 and ATPase6, or ND4L and ND4. The original names given to these bicistronic species were RNA14 and RNA7, respectively and have not been superseded. Although several of the original species 1–18 were identified as unprocessed molecules, it remains uncertain whether some of these species represent true precursors or products of imperfect cleavage. Such species include those of various lengths that contain the open reading frame for ND6 or MT-CO1+OriL but have very low steady state levels and so although present are rarely detected by standard methods [11]. It should, however, be noted that there are several persistent and uncleaved intermediates that are readily observable at steady state by simple northern analysis. Two common examples are the bicistronic RNA14, which is followed immediately by MT-CO3 with no intervening tRNA. The second, RNA19, contains uncleaved 16 S mt-rRNA, mt-tRNALeu(UUR) and MT-ND1.
3. How similar is the polyadenylation status of mt-mRNA species?
Upon processing, there is rapid constitutive polyadenylation of the 3′ terminus of mt-mRNAs [9,18]. Indeed, the first detectable mitochondrial transcripts following release from transcription inhibition all carry poly(A) 3′ extensions [20]. This also suggests that polyadenylation is a single processive event.
This maturation event captures the site of cleavage, which can be revealed by RT-PCR, cloning and sequencing. From these data, it is apparent that processing of several RNA species in cultured cells is reproducibly error prone. For example, in 20% of clones containing MT-CO2 and 5% of MT-ND3, the terminal mtDNA-encoded residue(s) were missing ([21] and RJT, unpublished observation). Circularisation assays were used to analyse mt-tRNAArg, which is immediately downstream of MT-ND3. This revealed additional residues that correspond to upstream nucleotides that are lost from the 3′ terminus of MT-ND3. The loss of the terminal 3′ residues of these mt-mRNAs, however, does not prevent normal polyadenylation. Further, when mt-mRNAs are endonucleolytically cleaved in vivo, the truncated species are immediately readenylated [22]. This is also consistent with a previous observation in a fibroblast cell line derived from a patient with a microdeletion at the 3′ terminus of RNA14 [20]. Although shortened on translation, the transcripts lacking termination codons were initially matured with poly(A) tails, making them indistinguishable from those seen in control cells.
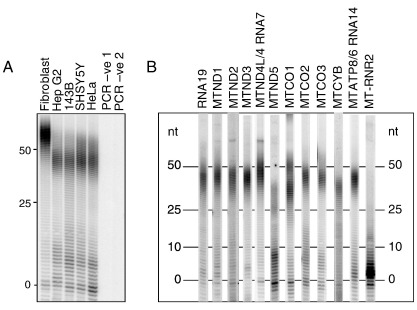
Mitochondrial poly(A) tail (MPAT) assays, as described in Ref. [20], have been devised for all human mitochondrial mRNAs with the exception of MT-ND6, for which a single consistent 3′ end has not be identified [16]. For the majority of the remaining mt-mRNA species there is a single major population that spans approximately 10 residues, with a median of around 45 nt (Table 1). There is however, a slight variation between cell types (Fig. 2A) as well as between transcripts within a cell type (Fig. 2B). On closer examination it is evident that poly(A) tail lengths of specific transcripts are not consistent between cell types. Moreover, certain mt-mRNAs bear 2 relatively abundant longer populations as seen on MT-CO1, which has medians of approximately 37 and 52 nt. A less dramatic example, MT-CO3, has median lengths of 42 and 57 nt (Fig. 2A). In Hep G2 and 143B cells the shorter tail dominates in contrast to fibroblasts where it is the longer form that is in the greatest abundance; species spanning both populations are visible in neuroblastoma (SHSY5Y) and HeLa cells. When such a bipartite distribution exists, this size difference of 15–17 nucleotides between the two populations is also not conserved. Perhaps the most strikingly divergent from the other human mt-mRNAs is that of MT-ND5. Although there are longer species averaging ∼ 25 or 35 nt, the major population is oligo- rather than polyadenylated, with an extension of between 0 and 10 nt in length, zero being the most frequently occurring (Fig. 2B). This is in line with the observations of Slomovic et al. [16] where they detect relatively little polyadenylation at this site. Although there is a broadly similar profile for MT-ND5 in HeLa, 143B and neuroblastoma cells, in HEK293 cells there is very little evidence of oligo-adenylated species and the dominant population has a median of 40 nt. Thus highlighting that there are differences in the length of poly(A) extensions both between transcripts within a cell type and also for the same transcript between cell types.
