Abstract
E7 is the major oncoprotein of high-risk human papillomaviruses (HPV) which causes cervical cancer. To date E7 oncoproteins have not been investigated in cervical adenocarcinoma. In this study we generated a rabbit monoclonal anti-HPV-16 E7 antibody, RabMab42-3, which recognizes a conformational epitope in the E7 carboxy-terminal zinc-finger resulting in a strong increase in the sensitivity for the detection of cell-associated HPV-16 E7 protein relative to conventional polyclonal anti-HPV-16 E7 antibodies. Using RabMab42-3, we show that the subcellular localization of endogenous HPV-16 E7 oncoprotein varies during the cell cycle in cervical cancer cells. Moreover, we demonstrate for the first time that the HPV-16 E7 oncoprotein is abundantly expressed in cervical adenocarcinoma in situ and adenocarcinoma, suggesting an important role of HPV-16 E7 for the development of these tumors. Our findings suggest that the HPV-16 E7 oncoprotein could be a useful marker for the detection of cervical adenocarcinoma and their precursors.
Keywords: Cervical cancer, Human papillomavirus, HPV-16 E7, Oncoprotein, Zinc-finger
Introduction
Persistent infections by human papillomaviruses (HPV) are the main etiologic factor for the development of cervical cancer, the second most frequent gynecological cancer worldwide (reviewed in zur Hausen, 2002). Approximately 85% of cervical cancers are squamous cell carcinomas (SCCs), whereas 15% are adenocarcinomas (ACs) (Vizcaino et al., 1998; Pirog et al., 2000; Munoz et al., 2003). About 40 HPV genotypes that can infect epithelial squamous and glandular cells in the cervical mucosa have been described (de Villiers et al., 2004). On the basis of epidemiological and biochemical data, these viruses are classified as high-risk HPVs associated with intraepithelial lesions with a high potential for progression to invasive carcinoma and low-risk HPVs associated with benign hyperplasia (zur Hausen, 1991; Muñoz et al., 1992, 2003; Bosch et al., 1995; Lacey et al., 2006). A subgroup of at least 15 high-risk HPVs is associated with cervical intraepithelial lesions with high potential for progression to carcinoma (Walboomers et al., 1999; Muñoz et al., 2003). Infections by high-risk genotypes are detected in virtually all cervical cancers (Bosch et al., 1995; Liaw et al., 1999; Wallin et al., 1999; Walboomers et al., 1999). HPV-16 is the predominant genotype worldwide in SCCs as well as in ACs with a prevalence of approximately 55% in SCCs and 50–70% in ACs (Pirog et al., 2000; Riethdorf et al., 2002; Muñoz et al., 2003).
Inactivation of cellular control functions in early cervical carcinogenesis permits deregulated transcription of the HPV genes E6 and E7, resulting in an overexpression of E6 and E7 (Dürst et al., 1985; Schwarz et al., 1985; Romanczuk and Howley, 1992; Stoler et al., 1992; Jeon and Lambert, 1995; Ziegert et al., 2003). This is consistent with the observed increase of the high-risk E7 oncoprotein levels during early steps of carcinogenesis in cells of the cervical squamous epithelium (Fiedler et al., 2004). Both oncoproteins cooperate to trigger reprogramming of cell proliferation, apoptosis, differentiation and metabolism and to induce epigenetic reorganization and genomic instability (reviewed in Zwerschke and Jansen-Dürr, 2000; McLaughlin-Drubin and Münger, 2009). Accordingly, E7, in cooperation with E6, can efficiently immortalize human keratinocytes (Münger et al., 1989; Hawley-Nelson et al., 1989; Hudson et al., 1990); and the consistent overexpression of these two oncogenes is required to induce and to maintain the transformed phenotype of cervical cancer cells (reviewed in Münger and Howley, 2002). E7 is the major HPV oncoprotein and its expression is sufficient to immortalize primary human epithelial cells at a low frequency (Halbert et al., 1991; Reznikoff et al., 1994; Wazer et al., 1995) and to induce cervical cancers in transgenic mice treated with low doses of estrogens (Riley et al., 2003). Work of the last two decades has demonstrated that E7 is an integral part of different cellular protein complexes in the cytoplasm as well as in the nucleus and that E7 has multiple biochemical functions in the deregulation of pathways necessary for the oncogenic potential of the virus (reviewed in Zwerschke and Jansen-Dürr, 2000; Narisawa-Saito and Kiyono, 2007; McLaughlin-Drubin and Münger, 2009; Ghittoni et al., 2010; Moody and Laimins, 2010; Pim and Banks, 2010).
The incidence of cervical ACs is increasing (Peters et al., 1986; Schwartz and Weiss, 1986; Chilvers et al., 1987; Eide, 1987; Zheng et al., 1996; Vizcaino et al., 1998; Altekruse et al., 2003; Wang et al., 2004; Bray et al., 2005). This disease is often at a more advanced stage when detected resulting in worse curability and survival (Shingleton et al., 1995). Moreover, adenocarcinoma in situ (ACIS) progress faster to cervical glandular carcinoma compared to high grade squamous intraepithelial lesions (Plaxe and Saltzstein, 1999), suggesting that improved cervical cancer screening procedures are particularly required for better detection of ACs and ACIS. Whereas high-risk HPV E7 proteins are regularly expressed in SCCs (Fiedler et al., 2004; Ressler et al., 2007), the levels of high-risk E7 oncoprotein in cervical ACs and their precursors are unknown. Therefore we set out to analyze the E7 oncoprotein levels in ACIS and AC and to evaluate whether high-risk HPV E7 oncoprotein is an adequate marker for the detection of cervical AC and ACIS, using a novel rabbit monoclonal anti-HPV-16 E7 antibody.
Results
Detection of the HPV-16 E7 oncoprotein in cervical cancer cells
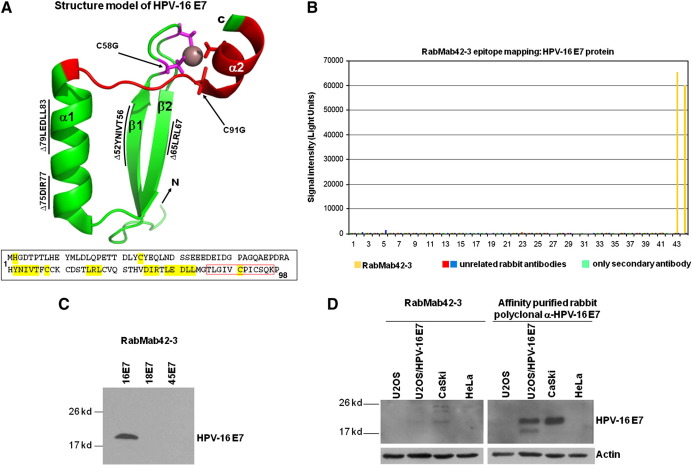
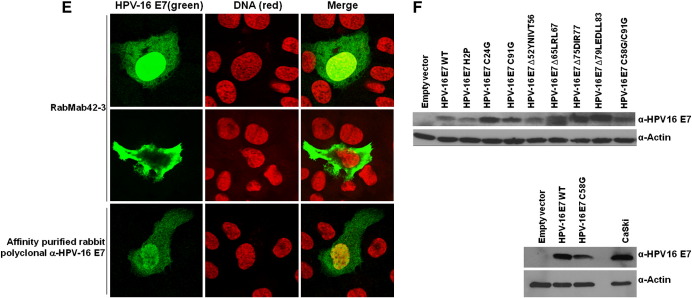
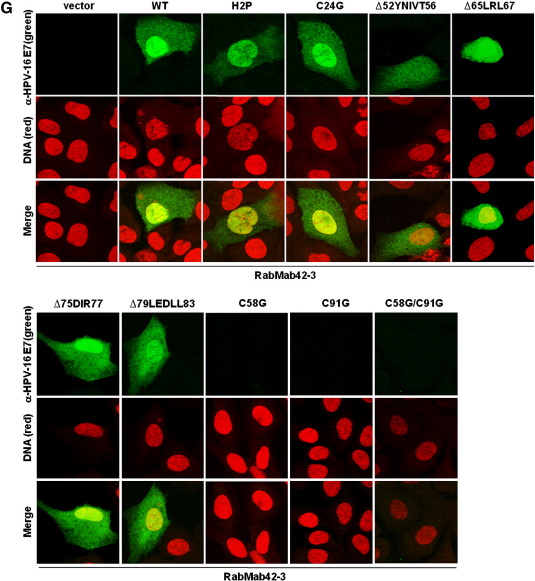
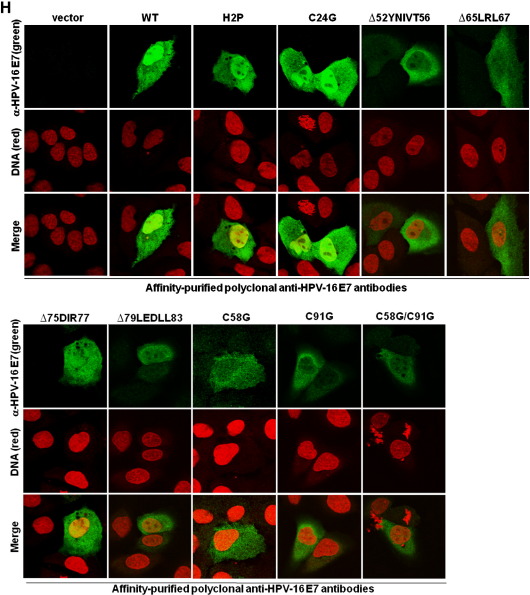
