Abstract
Mammalian cells depend on growth factor signaling to take up nutrients; however, coordination of glucose and glutamine uptake has been a mystery. In this issue of Genes & Development, Wellen and colleagues (pp. 2784–2799) show that glucose flux through the hexosamine biosynthesis pathway regulates growth factor receptor glycosylation and enables glutamine consumption. This mechanism ensures that cells do not engage in anabolic metabolism when nutrients are limiting, and highlights how substrate availability for protein modifications can modulate cell signaling.
Keywords: Metabolism, glucose, glutamine, hexosamine, growth factor signaling, glycosylation
Throughout the 20th century, biochemists systematically worked to outline the pathways cells use to capture energy as ATP and transform organic molecules to synthesize biomass from various substrates. Often working enzyme by enzyme, these studies provided the foundation for our mechanistic understanding of metabolism, or “biochemical networks,” in prokaryotes and higher organisms. More recently, analogous efforts by molecular biologists have elucidated signal transduction pathways, or “protein networks,” which control a multitude of cellular functions related to growth, differentiation, cell death, and metabolism. Recent work has found that many of the same signaling pathways that control growth also influence cellular metabolism (Kim and Dang 2006). For example, receptor-mediated signaling cascades initiated by growth factors stimulate nutrient uptake, cell growth, and cell cycle progression (DeBerardinis et al. 2008). In mammals, the two major nutrients taken up to supply carbon and nitrogen for cell proliferation are glucose and glutamine. Because glucose and glutamine provide biosynthetic precursors through distinct metabolic pathways, their consumption must be coordinated by cells. Nevertheless, despite a growing understanding of how signaling pathways regulate metabolism, much less is known about how flux through specific pathways is “sensed” to provide feedback control on signal transduction networks so that metabolic activity is coordinated with cell growth and proliferation.
In this issue of Genes & Development, Wellen et al. (2010) demonstrate that flux through the hexosamine biosynthesis pathway coordinates glucose and glutamine metabolism by regulating surface expression of growth factor receptors. Using a cellular system that decouples apoptosis from nutrient deprivation, Thompson and colleagues (Wellen et al. 2010) observed that glucose-deprived hematopoietic cells do not switch to glutamine metabolism despite the presence of the growth factor IL-3 and a plentiful extracellular supply of the amino acid. Interestingly, glucose-dependent glutamine uptake is restored in growth factor-replete cells by supplying N-acetylglucosamine (GlcNAc). GlcNAc is unable to enter glycolysis or other pathways of central carbon metabolism and is therefore not available to cells as a fuel source. Rather, GlcNAc supplementation replenishes the supply of precursors necessary for glycosylation of the IL-3 receptor α subunit (IL-3Rα), leading to proper receptor folding and trafficking to the cell surface. Thus, GlcNAc-mediated rescue of glucose-deprived cells restores surface expression of IL-3Rα and allows growth factor-dependent signal transduction, leading to glutamine consumption and cell growth.
The finding that cells are unable to consume a readily available supply of extracellular nutrients when glucose is absent is surprising. The Thompson group (Rathmell et al. 2000; Lum et al. 2005) has reported previously that the ability to initiate glucose uptake is a critical function of growth factor signaling to promote cell survival (Rathmell et al. 2000), and that apoptosis-resistant cells survive by autophagy in the absence of growth factor (Lum et al. 2005). Wellen et al. (2010) show that pools of metabolites involved in central carbon metabolism drop following glucose withdrawal. Nevertheless, these cells still require ATP generation to maintain cellular homeostasis irrespective of their inability to undergo programmed cell death. In the absence of glutamine or other amino acid uptake to support bioenergetics, it is likely that these cells generate ATP by autophagy. This suggests that the signals used by cells to initiate catabolism of endogenous cellular components to maintain survival are distinct from those used to metabolize nutrients in the extracellular environment. These findings also suggest that cells are wired to avoid consuming extracellular resources unless all the metabolic components are present along with a growth signal to support biosynthesis and cell proliferation.
