Abstract
Argonaute/Piwi proteins associate with small RNAs that typically provide sequence specificity for RNP function in gene and genome regulation. Here we show that Twi12, a Tetrahymena Piwi protein essential for growth, is loaded with mature tRNA fragments. The tightly bound ∼18- to 22-nucleotide tRNA 3′ fragments are biochemically distinct from the tRNA halves produced transiently in response to stress. Notably, the end positions of Twi12-bound tRNA 3′ fragments precisely match RNAs detected in total small RNA of mouse embryonic stem cells and human cancer cells. Our studies demonstrate unanticipated evolutionary conservation of mature tRNA processing to tRNA fragment small RNAs.
Keywords: Argonaute/Piwi RNP, tRNA cleavage, small RNA, Tetrahymena thermophila
New noncoding RNA populations continue to be discovered (Mercer et al. 2009; Siomi and Siomi 2009), implying that there are biological events of RNA-mediated regulation and RNA processing that remain to be appreciated. Beyond the microRNAs, siRNAs, and germline Piwi-interacting RNAs that guide the specificity of Argonaute/Piwi (Ago/Piwi) RNP function (Tolia and Joshua-Tor 2007), the search for novel small RNAs (sRNAs) has uncovered fragments of mature tRNAs that are poorly, nonpreferentially, or not specifically associated with Ago proteins (Cole et al. 2009; Thompson and Parker 2009a; Haussecker et al. 2010). Abundant tRNA fragments resulting from conditionally induced cleavage of the anti-codon loop were first reported in the ciliated protozoan Tetrahymena, and similar tRNA cleavage phenomena have been revealed as a broadly conserved prokaryotic and eukaryotic response to stress or change in developmental state (Lee and Collins 2005; Thompson and Parker 2009a; Garcia-Silva et al. 2010). Stress-induced tRNA cleavage involves a nuclease from the RNase T2 or RNase A family in budding yeast or human cells, respectively, not an RNase III family Dicer enzyme, and therefore generates a 5′ hydroxyl rather than 5′ monophosphate product (Fu et al. 2009; Thompson and Parker 2009b; Yamasaki et al. 2009). Libraries of RNA sequences obtained following Ago/Piwi protein enrichment or size selection of total RNA typically contain a minor fraction of tRNA fragments, which are expected contamination based on high tRNA abundance and the multiple pathways of tRNA degradation (Phizicky and Hopper 2010). Some examples of precursor or mature tRNA processing by Dicer have been reported, one of which occurs by Dicer recognition of an alternative short-hairpin-like fold of the primary transcript (Babiarz et al. 2008; Cole et al. 2009). Recent studies also describe Dicer-independent accumulation of the tRNA 3′ trailer (a primary transcript segment between the tRNA 3′ end and the transcription termination signal), creating sRNAs proposed to regulate cellular proliferation and/or the homeostasis of RNA silencing (Lee et al. 2009; Haussecker et al. 2010).
We previously purified tagged versions of eight distinct Tetrahymena Piwi family (Twi) proteins for analysis of sRNAs generated by the Tetrahymena Dicer enzymes Dcl1 and Dcr2. Dcl1 produces ∼28- to 29-nucleotide (nt) sRNAs that mediate heterochromatin formation and DNA elimination in the sexual cycle of reproduction (Malone et al. 2005; Mochizuki and Gorovsky 2005), while Dcr2 produces ∼23- to 24-nt sRNAs involved in gene regulation during asexual growth (Howard-Till and Yao 2006; Lee and Collins 2006, 2007; Couvillion et al. 2009). Of the eight distinct Twi proteins, only Twi12 failed to enrich a profile of sRNAs consistent with the size range for products of Dcl1 or Dcr2 (Couvillion et al. 2009). Surprisingly, among the TWI genes expressed in growing cells, only TWI12 is individually essential (Couvillion et al. 2009; additional data not shown). Because the heterogeneously sized sRNAs bound to Twi12 were previously isolated using Twi12 overexpressed in the presence of competing endogenous protein, they were of uncertain physiological specificity. Here we investigate the specificity of Twi12 sRNA loading.