Table 1.
Features of human mitochondrial transcripts.
| Protein | 5′ Leader (nt) | Open reading framea | 3′ Trailer (nt) | Initiation codon | Termination codon | “A” additions required for stop codon | Poly(A) tail lengthd |
|---|---|---|---|---|---|---|---|
| ND5 | 0 | 1811 | 568 | AUA | UAA | 0 | 0/8 |
| COX1 | 3 | 1541 | 72 | AUG | UAGc | 0 | 37 |
| ND4 | 296 | 1377 | 0 | AUG | UAA | 2 | 48 |
| CYTB | 0 | 1140 | 0 | AUG | UAA | 2 | 40 |
| ND2 | 0 | 1041 | 0 | AUU | UAA | 2 | 43 |
| ND1 | 2 | 955 | 0 | AUA | UAA | 1 | 45 |
| COX3 | 0 | 783 | 0 | AUG | UAA | 2 | 43 |
| COX2 | 0 | 708 | 24 (15)b | AUG | UAG | 0 | 45 |
| ATP6 | 161 | 679 | 0 | AUG | UAA | 1 | 45 |
| ND6 | 0 | 524 | ?e | AUG | UAGc | 0 | - |
| ND3 | 0 | 345 | 0 | AUA | UAA | 2 | 44 |
| ND4L | 0 | 296 | 1371 | AUG | UAA | 0 | 48 |
| ATP8 | 1 | 206 | 634 | AUG | UAG | 0 | 45 |
Protein coding sequences are ranked by length of open reading frame. Sequences taken from Ref. [1].
The 3′ UTR of MTCO2 contains a deletion of 9 residues in HepG2 cells.
As re-assigned in Temperley et al. [18].
Poly(A) tail lengths were determined by MPAT in Hep G2 cells.
There is no current consensus on the defined end of transcript MTND6[16].
Fig. 2.
Poly(A) profiles vary between cultured cell types and between transcripts. Panel A shows an MPAT assay for MT-CO3 transcripts in a variety of cell types including primary fibroblasts, Hep G2, 143B osteosarcoma, neuroblastoma (SHSY5Y) and HeLa. Negative controls for the first and second PCR steps are always performed. Panel B indicates the variation in poly(A) tail length between different human mt-transcripts at steady state in Hep G2 cells. Transcripts are aligned at the zero 3′ extension (0); nucleotide positions of 0, 10, 25 and 50 3′ extensions are indicated. The MPAT methodology was performed as described in Ref. [20].
The 3′ termini of the 12S and 16S mt-rRNA species are discernibly different from the mt-mRNAs. Sequence analysis of clones generated by ligation-mediated RT-PCR reveals that in the majority of cases there is no evidence for the addition of more than 3–4 nt to 12 S rRNA in Hep G2 cells or fibroblasts (RJT, unpublished observation). The 3′ cleavage site of 12 S mt-rRNA is as predicted by the mtDNA sequence, although marginally truncated forms have been observed. Unexpectedly, the majority of 16 S clones carry residues from the downstream mt-tRNALeu(UUR), this is perhaps as a consequence of transcription termination at this point rather than mis-processing. Given reports by other authors, it seems likely that sub-populations of more extensively polyadenylated mt-rRNAs do or can exist but this does not appear to be the predominant form under normal growth conditions in cultured cells [16].
4. Do mt-mRNAs contain untranslated regions and do these have known functions?
Human mt-mRNAs are generally described as having essentially no untranslated regions (UTR). However, there are only 4 RNA species that have no flanking untranslated regions of any kind in humans (Table 1); MT-CO3, MT-CYB, MT-ND2, and MT-ND3. Thus, UTRs do exist on human mt-mRNAs but can be as short as 1, 2 or 3 nucleotides, as seen in the 5′UTRs of RNA14, MT-ND1 and MT-CO1 respectively. There are two 5′UTRs that are found preceding either the ORF encoding ND4 or the ORF for ATPase6. Both of these ORFs constitute the downstream part of bicistronic transcripts RNA14 and RNA7, respectively, and hence the 5′UTR is in fact the out of frame sequence encoding each of the upstream ORFs. In reciprocal fashion, part of the downstream ORF in each case constitutes the 3′UTR for these two bicistronic RNA units. MT-CO2 also possesses a 3′UTR but this is not fully conserved between species and is the only UTR longer than three residues to consist of sequence with no known additional purpose. The remaining 3′UTRs consist of either anti-sense of coding sequences (following MT-CO1, MT-ND6 or MT-ND5) or overlapping open reading frames (RNA7, RNA14 as described above). This is significant as it suggests that any elements required to stimulate cleavage, polyadenylation or translation would have to be contained within coding sequences (sense or anti-sense) and have to be reconciled with mt-tRNA structures or the amino acid content of proteins.