Structural analysis showed that the N-terminal part of E7 is intrinsically unstructured, whereas the C-terminal domain forms a highly structured zinc-finger loop (Liu et al., 2006; Ohlenschläger et al., 2006; Fig. 1A). We immunized rabbits with purified native HPV-16 E7 protein (Fiedler et al., 2006) and screened the derived hybridomas for suitable antibodies. Epitope mapping using microarrays with HPV-16 E7 derived synthetic peptides (Fig. 1B and Table 1) identified one rabbit monoclonal anti-HPV-16 E7 antibody, RabMab42-3, which detected a specific epitope (86TLGIVCPICSQK97) in the carboxyl-terminal domain of E7 containing two of the cysteine residues known as key structuring elements of the E7 zinc-finger (Fig. 1A). Although conformation-specific antibodies as a rule hardly work in peptide microarray-based epitope mapping assays (Harlow and Lane, 1998), data presented here suggests that the RabMab42-3-epitope is most likely present in a rigid—the zinc ion comprising—structure in the native protein. In keeping with our assignment as a conformation-specific antibody, RabMab42-3 had relatively low affinity to HPV-16 E7 in Western blot experiments where the protein was denatured by SDS. Although RabMab42-3, when used in concentrations of 15 μg/ml for Western blotting, recognized 10 ng of purified HPV-16 E7 protein (Fig. 1C) as well as HPV-16 E7 protein in lysates of HPV-16 E7 expressing cells (Fig. 1D), our affinity-purified rabbit polyclonal anti-HPV-16 E7 antibodies (Fiedler et al., 2004) recognized the denatured HPV-16 E7 protein much better even when used at much (> 25fold) lower concentration (Fig. 1D). In contrast, RabMab42-3 had a high affinity to native cell-associated HPV-16 E7 protein upon detection by indirect immunofluorescence. U-2OS cells transiently expressing HPV-16 E7 were stained with RabMab42-3 (Fig. 1E, upper and middle panel), and viewed using a confocal laser-scanning microscope. In the vast majority of the transiently transfected cells both cytoplasmic and nuclear HPV-16 E7 staining was detected (Fig. 1E, upper panel), while in a small number of the transiently transfected cells E7 was predominantly localized in the cytoplasm (Fig. 1E, middle panel). The signal generated by the affinity-purified rabbit polyclonal anti-HPV-16 E7 antibodies, when used in the same concentration, was less intense (Fig. 1E). To further investigate the specificity of RabMab42-3, U-2 OS cells were transiently transfected with expression vectors for HPV-16 E7 mutants addressing the unstructured N-terminus as well as the major structure determining elements in the C-terminal domain (Fig. 1A). Western blot analysis using polyclonal antibodies showed that the protein levels of all E7 mutants were either similar or higher as the level of the HPV-16 E7 wild type protein (Fig. 1F). In the immunofluorescence assays RabMab42-3 detected the two N-terminal E7 mutants, H2P and C24G, as well as carboxyl-terminal mutants containing deletions in β1-sheet (∆52YNIVT56), β2-sheet (∆65LRL67), and helix α1 (∆75DIR77 and ∆79LEDLL83) (Fig. 1G). However, RabMab42-3 could neither detect HPV-16 E7 C91G nor C58G which both contain single point mutations in the zinc coordination site. As expected from these results, the zinc-finger double cysteine mutant HPV-16 E7 C58G/C91G was also not detectable by RabMab42-3. This mutational analysis indicates that the intact zinc-finger structure, which is coordinated by the two CXXC motifs and the zinc ion (Liu et al., 2006; Ohlenschläger et al., 2006; see also above Fig. 1A), is necessary for an efficient recognition of the HPV-16 E7 protein by RabMab42-3. In contrast, the affinity-purified rabbit polyclonal anti-HPV-16 E7 antibodies could readily detect all HPV-16 E7 mutants, including the CXXC mutants, in U-2 OS cells by immunofluorescence analysis (Fig. 1H). Thus our mutational analysis indicates that RabMab42-3 specifically recognizes the epitope 86TLGIVCPICSQK97 and suggests that the binding of the monoclonal antibody depends on the intact conformation of the HPV-16 E7 zinc-finger.
Fig. 1.
RabMab42-3 detects a conformational epitope in the HPV-16 E7 protein. (A) Model of the structure of the HPV-16 E7 protein deduced from the NMR structure of HPV-45 E7 (Ohlenschläger et al., 2006) and the alignment of E7 sequences of HPV-45 and -16 (Morandell et al., 2008, see also Fig. 3B). (Upper panel) The carboxyl-terminal domains containing the β1β2α1α2 secondary structure are shown. The side chains of the four cysteines (C58, C61, C91, C94) in the two CXXC motifs coordinating the zinc ion (brown) located in the turn connecting β1 and β2 and in the C-terminal α2-helix are indicated. The epitope 86TLGIVCPICSQK97 in the carboxyl-terminal E7 zinc-finger recognized by RabMab42-3 is indicated in red. Moreover, the localization of the C-terminal mutations ∆52YNIVT56, ∆65LRL67, ∆75DIR77, ∆79LEDLL83, C58G and C91G are indicated. Note the unstructured N-terminus and the mutations H2P and C24G are not indicated. (Lower panel) Amino acid sequence of the HPV-16 E7 protein. The sequences selected for mutations are indicated in yellow and the epitope recognized by RabMab42-3 is framed by the red box. (B) RabMab42-3 epitope mapping: HPV-16 E7 peptides (see Table 1) were spotted on microarrays and incubated with RabMab42-3 and unrelated rabbit antibodies followed by a fluorescently labeled secondary antibody (anti-rabbit-Cy5). The results of the HPV 16 E7 antigen peptide arrays with RabMab42-3, two unrelated rabbit antibodies and “only secondary antibodies” are shown. (C) Ten nanograms of purified recombinant HPV-16 E7, HPV-18 E7 and HPV-45 E7 proteins were separated by SDS PAGE and the E7 proteins detected by Western blotting using 15 μg/ml RabMab42-3. (D) Lysates of U-2 OS cells transiently transfected with an expression vector for HPV-16 E7 or empty vector and the cervical cancer cell lines CaSki (HPV-16 positive) and HeLa (HPV-18 positive) were separated by SDS PAGE and blotted onto the same PVDF membrane. The HPV-16 E7 protein was detected by Western blotting using either 15 μg/ml RabMab42-3 (left panel) or 0.625 μg/ml affinity-purified polyclonal anti-HPV-16 E7 antibodies (right panel). (E) U-2 OS cells transiently transfected with an HPV-16 E7 expression vector were stained in indirect immunofluorescence experiments either with 200 pg/μl RabMab42-3 or 200 pg/μl affinity-purified polyclonal anti-HPV-16 E7 antibodies (positive control), as indicated, followed by application of AlexaFluor488-labeled secondary antibodies. (F) Lysates of U-2 OS cells transiently expressing HPV-16 E7 wild type and mutants as indicated were separated by SDS PAGE and the HPV-16 E7 protein was detected by Western blotting using 0.625 μg/ml affinity-purified polyclonal anti-HPV-16 E7 antibodies. (G) U-2 OS cells transiently expressing HPV-16 E7 wild type and mutants as indicated were stained in indirect immunofluorescence experiments with 2 ng/μl RabMab42-3 followed by application of AlexaFluor488-labeled secondary antibodies. (H) U-2 OS cells transiently expressing HPV-16 E7 wild type and mutants as indicated were stained in indirect immunofluorescence experiments with 2 ng/μl affinity-purified polyclonal anti-HPV-16 E7 antibodies followed by application of AlexaFluor488-labeled secondary antibodies.
Table 1.
Sequence of the HPV-16 E7 derived 13mer peptides used in the peptide microarrays.
