Abstract
Energy homeostasis depends on insulin signaling in metazoans. Insulin levels reflect the nutritional status of the animal to control levels of circulating sugar and regulate storage of resources in the form of glycogen and fat. Over the past several years, evidence has begun to accumulate that insulin production and secretion, as well as cellular responsiveness to insulin, are subject to regulation by microRNAs. Here we present evidence that miR-14 acts in the insulin-producing neurosecretory cells in the adult Drosophila brain to control metabolism. miR-14 acts in these cells through its direct target, sugarbabe. sugarbabe encodes a predicted zinc finger protein that regulates insulin gene expression in the neurosecretory cells. Regulation of sugarbabe levels by nutrients and by miR-14 combines to allow the fly to manage resource mobilization in a nutritionally variable environment.
Keywords: MicroRNA, metabolism, obesity, gene expression, neurosecretory cell
In mice, miR-375 affects metabolism by regulating insulin secretion in pancreatic β cells (Poy et al. 2004). Mutant mice lacking miR-375 show defects in glucose homeostasis, apparently due to reduced pancreatic β-cell mass (Poy et al. 2009). Mammalian microRNAs (miRNAs) also regulate insulin levels by other means. miR-9 and miR-96 have been implicated in insulin release through up-regulation of granuphilin, a negative regulator of secretion involved in vesicle docking (Lovis et al. 2008).
In Drosophila, miRNAs have been implicated in regulating insulin responsiveness in a metabolically important organ called the fat body. The fat body combines the functions of liver and adipose tissue and plays a central role in coordinating metabolism and growth of the organism during development. miR-278 mutant flies show insulin resistance in most tissues, but most strongly in the fat body (Teleman et al. 2006). miR-278 acts by regulating expression of the expanded gene. Expanded is best known as a membrane-associated FERM domain protein, which negatively regulates the Hippo signaling pathway (Cho et al. 2006; Hamaratoglu et al. 2006). However, the Hippo pathway does not appear to be involved in this context. A second miRNA, miR-8, also acts in fat body to control insulin signaling (Hyun et al. 2009). miR-8 and its vertebrate ortholog, miR-200, indirectly activate phosphatidylinositol 3-kinase (PI3K) by repressing the expression of their conserved targets, U-shaped/FOG2. FOG2 protein binds to the regulatory subunit of PI3K and prevents formation of the active enzyme complex. Thus, loss of mir-8 leads to reduced insulin sensitivity in the fat body, with effects on growth and metabolism.
In Drosophila, metabolic control by insulin production depends on insulin gene expression in a set of 14 neurosecretory cells in the brain (Brogiolo et al. 2001; Cao and Brown 2001; Ikeya et al. 2002). The developmental origin of these insulin-producing cells (IPCs) suggests an evolutionary relationship to mammalian pancreatic β cells (Wang et al. 2007; Clements et al. 2008). Flies lacking IPCs have elevated carbohydrate and fat levels (Rulifson et al. 2002; Broughton et al. 2005), indicating a role for IPCs in metabolic control. Previous reports have shown that miR-14 mutant flies present defects related to apoptosis, stress response, survival, and metabolism (Xu et al. 2003). Misregulation of the Ecdysone receptor (EcR) was shown to be the cause of the pupal stage survival and metamorphosis defects and the reduced adult life span, but was not responsible for the obesity observed in the mutant adult flies (Varghese and Cohen 2007). In this study, we provide evidence that the metabolic function of miR-14 depends on its activity in the IPCs, and explore its mechanism of action.