Results and Discussion
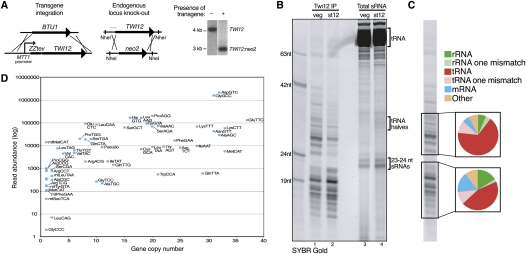
We first established the biological function of N-terminally tagged Twi12 by disruption of endogenous TWI12 in the presence of a tagged protein transgene integrated at the nonessential BTU1 locus (Fig. 1A). The MTT1 promoter used to drive transgene expression has low basal expression but is rapidly induced to high expression by cadmium addition to media (Shang et al. 2002). The transgene ORF encodes Twi12 fused to tandem Protein A domains with an intervening protease cleavage site (ZZtev) (Fig. 1A, left). Wild-type cells lacking the integrated transgene were not able to replace the endogenous TWI12 locus with a neo2 selection cassette, while cells with the transgene could fully replace TWI12 even when selected in the absence of cadmium (Fig. 1A, right). The uninduced level of transgene mRNA expression was only slightly higher than the level of endogenous TWI12 mRNA, while cadmium induction gave >10-fold overexpression (Supplemental Fig. S1). Cells with an uninduced level of transgene expression that also lacked endogenous Twi12 were used to purify Twi12 and associated sRNAs from the rapidly dividing cells of a vegetative growth culture (veg) or from cultures of nondividing cells harvested after 12 h of nutrient starvation (st12). Copurified RNA was resolved by denaturing polyacrylamide gel electrophoresis and directly stained with SYBR Gold (Fig. 1B, lanes 1,2). Comparison with total RNA size-enriched for smaller RNAs (<100 nt) revealed that the Twi12-bound sRNAs were distinct from any abundant sRNA population, including the constitutively accumulated ∼23- to 24-nt sRNA products of Dcr2 and the starvation-induced ∼30- to 35-nt halves of tRNA cleaved in the anti-codon loop (Fig. 1B, lanes 3,4). Parallel mock purifications using wild-type cell extract lacking tagged Twi12 failed to enrich sRNAs (Supplemental Fig. S2).
Figure 1.
Twi12 association with tRNA fragments. (A) The schematics show the strategy of ZZTwi12 transgene knock-in and endogenous TWI12 locus knockout; the latter is evidenced with Southern blot data using NheI-digested genomic DNA at right. The position of the Southern blot probe is indicated in the TWI12 locus schematic with a thin gray line. (B) RNAs copurified with Twi12 in vegetative growth (veg) or after 12 h of starvation (st12) were stained directly with SYBR Gold in comparison with size-selected total RNA from the same cultures. (C) The composition of sRNA library sequencing reads is depicted for reads with either a perfect match to the genome or a single internal mismatch as indicated. (D) Read numbers for tRNA fragments copurified with Twi12 were plotted relative to tRNA gene copy number as an approximation of full-length tRNA abundance. The labels of some data points at left were removed for clarity: mtGluTCC, SecTCA, ThrCGT, mtHisGTG. Pseudo indicates combined reads from the 11 annotated tRNA pseudogenes, and mt indicates mitochondrial.
Twi12-bound sRNAs from vegetative growth were used for library construction and deep sequencing. Surprisingly, the majority of sequences from both Twi12-associated sRNA size classes mapped to tRNA genes (Fig. 1C, with dark shading for perfect sequence matches or light shading for allowing a single internal mismatch). Given that conserved post-transcriptional tRNA modifications such as N1-methyl-adenosine at position 58 greatly reduce the efficiency of the reverse transcription step required for library construction (Saikia et al. 2010), the observed predominance of tRNA sequence reads is likely to be a dramatic underestimate. Indeed, we were unable to detect the presence of any non-tRNA sequence read in the Twi12-enriched sRNA pool by direct hybridization (see below; additional data not shown). Read numbers for each tRNA sequence were plotted against gene copy number as an approximation of tRNA abundance, with the caveat that modification differences between tRNAs would bias relative representation of tRNA reads in the library. This analysis suggests a general trend for more abundant tRNAs to generate more tRNA fragment reads (Fig. 1D; Supplemental Table 1 provides an updated and curated inventory of Tetrahymena thermophila tRNA gene sequences and copy numbers). We conclude that Twi12 carries sRNAs broadly representative of total cellular tRNA.