| HPV-16 E7 protein (peptide number) | Amino acid position | Sequence |
|---|---|---|
| 1 | 1 | MHGDTPTLHEYML |
| 2 | 3 | GDTPTLHEYMLDL |
| 3 | 5 | TPTLHEYMLDLQP |
| 4 | 7 | TLHEYMLDLQPET |
| 5 | 9 | HEYMLDLQPETTD |
| 6 | 11 | YMLDLQPETTDLY |
| 7 | 13 | LDLQPETTDLYSY |
| 8 | 15 | LQPETTDLYSYEQ |
| 9 | 17 | PETTDLYSYEQLN |
| 10 | 19 | TTDLYSYEQLNDS |
| 11 | 21 | DLYSYEQLNDSSE |
| 12 | 23 | YSYEQLNDSSEEE |
| 13 | 25 | YEQLNDSSEEEDE |
| 14 | 27 | QLNDSSEEEDEID |
| 15 | 29 | NDSSEEEDEIDGP |
| 16 | 31 | SSEEEDEIDGPAG |
| 17 | 33 | EEEDEIDGPAGQA |
| 18 | 35 | EDEIDGPAGQAEP |
| 19 | 37 | EIDGPAGQAEPDR |
| 20 | 39 | DGPAGQAEPDRAH |
| 21 | 41 | PAGQAEPDRAHYN |
| 22 | 43 | GQAEPDRAHYNIV |
| 23 | 45 | AEPDRAHYNIVTF |
| 24 | 47 | PDRAHYNIVTFSS |
| 25 | 49 | RAHYNIVTFSSKS |
| 26 | 51 | HYNIVTFSSKSDS |
| 27 | 53 | NIVTFSSKSDSTL |
| 28 | 55 | VTFSSKSDSTLRL |
| 29 | 57 | FSSKSDSTLRLSV |
| 30 | 59 | SKSDSTLRLSVQS |
| 31 | 61 | SDSTLRLSVQSTH |
| 32 | 63 | STLRLSVQSTHVD |
| 33 | 65 | LRLSVQSTHVDIR |
| 34 | 67 | LSVQSTHVDIRTL |
| 35 | 69 | VQSTHVDIRTLED |
| 36 | 71 | STHVDIRTLEDLL |
| 37 | 73 | HVDIRTLEDLLMG |
| 38 | 75 | DIRTLEDLLMGTL |
| 39 | 77 | RTLEDLLMGTLGI |
| 40 | 79 | LEDLLMGTLGIVS |
| 41 | 81 | DLLMGTLGIVSPI |
| 42 | 83 | LMGTLGIVSPISS |
| 43 | 85 | GTLGIVSPISSQK |
| 44 | 86 | TLGIVSPISSQKP |
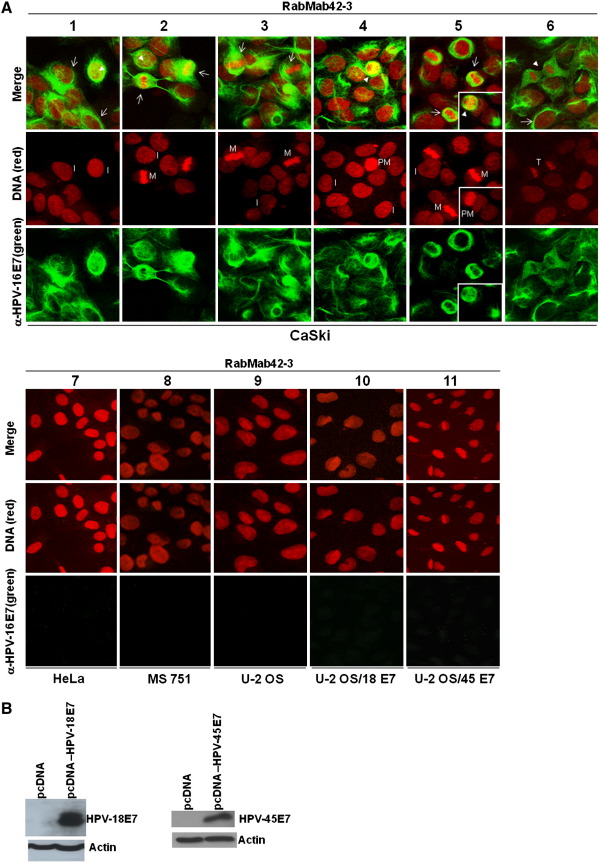
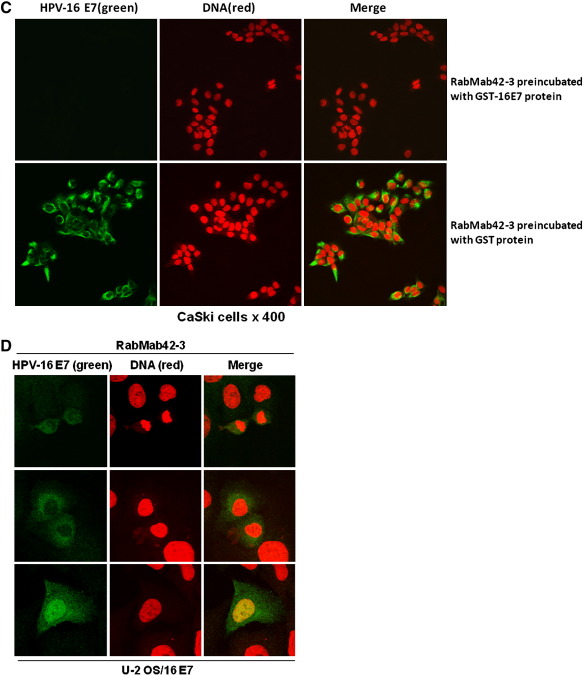
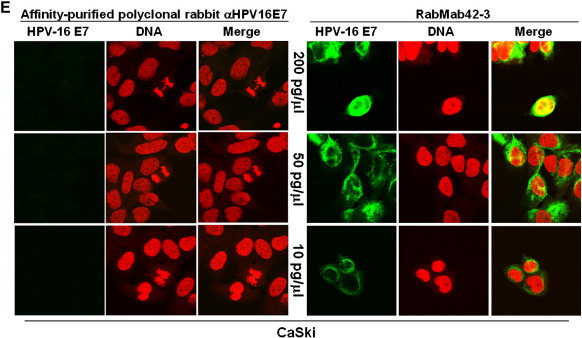
Next RabMab42-3 was employed for immunofluorescence based detection of endogenous E7 protein in cervical cancer cells. We found that RabMab42-3 strongly detected the E7 protein in the HPV-16 positive CaSki cells by immunofluorescence (Fig. 2A, panels 1–6). In contrast, no signal was observed in HPV-18 positive HeLa, HPV-45 positive MS 751 cervical cancer cells and in unrelated U-2OS osteosarcoma cells (Fig. 2A, panels 7–9). Moreover, RabMab42-3 did not cross react with HPV-18 E7 and HPV-45 E7 proteins transiently overexpressed in U-2 OS cells (Fig. 2A, panels 10–11) despite the fact that both oncoproteins were strongly expressed in the U-2 OS cells, as demonstrated in Western blot experiments using affinity-purified polyclonal anti-HPV-18 E7 and anti-HPV-45 E7 antibodies (Fig. 2B). Finally, the staining by RabMab42-3 could be blocked by preincubation of the monoclonal antibody with purified HPV-16 E7 antigen (Fig. 2C). These results establish the specificity of RabMab42-3. Confocal microscopy analysis demonstrated E7 microstructures localized in both the cytoplasmic and nuclear compartments of CaSki cells. While many of the cells in interphase showed a predominantly diffuse cytoplasmic E7-staining with a ring structure surrounding the nucleus and faint nuclear E7 microstructures (Fig. 2A, arrows in panels 1 and 6), some of these cells (~ 1%) showed a strong nuclear E7 staining (Fig. 2A, arrowheads in panels 1 and 2, see also below, Fig. 2E). A thorough analysis of U-2 OS cells transiently overexpressing HPV-16 E7 demonstrated that most of the cells with predominantly cytoplasmic HPV-16 E7 occurred shortly after cytokinesis/G1 transition in the early G1 phase (Fig. 2D, upper and middle panels), while the vast majority of the interphase cells had predominantly nuclear E7 (Fig. 2D, lower panel). In CaSki cells undergoing mitosis, RabMab42-3 stained various structures. One conspicuous figure was formed by ligaments annulated around the nucleus in prometaphase cells (Fig. 2A, arrowheads in panels 4 and 5). In metaphase (Fig. 2A, arrows in panels 2, 3 and 5) and telophase cells (Fig. 2A, arrowhead in panel 6) RabMab42-3 stained large structures encompassing the chromosomes. These results demonstrate that the subcellular localization of endogenous HPV-16 E7 oncoprotein varies during the cell cycle. Strikingly, RabMab42-3 yielded high quality immunofluorescence E7 signals in CaSki cells at very low antibody concentration (10 pg/μl), as compared to affinity-purified rabbit polyclonal anti-HPV-16 E7 antibodies (Fig. 2E), suggesting that the sensitivity of RabMab42-3 is at least 20 times higher in immunofluorescence experiments, with no apparent loss of specificity.
Fig. 2.
Detection of endogenous HPV-16 E7 protein in CaSki cells. (A) Cells were stained with RabMab42-3 as indicated, followed by application of AlexaFluor488-labeled secondary antibodies. CaSki cells (panels 1–6). HeLa (panel 7), MS 751 (panel 8), U-2 OS (panel 9), U-2 OS/HPV-18 E7 (panel 10) and U-2 OS/HPV-45 E7 (panel 11) cells served as negative controls. I = interphase, PM = prometaphase, M = metaphase, T = telophase. (B) U-2 OS cells transiently expressing HPV-18 E7 or HPV-45 E7 wild type as indicated were separated by SDS PAGE and the E7 proteins detected by Western blotting using affinity-purified polyclonal antibodies against HPV-18 E7 and HPV-45 E7 (Fiedler et al., 2005), respectively. (C) Immunofluorescence staining by RabMab42-3 was blocked by preincubation of the antibody with purified immobilized GST-HPV-16 E7 antigen (1 μg/μl) but not GST protein (1 μg/μl). (D) U-2 OS cells transiently overexpressing HPV-16 E7 were stained in indirect immunofluorescence experiments with RabMab42-3 followed by application of AlexaFluor488-labeled secondary antibodies. (Upper and middle panel) HPV-16 E7 expressing cells shortly after cytokinesis/G1 transition are shown. (Lower panel) An interphase cell with predominantly nuclear HPV-16 E7 staining is shown. (E) Comparison of the sensitivity of RabMab42-3 and affinity-purified rabbit polyclonal α-HPV-16 E7 antibodies for the detection of cell-associated endogenous HPV-16 E7 protein in indirect immunofluorescence experiments. For the detection of E7 protein, HPV16-positive CaSki cells were stained with decreasing concentrations (200–10 pg/μl) of either RabMab42-3 (right panel) or affinity purified polyclonal α-HPV16-E7 antibodies (left panel), as indicated, followed by application of AlexaFluor488-labeled secondary antibodies.