Results and Discussion
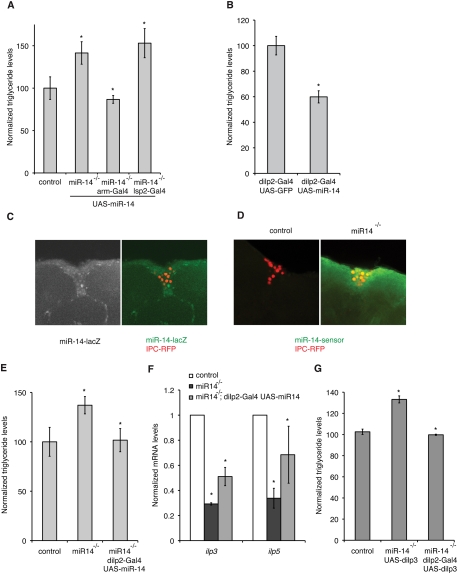
To explore the basis for the metabolic defect in miR-14 mutant flies, we sought to use tissue-specific rescue of the mutant phenotype as a means to determine where miR-14 expression is required. We first confirmed that expression of a UAS-miR-14 transgene under the control of a ubiquitously expressed Gal4-driver (armadillo-Gal4) could rescue the obesity phenotype. To assess obesity, we compared the ratio of total body triglyceride with total body protein. miR-14 mutant flies showed an elevated fat to protein ratio, but this was restored to normal in the rescued mutant (Fig. 1A). Interestingly, the mutant was not rescued by expressing UAS-miR-14 in the adipose tissue, using the fat body-specific driver lsp2-Gal4 (Fig. 1A). This finding suggested that miR-14 does not act in the adipose tissue to regulate triglyceride levels.
Figure 1.
miR-14 acts in the insulin-producing neurosecretory cells to control insulin production. (A,B,E,G) Histograms showing the ratio of total body triglyceride to total body protein (genotypes as indicated). Data are mean ± standard deviation (SD) based on three independent biological replicates. P-values were determined using Student's t-test. (A) Genetic rescue of miR-14 mutants by expression of a UAS-miR-14 transgene. P = 0.02 comparing control and miR-14 mutant; P = 0.01 comparing miR-14 mutant and miR-14 mutant, arm-Gal4-UAS-miR-14; P = 0.41 comparing miR-14 mutant and miR-14 mutant, lsp2-Gal4-UAS-miR-14. (B) Overexpression of miR-14 in the IPCs using dILP2-Gal4. P = 0.03 comparing dilp2-Gal4 UAS-GFP and dilp2-Gal4 UAS-miR-14. (C) miR-14 lacZ expression in the adult brain visualized using anti-β−Gal (green). IPCs labeled by dILP2-Gal4-directed expression of UAS-nuclear RFP (red). (D) miR-14 sensor GFP in control and miR-14 mutant adult brain (green). IPCs labeled by dILP2-Gal4 UAS-nRFP (red). (E) Rescue of the miR-14 mutant by IPC-specific expression of miR-14 using dILP2-Gal4. P = 0.007 comparing miR14 mutant with miR14 mutant, dilp2-Gal4 UAS-miR-14. (F) Insulin-like peptide mRNA levels measured by quantitative RT–PCR (qRT–PCR). Data represent three independent experiments, normalized to RP49 mRNA and then to the level in control flies. For ilp3, P = 2.4E-09 comparing control with miR-14 mutant; P = 0.001 comparing miR14 mutant with miR14 mutant, dilp2-Gal4 UAS-miR-14. For ilp5, P = 1.7E-05 comparing control with miR-14 mutant; P = 0.01 comparing miR14 mutant with miR-14 mutant, dilp2-Gal4 UAS-miR-14. (G) Rescue of the miR-14 mutant by IPC-specific expression of UAS-dilp3 using dILP2-Gal4. P-value = 0.0001 comparing control and miR-14 mutant, UAS-ilp3 (without the Gal4 driver, middle); P-value = 3.3E-05 comparing miR-14 mutant UAS-ilp3 with miR-14 mutant, dilp2-Gal4 UAS-ilp3 (with the Gal4 driver).
We made use of the observation that miR-14 overexpression can make flies lean (Xu et al. 2003) to identify tissues in which miR-14 activity can influence fat levels (Supplemental Fig. S1). Expression of miR-14 in the CNS using a pan-neuronal driver (Supplemental Fig. S1A)—or, more selectively, in the neurosecretory IPCs using dILP2-Gal4—produced lean flies (Fig. 1B). A miR-14 lac-Z reporter transgene showed expression in the IPCs and in most other cells of the brain (IPCs are identified by dilp-Gal4 UAS-nRFP) (Fig. 1C; Supplemental Fig. S2A,B). Figure 1D shows expression of a miR-14 sensor (a ubiquitously expressed GFP reporter transgene with perfect miR-14 sites in the 3′ untranslated region [UTR]). Use of perfect miR-14 sites allows for strong down-regulation of the sensor in miR-14-expressing cells. This is illustrated by comparing sensor levels in miR-14 mutant clones and neighboring miR-14/+ heterozygous tissue in larval wing discs (Supplemental Fig. S2C). Similarly, miR-14 sensor levels were very low in control brains, but increased considerably in miR-14 mutant brains, indicating that miR-14 is active in the IPCs as well as other cells of the brain (Fig. 1D). Restoring miR-14 expression selectively in the IPCs of otherwise miR-14 mutant flies proved to be sufficient to restore fat levels to normal (Fig. 1E). Although miR-14 is broadly expressed and active in the brain, this tissue-specific rescue identifies the IPCs as a key site of miR-14 function in the control of metabolism.