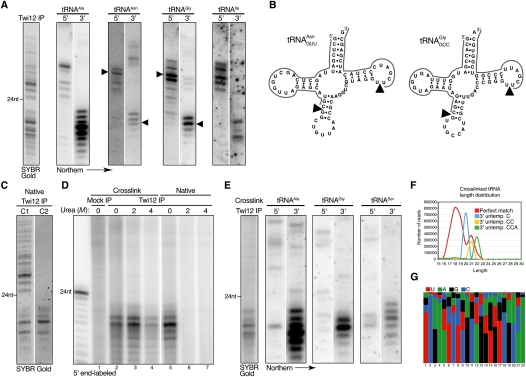
To verify the abundance of tRNA fragments as Twi12-associated sRNAs, we used oligonucleotides complementary to each end of several tRNAs to probe blots of Twi12-associated sRNAs. Consistently, Twi12-bound tRNA 5′ end fragments were predominantly ∼25- to 30 nt in length, while tRNA 3′ end fragments were predominantly <23 nt in length, matching the two size classes of Twi12-bound sRNAs detected by SYBR Gold (Fig. 2A; additional data not shown). These fragment sizes and sRNA sequence data establish that typical Twi12-bound tRNA 3′ fragments begin in the T-loop and end at variable positions of the mature tRNA 3′ CCA tail (see below), while typical Twi12-bound tRNA 5′ fragments begin at the mature tRNA 5′ end and end variably in the anti-codon stem (illustrated in Fig. 2B).
Figure 2.
Retention of the tRNA 3′ fragment as the tightly bound strand. (A) Blot hybridization of Twi12-bound sRNAs was performed using probes complementary to the 5′ or 3′ end of several abundant tRNAs. Direct SYBR Gold staining of Twi12-bound sRNAs is shown at left for reference. (B) Arrowheads and lines indicate the potential processing positions and detected sRNAs, respectively, for two of the tRNAs from A. The 3′ CCA nucleotides of a mature tRNA are not illustrated. (C) Twi12-associated sRNAs were directly stained after RNP isolation using two different gentle, nondenaturing buffer conditions. (D) Twi12-associated sRNAs were 5′ end-labeled after isolation using stringent purification conditions, with or without prior cross-linking; different concentrations of urea were used for washes prior to RNA extraction. (E–G) Twi12-cross-linked sRNAs washed with 2 M urea were used for blot hybridization to detect specific tRNA fragments (shown in E) and for library construction. The length distribution of sequencing reads (F) is shown for 3′ fragments that match the genome perfectly or have an untemplated 3′ C, CC, or CCA, with the combined nucleotide frequency at each position (G) revealing a 5′ sequence signature.
To investigate differential association of Twi12 with the 5′ versus 3′ tRNA fragments, we used more stringent RNP purification conditions. Under some conditions, Twi12 selectively enriched only the ∼18- to 22-nt sRNAs (Fig. 2C). Furthermore, when Twi12 was purified in the presence of high detergent concentration, with or without prior in vivo cross-linking with formaldehyde, only the ∼18- to 22-nt sRNA population was enriched (Fig. 2D, lanes 2,5). The sRNAs of cross-linked but not native complexes were resistant to dissociation by urea (Fig. 2D, cf. lanes 2–4 and 5–7), confirming the direct cross-linking, and thus close-range physical association, of Twi12 and the ∼18- to 22-nt sRNAs. Hybridization analysis confirmed that the sRNAs preferentially enriched with Twi12 after in vivo cross-linking are the tRNA 3′ fragments (Fig. 2E; additional data not shown). These findings suggest that, analogous to other Ago/Piwi proteins, Twi12 is initially loaded with an asymmetric RNA duplex, such that one guide strand is tightly bound (the strand containing the 3′ side of the acceptor stem), while the other passenger strand is readily released (the strand containing the 5′ side of the acceptor stem).
The 3′ CCA is added post-transcriptionally to eukaryotic tRNAs. To better define the termini of the tRNA 3′ fragments tightly bound to Twi12, we deep-sequenced a library from sRNAs enriched by cross-linking to Twi12 in vivo. This population of tRNA 3′ fragments was analyzed for sequence reads that mapped only to the genome-encoded portion of the tRNA or were extended at their 3′ end by addition of C, CC, or CCA (Fig. 2F). About one-third of the reads include at least one post-transcriptionally added nucleotide, indicating that the Twi12-associated tRNA fragments derive at least in part from matured tRNAs. A pronounced 5′ sequence signature was evident for the cross-linked Twi12 sRNAs (Fig. 2G). The 5′ ends of the tRNA fragments correspond to cleavage between the thymidine and pseudouridine (ψ) of the TψC motif, giving rise to a strong 5′ UC bias in the sequenced fragments. The overwhelming representation of cytidine at the second position suggests that tightly bound Twi12 sRNAs vary more in their extent of retained 3′ CCA than in their 5′ end position within the T-loop.