Wellen et al. (2010) found that surface expression of the IL-3Rα chain was dependent on the availability of UDP-GlcNAc for receptor N-glycosylation (Fig. 1). Production of UDP-GlcNAc under standard growth conditions requires glucose flux into the hexosamime biosynthesis pathway, glutamine availability to supply the nitrogen for the amino sugar, and acetyl-coenzyme A (AcCoA) to donate the acetyl moiety. Numerous surface proteins and growth factor receptors are modified by N-linked glycosylation. Indeed, receptors important for growth regulation—such as CTLA-4, GLUT4, IGFR, EGFR, HER2/ErbB2, and FGFR—are glycosylated, and the extent of glycosylation influences surface expression as well as cell growth or differentiation (Lau et al. 2007; Fang et al. 2010). This suggests that the availability of UDP-GlcNAc for protein glycosylation is a means by which cells detect glucose availability. Notably, physiologically relevant changes in glucose concentration increased IL-3Rα surface expression in a dose-dependent manner, indicating the sensitivity of this feedback regulation to glucose supply. Because glutamine and AcCoA are also needed to make UDP-GlcNAc, glutamine withdrawal or AcCoA limitation may result in decreased growth factor receptor expression via the same mechanism. Indeed, glucose uptake is also dependent on glutamine metabolism, as both glutamine and cell-permeable α-ketoglutarate (αKG) can repress the MondoA transcription factor, stimulating glucose consumption and cell proliferation (Kaadige et al. 2009). Another recent study identified an energetic checkpoint that only allowed effective receptor glycosylation and folding when ATP was in excess (Fang et al. 2010). Together, these studies demonstrate that cells require both ATP and biosynthetic precursors for proper folding and surface expression of growth factor receptors. The failure to effectively transduce cell growth signals when nutrients are limited may ensure that cells do not activate the cell growth machinery unless adequate substrates are present.
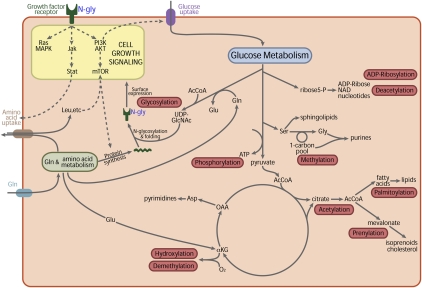
Figure 1.
Metabolic pathways provide substrates for post-translational modifications (PTMs) that influence cell signaling. Growth factor signaling induces uptake of nutrients such as glucose and glutamine that fuel bioenergetic and biosynthetic pathways in cells. In turn, the availability of UDP-GlcNAc generated by these pathways as a substrate for protein glycosylation coordinates growth factor signaling with nutrient availability. Glucose, glutamine, and AcCoA are required for N-linked glycosylation, folding, and trafficking of growth factor receptors to the cell surface. Surface expression of receptors is required for activation of downstream signals to drive nutrient uptake and cell growth. Limited availability of glucose short circuits this feedback loop, leading to loss of growth factor signaling, decreased uptake of amino acids, and growth arrest. Substrates for PTMs derived from other pathways can participate in similar feedback loops to couple metabolic flux information to the control of various cellular processes. Specific PTM reactions are highlighted alongside their associated metabolites and pathways in central carbon metabolism. Amino acids are listed with their three-letter designations. (OAA) Oxaloacetate; (αKG) α-ketoglutarate; (NAD) nicotinamide adenine dinucleotide; (MAPK) mitogen-activated protein kinase; (PI3K) phosphoinositide 3-kinase; (mTOR) mammalian target of rapamycin.
Remarkably, cells that are supplemented with GlcNAc in the absence of glucose are able to grow by consuming glutamine and other amino acids. Under these conditions, glutamine serves as an anaplerotic substrate to replenish tricarboxylic acid (TCA) cycle intermediates and drive oxidative ATP generation, provide carbon for lipid synthesis, and facilitate leucine uptake. Two key observations were made upon GlcNAc-mediated rescue of IL-3-dependent cell growth. First, glutamine and other amino acids did not allow cell division in the absence of glucose. The finding that cells grow but do not divide in the absence of glucose suggests another mechanism is present to sense glucose or a downstream metabolite. Do cells use unique metabolic checkpoints to advance through different stages of the cell cycle? Second, glutamine must have the ability to contribute to multiple metabolic pathways in order to mediate cell growth. Through what pathways is glutamine preferentially metabolized within cells, and under what physiological conditions is glutamine used as a carbon source to build biomass?