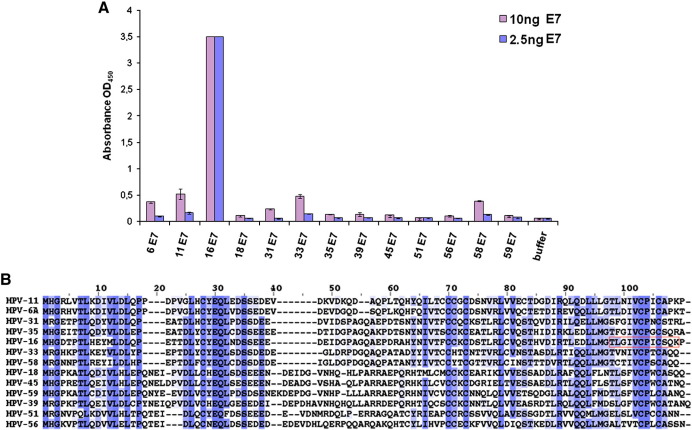
To more precisely characterize the performance of RabMab42-3 we established a direct ELISA using different amounts of randomly coated recombinant HPV E7 protein and analyzed its reactivity against HPV-16 E7, the E7 proteins of 10 other high-risk HPV genotypes and the 2 most common low risk viruses, HPV-6 and HPV-11. As shown in Fig. 3A, 2.5 ng HPV-16 E7 protein was readily detected using this assay. However no cross reactivity was found against the E7 proteins of the other HPV types. As expected, at higher antibody/antigen ratio a moderate cross reactivity was detected against the E7 proteins of HPV-33 and HPV-58 which belong to HPV species 9 (HPV-16 family) and against the E7 proteins of HPV-6 and HPV-11. Almost no cross reactivity was found against the E7 proteins of HPV species 7 (HPV-18, -39, -45, -59). The alignment of the HPV E7 protein sequences (Fig. 3B) suggests that especially the amino acids upstream and downstream from the highly conserved 90VCPXC94 motif in the epitope 86TLGIVCPICSQK97 provide the specific recognition of the HPV-16 E7 protein by RabMab42-3. Note that the cross reactivity analysis was conducted with relatively high amounts of E7 proteins. In an appropriate sandwich ELISA, RabMab42-3 detects ~ 500 fg of recombinant HPV-16 E7 protein (Ehehalt et al., unpublished findings). In keeping with these results RabMab42-3 showed also no cross reactivity in immunofluorescence experiments using U-2 OS cells transiently overexpressing the corresponding HPV E7 proteins (Fig. 2A and data not shown). Moreover, RabMab42-3 showed no cross reactivity in peptide microarrays conducted with collections of 13mer peptides derived from the E7 proteins of the HPV genotypes tested in Fig. 3.
Fig. 3.
Specificity of RabMab42-3 in ELISA using randomly coated HPV E7 proteins. A) Direct ELISA using 2.5 and 10 ng of randomly coated E7 antigens as indicated and biotinylated RabMab42-3 (14 ng/100 μl) is shown. B) Alignment of HPV E7 protein sequences that were used for ELISA in Fig. 2A. Color coding: deep to light blue: high to low sequence conservation. RabMab42-3 epitope is boxed in red.
Detection of the HPV-16 E7 oncoprotein in cervical AC and ACIS
Analysis of cervical ACs from 30 patients using GP5+/GP6+ primer PCR and HPV typing by EIA identified infections with different HPV genotypes (Table 2). Twenty-six (87%) ACs were HPV-DNA positive, 14 (47%) contained mixed infections by two or more genotypes, 23 (77%) were HPV-16 positive, 3 (10%) contained only HPV-18 and in 4 ACs the HPV-type was apparently not detectable by PCR. Among the 6 ACIS were 2 HPV-16 positive, 1 contained HPV-16/58, 2 HPV-16/18 and others, and 1 HPV-18 (Table 3). As negative controls, we used 22 cervical biopsies containing normal squamous and glandular epithelia from hysterectomy specimens with multiple leiomyomas and without cervical lesions, which were HPV-negative in the PCR analysis (Ressler et al., 2007).
Table 2.
HPV-typing of 30 cervical adenocarcinomas (AC) by GP5+/6+ primer PCR and EIA.
| HPV type | AC |
|---|---|
| HPV-16 | 9 (30%) |
| HPV-18 | 3 (10%) |
| HPV-16, -18 | 8 (27%) |
| HPV-16, -18, -31 | 1 (3%) |
| HPV-16, -18, -58 | 3 (10%) |
| HPV-16, 58 | 2 (7%) |
| not detected | 4 (13%) |
| Total | 30 (100%) |
Table 3.
HPV-typing of six adenocarcinoma in situ (ACIS) by GP5+/6+ primer PCR and EIA.
| HPV type | ACIS |
|---|---|
| HPV-16 | 2 (33%) |
| HPV-16, -58 | 1 (17%) |
| HPV-16, -58, -18 | 1 (17%) |
| HPV-16, -58, -18, -33 | 1 (17%) |
| HPV-18 | 1 (17%) |
| Total | 6 (100%) |
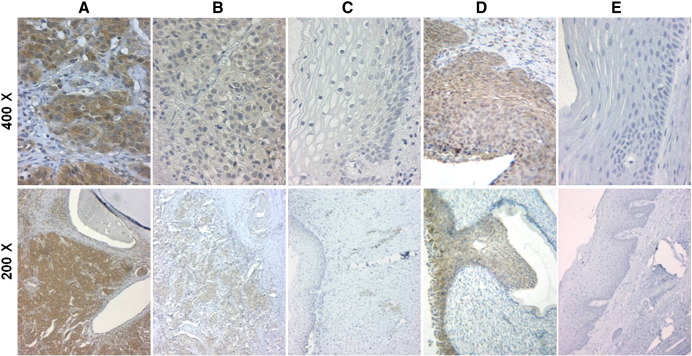
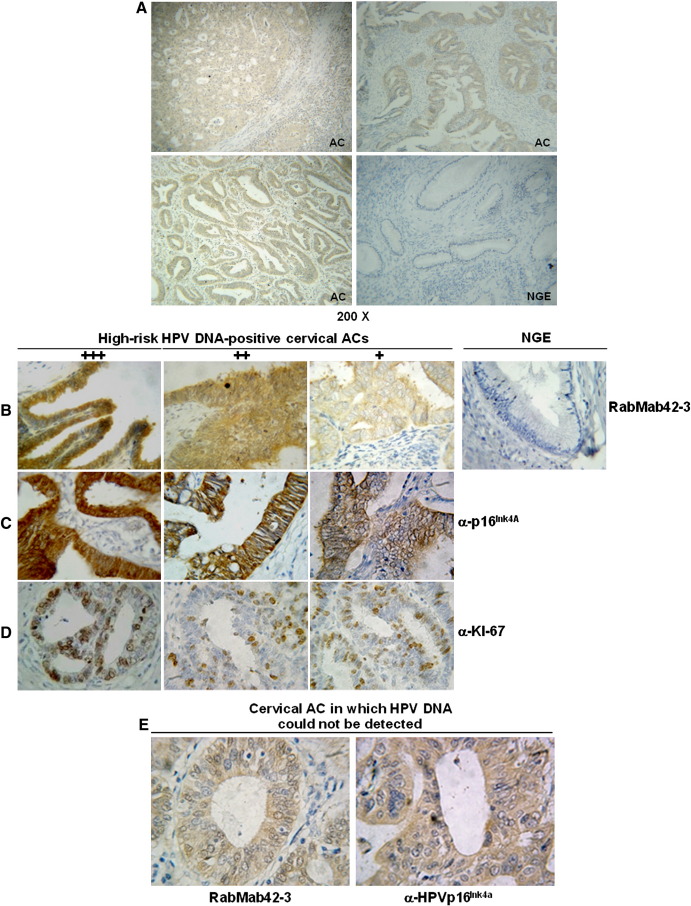
Like affinity-purified polyclonal anti-HPV-16 E7 antibodies (Fiedler et al., 2004; Ressler et al., 2007) RabMab42-3 recognized the HPV-16 E7 protein in paraffin sections of HPV-16 positive cervical SCCs and CIN IIIs but generated no signal in HPV-negative normal cervical squamous epithelia (Fig. 4). RabMab42-3 immunohistochemically detected the E7 oncoprotein in paraffin sections of all 23 HPV-16 positive ACs (Fig. 5A). Almost all epithelial tumor cells within the tumor islets were stained but not the cells in adjacent connective tissues (Fig. 5B). A detailed analysis showed variations in the E7 immunoreactivity across the individual biopsies. Using an arbitrary scoring system, we classified the E7 protein expression levels by semi-quantitative analysis in three categories (Fig. 5B): 3 ACs were classified as + 3, 2 as + 2 and 18 as + 1. No E7 proteins were detected in normal cervical specimens, neither in connective tissue and cervical glandular epithelia (Fig. 5B, right panel) nor in cervical squamous epithelia (Fig. 4). Both glandular cells and squamous epithelial cells in non-neoplastic areas of cancer biopsies were not stained.
Fig. 4.
Detection of high-risk HPV E7 oncoproteins in cervical SCC and CIN III. (A and B) Detection of HPV-16 E7 protein in paraffin sections of a HPV-16 DNA positive cervical SCC by RabMab42-3 using an immunoperoxidase staining protocol. (C) Epithelium and connective tissue adjacent to tumor islets in the HPV-16 DNA positive cervical SCC shown in A and B. (D) Detection of HPV-16 E7 protein in paraffin sections of a HPV-16 DNA positive CIN III by RabMab42-3 using an immunoperoxidase staining protocol. (E) HPV DNA negative normal cervical squamous epithelium served as negative control.
Fig. 5.
Detection of high-risk HPV E7 oncoproteins in cervical ACs. (A) Detection of HPV-16 E7 protein in paraffin sections of different lesions of HPV-16 DNA positive cervical ACs by RabMab42-3 using an immunoperoxidase staining protocol. HPV DNA negative normal cervical glandular epithelium (NGE) served as negative control (Magnifications 200×). (B) (Left panels) Immunoperoxidase stainings of paraffin sections from different lesions of HPV-16 DNA positive ACs with RabMab42-3. An arbitrary scoring system to grade the E7 protein staining intensity in 3 categories was used. The strongest staining intensity was set 100% and the staining intensity rated as follows: (+ 3) 71–100% +++; (+ 2) 41–70% ++; (+ 1) 10–40% +. (Right panel) Normal cervical glandular epithelium (NGE) from a HPV-DNA negative biopsy stained with RabMab42-3 (Magnification 400×). (C) Histological sections of HPV-16 positive ACs stained by anti-p16INK4a antibodies (Magnification 400×). (D) Histological sections of HPV-16 positive ACs stained by anti-Ki-67 antibodies (Magnification 400×). (E) Cervical AC biopsy in which HPV DNA could not be detected by PCR analysis, stained by RabMab42-3 and anti-p16Ink4a antibodies (Magnification 400×).