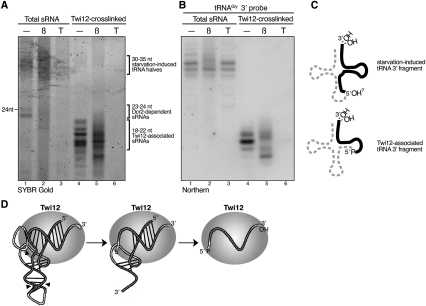
The Twi12-bound tRNA fragments differ from the starvation or other stress-induced tRNA halves (Lee and Collins 2005) in their constitutive accumulation, shorter length, and lower abundance. Notably, the starvation-induced tRNA halves were not detectably associated with Twi12 (Fig. 1B), even if Twi12 was dramatically overexpressed by cadmium induction (data not shown). To investigate potential biochemical differences between the Twi12-bound and starvation-induced tRNA 3′ fragments, we compared the phosphorylation and modification status of the fragment ends. Size-enriched total RNA from starved cells containing predominantly ∼30- to 35-nt tRNA halves and ∼23- to 24-nt products of Dcr2 (Fig. 3A, lane 1) or Twi12-bound sRNAs enriched by affinity purification after in vivo cross-linking (Fig. 3A, lane 4) were subject to nuclease or chemical treatment. The entire pool of RNAs was visualized by direct staining with SYBR Gold (Fig. 3A), and specific tRNA 3′ fragments were detected by blot hybridization (Fig. 3B; additional data not shown). The 5′ monophosphate-dependent nuclease Terminator degraded the ∼23- to 24-nt sRNA products of Dcr2 but not the starvation-induced tRNA 3′ halves (Fig. 3A,B, lanes 3), consistent with the known specificities of Dicer and the nucleases responsible for stress-induced tRNA cleavage in yeast and mammals (Fu et al. 2009; Thompson and Parker 2009b; Yamasaki et al. 2009). Unlike the starvation-induced tRNA 3′ halves, the Twi12-bound tRNA 3′ fragments were degraded by Terminator treatment (Fig. 3A,B, lanes 6). This result establishes that the tightly bound Twi12 sRNAs possess a 5′ monophosphate. A substantial fraction of the ∼23- to 24-nt Dcr2 products, ∼30- to 35-nt starvation-induced tRNA 3′ halves, and ∼18- to 22-nt Twi12-bound tRNA 3′ fragments were reactive to β elimination, indicating the presence of both 2′ and 3′ hydroxyl groups at the RNA 3′ end (Fig. 3A,B, lanes 2,5).
Figure 3.
Distinct biochemical features of the Twi12-bound and starvation-induced tRNA 3′ fragments. (A–C) Size-enriched total RNA from cells harvested 3 h after initiation of starvation (lanes 1–3) and cross-linked Twi12 sRNAs from vegetative growth (lanes 4–6) were subjected to β elimination or Terminator exonuclease treatment followed by direct staining (A) or blot hybridization (B) to detect a specific tRNA 3′ fragment. The tRNAGly 3′ fragment detected by blot hybridization is depicted in C as a solid black line against the dashed full-length tRNA. (D) A speculative model is shown for Twi12 tRNA fragment loading and release. The line drawing is based on previous illustrations of tRNA tertiary structure (Purves et al. 1995).
These assays demonstrate that Twi12-bound tRNA 3′ fragments have biochemical features distinct from the starvation-induced tRNA 3′ halves (summarized in Fig. 3C), including the presence of a 5′ monophosphate characteristic of most Ago/Piwi-bound sRNAs. Consistent with known principles of Ago/Piwi sRNA strand incorporation and displacement (Tolia and Joshua-Tor 2007; Kawamata et al. 2009; Siomi and Siomi 2009), we envision that Twi12 binds initially to full-length or endonucleolytically cleaved tRNAs retaining most of the tRNA secondary structure (Fig. 3D). The tRNA fold stacks the acceptor and T-loop stems, presenting a relatively canonical length of sRNA duplex for Ago/Piwi interaction. The cleavage reactions that generate a 5′ monophosphate terminus at the ψ of the TψC motif may provide an optimal 5′ end for recognition by Twi12. The 3′ end of the tightly bound sRNA is typically not a complete CCA, which could reflect endogenous turnover of this sequence prior to Twi12 association or the trimming of a bound tRNA fragment to optimal length for end protection. The tRNA 5′ fragment initially base-paired to the 3′ fragment would have less-protected ends, allowing nuclease nibbling and, ultimately, loss of the 5′ fragment via nuclease degradation, helicase unwinding, or passive dissociation.
