Abstract
The positive link between the SWI/SNF and the Gcn5 histone acetyltransferase in transcriptional activation has been well described. Here we report an inhibitory role for Gcn5 in SWI/SNF targeting. We demonstrate that Gcn5-containing complexes directly acetylate the Snf2 subunit of the SWI/SNF complex in vitro, as well as in vivo. Moreover, the acetylation of Snf2 facilitates the dissociation of the SWI/SNF complex from acetylated histones, and reduces its association with promoters in vivo. These data reveal a novel mechanism by which Gcn5 modulates chromatin structure not only through the acetylation of histones, but also by directly acetylating Snf2.
Keywords: Acetylation, Gcn5, SWI/SNF
Yeast SWI/SNF is a 1.2-MDa chromatin remodeling complex composed of 12 different subunits, including the catalytic subunit Swi2/Snf2 (Lee et al. 2004). SWI/SNF uses ATP to alter nucleosome structure, which is necessary to regulate transcription of a subset of yeast genes (Cote et al. 1994; Sudarsanam et al. 2000). SWI/SNF is targeted to the promoter of specific genes by sequence-specific DNA-binding transcription factors that interact with Swi1 and Snf5 subunits (Prochasson et al. 2003).
Gcn5 (KAT2) is the catalytic core of three different HAT complexes (SAGA, SLIK, and ADA) that are important coactivators for a number of genes (Brownell et al. 1996; Lee and Workman 2007). Recently, it was reported that Gcn5 acetylated nonhistone proteins, including Rsc4 and Cdc6 in yeast and proliferator γ coactivator 1 (PGC-1) in mammals (Lerin et al. 2006; VanDemark et al. 2007; Paolinelli et al. 2009).
Genetic and biochemical evidence suggests a functional link between SWI/SNF and Gcn5 (Roberts and Winston 1997; Hassan et al. 2001). For a subset of genes that include PHO8, SUC2, and HO, both Gcn5 and SWI/SNF are required for maximal gene expression (Cosma et al. 1999; Reinke et al. 2001; Mitra et al. 2006). Upon induction of these genes, the targeted Gcn5 complex transiently acetylates promoter-flanking nucleosomes. SWI/SNF is then subsequently or concurrently recruited to the promoter, where it either slides the acetylated nucleosomes or evicts the histones (Reinke et al. 2001; Chandy et al. 2006).
The acetylation of histones by Gcn5 also influences the duration of time for which the SWI/SNF and Gcn5 complexes remain on chromatin. Gcn5 and the Snf2 subunit of the SWI/SNF complex contain bromodomains that recognize and associate with acetylated lysine residues (Dhalluin et al. 1999; Ornaghi et al. 1999; Hudson et al. 2000; Hassan et al. 2002). Indeed, the bromodomain of Snf2 is required for not only the retention of SWI/SNF on acetylated chromatin, but also the removal of acetylated histones from chromatin (Chandy et al. 2006; Hassan et al. 2006). Although the sequential mechanism by which SWI/SNF and Gcn5 activate genes has been demonstrated previously (Hassan et al. 2001; Reinke et al. 2001; Agalioti et al. 2002), it is still unknown how SWI/SNF dissociates from chromatin after remodeling nucleosomes at promoters.
In this study, we demonstrate that Gcn5 acetylates the Snf2 subunit of the SWI/SNF complex. We identify two acetylation sites on Snf2 that are located between tandem AT hook domains. Peptide pull-down assays reveal that acetylation of Snf2 protein inhibits the interaction between its bromodomain and acetylated histones. Furthermore, the bromodomain of Snf2 recognizes the two acetylated lysine residues located between the AT hook domains, suggesting that an intramolecular interaction facilitates dissociation of SWI/SNF from acetylated chromatin. Indeed, chromatin immunoprecipitation (ChIP) assays show that these acetylations regulate the period of retention of SWI/SNF on chromatin in vivo. Overall, our studies demonstrate that Gcn5 modulates the retention of the SWI/SNF complex on chromatin both positively through histone acetylation and negatively through acetylation of the Snf2 subunit. This feedback loop helps explain how chromatin-bound SWI/SNF is able to dissociate from acetylated nucleosomes.
Results and Discussion
Snf2 is acetylated by Gcn5 and is deacetylated by Hst2 and Rpd3
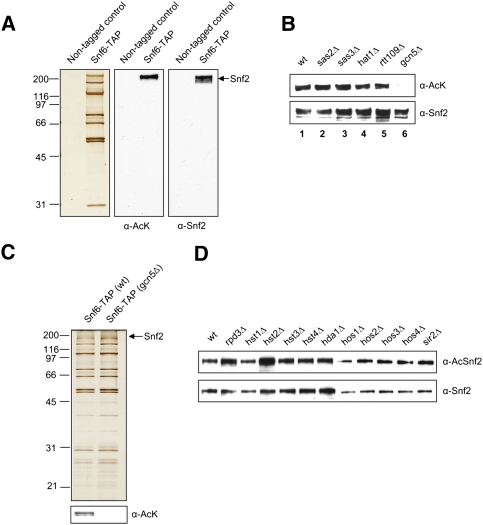
Subunits of chromatin remodeling complexes have been shown to undergo post-translational modifications (PTMs) (Morrison et al. 2007; VanDemark et al. 2007). We investigated whether components of the SWI/SNF complex were post-translationally modified. First, SWI/SNF complexes purified from a Snf6-TAP-tagged strain or untagged strain were probed with anti-acetyl lysine antibody (α-AcK) (Fig. 1A). We observed that the catalytic subunit Snf2 was acetylated (Fig. 1A). We next deleted nonessential histone acetyltransferases (HATs) from the Snf6-TAP-tagged strain to determine the enzyme responsible for Snf2 acetylation. SWI/SNF was immunoprecipitated from equal quantities of whole-cells extracts using IgG-coupled sepharose and was analyzed by Western blotting. Although the levels of Snf2 protein were similar, we did not observe the acetylation of Snf2 in gcn5Δ cells, demonstrating that Gcn5 is required for Snf2 acetylation (Fig. 1B, lane 6).