The cyclin-dependent kinase inhibitor p16INK4a is frequently upregulated in HPV-positive cervical precancers and cancers (Klaes et al., 2001; Tsoumpou et al., 2009). Although a few cervical cancers are p16INK4a negative (Tsoumpou et al., 2009), some other tumors are p16INK4a positive via non-HPV related mechanisms and p16INK4a was detected in normal epithelia (Riethdorf et al., 2002; Ressler et al., 2006), p16INK4a is applied as surrogate marker of high-risk or integrated HPV infection in cervical specimens which could act as an adjunct to current cytological and histological assessment of cervical smears and biopsies. In our study all HPV-16 E7 positive ACs stained positive by the anti-p16INK4a antibodies (Fig. 5C), underlining that p16INK4a is frequently expressed in cervical ACs. The Ki-67 antibodies, which were employed as a control for proliferating cells (Gore et al., 1995), stained the same tumor cell islets as RabMab42-3 and the anti-p16INK4a antibodies (Fig. 5D). In accordance with previous studies approximately 50% of the tumor cells stained positive for Ki-67.
RabMab42-3 stained the 4 ACs in which HPV-DNA was undetectable by PCR analysis in our hands (Fig. 5E, left panel), suggesting that these biopsies do contain HPV-16. These findings are underlined by the positive anti-p16INK4a staining (Fig. 5E, right panel).
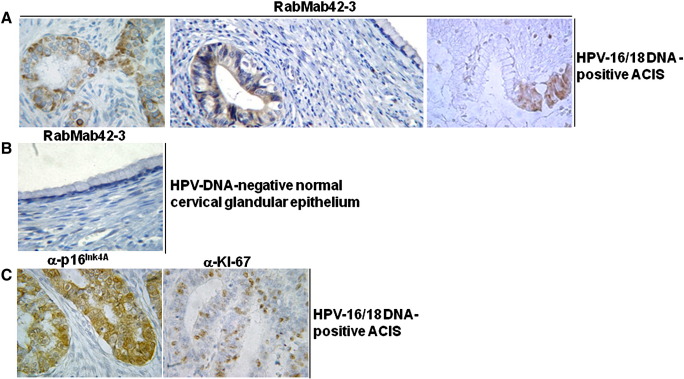
We analyzed whether the HPV-16 E7 oncoprotein is detectable in ACIS. The dysplastic cells in the 5 HPV-16 positive ACIS stained positive with RabMab42-3 (Fig. 6A). The intensity of the E7 staining in the tumor cells of all five specimens were classified as strong (+ 3) by semi-quantitative analysis using the same arbitrary scoring system as for the ACs. This suggests that high HPV-16 E7 oncoprotein levels correlate with ACIS and that the E7 levels in ACIS are sufficient for malignant progression. Similar to the results obtained by the immunofluorescence experiments with the HPV-16 positive CaSki cells (Fig. 2A, C and E), the subcellular localization of E7 in most of the tumor cells in the ACIS was predominantly cytoplasmic, whereas a few cells had a predominantly nuclear E7 staining. Normal glandular epithelial cells adjacent to dysplastic epithelial cells in a gland of the same ACIS were not stained (Fig. 6A, right panel), and no staining was observed in any cell type in HPV-DNA negative biopsies of normal tissue (Fig. 6B). Anti-p16INK4a antibodies stained all tumor cells in the ACIS (Fig. 6C, left panel) and the proliferation marker Ki-67 was detected in 30–50% of the cells in the tumor islets (Fig. 6C, right panel).
Fig. 6.
Detection of HPV E7 oncoproteins in cervical ACIS. (A) Detection of the HPV-16 E7 oncoprotein in three different mixed infected HPV 16/18 DNA positive ACIS using RabMab42-3 (Magnification 400×). (B) No staining of normal cervical glandular epithelium and connective tissue from a HPV-DNA negative biopsy with the anti-HPV-16 E7 RabMab (Magnification 400×). (C) Staining of a HPV-16/18 DNA-positive ACIS by anti-p16INK4a and anti-Ki-67 antibodies, as indicated (Magnification 400×).
Discussion
High-risk HPV E7 oncoproteins have great potential as markers for the detection of cervical precancers and cancers (Doorbar, 2007; Ressler et al., 2007; zur Hausen, 2002). However, HPV E7 oncoproteins are little investigated in cervical precancers and cancers in humans due to the lack of antibodies sufficient in specificity and sensitivity for the detection of E7. In this study, we generated highly specific and sensitive anti-HPV-16 E7 RabMabs and showed for the first time that the HPV-16 E7 oncoprotein is abundantly expressed in cervical ACs and ACIS but not in normal glandular cervical epithelium. This indicates the importance of the E7 oncoprotein for the development of ACIS as well as progression to cervical AC. These data, together with studies detecting E7 oncoproteins in cervical SCCs and high-grade CINs (Fiedler et al., 2004, 2005; Ressler et al., 2007) suggest that E7 oncoproteins can be considered as markers for premalignant and malignant cervical tumors.
The role of high-risk HPV E7 as oncoprotein in cervical carcinogenesis is established (reviewed in McLaughlin-Drubin and Münger, 2009). However, earlier attempts to detect E7 oncoproteins in cervical specimens were hampered by the fact that HPV E7 oncoproteins display low immunogenicity. The recent description of a solution structure for HPV E7 proteins indicates that the highly conserved N-terminal part of E7 proteins is intrinsically unstructured (Liu et al., 2006; Ohlenschläger et al., 2006), providing a potential explanation for the low immunogenicity. Whereas E7 proteins contain a rigid zinc-finger structure in the C-terminal domain, no antibodies were described so far that would specifically recognize this domain. We produced polyclonal anti-HPV-16 E7 antisera in both rabbits and goats which are of sufficient sensitivity and specificity to detect E7 proteins in cervical SCCs (Fiedler et al., 2004, 2005, 2006; Ressler et al., 2007). These antisera recognize epitopes in both the N-terminal and C-terminal part of E7 with varying affinity (unpublished results). This is probably due to the presence of antibodies of lower affinity and/or specificity recognizing linear epitopes in the N-terminus and antibodies of higher affinity and/or specificity recognizing epitopes in the C-terminus of E7. While the lack of conformational diversity in a rigid structure, such as the E7 zinc-finger, lowers the energy needed for protein binding, it is conceivable that this leads to a high affinity to target proteins (Colas, 2000). This suggests that the affinity of specific monoclonal antibodies to the E7 zinc-finger might be higher than to the unstructured E7 N-terminus. Such antibodies can be selected by screening of the given hybridomas (Harlow and Lane, 1998). In fact, when monoclonal anti-HPV-16 E7 antibodies were generated from rabbits, one hybridoma (RabMab42-3) was derived that produced an antibody against a C-terminal epitope of E7 within the C-terminal zinc-finger structure. This epitope differs from the epitopes recognized by our previously used polyclonal anti-HPV-16 E7 antibodies (Fiedler et al., 2004; Ressler et al., 2007; unpublished results). Employing several independent techniques, epitope mapping using peptide microarrays, Western blot analysis, mutational analysis of the RabMab42-3/HPV-16 E7 interaction using immunofluorescence assays, ELISA, competition experiments with purified HPV-16 E7 protein and immunohistochemistry experiments using paraffin embedded cervical cancer biopsies, we provided evidence that the monoclonal rabbit anti-HPV-16 E7 antibody RabMab42-3 specifically recognizes a conformational epitope (86TLGIVCPICSQK97) in the carboxyl-terminal domain of the HPV-16 E7 protein containing half of the zinc coordinating motif in its center (Fig. 1A). Of note, we demonstrated that the rabbit polyclonal antiserum recognized the HPV-16 E7 mutants C58G, C91G and C58G/C91G, which contain point mutations in the major structuring elements of the E7 zinc-finger, in Western blot as well as immunofluorescence experiments; however, the HPV-16 E7 CXXC mutants were not recognized by RabMab42-3, suggesting that the RabMab42-3/HPV-16 E7 interaction strictly depends on the intact spatial structure of the HPV-16 E7 zinc-finger. The specificity of RabMab42-3 is further underscored by the fact that RabMab42-3 showed almost no cross reactivity against the E7 proteins of diverse HPV genotypes.
RabMab42-3 specifically detected endogenous E7 protein in CaSki cells. In comparison to the E7 pattern recognized by affinity-purified polyclonal anti-HPV-16 E7 antibodies (Fiedler et al., 2004; Ressler et al., 2007), the cytoplasmic as well as nuclear E7 structures recognized by RabMab42-3 were less homogenously distributed throughout the cells. It is likely that this result is due to the differences in epitope-recognition between the given antibodies and to the markedly higher sensitivity of RabMab42-3 in the immunofluorescence experiments detecting the cell-associated native HPV-16 E7 protein. Nevertheless, both antibodies detected endogenous E7 protein in cytoplasmic as well as nuclear sites in CaSki cells (Fiedler et al., 2004; Ressler et al., 2007; this study). The microstructures stained by RabMab42-3 in the confocal laser-scanning microscopy experiments (layer thickness ≤ 1.3 micrometer) varied among the individual cells in the CaSki population, which may reflect a differential subcellular localization of E7 during the cell cycle. In mitotic CaSki cells, RabMab42-3 detected structures resembling components of the cytoskeleton and in part of the spindle apparatus. This supports conclusions from previous studies showing a functional interaction of E7 with centrosomes (Duensing et al., 2000; Nguyen et al., 2007). However, the clarification of the cytoplasmic structures stained by RabMab42-3 warrants further studies. A small subset of interphase CaSki cells had a strong nuclear E7 staining, whereas others showed faint E7 structures pervading the nucleus. It is likely that the nuclear E7 structures reflect well established nuclear E7 functions, such as deregulation of the p16Ink4A/pRb pathway (reviewed in Howley et al., 1991; Zwerschke and Jansen-Dürr, 2000; zur Hausen, 2002; Narisawa-Saito and Kiyono, 2007; McLaughlin-Drubin and Münger, 2009; Ghittoni et al., 2010; Moody and Laimins, 2010; Pim and Banks, 2010). In the vast majority of the interphase cells, RabMab42-3 produced predominantly cytoplasmic staining, probably reflecting the well described cytoplasmic E7 functions (reviewed in Zwerschke and Jansen-Dürr, 2000; McLaughlin-Drubin and Münger, 2009). Similar to our results in CaSki cells, HPV-16 E7 was detected in both the cytoplasm and nucleus of the tumor cells in both ACIS and AC, whereby the cytoplasmic localization of HPV E7 preponderated in all biopsies. These data are in agreement with results previously demonstrated in cervical SCCs and CINIIIs (Fiedler et al., 2004, 2005; Ressler et al., 2007). The subcellular localizations of E7 found in the present study are in agreement with biochemical, indirect immunofluorescence, immunohistochemical and electron microscopy studies, conducted in cervical cancer biopsies in situ, cervical cancer cell lines and experimental cells transiently overexpressing E7, which have demonstrated that HPV-16 E7 is located in both the cytoplasm (Smotkin and Wettstein, 1987; Zatsepina et al., 1997; Zwerschke et al., 2000; Fiedler et al., 2004; Huh et al., 2005; Nguyen et al., 2007; Ressler et al., 2007) and the nucleus (Sato et al., 1989; Greenfield et al., 1991; Zatsepina et al., 1997; Smith-McCune et al., 1999; Fiedler et al., 2004; Ressler et al., 2007). In keeping with these findings it was shown that HPV-16 E7 can be actively transported into the nucleus (Angeline et al., 2003), contains a nuclear export sequence and can shuttle between the nucleus and cytoplasm (Knapp et al., 2009).