How does miR-14 expression in the IPCs affect fat levels? Previous work has shown that systemic insensitivity to insulin leads to an obese phenotype (Böhni et al. 1999; Tatar et al. 2001). Flies lacking IPCs or with reduced insulin-like peptide (ILP) levels store excess fat (Hwangbo et al. 2004; Broughton et al. 2005; Pospisilik et al. 2010). We therefore examined expression of the genes encoding IPC-specific ILPs in the miR-14 mutant. The nutrient-sensitive ilp (ilp3 and ilp5) (Ikeya et al. 2002) mRNA levels were reduced (Fig. 1F). This was partially rescued by miR-14 expression in the mutant IPCs (Fig. 1F). The level of the non-nutrient-sensitive ilp2 mRNA was also reduced, but to a lesser extent (Supplemental Fig. S1B). Interestingly, restoring ilp3 expression in the IPCs by expression of a UAS-ilp3 transgene was sufficient to restore normal fat levels in the miR-14 mutant (Fig. 1G). This suggests that reduced ilp gene expression in the IPCs is sufficient to explain the metabolic effects of removing miR-14.
The observation that IPC-specific expression was sufficient to rescue the miR-14 phenotype provided a means to search for miR-14 targets that are functionally relevant in these cells. Expression profiling was performed using RNA isolated from adult heads of control flies, miR-14 mutants, and IPC-rescued miR-14 mutants. Of 810 mRNAs up-regulated by >1.5-fold (P < 0.05) in the mutant, 165 were reduced by >50% in the mutant with miR-14 expression restored selectively in the IPCs. Forty-six of these had predicted miR-14 target sites in their 3′UTRs. Ten of the 46 were among mRNAs enriched by Ago1 immunoprecipitation from S2 cells (Hong et al. 2009), where miR-14 is abundantly expressed. These 10 mRNAs were selected for further analysis (Supplemental Table S1).
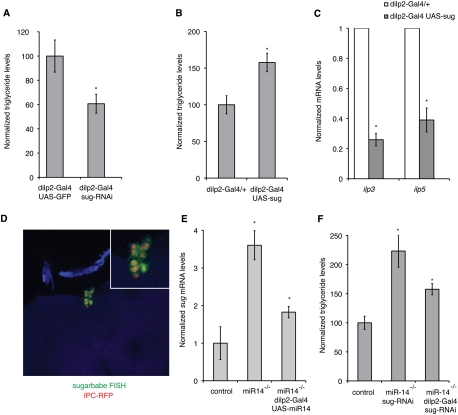
dILP2-Gal4 was used to express UAS-RNAi transgenes to reduce expression of the 10 candidates in the IPCs. This should mimic the effects of miR-14 overexpression, which makes flies lean. Two of the candidates, sugarbabe and fuzzy, produced a lean phenotype when depleted by RNAi (Fig. 2A; Supplemental Table S1). As a second test, we asked whether overexpression in the IPCs could mimic the miRNA mutant phenotype. Overexpression of fuzzy had no effect, but sugarbabe overexpression in the IPCs led to elevated fat levels (Fig. 2B; Supplemental Table S1) and reduced levels of ilp transcripts (Fig. 2C; Supplemental Fig. S3A). Similar to what was observed in the miR-14 mutant flies, the effect of sugarbabe overexpression was more pronounced on the levels of the two nutrient-sensitive ilp transcripts.
Figure 2.