5. Codon usage to start and stop translation
In 8 of the 13 ORFs, the initiation codon starts at the very 5′ terminal nucleotide of the mt-mRNA, in 3 cases translation initiates a few (< 4) nucleotides down from the 5′ terminal nucleotide and as described above, the remaining 2 ORFs are embedded in the mature mt-mRNA as they constitute the downstream coding sequences of the bicistronic transcripts. While AUG is the most common initiation codon, occurring in nine instances, AUA and AUU are also used (Table 1). Hence, it appears likely that there must be recognisable and distinctive features that facilitate loading of the mitoribosomes to the correct start site for translation initiation. Spremulli and colleagues [23] used chemical characterisation of the 5′ sequences and identified a lack of secondary structure that could more easily permit mitochondrial ribosome loading. No sequences or structures in UTRs or ORFs have been identified as having a role in recruitment or assembly of mitochondrial ribosomes, however it is likely such elements must exist, given the variation in initiation codons and their positions. Such an association is suggested for TACO1, the first translational activator for a human mt-mRNA, where mutations in this protein cause specific reduction in the protein synthesis of COX1 [24].
There is no correlation between use of an unconventional initiation codon and possession of a 5′ UTR. In contrast, non-UAA termination codons are always followed by a 3′ UTR in human mt-mRNA. Until recently, no functional significance has been attached to this. In humans, UAA terminates nine open reading frames. Of the remaining four, two end in UAG. AGA/AGG were also designated as stops in the 1981 sequencing of the human mitochondrial genome [1]. Recent data have now shown that the AGA/AGG triplets are not classical stop codons that are recognised by the mitochondrial release factor mtRF1a, but instead contribute to a −1 frameshift. This repositioning of the mitochondrial ribosome on the mt-mRNA results in a UAG occupying the A-site, which then acts as a termination signal in these two cases [22]. For this −1 frameshift to transpire, the 3′UTR plays a crucial role. The downstream 3′UTR sequence forms a strong secondary structure that inhibits any potential readthrough. Codons AGA or AGG in the A site would not be recognised by the mitochondrial release factor or any mt-tRNA. The secondary structure generated within the 3′UTR immediately downstream of the AGA/G therefore acts in conjunction with the “unrecognised” AGA/G forcing the ribosome to reposition and allow the standard UAG stop codon to be recognised by the mitochondrial release factor, mtRF1a [22,25].
In summary, each mt-transcription unit must contain cis-acting elements that promote cleavage, allow subsequent maturation and for mt-mRNA, translation. The specific nature of the elements that recruit the required trans-acting factor(s) or indeed many of the trans-acting factors themselves have yet to be fully characterised. The assortment of initiation codons and variety of their locations within RNA molecules suggests a role for additional factors that can steer mitochondrial ribosomes to appropriate locations. Although not discussed above, elements necessary for maintenance and replication of mtDNA, or coupling transcription to translation may also be encrypted in the transcript [26]. The scarcity of genuinely untranslated regions in mt-mRNAs necessitates an overlayering of functions to a single sequence, which would enforce constraints on a genome generally perceived to be relatively permissive to mutation.
6. Concluding remarks
The apparent simplicity of the human mitochondrial genetic system may lead one to suspect that there are generic mechanisms of RNA cleavage, maturation, translation and degradation. At a mechanistic level this may be true, however, the cis-acting elements that direct and promote these processes appear to vary. The transcript descriptions given here are not comprehensive, but it is clear that there is a lack of homogeneity between human mt-mRNAs and an absence of obvious features carried by all species. Not even poly(A) tails can be shown to be present on all mRNAs; where present they vary substantially in length, for reasons that remain to be determined, and can also be found on molecules where stability or translation would not appear to be desirable. Recent evidence of polyuridylation of transcripts further complicates any simplistic interpretation [27]. Hence, it appears that human mitochondrial mRNAs, like members of all families, are similar but different.
Acknowledgments
RNL and ZMACL would like to thank The Wellcome Trust [074454/Z/04/Z] and the BBSRC [BB/F011520/1] for continuing support.