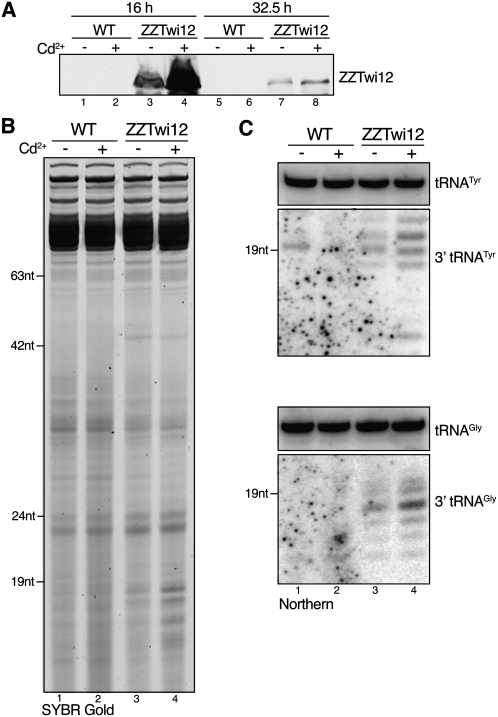
Selective retention of tRNA 3′ fragments as the guide strands of Twi12 RNPs predicts that steady-state accumulation of this specific tRNA fragment population should depend on the cellular level of Twi12. Following unsuccessful attempts to engineer inducible TWI12 genetic depletion, we addressed whether Twi12-associated sRNAs were altered in steady-state accumulation by transient protein overexpression. Cadmium addition results in a rapid increase in expression from the MTT1 promoter, followed by a decline as cadmium is chelated by induced metallothionein proteins. Transgene-encoded ZZTwi12 was robustly induced from its basal expression level by cadmium addition during vegetative growth (Fig. 4A, lanes 1–4) and then declined in level as cells continued to divide, until reaching the maximal density of stationary phase (Fig. 4A, lanes 5–8). Notably, Twi12 overexpression increased the accumulation of RNAs smaller than the ∼23- to 24-nt sRNA products of Dcr2 (Fig. 4B), which were detected as tRNA 3′ fragments by blot hybridization (Fig. 4C). The increased steady-state accumulation of tRNA 3′ fragments was not accompanied by an increase in accumulation of the 5′ fragments or the stress-induced tRNA halves (Supplemental Fig. S3), consistent with selective stabilization of the 3′ fragments in Twi12 RNPs.
Figure 4.
Increased tRNA 3′ fragment accumulation upon Twi12 overexpression. (A) ZZTwi12 accumulation in wild-type or transgene-containing cells grown with or without cadmium (Cd2+) was determined by immunoblot against the epitope tag at the indicated time points of continuous growth. Cadmium was first administered at time 0 (0.75 μg/mL), and was supplemented again after 12 h (0.3 μg/mL) and 21.5 h (0.5 μg/mL), followed by overnight culture to the maximal cell density of stationary phase. (B,C) Total RNA was harvested and size-enriched from cells in stationary phase (32.5 h), followed by direct staining (B) or blot hybridization (C). Similar results were observed using samples from cells harvested prior to reaching maximum density (data not shown), but stationary-phase culture sRNAs are shown here to allow comparison of Twi12-bound tRNA 3′ fragments and all other tRNA fragment populations, including tRNA halves (see Supplemental Fig. S3).