Genetic evidence that miR-14 acts via sugarbabe. (A,B,F) Ratio of total body triglyceride to total body protein. (C,E) mRNA levels measured in flies of the indicated genotypes. Data are presented as mean ± SD based on three independent biological replicates. P-values determined using Student's t-test. (A) Depletion of sugarbabe by expression of a UAS-RNAi transgene in IPCs reduced fat levels. P = 0.006 comparing dilp2-Gal4 UAS-GFP with dilp2-Gal4 UAS-sug-RNAi. (B) Overexpression of sugarbabe in IPCs increased fat levels. P = 0.002 comparing dilp2-Gal4/+ with dilp2-Gal4 UAS-sug. (C) Overexpression of sugarbabe reduced expression of ilp mRNAs. P = 1.2E-08 for ilp3 mRNA; P = 2.8E-05 for ilp5 mRNA. (D) sugarbabe expression in the adult IPCs using FISH (green). IPCs marked by dILP2-Gal4 nRFP (red). (E) sugarbabe mRNA levels from expression-profiling data. P = 0.0013 comparing control with miR-14 mutant; P = 0.0052 comparing miR-14 mutant with rescue-miR-14 mutant, dilp2-Gal4 UAS-miR-14. (F) Partial suppression of miR-14 mutant phenotype by IPC-specific depletion of sugarbabe. P = 1.96E-05 comparing control with miR-14 mutant, UAS-sug-RNAi (without the Gal4 driver, middle); P = 0.001 comparing miR-14 mutant, UAS-sug-RNAi with miR-14 mutant, dilp2-Gal4 UAS-sug-RNAi (with the Gal4 driver).
These experiments suggest that sugarbabe might be a biologically relevant target of miR-14 activity in the IPCs. Fluorescent in situ hybridization (FISH) was used to visualize sugarbabe mRNA expression in the adult brain. sugarbabe mRNA was present in the IPC cells (identified by dILP2-Gal4-directed expression of a UAS-nRFP transgene) (Fig. 2D). Little or no sugarbabe mRNA was detected in other parts of the brain.
As a more stringent test of sugarbabe function, we asked whether limiting its overexpression selectively in the IPCs would reduce the severity of the miR-14 mutant phenotype. IPC-specific expression of a sugarbabe UAS-RNAi transgene reduced sugarbabe mRNA levels in the adult head by ∼30% and resulted in increased ilp transcript levels (Supplemental Fig. S3B). sugarbabe transcript levels increased approximately threefold in miR-14 mutant head RNA (Fig. 2E; Supplemental Fig. S3C), and this was partially offset by IPC-specific expression of the UAS-RNAi transgene (Supplemental Fig. S3C). This also lowered fat levels (Fig. 2F). These assays point to sugarbabe as a functionally important miR-14 target in the IPCs.
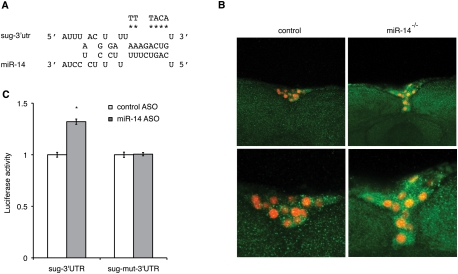
There is one predicted miR-14 site in the sugarbabe 3′UTR (Fig. 3A). This site is conserved in the eight species of the Drosophilia melanogaster and obscura groups. To test its function in vivo, we prepared transgenic flies expressing a GFP reporter with the intact sugarbabe 3′UTR. GFP expression levels were higher in the IPCs and in other brain cells in miR-14 mutant flies compared with wild-type controls (Fig. 3B). This indicates that endogenous levels of miR-14 are sufficient to negatively regulate gene expression via the sugarbabe 3′UTR in the IPCs in vivo. As a further test, we made use of a luciferase reporter with the intact sugarbabe 3′UTR and a second version in which the site was altered to disrupt pairing to the miRNA seed (Fig. 3A). Because S2 cells endogenously express miR-14, we asked whether depleting miR-14 would lead to elevated expression of the luciferase reporters. The reporter with the mutated site was unchanged, but the reporter with the intact miR-14 site showed a 40% increase in activity in miR-14-depleted cells (Fig. 3C). Reciprocally, overexpression of miR-14 caused reduced luciferase activity from the intact reporter (Supplemental Fig. S3D). These results suggest that miR-14 can act directly via the predicted target site to regulate sugarbabe levels.