Glutamine enters the cell through amino acid transporters and acts as a critical source of nitrogen for cells. The amido-nitrogen of glutamine (rather than the nitrogen that remains on glutamate) is essential for synthesis of nucleotides and hexosamines, and glutamine can also be deaminated by glutaminase to generate free ammonia in cells. Each of these reactions produces glutamate, one of the most abundant metabolites within cells (Munger et al. 2008). Glutamate can be converted to αKG via reactions catalyzed by glutamate dehydrogenase or aminotransferases. These aminotransferases are important nitrogen donors for biosynthesis and are highly active within cells. In fact, 15N-amino-labeled glutamine can even transfer nitrogen to essential amino acids such as leucine, isoleucine, and valine, presumably through reversible activity of branched chain aminotransferases (Hiller et al. 2010).
At least in cultured tumor cell lines, glutamine is a major contributor to the glutamate and αKG pool, as evidenced by the contribution of 13C-labeled glutamine atoms to TCA cycle metabolites (DeBerardinis et al. 2007; Hiller et al. 2010; Ward et al. 2010; Wellen et al. 2010). Glutamine is converted to lactate at high rates in some cell lines, and can also supply carbon for lipogenesis (DeBerardinis et al. 2007; Wellen et al. 2010). This latter function is crucial, as evidenced by the rescue of cell growth by cell-permeable αKG upon acute glutamine withdrawal (Wise et al. 2008; Weinberg et al. 2010). AcCoA is the building block for lipid synthesis, and glutamine can only contribute carbon to AcCoA through two pathways: glutaminolysis and reductive carboxylation. Glutaminolysis involves the oxidative conversion of glutamine carbon to malate in the TCA cycle and subsequent generation of pyruvate via malic enzyme. Pyruvate dehydrogenase activity then produces AcCoA that is exported to the cytosol for lipid synthesis through the citrate shuttle. Deberardinis et al. (2007) have demonstrated the definitive use of this pathway for lipid synthesis using [3-13C]glutamine (i.e., glutamine labeled with 13C on the 3-carbon) in glioblastoma cells. Alternatively, cells can reductively metabolize αKG to form citrate via NADP+-dependent isocitrate dehydrogenase (IDH) and aconitase (ACO) enzymes. Significant activity of this pathway has been demonstrated in hepatoma and differentiated brown fat adipocyte cell lines using [5-14C] and [5-13C]glutamine tracers (Holleran et al. 1995; Yoo et al. 2008), and reversibility of the IDH and ACO reactions has been detected in glioblastoma and lung adenocarcinoma cell lines (Metallo et al. 2009; Ward et al. 2010). How cells regulate flux through either pathway remains to be determined.
Wellen et al. (2010) speculate that, because neither GlcNAc nor glutamine can enter the glycolytic pathway, GlcNAc-mediated rescue of glutamine flux is unable to support cell proliferation. Their finding that glucosamine, which can enter glycolysis and the hexosamine pathway, is able to rescue proliferation of glucose-deprived cells supports this hypothesis. Few physiologically relevant substrates can replace the essential role of glucose in driving glycolysis. Pentose phosphates are synthesized directly from glucose, and the lack of ribose moieties presumably prevents nucleotide synthesis and entry into S phase of the cell cycle. Furthermore, this possibility suggests that cells have mechanisms of sensing other essential metabolites to influence checkpoints for proliferation apart from those used to control cell growth. The identification of these checkpoints will provide crucial insight into how cells progress through the cell cycle or bypass these controls during tumorigenesis.
In addition to flux through the hexosamine pathways, evidence exists that sensing of other nutrients can influence signal transduction, leading to activation of the cell growth machinery. Amino acid uptake from the extracellular space often occurs via cotransporters, and mechanisms have been proposed for how subsets of amino acids might influence growth control. For instance, glutamine efflux coupled to leucine uptake through the Slc7a5 transporter can influence mTOR (mammalian target of rapamycin) signaling (Nicklin et al. 2009). Absence of a number of amino acids, including leucine, results in decreased mTOR activity even in the presence of an active upstream growth signal. This regulation occurs at least in part by subcellular localization of the mTOR signaling complex (Sancak and Sabatini 2009); however, how the amino acids are “sensed” remains a mystery.