All HPV DNA-positive cervical ACs and ACIS investigated were either HPV-16 and/or HPV-18 positive and the 4 ACs in which the HPV-type was not detectable in the paraffin embedded specimens by PCR in our hands stained HPV-16 E7 protein as well as p16INK4a positive, reflecting the high prevalence of HPV-16 and HPV-18 in cervical AC (Pirog et al., 2000; Riethdorf et al., 2002). HPV-18 E7 could also be detected in cervical ACs using RabMabs to HPV-18 E7 (data not shown); however, available antibodies to HPV-18 E7 recognized linear epitopes in the N-terminus of E7 and were therefore not included in this study.
Using RabMab42-3, we detected high levels of HPV-16 E7 associated with ACs as well as ACIS relative to normal cervical epithelium. Although larger studies are necessary, this initial work suggests an important role of E7 in precancerous and cancerous progression. In most of the ACs analyzed, the E7 protein levels were at a similar level or lower as in ACIS, suggesting that the E7 levels in ACIS are sufficient for malignant progression. Tumor size, depth of invasion, and grading of the invasive tumor front are main prognostic factors in early cervical AC (El-Ghobashy et al., 2005). We compared the E7 oncoprotein levels in the cervical AC with the histopathologic staging and grading of these lesions according to the criteria defined by the tumor-node-metastasis system of the Union Internationale Contre le Cancer (Sobin and Wittekind, 2002). However, whereas the detection of the E7 oncoprotein always coincided with the presence of an invasive cervical adenocarcinoma, there was no correlation between the E7 protein levels and the pathologic prognostic variables (tumor size, lymph node status, metastases at distance, or histologic grading).
Our results are in agreement with the regular detection of HPV E7 oncoproteins in human cervical SCCs and high-grade CINs (Fiedler et al., 2004, Fiedler et al., 2005; Ressler et al., 2007) as well as the high oncogenic potential of high-risk HPV E7 in vitro and in animal models (reviewed in Zwerschke and Jansen-Dürr, 2000; zur Hausen, 2002; Narisawa-Saito and Kiyono, 2007; McLaughlin-Drubin and Münger, 2009; Ghittoni et al., 2010; Moody and Laimins, 2010; Pim and Banks, 2010). Together the data suggest that high-risk HPV E7 proteins could be useful as specific markers for the detection of cervical cancers and high-grade precancers of squamous and glandular origin.
Materials and methods
Patients
Paraffin-embedded conization specimens from women with cervical AC, ACIS, SSC, and CIN III were diagnosed according to the WHO-classification of tumors of female genital organs (Wells et al., 2003) by the division of pathology and collected by the National Tumour Registry of the National Health Laboratory, Luxembourg or by the departments of Obstetrics and Gynecology, Medical University Innsbruck, Austria. Twenty-two normal cervical squamous and glandular epithelia were obtained from the division of Pathology, National Health Laboratory, Luxembourg and the departments of Obstetrics and Gynecology, Medical University Innsbruck, Austria (Ressler et al., 2007).
HPV-typing
Preparation of HPV DNA and HPV typing was conducted as described (Jacobs et al., 1997).
Generation and characterization of rabbit monoclonal anti-HPV-16 E7 antibodies
HPV-16 E7 protein was purified as described (Fiedler et al., 2006) and used to generate rabbit monoclonal antibodies in collaboration with Epitomics Inc., (Burlingame, CA, USA). Hybridome subclones were characterized by ELISA, Western blot, and immunofluorescence. E7 epitopes were analyzed by JPT Peptide Technologies GmbH (Berlin, Germany) using peptide microarrays. To do this, collections of HPV E7 derived 13mer peptides displayed on peptide microarrays were incubated with RabMab42-3 and unrelated rabbit control antibodies. The determination of peptide-antibody binding was performed by RepliTope-analysis where the peptide microarray was incubated with the primary antibody followed by a fluorescently labeled secondary antibody (anti-rabbit-Cy5). After washing the peptide microarrays were dried using a microarray centrifuge and scanned in a high resolution microarray scanning system with appropriate wavelength settings.
Cell culture and transfection
The human cervical cancer cell lines CaSki (German Cancer Research Center, Heidelberg, Germany), MS751 (Geisbill et al., 1997) and HeLa (ATCC-LGC, Manassas, USA) and the human osteosarcoma cell line U-2OS were cultured in DMEM plus 10% FCS (Fiedler et al., 2004). Cells were transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). HPV-16 E7 wild type and mutants were overexpressed using pJ4W plasmids (Massimi et al., 1997; Mannhardt et al., 2000; Prathapam et al., 2001).
Western blot analysis
Western blot analysis was performed as described (Fiedler et al., 2004).
Indirect immunofluorescence experiments
Cells were fixed with 4% (w/v) PFA/1× PBS, permeabilized with 0.1% (w/v) Na-Citrate/0.3% (v/v) Triton-X-100, blocked with 1× PBS/1%BSA and incubated for 1 h at 37 °C with anti-HPV-16 E7 antibodies in 1× PBS/1%BSA. After washing in 1× PBS and staining with secondary IgGs (DAKOCytomation, Hamburg), cells were processed for indirect immunofluorescence microscopy and viewed using a confocal laser-scanning system (Mannhardt et al., 2000).
Immunohistochemical detection of HPV-16 E7, Ki-67 and p16INK4a
Paraffin-embedded tissue-sections (2 μM) were deparaffinized in xylene and incubated for 5 min each in 100%, 90%, 80%, 70% and 50% isopropanol. Probes were cooked in a steamer, 30 min in 10 mM Citrate buffer pH 6.0 for p16INK4a and Ki-67 staining or 1 h in DAKO retrieval solution pH 6.1 (S1700) for HPV E7 staining. Peroxidase was blocked with 20% H2O2 for 15 min. Note since RabMab42-3 is a conformation-specific antibody the initial fixing of the tissues and the antigen retrieval procedure is critical. After washing in H2O the samples were blocked for 15 min in serum (goat serum for RabMab42-3 and anti-Ki-67 antibody staining; rabbit serum for anti-p16INK4a antibody staining) diluted 1:10 in TBS/BSA (TBS = 7.75 g Tris–HCl pH 7.5, 8.78 g NaCl ad 1 l H2O; TBS/BSA = 5% BSA in TBS). The sections were then incubated for 1 h at RT in TBS/BSA either with biotinylated anti-HPV-16 E7 RabMab42-3 (100–250 ng/μl), anti-p16INK4a (Neomarkers, Vienna) or anti-Ki-67 antibodies (Neomarkers). After washing in TBS/0.1% (v/v) Tween 20 bound antibodies were detected with biotin/streptavidin peroxidase conjugates, visualized with DAB solution, counterstained with Hemalaun, dehydrated and mounted as described (Ressler et al., 2007).
ELISA procedure
Wells of microtiter plates (Maxisorp F, Nunc, Vienna) were coated overnight (4 °C) with different amounts of recombinant bacterial produced untagged HPV E7 proteins (Fiedler et al., 2006) in 100 μl of coating buffer (0.1 M NaHCO3, pH 9.6). After washing three times in PBS, pH 7.4, containing 0.05% Tween20, wells were blocked with 300 μl Universal Casein Diluent/Blocker (UCDB, SDT, Baesweiler, Germany) for 2 h at room temperature. Wells were washed three times. A 100 μl biotinylated primary antibody RabMab42-3 (appropriate dilution in UCDB) was added to each well and incubated for 1 h at room temperature. After three washing steps, 100 μl Streptavidin-PolyHRP40 conjugate (SDT, 0.2 μg/ml in UCDB) was added to each well, followed by 1 h incubation at room temperature. After washing six times, successful binding of the antibody was visualized by the addition of 100 μl chromogenic substrate (es(HS)TMB, SDT) to each well and follow-up incubation for 30 min in the dark at room temperature. The reaction was stopped by the addition of 50 μl 2 N H2SO4 and quantified by absorbance measurement (450 nm) in a multilabel plate reader (VICTOR TM X5, Perkin Elmer, Vienna, Austria).
Disclosure Statement
K. Dreier, D. Ehehalt, B. Lener, H. Pircher, P. Jansen-Dürr and W. Zwerschke declare that they are listed as inventors on a patent application related to anti high-risk HPV E7 RabMabs submitted by the Austrian Academy of Science (ÖAW) and AWS-Austria Wirtschaftsservice.