Figure 3.
miR-14 regulates sugarbabe expression. (A) Predicted miR-14 target site in the sugarbabe 3′UTR. Asterisks indicate residues changed in the target site mutant. (B) sugarbabe 3′UTR reporter transgene showing GFP expression in control and miR-14 mutant brains (green). IPCs marked by dILP2-Gal4 nRFP (red). (C) Luciferase assays showing regulation of a sugarbabe 3′UTR reporter. S2 cells were transfected to express a firefly luciferase sugarbabe 3′UTR reporter or a version of the reporter with the site mutated. Cells were cotransfected with a renilla luciferase reporter as a control for transfection efficiency. Data show the ratio of firefly to Renilla luciferase activity. Cells were treated with antisense oligonucleotide to deplete miR-14 or with a control oligonucleotide. Data represent mean ± SD for three independent biological replicates. P = 4.9E-05 (Student's t-test).
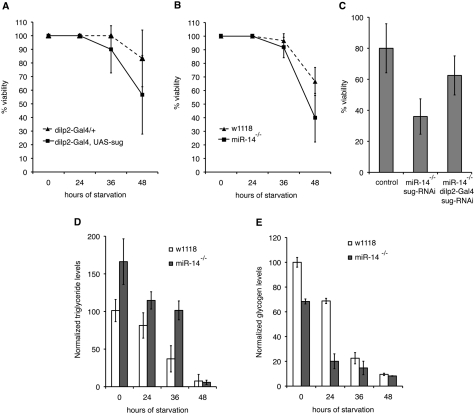
In the laboratory, flies are reared on a nutritionally rich medium containing protein, amino acids, fats, and complex sugars. Natural environments offer a more variable nutrient supply, and the ability to adapt to changing conditions may confer an advantage. Like other animals, flies store energy in the form of glycogen and fat. We examined the contribution of miR-14 and sugarbabe to their response to nutrient stress. For these experiments, larvae were reared under controlled growth conditions on rich medium, and adult flies were aged on this medium for 4 d before being deprived of nutrients. miR-14 mutant flies were more sensitive to nutrient deprivation, as were flies selectively overexpressing sugarbabe in the IPCs, which phenocopies the miR-14 mutant (Fig. 4A,B). We confirmed that the reduced survival of the miR-14 mutants under these conditions was due to elevated sugarbabe levels by selectively expressing the sugarbabe RNAi transgene in the IPC cells of the miR-14 mutant. Partially compensating for the elevated sugarbabe mRNA level improved survival of the mutant flies (Fig. 4C)
Figure 4.
miR-14 effects on nutrient deprivation. (A,B) Survival curves of adult male flies during nutrient deprivation. Flies were reared and aged under controlled conditions and deprived of nutrients at t = 0. Data represent three independent biological replicates. (A) P = 0.105 (36 h) and P = 0.005 (48 h) comparing dilp-Gal4 with dilp-Gal4 UAS-sug. (B) P = 0.045 (36 h) and P = 0.13 (48 h) comparing control w1118 with miR-14 mutant. (C) Partial suppression of the reduced viability of miR-14 mutants by IPC-specific depletion of sugarbabe at 48 h. P = 0.0005 control versus miR-14, UAS-sug-RNAi (without the Gal4 driver, middle); P = 0.001 comparing miR-14 mutant, UAS-sug-RNAi with miR-14 mutant, dilp2-Gal4 UAS-sug-RNAi (with the Gal4 driver). (D) Ratio of triglyceride to protein on nutrient-deprived flies, reared as in B. P = 0.02 (0 h), P = 0.02 (24 h), P = 0.003 (36 h), and P = 0.38 (48 h) comparing control w1118 with miR-14 mutant. (E) Glycogen levels of nutrient-deprived flies, reared as in B. P = 0.001 (0 h), P = 9.28E-05 (24 h), P = 0.06 (36 h), and P = 0.04 (48 h) comparing control w1118 with miR-14 mutant.
At the start of the experiment, before nutrient deprivation, fat levels were higher in the mutant (Fig. 4D), but glycogen levels were lower—an expected outcome of reduced insulin signaling (Fig. 4E). During nutrient deprivation, glycogen drops more quickly than in control flies, but reaches similar low levels by 36 h. Fat reserves were initially mobilized more slowly, but then dropped sharply at the stage when the mutant flies begin to die (Fig. 4D). Thus, there is an initial metabolic imbalance in the miR-14 mutants, with nutrient reserves shifted more toward fat storage, perhaps at the expense of glycogen. The resulting imbalance in nutrient mobilization may be responsible for the impaired survival of the mutant flies when nutrient-deprived.