It is intriguing to speculate how perturbations in metabolism might affect signal transduction. After transcription and translation, cells employ various mechanisms to control protein function. Metabolites may bind directly to proteins and allosterically regulate function. This type of regulation is well described for many metabolic enzymes, but can also control signaling proteins. Cells respond to energy stress by activating AMPK to decrease ATP-consuming processes and increase ATP production, and activation of this kinase is driven by AMP binding (Xiao et al. 2007). Protein function can also be influenced by post-translational modification (PTM). Many PTMs involve covalent modification of specific residues using metabolic substrates that may ultimately change the localization, activity, or stability of a given protein. The chemical reactions leading to PTMs have been reviewed in detail, and examples include phosphorylation, glycosylation, acetylation, methylation, prenylation, and hydroxylation (Walsh et al. 2005). However, an emerging concept is that the metabolic pathways that generate precursor molecules for PTMs can be limiting in cells (Fig. 1). Thompson and colleagues (Wellen et al. 2009, 2010) have identified two such metabolic pathways, hexosamine biosynthesis and AcCoA generation, which can exert control over growth factor receptor glycosylation and histone acetylation, respectively. Such relationships are numerous within cells, and may in fact play a central role in the progression of diseases.
Histone modification is a high-level regulatory process capable of controlling global gene expression in the nucleus, and such epigenetic switches are mediated by protein modifications involving methyl and acetyl groups. The ATP citrate lyase (ACL) enzyme is responsible for generating AcCoA from citrate for lipid biosynthesis in the cytosol (Hatzivassiliou et al. 2005), though more recent efforts have demonstrated its presence in the nucleus (Wellen et al. 2009). ACL activity and glucose availability to supply AcCoA are both required for growth factor or differentiation-induced histone acetylation. Although other substrates, such as acetate, can also supply carbon for acetylation, these findings establish a role for AcCoA availability in regulating the activity of histone acetyltransferases. Acetylation is a dynamic and ubiquitous process that affects nonhistone proteins in the nucleus, cytoplasm, and mitochondria. Indeed, virtually all enzymes in central carbon metabolism are acetylated on lysine residues, and the activity of many proteins changes in response to acetylation status (Zhao et al. 2010). While we do not understand how acetylation regulates metabolism at the systems level, the AcCoA concentration within a cell or subcellular compartment represents an important and potentially direct method of regulating metabolic activity. It also provides a means to provide metabolic input into the regulation of gene expression, and potentially other processes important for cell physiology.
Protein acetylation is reversible, and the removal of these modifications is catalyzed by histone deacetylases (HDACs). Sirtuins (class III HDACs) require nicotinamide adenine dinucleotide (NAD+) for activity, and are therefore responsive to the redox state within cellular compartments (Schwer and Verdin 2008). These enzymes regulate signaling proteins such as p53, and they can directly control metabolic activity of enzymes like IDH2 and GDH (Schlicker et al. 2008; van Leeuwen and Lain 2009). In these deacetylase reactions, NAD+ is converted to O-acetyl-ADP-ribose, which is hydrolyzed to form ADP-ribose. Interestingly, some sirtuins also exhibit ADP-ribosyl transferase activity while consuming NAD+ in a manner similar to poly-ADP-ribose polymerases (PARPs). ADP-ribosylation itself is a unique PTM responsible for regulating various cellular functions, in particular DNA damage (de Murcia et al. 1997). Glucose metabolism in the pentose phosphate pathway is the primary source of ribose for nucleotide synthesis. Might ribose availability or various salvage pathways that recycle these cofactors affect the activity of sirtuins or PARPs by limiting the supply of NAD+ and/or ADP-ribose? Similarly, changes in the redox state and ratio of NAD+ to that of the reduced form (NADH) may also influence the activity of these enzymes and afford the cell a method of sensing its metabolic state.