What is the biological function of Twi12 RNPs? Twi12-faciliated tRNA turnover may be important in tRNA quality control or in balancing tRNA levels to improve translational fidelity (Dittmar et al. 2006; Kramer and Farabaugh 2007; Phizicky and Hopper 2010). Also, although Twi12 does not conserve the active site residues of some Ago/Piwi proteins, the tRNA fragments could provide sequence specificity for target RNA or DNA regulation. Nonabundant sRNA populations notably similar to the Twi12-bound sRNAs have been detected in total RNA of rapidly dividing mouse and human cell lines. In mouse embryonic stem cells, beneath the abundant reads from Dicer processing of a misfolded tRNA precursor, some Dicer-independent tRNA 5′ and 3′ end fragments are similar in length to the Twi12-bound sRNAs (Babiarz et al. 2008). Also, among the tRNA fragment populations sequenced from total RNA of human liver carcinoma or prostate cancer cell lines, there are tRNA 3′ fragments with the same length, 5′ end position, 3′ CCA residues, and 5′ monophosphate terminus (when possible to infer from the cloning method) as the Twi12-bound sRNAs (Kawaji et al. 2008; Lee et al. 2009). Thus, the unique loading specificity and function of Twi12 may represent a ciliate evolutionary adaptation for handling the products of a conserved tRNA endonucleolytic processing reaction. Whether animal Piwi proteins are also loaded with ∼20-nt tRNA 3′ fragments remains to be investigated in a suitable physiological context.
Materials and methods
Strains and extracts
Strain construction and culture growth were performed as described previously (Couvillion et al. 2009) in the CU522 strain background. Transgene-encoded Twi12 is expressed from the endogenous TWI12 ORF. Purifications under condition 1 (C1) were performed as described previously (Lee and Collins 2007). For condition 2 (C2), extract preparation was modified by cell lysis in a volume ∼10-fold that of the packed cell pellet, addition of 0.1% Triton X-100, doubled monovalent ion concentration (100 mM NaCl), omission of β-mercaptoethanol, and clearing by centrifugation at 16,000g for 15 min. For cross-linking, cells were washed in 1× PBS, resuspended to 5 × 105 cells per milliliter, and swirled with 0.75% formaldehyde for 10 min. The reaction was quenched by addition of 125 mM Tris-HCl (pH 7.5) for 5 min. Cells were then washed twice in 1× PBS, resuspended to 5 × 106 cells per milliliter in RIPA (50 mM Tris-HCl at pH 7.5, 1% Igepal, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl) plus protease inhibitors, and sonicated in 15-sec pulses at 20% amplitude until fully lysed (2–2.5 min). Lysate from cross-linked cells was cleared at 16,000g. After Twi12 binding to IgG agarose, resin was washed three times for 10 min each at room temperature in RIPA-HS (RIPA with 500 mM NaCl, 1% sodium deoxycholate, 0.1% SDS) with the indicated final concentration of urea.
Nucleic acid analyses
DNA blots were hybridized with hexamer-primed probes. RNA blots were hybridized with 5′ end-labeled 20- to 22-nt oligonucleotides perfectly complementary to the indicated tRNA, with an endpoint at the mature tRNA 5′ or 3′ end (excluding the 3′ CCA). Total RNA was isolated and size-enriched as described previously (Lee and Collins 2006). Terminator exonuclease (Epicentre) was used based on the manufacturer's protocol, and β elimination was performed as described (Couvillion et al. 2009). Blotting to detect the ZZ tag was performed using whole-cell lysates (Couvillion et al. 2009). Library preparation, deep sequencing, and annotation were performed largely as described (Couvillion et al. 2009), with additional consideration for tRNA-specific features (see the Supplemental Material). Reads from the sRNA libraries are deposited at Gene Expression Omnibus (accession no. GSE24916, data sets GSM612779–GSM612780).
Acknowledgments
We thank Leath Tonkin of the Vincent J. Coates Genomics Sequencing Laboratory for sRNA library sequencing.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1996210.
Supplemental material is available for this article.
References
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K 2009. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev 23: 2016–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T 2006. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2: e221 doi: 10.1371/journal.pgen.0020221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442 [DOI] [PubMed] [Google Scholar]
- Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A 2010. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 171: 64–73 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16: 673–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till RA, Yao MC 2006. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol Cell Biol 26: 8731–8742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. 2008. Hidden layers of human small RNAs. BMC Genomics 9: 157 doi: 10.1186/1471-2164-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y 2009. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16: 953–960 [DOI] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749 [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K 2006. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev 20: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol 14: 604–610 [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL 2005. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol 25: 9151–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2005. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev 19: 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves WK, Orians GH, Heller HC 1995. Life, the science of biology. W.H. Freeman, Salt Lake City, UT [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T 2010. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci 99: 3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2009. On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2009a. Stressing out over tRNA cleavage. Cell 138: 215–219 [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2009b. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L 2007. Slicer and the argonautes. Nat Chem Biol 3: 36–43 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]