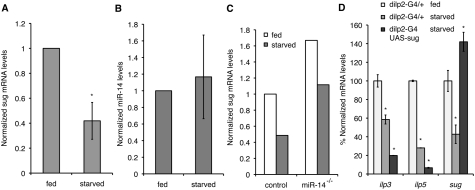
Overexpression of sugarbabe in the IPCs is sufficient to mimic the effects of the miR-14 mutant on fat storage, expression of the nutritionally sensitive ilp transcripts, and starvation sensitivity. To further explore their function in this context, we asked whether their expression in the adult head was nutrient-responsive. sugarbabe has been shown previously to be up-regulated in the gut, malphigian tubules, and fat body tissues of larvae fed on a high-sugar diet (Zinke et al. 2002). We found that sugarbabe mRNA levels decreased by >50% in RNA from adult heads (Fig. 5A), whereas miR-14 levels were not affected significantly (Fig. 5B). The change in sugarbabe levels was also seen in nutrient-deprived miR-14 mutants (Fig. 5C), showing a miR-14-independent change of sugarbabe during starvation. Thus, sugarbabe expression is controlled in the IPCs by two independent means.
Figure 5.
Responses to nutrient deprivation. (A) sugarbabe mRNA levels measured by qRT–PCR in w1118 control flies reared under controlled conditions. “Starved” indicates flies deprived of nutrients for 24 h. Data represent three independent biological replicates. P = 0.0002. (B) miR-14 miRNA levels measured by qPCR. P = 0.3. (C) sugarbabe mRNA levels compared in control and miR-14 mutants under fed and nutrient-deprived conditions. (D) ilp3, ilp5, and sugarbabe mRNA levels measured by qRT–PCR in dilp2-Gal4/+ control flies and dilp2-Gal4 UAS-sugarbabe (Gal4-directed sugarbabe expression in IPCs) reared and treated as in A. Data represent three independent biological replicates. For ilp3 levels, P = 0.018 comparing fed versus starved control flies; P = 0.007 comparing starved control and sugarbabe-overexpressing flies. For ilp5 levels, P = 0.003 comparing fed and starved control flies; P = 0.0002 comparing starved control and sugarbabe overexpression flies. For sugarbabe levels, P = 0.03 comparing fed and starved control flies; P = 0.01 comparing starved control and sugarbabe-overexpressing flies.
Sugarbabe encodes a predicted zinc finger protein (Zinke et al. 2002). Our findings suggest that sugarbabe levels in the IPC are under nutritional regulation, and that this in turn controls the levels of ilp mRNAs. Sugarbabe might act as a transcription factor to regulate ilp gene expression, but we do not exclude the possibility that its effects are post-transcriptional, affecting RNA processing or stability.
How do the nutrition-dependent changes in sugarbabe expression correlate with regulation of ilp transcript levels? Based on the data presented so far, the observed reduction in sugarbabe levels upon nutrient deprivation is expected to elevate ilp transcript levels. Yet, ilp levels decrease under these conditions. To address this issue, we examined the effects of offsetting the decrease in sugarbabe levels by expressing UAS- sugarbabe in the IPCs under dilp2-Gal4 control. Under these conditions, the levels of ilp3 and ilp5 transcripts decreased by considerably more than in the nutrient-deprived controls (Fig. 5D). This indicates that down-regulation of sugarbabe during nutrient deprivation serves to limit the effects of nutrient deprivation on ilp gene expression.
Our findings highlight the importance of sugarbabe levels in the IPCs as a regulator of energy balance. Under normal circumstances, elevated fat levels can confer resistance to starvation (Hader et al. 2003; Gronke et al. 2007). However, flies with defects in fat mobilization can be hypersensitive to starvation despite being obese (Gronke et al. 2007). Impairing insulin signaling has been shown to enhance fat storage (Böhni et al. 1999; Tatar et al. 2001; Hwangbo et al. 2004; Broughton et al. 2005), but it can also sensitize flies to starvation in some contexts (Clancy et al. 2001). miR-14 mutants show a similar syndrome of low insulin and obesity coupled with starvation sensitivity. Our findings suggest that nutritional regulation of sugarbabe might serve to limit the reduction of insulin expression during starvation. We speculate that maintaining a sufficient level of insulin activity might be important to allow mobilization of nutrient reserves from fat stores. The finding that sugarbabe is nutritionally controlled, while miR-14 levels are not, provides the opportunity for an interplay between nutritionally dependent and independent regulation of insulin production.