Acknowledgments
We thank Daniela Köttner and Brigitte Jenewein for excellent technical assistance. This work was supported by the Austrian Cancer Society-Tyrol, BMBWK (BMBWK-651.048/0001-VI/2/2006) and European Union (INCA project LSHC-CT-2005-018704) to W.Z. and by the Austrian Science Funds (FWF) to P.J.D.
References
- Altekruse S.F., Lacey J.V., Jr., Brinton L.A., Gravitt P.E., Silverberg S.G., Barnes W.A., Jr., Greenberg M.D., Hadjimichael O.C., McGowan L., Mortel R., Schwartz P.E., Hildesheim A. Comparison of human papillomavirus genotypes, sexual, and reproductive risk factors of cervical adenocarcinoma and squamous cell carcinoma: Northeastern United States. Am. J. Obstet. Gynecol. 2003;188:657–663. doi: 10.1067/mob.2003.132. [DOI] [PubMed] [Google Scholar]
- Angeline M., Merle E., Moroianu J. The E7 oncoprotein of high-risk human papillomavirus type 16 enters the nucleus via a nonclassical Ran-dependent pathway. Virology. 2003;317:13–23. doi: 10.1016/j.virol.2003.08.024. [DOI] [PubMed] [Google Scholar]
- Bosch F.X., Manos M.M., Muñoz N., Sherman M., Jansen A.M., Peto J., Schiffman M.H., Moreno V., Kurman R., Shah K.V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J. Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- Bray F., Carstensen B., Møller H., Zappa M., Zakelj M.P., Lawrence G., Hakama M., Weiderpass E. Incidence trends of adenocarcinoma of the cervix in 13 European countries. Cancer Epidemiol. Biomark. Prev. 2005;14:2191–2199. doi: 10.1158/1055-9965.EPI-05-0231. [DOI] [PubMed] [Google Scholar]
- Chilvers C., Mant D., Pike M.C. Cervical cancer adenocarcinoma and oral contraceptives. Br. Med. J. Clin. Res. 1987;295:1446–1447. doi: 10.1136/bmj.295.6611.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas P. Combinatorial protein reagents to manipulate protein function. Curr. Opin. Chem. Biol. 2000;4:54–59. doi: 10.1016/s1367-5931(99)00051-4. [DOI] [PubMed] [Google Scholar]
- de Villiers E.M., Fauquet C., Broker T.R., Bernard H.U., zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Doorbar J. Papillomavirus life cycle organization and biomarker selection. Dis. Markers. 2007;23:297–313. doi: 10.1155/2007/613150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing S., Lee L.Y., Duensing A., Basile J., Piboonniyom S., Gonzalez S., Crum C.P., Münger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl Acad. Sci. USA. 2000;97:10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürst M., Kleinheinz A., Hotz M., Gissman L. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumors. J. Gen. Virol. 1985;66:1515–1522. doi: 10.1099/0022-1317-66-7-1515. [DOI] [PubMed] [Google Scholar]
- Eide T.J. Cancer of the uterine cervix in Norway by histological type 1970–1984. J. Natl Cancer Inst. 1987;79:199–205. [PubMed] [Google Scholar]
- El-Ghobashy A.A., Shaaban A.M., Herod J., Herrington C.S. The pathology and management of endocervical glandular neoplasia. Int. J. Gynecol. Cancer. 2005;15:583–592. doi: 10.1111/j.1525-1438.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Fiedler M., Müller-Holzner E., Viertler H.P., Widschwendter A., Laich A., Pfister G., Spoden G.A., Jansen-Dürr P., Zwerschke W. High level HPV-16 E7 oncoprotein expression correlates with reduced pRb-levels in cervical biopsies. FASEB J. 2004;18:1120–1122. doi: 10.1096/fj.03-1332fje. [DOI] [PubMed] [Google Scholar]
- Fiedler M., Ressler S., Campo-Fernández B., Laich A., Jansen L., Widschwendter A., Viertler H.P., Bacher N., Morandell D., Müller-Holzner E., Dürst M., Jansen-Dürr P., Zwerschke W. Expression of the high-risk human papillomavirus type 18 and 45 E7 oncoproteins in cervical carcinoma biopsies. J. Gen. Virol. 2005;86:3235–3241. doi: 10.1099/vir.0.81390-0. [DOI] [PubMed] [Google Scholar]
- Fiedler M., Campo-Fernández B., Laich A., Moser B., Stöckl P., Jansen-Dürr P., Zwerschke W. Purification and characterisation of the E7 oncoproteins of the high-risk human papillomavirus types 16 and 18. J. Virol. Meth. 2006;134:30–35. doi: 10.1016/j.jviromet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Geisbill J., Osmers U., Dürst M. Detection and characterization of human papillomavirus type 45 DNA in the cervical carcinoma cell line MS751. J. Gen. Virol. 1997;78:655–658. doi: 10.1099/0022-1317-78-3-655. [DOI] [PubMed] [Google Scholar]
- Ghittoni R., Accardi R., Hasan U., Gheit T., Sylla B., Tommasino M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes. 2010;40:1–13. doi: 10.1007/s11262-009-0412-8. [DOI] [PubMed] [Google Scholar]
- Gore S.D., Weng L.J., Jones R.J., Cowan K., Zilcha M., Piantadosi S., Burke P.J. Impact of in vivo administration of interleukin 3 on proliferation, differentiation, and chemosensitivity of acute myeloid leukemia. Clin. Cancer Res. 1995;1:295–303. [PubMed] [Google Scholar]
- Greenfield I., Nickerson J., Penman S., Stanley M. Human papillomavirus 16 E7 protein is associated with the nuclear matrix. Proc. Natl Acad. Sci. USA. 1991;88:11217–11221. doi: 10.1073/pnas.88.24.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert C.L., Demers G.W., Galloway D.A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. Coldspring Harbor Laboratory Press; Cold Spring Harbor, New York: 1998. Using Antibodies: A Laboratory Manual. [Google Scholar]
- Hawley-Nelson P., Vousden K.H., Hubbert N.L., Lowy D.R., Schiller J.T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley P.M., Münger K., Romanczuk H., Scheffner M., Huibregtse J.M. Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses. Princess Takamatsu Symp. 1991;22:239–248. [PubMed] [Google Scholar]
- Hudson J.B., Bedell M.A., McCance D.J., Laiminis L.A. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J. Virol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh K.W., DeMasi J., Ogawa H., Nakatani Y., Howley P.M., Münger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl Acad. Sci. USA. 2005;102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M.V., Snijders P.J., van den Brule A.J., Helmerhorst T.J., Meijer C.J., Walboomers J.M. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 1997;35:791–795. doi: 10.1128/jcm.35.3.791-795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S., Lambert P.F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl Acad. Sci. USA. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaes R., Friedrich T., Spitkovsky D., Ridder R., Rudy W., Petry U., Dallenbach-Hellweg G., Schmidt D., von Knebel Doeberitz M. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int. J. Cancer. 2001;92:276–284. doi: 10.1002/ijc.1174. [DOI] [PubMed] [Google Scholar]
- Knapp A.A., McManus P.M., Bockstall K., Moroianu J. Identification of the nuclear localization and export signals of high-risk HPV16 E7 oncoprotein. Virology. 2009;383:60–68. doi: 10.1016/j.virol.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey C.J., Lowndes C.M., Shah K.V. Chapter 4: Burden and management of non-cancerous HPV-related conditions: HPV-6/11 disease. Vaccine. 2006;24(Suppl 3):S3/35-41. doi: 10.1016/j.vaccine.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Liaw K.L., Glass A.G., Manos M.M., Greer C.E., Scott D.R., Sherman M., Burk R.D., Kurman R.J., Wacholder S., Rush B.B., Cadell D.M., Lawler P., Tabor D., Schiffman M. Detection of human papillomavirus DNA in cytologically normal women and subsequent cervical squamous intraepithelial lesions. J. Natl Cancer Inst. 1999;91:954–960. doi: 10.1093/jnci/91.11.954. [DOI] [PubMed] [Google Scholar]
- Liu X., Clements A., Zhao K., Marmorstein R. Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem. 2006;281:578–586. doi: 10.1074/jbc.M508455200. [DOI] [PubMed] [Google Scholar]
- Mannhardt B., Weinzimer S.A., Wagner M., Fiedler M., Cohen P., Jansen-Durr P., Zwerschke W. Human papillomavirus type 16 E7 oncoprotein binds and inactivates growth-inhibitory insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2000;20:6483–6495. doi: 10.1128/mcb.20.17.6483-6495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimi P., Pim D., Banks L. Human papillomavirus type 16 E7 binds to the conserved carboxy-terminal region of the TATA box binding protein and this contributes to E7 transforming activity. J. Gen. Virol. 1997;78:2607–2613. doi: 10.1099/0022-1317-78-10-2607. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin M.E., Münger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384:335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Morandell D., Rostek U., Bouvard V., Campo-Fernández B., Fiedler M., Jansen-Dürr P., Zwerschke W. Human papillomavirus type 45 E7 is a transforming protein inducing retinoblastoma protein degradation and anchorage-independent cell cycle progression. Virology. 2008;379:20–29. doi: 10.1016/j.virol.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Münger K., Howley P.M. Human papillomavirus immortalization and transformation functions. Virus Res. 2002;89:213–228. doi: 10.1016/s0168-1702(02)00190-9. [DOI] [PubMed] [Google Scholar]
- Münger K., Phelps W.C., Bubb V., Howley P.M., Schlegel R. The E6 and E7 genes of human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz N., Bosch F.X., de Sanjosé S., Tafur L., Izarzugaza I., Gili M., Viladiu P., Navarro C., Martos C., Ascunce N. The causal link between human papillomavirus and invasive cervical cancer: a population-based case–control study in Colombia and Spain. Int. J. Cancer. 1992;52:743–749. doi: 10.1002/ijc.2910520513. [DOI] [PubMed] [Google Scholar]