AcCoA is the precursor for fatty acids and cholesterol, and metabolites along these synthesis pathways can serve as substrates for acylation and prenylation. In fact, many signaling ligands are lipid-modified, including members of the Wnt, Hedgehog (Hh), and epidermal growth factor receptor (EGFR) ligand families (Miura and Treisman 2006). Well-conserved cysteine residues in secreted Wnt proteins are palmitoylated, and this PTM is necessary for signaling activity and secretion (van den Heuvel et al. 1993; Willert et al. 2003). Palmitoylation and myristoylation can also affect the secretion and function of Hh and the EGFR ligand Spitz (Chamoun et al. 2001; Miura et al. 2006). Similarly, farnesyl and geranylgeranyl diphosphate, metabolites of the isoprenoid pathway, are used to modify cysteine thiolate side chains on some proteins (Walsh et al. 2005). Ras family GTPases are well-known targets of farnesyltransferase and geranylgeranyltransferase enzymes (Sebti 2005). While the dynamic role of these PTMs in regulating cellular signaling has been established, the sensitivity of acyltransferases to physiological levels of substrate has not been well studied.
Although most extensively explored with respect to histones, protein methylation is another PTM regulated by “metabolically sensitive” enzymes (see Teperino et al. 2010 for a detailed discussion of these phenomena). Virtually all methyltransferases employ S-adenosyl-methionine (SAM) as the methyl donor and generate S-adenosyl-homocysteine. SAM is ultimately regenerated via one carbon metabolism after recycling of homocysteine to methionine. As a result, methyltransferases are dependent on a supply of methionine or carbon flux into the folate pool.
Two distinct families of demethylases have been identified, and the activities of both are influenced by metabolite levels. Lysine-specific demethylase 1 (LSD1) and homologous protein activity is dependent on flavin adenine dinucleotide (FAD) reduction to FADH2 (Culhane and Cole 2007). Therefore, like the Sirtuins and PARPs, LSD1 demethylase activity is sensitive to the local redox status. Jumonji C (JmjC) domain-containing proteins exhibit demethylase activity on mono-, di-, and trimethylated lysine residues and are members of the large αKG- and Fe2+-dependent dioxygenase family (Tsukada et al. 2006). These proteins consume oxygen, convert αKG to succinate, and require ascorbate to regenerate the Fe2+ cofactor as part of their enzymatic activity. Thus, altered concentrations of any of these molecules may affect demethylase activity. Other similarly regulated dioxygenases instead catalyze proline or asparagine hydroxylation rather than demethylation. Owing to their oxygen dependence, the Egl nine-homolog proteins (EGLN2 in particular) are well-described components of the cellular oxygen-sensing machinery and influence stability of the hypoxia-inducible transcription factors (HIFs) that regulate metabolic enzyme expression and neo-vascularization (Epstein et al. 2001). Intriguingly, hypoxic microenvironments and HIF2 expression are both associated with stem/progenitor cells (Covello et al. 2006), and low oxygen tension can improve the generation of induced pluripotent stem cells (Yoshida et al. 2009). Substrate availability may regulate both hydroxylating and demethylating dioxygenases to influence the gene expression or epigenetic changes that control cell fate.
PTMs provide a mechanism for cells to sense metabolite levels, allowing specific metabolic fluxes to influence signal transduction pathways regulating cellular processes such as growth and proliferation. However, a quantitative approach is required to characterize metabolic pathways and explore how changes in flux might influence cell signaling. Over the last decade, significant advances have been made in our ability to computationally and experimentally estimate metabolic fluxes in eukaryotic cells (Zamboni 2010). Mass spectrometry and nuclear magnetic resonance-based techniques now enable quantification of labeling in metabolite pools from isotopic tracers at extraordinarily high resolution. In turn, this information can be analyzed in simple or complex networks to reliably estimate fluxes and confidence intervals using various methods (Antoniewicz et al. 2006; Young et al. 2008; Hiller et al. 2010). Applying these analytical techniques in tandem with analyses of PTMs and signal transduction is needed to fully elucidate how metabolic flux controls these cellular processes.
In their description of flux-mediated regulation of glycosylation and acetylation, Wellen et al. (2009, 2010) acutely withdrew glucose or knocked down ACL protein levels. What cellular conditions might limit intracellular metabolite availability? While not all PTM substrates are present at rate-limiting concentrations for these reactions, changes in the in vivo microenvironment may induce significant fluctuations of metabolite levels. Metabolic fluxes change in response to differentiation, proximity to blood vessels, and cell growth signals, and even during different phases of the cell cycle. For example, protein and lipid synthesis rates are thought to be highest in G1 and G2 phases, while nucleotide synthesis is maximal during S phase (Tu et al. 2007). Such temporal compartmentalization requires coordinated regulation of metabolic pathways. Mechanisms are also required to inform the signaling machinery orchestrating these complex processes to ensure that metabolic flux is adequate to complete the task. By identifying the coupled regulation of glucose and glutamine metabolism through IL-3Rα glycosylation, Wellen et al. (2010) have added to the growing body of evidence that metabolism is not a static bystander, but plays an active role in coordinating signal transduction in mammalian cells.