Materials and methods
Reagents and fly strains
Reagents and fly strains used were as follows: miR-14 mutant and P{lacW}miR-14k10213 (Xu et al. 2003); UAS-miR-14 and tub-miR-14 (Varghese and Cohen 2007); UAS-sugarbabe (Zinke et al. 2002); dilp2-Gal4 and UAS-dilp3 (Ikeya et al. 2002). armadillo-Gal4, elav-Gal4, and UAS-GFP were from the Bloomington Stock Center (http://flybase.bio.indiana.edu). lsp2-Gal4 was provided by Bassem Hassan. UAS-sugarbabe-RNAi (Dietzl et al. 2007). Other RNAi lines were from Vienna Drosophila RNAi Center (VDRC), National Institute of Genetics (NIG) RNAi Stock Center, and Bloomington Stock Center. miR-14-sensor GFP was generated as described (Brennecke et al. 2003). sug-3′UTR GFP and luciferase reporters were generated as described (Varghese and Cohen 2007).
Conditions for phenotype analysis
Controlled growth conditions were used for all experiments related to metabolism. First instar larvae were collected within 2–3 h of hatching. GFP balancers were used to allow genotyping. Fifty larvae were placed in fresh vials to maintain uncrowded conditions. Adult males were picked within 6 h of emergence and aged for 5 d (10 per vial) before triglyceride measurement or quantitative real-time PCR assays. For starvation, flies were collected from similar conditions. They were then transferred to normal fly food vials or vials containing only 1.5% agar and collected at the indicated time points.
Triglyceride and glycogen measurements
Batches of 10 flies were homogenized using Sartorius Potter-S tissue homogenizer. After 5 min of heat inactivation at 70°C and centrifugation at 13000 rpm for 3 min, triglyceride and protein levels were measured using the Sigma Triglyceride kit and Bio-Rad protein assay reagent. Colorimetric analysis was performed using a TECAN microplate reader in 96-well format. Sample preparation for glycogen measurement was similar to triglycerides, following the manufacturer's protocol (BioVision). Data were normalized to total protein.
Quantitative PCR (qPCR)
Batches of 20 flies were collected, snap-frozen in liquid nitrogen, and shaken to separate head and body. Total RNA was extracted from heads using Trizol. RNA samples were treated with DNase1 (Qiagen RNeazy kit). Reverse transcription used oligo-dT primers and SuperScript RT-III (Invitrogen) and the cDNAs used for qPCR with Power SYBR Green (Applied Biosystems) and ABI-7500 Fast qPCR machine.
The following primers were used: dilp2 FP, 5′-GGCCAGCTCCACAGTGAAGT-3′; dilp2 RP, 5′-TCGCTGTCGGCACCGGGCAT-3′; dilp3 FP, 5′-CCAGGCCACCATGAAGTTGT-3′; dilp3 RP, 5′-TTGAAGTTCACGGGGTCCAA-3′; dilp5 FP, 5′-TCCGCCCAGGCCGCAAACTC-3′; dilp5 RP, 5′-TAATCGAATAGGCCCAAGGT-3′; sugarbabe FP, 5′-CCGCCAGCGATTTCGTATG-3′; sugarbabe RP, 5′-GCCAGTGCATCCAAGGTGTC-3′.
Transfection and antisense oligonucleotide-mediated depletion of miR-14 in S2 cells
S2 cells were transfected in 24-well plates using 250 ng of tubulin-miR14 plasmid DNA or the empty vector control, 25 ng of firefly luciferase DNA or sug-3′UTR or mutant-3′UTR luciferase reporter DNA, and 25 ng of Renilla luciferase DNA as a transfection control. Dual-luciferase assays were performed 60 h post-transfection according to the manufacturer's protocol (Promega). Antisense oligos were designed and S2 cells were transfected according to Horwich and Zamore (2008): miR-14 ASO, 5′-AGAGAUAGGAGAGAGAAAAAGACUGAAGGAU-3′; control ASO (miR-14 shuffle), 5′-AGAGAUAAGUAGGAGUGAGAAGAGAAAGGAU-3′. ASOs incorporated 2′-O-methyl-modified nucleotides and had a 3′-cholesterol modification (Dharmacon). ASOs and luciferase reporter constructs were transfected using Dharmafect 4. Cells were harvested 72 h later, and dual-luciferase assays were performed.