- Muñoz N., Bosch F.X., de Sanjosé S., Herrero R., Castellsagué X., Shah K.V., Snijders P.J., Meijer C.J., International Agency for Research on Cancer Multicenter Cervical Cancer Study Group Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J. Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Narisawa-Saito M., Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–1511. doi: 10.1111/j.1349-7006.2007.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C.L., Eichwald C., Nibert M.L., Münger K. Human papillomavirus type 16 E7 oncoprotein associates with the centrosomal component gamma-tubulin. J. Virol. 2007;81:13533–13543. doi: 10.1128/JVI.01669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlenschläger O., Seiboth T., Zengerling H., Briese L., Marchanka A., Ramachandran R., Baum M., Korbas M., Meyer-Klaucke W., Dürst M., Görlach M. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene. 2006;25:5953–5959. doi: 10.1038/sj.onc.1209584. [DOI] [PubMed] [Google Scholar]
- Peters R.K., Chao A., Mack T.M., Thomas D., Bernstein L., Henderson B.E. Increased frequency of adenocarcinomas of the uterine cervix in young women in Los Angeles County. J. Natl Cancer Inst. 1986;76:423–428. [PubMed] [Google Scholar]
- Pim D., Banks L. Interaction of viral oncoproteins with cellular target molecules: infection with high-risk vs low-risk human papillomaviruses. APMIS. 2010;118:471–493. doi: 10.1111/j.1600-0463.2010.02618.x. [DOI] [PubMed] [Google Scholar]
- Pirog E.C., Kleter B., Olgac S., Bobkiewicz P., Lindeman J., Quint W.G., Richart R.M., Isacson C. Prevalence of human papillomavirus DNA in different histological subtypes of cervical adenocarcinoma. Am. J. Pathol. 2000;157:1055–1062. doi: 10.1016/S0002-9440(10)64619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxe S.C., Saltzstein S.L. Estimation of the duration of the preclinical phase of cervical adenocarcinoma suggests that there is ample opportunity for screening. Gynecol. Oncol. 1999;75:55–61. doi: 10.1006/gyno.1999.5524. [DOI] [PubMed] [Google Scholar]
- Prathapam T., Kühne C., Banks L. The HPV-16 E7 oncoprotein binds Skip and suppresses its transcriptional activity. Oncogene. 2001;20:7677–7685. doi: 10.1038/sj.onc.1204960. [DOI] [PubMed] [Google Scholar]
- Ressler S., Bartkova J., Niederegger H., Bartek J., Scharffetter-Kochanek K., Jansen-Dürr P., Wlaschek M. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5:379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- Ressler S., Scheiden R., Dreier K., Laich A., Müller-Holzner E., Pircher H., Morandell D., Stein I., Viertler H.P., Santer F.R., Widschwendter A., Even J., Jansen-Dürr P., Capesius C., Zwerschke W. High-risk HPV E7 oncoprotein expression in cervical squamous cell carcinoma. Clin. Cancer Res. 2007;13:7067–7072. doi: 10.1158/1078-0432.CCR-07-1222. [DOI] [PubMed] [Google Scholar]
- Reznikoff C.A., Belair C., Savelieva E., Zhai Y., Pfeifer K., Yeager T., Thompson K.J., DeVries S., Bindley C., Newton M.A., Sekhon G., Waldman F. Long-term genome stability and minimal genotypic and phenotypic alterations in HPV16 E7-, but not E6-, immortalized human uroepithelial cells. Genes Dev. 1994;8:2227–2240. doi: 10.1101/gad.8.18.2227. [DOI] [PubMed] [Google Scholar]
- Riethdorf L., Riethdorf S., Lee K.R., Cviko A., Löning T., Crum C.P. Human papillomaviruses, expression of p16, and early endocervical glandular neoplasia. Hum. Pathol. 2002;33:899–904. doi: 10.1053/hupa.2002.127439. [DOI] [PubMed] [Google Scholar]
- Riley R.R., Duensing S., Brake T., Münger K., Lambert P.F., Arbeit J.M. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 2003;63:4862–4871. [PubMed] [Google Scholar]
- Romanczuk H., Howley P.M. Disruption of either the E1or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl Acad. Sci. USA. 1992;89:3159–3163. doi: 10.1073/pnas.89.7.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Watanabe S., Furuno A., Yoshiike K. Human papillomavirus type 16 E7 protein expressed in Escherichia coli and monkey COS-1 cells: immunofluorescence detection of the nuclear E7 protein. Virology. 1989;170:311–315. doi: 10.1016/0042-6822(89)90386-3. [DOI] [PubMed] [Google Scholar]
- Schwartz S.M., Weiss N.S. Increased incidence of adenocarcinoma of the cervix in young women in the United States. Am. J. Epidemiol. 1986;124:1045–1047. doi: 10.1093/oxfordjournals.aje.a114474. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Freese U.K., Gissmann L., Mayer W., Roggenbuck B., Stremlau A., zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- Shingleton H.M., Bell M.C., Fremgen A., Chmiel J.S., Russell A.H., Jones W.B., Winchester D.P., Clive R.E. Is there really a difference in survival of women with squamous cell carcinoma, adenocarcinoma, and adenosquamous cell carcinoma of the cervix? Cancer. 1995;76:1948–1955. doi: 10.1002/1097-0142(19951115)76:10+<1948::aid-cncr2820761311>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Smith-McCune K., Kalman D., Robbins C., Shivakumar S., Yuschenkoff L., Bishop J.M. Intranuclear localization of human papillomavirus 16 E7 during transformation and preferential binding of E7 to the Rb family member p130. Proc. Natl Acad. Sci. USA. 1999;96:6999–7004. doi: 10.1073/pnas.96.12.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smotkin D., Wettstein F.O. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein. J. Virol. 1987;61:1686–1689. doi: 10.1128/jvi.61.5.1686-1689.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin L.H., Wittekind C.H. Wiley-Liss; New York: 2002. TNM Classification of Malignant Tumours, sixth edition, UICC. [Google Scholar]
- Stoler M.H., Rhodes C.R., Whitbeck A., Wolinsky S.M., Chow L.T., Broker T.R. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol. 1992;23:117–128. doi: 10.1016/0046-8177(92)90232-r. [DOI] [PubMed] [Google Scholar]
- Tsoumpou I., Arbyn M., Kyrgiou M., Wentzensen N., Koliopoulos G., Martin-Hirsch P., Malamou-Mitsi V., Paraskevaidis E. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat. Rev. 2009;35:210–220. doi: 10.1016/j.ctrv.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino A.P., Moreno V., Bosch F.X., Muñoz N., Barros-Dios X.M., Parkin D.M. International trends in the incidence of cervical cancer: I. Adenocarcinoma and adenosquamous cell carcinomas. Int. J. Cancer. 1998;75:536–545. doi: 10.1002/(sici)1097-0215(19980209)75:4<536::aid-ijc8>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Wallin K.L., Wiklund F., Angstrom T., Bergman F., Stendahl U., Wadell G., Hallmans G., Dillner J. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N Engl J. Med. 1999;341:1633–1638. doi: 10.1056/NEJM199911253412201. [DOI] [PubMed] [Google Scholar]
- Wang S.S., Sherman M.E., Hildesheim A., Lacey J.V., Jr., Devesa S. Cervical adenocarcinoma and squamous cell carcinoma incidence trends among white women and black women in the United States for 1976–2000. Cancer. 2004;100:1035–1044. doi: 10.1002/cncr.20064. [DOI] [PubMed] [Google Scholar]
- Wazer D.E., Liu X.L., Chu Q., Gao Q., Band V. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc. Natl Acad. Sci. USA. 1995;92:3687–3691. doi: 10.1073/pnas.92.9.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M., Östör A.G., Crum C.P., Franceschi S., Tommasino M., Nesland J.M., Goodman A.K., Sankaranarayanan R., Hanselaar A.G., Albores-Saavedra J. Tumors of the uterine cervix. Epithelial tumors. In: Tavassoli F.A., Devilee P., editors. WHO Classification of Tumors, Pathology & Genetics. Tumors of the Breast and Female Genital Organs. IARC-Press; Lyon: 2003. pp. 269–276. [Google Scholar]
- Zatsepina O., Braspenning J., Robberson D., Hajibagheri M.A., Blight K.J., Ely S., Hibma M., Spitkovsky D., Trendelenburg M., Crawford L., Tommasino M. The human papillomavirus type 16 E7 protein is associated with the nucleolus in mammalian and yeast cells. Oncogene. 1997;14:1137–1145. doi: 10.1038/sj.onc.1200946. [DOI] [PubMed] [Google Scholar]
- Zheng T., Holford T.R., Ma Z., Chen Y., Liu W., Ward B.A., Boyle P. The continuing increase in adenocarcinoma of the uterine cervix: a birth cohort phenomenon. Int. J. Epidemiol. 1996;25:252–258. doi: 10.1093/ije/25.2.252. [DOI] [PubMed] [Google Scholar]
- Ziegert C., Wentzensen N., Vinokurova S., Kisseljov F., Einenkel J., Hoeckel M., von Knebel Doeberitz M. A comprehensive analysis of HPV integration loci in anogenital lesions combining transcript and genome-based amplification techniques. Oncogene. 2003;22:3977–3984. doi: 10.1038/sj.onc.1206629. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- Zwerschke W., Jansen-Dürr P. Cell transformation by the E7 oncoprotein of human papillomavirus type 16: interactions with nuclear and cytoplasmic target proteins. Adv. Cancer Res. 2000;78:1–29. doi: 10.1016/s0065-230x(08)61022-2. [DOI] [PubMed] [Google Scholar]
- Zwerschke W., Mannhardt B., Massimi P., Nauenburg S., Pim D., Nickel W., Banks L., Reuser A.J., Jansen-Dürr P. Allosteric activation of acid alpha-glucosidase by the human papillomavirus E7 protein. J. Biol. Chem. 2000;275:9534–9541. doi: 10.1074/jbc.275.13.9534. [DOI] [PubMed] [Google Scholar]