Acknowledgments
We thank Brooke Bevis for help generating the figure.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2010510.
References
- Antoniewicz MR, Kelleher JK, Stephanopoulos G 2006. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng 8: 324–337 [DOI] [PubMed] [Google Scholar]
- Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, Basler K 2001. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293: 2080–2084 [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B 2006. HIF-2α regulates Oct-4: Effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev 20: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane JC, Cole PA 2007. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol 11: 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB 2007. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci 104: 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB 2008. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20 [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci 94: 7303–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Fang M, Zhirong S, Huang S, Zhao L, Chen S, Mak TM, Wang X 2010. The endoplasmic reticulum UDPase ENTPD5 promotes protein N-glycosylation and Warburg effect in the PI3K/PTEN pathway. Cell doi: 10.1016/j.cell.2010.10.010 [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB 2005. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8: 311–321 [DOI] [PubMed] [Google Scholar]
- Hiller K, Metallo CM, Kelleher JK, Stephanopoulos G 2010. Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal Chem 82: 6621–6628 [DOI] [PubMed] [Google Scholar]
- Holleran AL, Briscoe DA, Fiskum G, Kelleher JK 1995. Glutamine metabolism in AS-30D hepatoma cells. Evidence for its conversion into lipids via reductive carboxylation. Mol Cell Biochem 152: 95–101 [DOI] [PubMed] [Google Scholar]
- Kaadige MR, Looper RE, Kamalanaadhan S, Ayer DE 2009. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc Natl Acad Sci 106: 14878–14883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Dang CV 2006. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res 66: 8927–8930 [DOI] [PubMed] [Google Scholar]
- Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW 2007. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129: 123–134 [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120: 237–248 [DOI] [PubMed] [Google Scholar]
- Metallo CM, Walther JL, Stephanopoulos G 2009. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol 144: 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura GI, Treisman JE 2006. Lipid modification of secreted signaling proteins. Cell Cycle 5: 1184–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura GI, Buglino J, Alvarado D, Lemmon MA, Resh MD, Treisman JE 2006. Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev Cell 10: 167–176 [DOI] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB 2000. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol Cell 6: 683–692 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Sabatini DM 2009. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochem Soc Trans 37: 289–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C 2008. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 382: 790–801 [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E 2008. Conserved metabolic regulatory functions of sirtuins. Cell Metab 7: 104–112 [DOI] [PubMed] [Google Scholar]
- Sebti SM 2005. Protein farnesylation: Implications for normal physiology, malignant transformation, and cancer therapy. Cancer Cell 7: 297–300 [DOI] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J 2010. Histone methyl transferases and demethylases; Can they link metabolism and transcription? Cell Metab 12: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL 2007. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci 104: 16886–16891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Harryman-Samos C, Klingensmith J, Perrimon N, Nusse R 1993. Mutations in the segment polarity genes wingless and porcupine impair secretion of the wingless protein. EMBO J 12: 5293–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen I, Lain S 2009. Sirtuins and p53. Adv Cancer Res 102: 171–195 [DOI] [PubMed] [Google Scholar]
- Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr 2005. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed Engl 44: 7342–7372 [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS 2010. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci 107: 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Scarino JMS, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB 2010. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev (this issue). doi: 10.1101/gad.1985910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR III, Nusse R 2003. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423: 448–452 [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, et al. 2007. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 449: 496–500 [DOI] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK 2008. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 283: 20621–20627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S 2009. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 5: 237–241 [DOI] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G 2008. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng 99: 686–699 [DOI] [PubMed] [Google Scholar]
- Zamboni N 2010. 13C metabolic flux analysis in complex systems. Curr Opin Biotechnol. doi: 10.1016.copbio.2010.08.009 [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]