Microarray analysis
Microarray profiling was performed by the EMBL Gene Core using Affymetrix Drosophila 2.0 arrays. RNA was amplified and labeled using standard protocols for hybridization, washing, and data acquisition (Fluidics Station 400, GeneArray 2500 scanner, Affymetrix Microarray suite version 5.1). Data were normalized using CARMAWeb version 1.4.
FISH and antibody labeling
The sugarbabe antisense RNA probe was synthesized using the DIG-labeling kit (Roche). Anti-DIG-POD primary antibody was used and detected using the Tyramide Signal Amplification kit from Perkin Elmer following standard protocols. Rabbit anti-β-galactosidase antibody was from Cappel. Mouse anti-GFP antibody was from Molecular Probes.
Acknowledgments
We are grateful to V. Benes and T. Bähr-Ivacevic of the EMBL GeneCore for expression profiling, and to Neeti Birla for help with the IPC screen for target genes. This work was supported in part by the Singapore National Research Foundation under CRP Award number NRF-CRP3-2008-03, by the Singapore Millennium Foundation through core funding to the Temasek Life Sciences Laboratory, by IMCB, and by EU-FP6 grant “Sirocco” LSHG-CT-2006-037900.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1995910.
Supplemental material is available for this article.
References
- Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 97: 865–875 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the pro-apoptotic gene hid in Drosophila . Cell 113: 25–36 [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11: 213–221 [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci 102: 3105–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Brown MR 2001. Localization of an insulin-like peptide in brains of two flies. Cell Tissue Res 304: 317–321 [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD 2006. Delineation of a Fat tumor suppressor pathway. Nat Genet 38: 1142–1150 [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104–106 [DOI] [PubMed] [Google Scholar]
- Clements J, Hens K, Francis C, Schellens A, Callaerts P 2008. Conserved role for the Drosophila Pax6 homolog Eyeless in differentiation and function of insulin-producing neurons. Proc Natl Acad Sci 105: 16183–16188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, Kuhnlein RP 2007. Dual lipolytic control of body fat storage and mobilization in Drosophila . PLoS Biol 5: e137 doi: 10.1371/journal.pbio.0050137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hader T, Muller S, Aguilera M, Eulenberg KG, Steuernagel A, Ciossek T, Kuhnlein RP, Lemaire L, Fritsch R, Dohrmann C, et al. 2003. Control of triglyceride storage by a WD40/TPR-domain protein. EMBO Rep 4: 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol 8: 27–36 [DOI] [PubMed] [Google Scholar]
- Hong X, Hammell M, Ambros V, Cohen SM 2009. Immunopurification of Ago1 miRNPs selects for a distinct class of microRNA targets. Proc Natl Acad Sci 106: 15085–15090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Zamore PD 2008. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat Protoc 3: 1537–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo DS, Gersham B, Tu MP, Palmer M, Tatar M 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562–566 [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN 2009. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139: 1096–1108 [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E 2002. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila . Curr Biol 12: 1293–1300 [DOI] [PubMed] [Google Scholar]
- Lovis P, Gattesco S, Regazzi R 2008. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem 389: 305–312 [DOI] [PubMed] [Google Scholar]
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, et al. 2010. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell 140: 148–160 [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230 [DOI] [PubMed] [Google Scholar]
- Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M 2009. miR-375 maintains normal pancreatic α- and β-cell mass. Proc Natl Acad Sci 106: 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R 2002. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 296: 1118–1120 [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107–110 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Maitra S, Cohen SM 2006. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev 20: 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J, Cohen SM 2007. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila . Genes Dev 21: 2277–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tulina N, Carlin DL, Rulifson EJ 2007. The origin of islet-like cells in Drosophila identifies parallels to the vertebrate endocrine axis. Proc Natl Acad Sci 104: 19873–19878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA 2003. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13: 790–795 [DOI] [PubMed] [Google Scholar]
- Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ 2002. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J 21: 6162–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]