References
- 1.Anderson S., Bankier A.T., Barrell B.G., De Bruijn M.H.L., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., Schreier P.H., Smith A.J.H., Staden R., Y.I.J. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Falkenberg M., Larsson N.-G., Gustafsson C.M. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 3.Gelfand R., Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol. Cell. Biol. 1981;1:497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montoya J., Christianson T., Levens D., Rabinowitz M., Attardi G. Identification of initiation sites for heavy strand and light strand transcription in human mitochondrial DNA. Proc. Natl. Acad. Sci. U. S. A. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linder T., Park C.B., Asin-Cayuela J., Pellegrini M., Larsson N.G., Falkenberg M., Samuelsson T., Gustafsson C.M. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005;48:265–269. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- 6.Hyvärinen A.K., Pohjoismäki J.L., Reyes A., Wanrooij S., Yasukawa T., Karhunen P.J., Spelbrink J.N., Holt I.J., Jacobs H.T. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin M., Cho J., Cesare A.J., Griffith J.D., Attardi G. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 8.Ekstrand M.I., Falkenberg M., Rantanen A., Park C., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.G. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 9.Ojala D., Montoya J., Attardi G. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 10.Bindoff L.A., Howell N., Poulton J., McCullough D., Morten K.J., Lightowlers R.N., Turnbull D.M., Weber K. J. Biol. Chem. 1993;268:19559–19564. [PubMed] [Google Scholar]
- 11.Szczesny R.J., Borowski L.S., Brzezniak L.K., Dmochowska A., Gewartowski K., Bartnik E., Stepien P.P. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selwood S.P., McGregor A., Lightowlers R.N., Chrzanowska-Lightowlers Z.M. FEBS Lett. 2001;494:186–191. doi: 10.1016/s0014-5793(01)02345-6. [DOI] [PubMed] [Google Scholar]
- 13.Guan M.X., Enriquez J.A., Fischel-Ghodsian N., Puranam R.S., Lin C.P., Maw M.A., Attardi G. Mol. Cell. Biol. 1998;18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sbisa E., Nardelli M., Tanzariello F., Tullo A., Saccone C. Curr. Genet. 1990;17:247–253. doi: 10.1007/BF00312616. [DOI] [PubMed] [Google Scholar]
- 15.Sbisà E., Tullo A., Nardelli M., Tanzariello F., Saccone C. FEBS Lett. 1992;296:311–316. doi: 10.1016/0014-5793(92)80311-4. [DOI] [PubMed] [Google Scholar]
- 16.Slomovic S., Laufer D., Geiger D., Schuster G. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holzmann J., Frank P., Löffler E., Bennett K.L., Gerner C., Rossmanith W. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Dubrovsky E.B., Dubrovskaya V.A., Levinger L., Schiffer S., Marchfelder Nucleic A. Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoya J., Ojala D., Attardi G. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 20.Temperley R.J., Seneca S.H., Tonska K., Bartnik E., Bindoff L.A., Lightowlers R.N., Chrzanowska-Lightowlers Z.M. Hum. Mol. Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 21.M. Wydro, A. Bobrowicz, R.J. Temperley, R.N. Lightowlers, Z.M. Chrzanowska-Lightowlers. Nucleic Acids Res. (in press). doi:10.1093/nar/gkq068. [DOI] [PMC free article] [PubMed]
- 22.Temperley R., Richter R., Dennerlein S., Lightowlers R.N., Chrzanowska-Lightowlers Z.M.A. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- 23.Jones C.N., Wilkinson K.A., Hung K.T., Weeks K.M., Spremull L.L. RNA. 2008;14:862–871. doi: 10.1261/rna.909208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J.E., Lochmüller H., Chevrette M., Kaufman B.A., Horvath R., Shoubridge E.A. Nat. Genet. 2009;41:833–837. doi: 10.1038/ng.390. [DOI] [PubMed] [Google Scholar]
- 25.Soleimanpour-Lichaei H.R., Kuhl I., Gaisne M., Passos J.F., Wydro M., Rorbach J., Temperley R., Bonnefoy N., Tate W., Lightowlers R., Chrzanowska-Lightowlers Z. Mol. Cell. 2007;27:745–757. doi: 10.1016/j.molcel.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welter C., Dooley S., Zang K.D., Blin N. Nucleic Acids Res. 1989;17:6077–6086. doi: 10.1093/nar/17.15.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slomovic S., Schuster G. RNA. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]