Figure 1.
Snf2 is acetylated by Gcn5 and deacetylated by Hst2 and Rpd3. (A) TAP-purified proteins from Snf6-TAP-tagged strain (Snf6-TAP) or untagged strain (nontagged control) were resolved with SDS-PAGE and analyzed by immunoblotting with acetyl lysine (α-AcK) and Snf2 (α-Snf2) antibodies. (B) Immunoprecipitated SWI/SNF from either wild-type or mutant cells was analyzed by immunoblotting with α-AcK and α-Snf2. (C) SWI/SNF complexes from wild type or gcn5Δ were resolved with SDS-PAGE and analyzed by silver staining and Western blot using α-AcK. (D) SWI/SNF was immunoprecipitated with Flag agarose from wild type or a series of HDAC deletion mutants. Immunoprecipitated complexes were analyzed with acetylated Snf2 antibody (α-AcSnf2) and α-Snf2.
Then, we tested whether acetylation of Snf2 affects complex integrity. SWI/SNF purified from wild-type or gcn5Δ cells was visualized by silver staining (Fig. 1C). SWI/SNF from gcn5Δ and SWI/SNF from wild type were indistinguishable, suggesting that the complex is intact in the absence of acetylation. It was further confirmed by mass spectrometry analysis (data not shown). Moreover, in order to identify which histone deacetylase (HDAC) is responsible for deacetylation of Snf2, the levels of Snf2 acetylation were examined in a series of HDAC deletion mutants. As shown in Figure 1D, levels of Snf2 acetylation increased in hst2Δ and rpd3Δ, indicating that Rpd3 and Hst2 HDACs are involved in deacetylation of Snf2.
Gcn5 acetylates two lysine residues located between the AT hook domains of Snf2, both in vivo and in vitro
In order to identify the Gcn5 acetylation sites on Snf2, we analyzed PTMs of the SWI/SNF complex purified from either wild type or gcn5Δ by mass spectrometry analysis. Both Lys 1493 (K1493) and Lys 1497 (K1497), located between the Snf2 AT hook domains, were acetylated in the wild type. Neither K1493 nor K1497 acetylation was detected on Snf2 that was purified from gcn5Δ (data not shown).
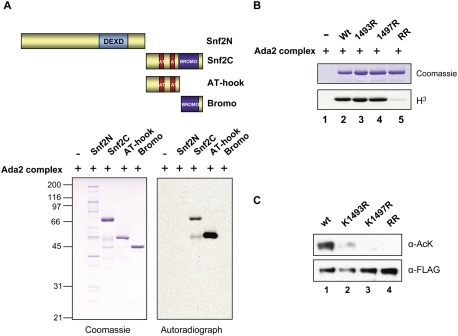
Yeast Snf2 is composed of a DEXD domain in the middle of the protein, followed by two AT hook domains and bromodomain at the C terminus. Snf2 cDNA was chopped into four different pieces, and each piece was subcloned into an N-terminal GST-tagged vector. The four different proteins were then expressed in Escherichia coli and purified using glutathione sepharose (Fig. 2A). Acetylation of the purified proteins was carried out in vitro using TAP-purified Ada2 (Ada2 complex). We used the Ada2 complex, which contained all three Gcn5 complexes, for this assay because we found that all three complexes acetylated Snf2 indistinguishably (Supplemental Fig. S1). Specifically, the two Snf2 segments harboring AT hook domains (Snf2C and AT hook) were acetylated by the Ada2 complex (Fig. 2A). This finding implied that the acetylation sites were localized between the AT hook domains of Snf2. Indeed, this region contained the K1493 and K1497 residues identified earlier by mass spectrometry. Note that these lysine residues are not conserved in other organisms (Supplemental Fig. S2).
Figure 2.
Gcn5 acetylates two lysine residues located between the AT hook domains of Snf2, both in vivo and in vitro. (A) Schematic diagram of Snf2 fragments: Snf2N (1–1383 amino acids), Snf2C (1384–1703 amino acids), AT hook (1384–1546 amino acids), and bromodomain (1547–1703 amino acids). GST-Snf2 fragments acetylated by Ada2 complexes were subjected to SDS-PAGE and visualized by autoradiograph. (B) Wild-type and mutant AT hook proteins acetylated by Ada2 complexes in vitro were subjected to SDS-PAGE and visualized by autoradiograph (3H). (C) SWI/SNF was immunoprecipitated with Flag agarose from wild type or mutants expressing K1493R, K1497R, or K1493R and K1497R (RR) Snf2 under native promoter. Immunoprecipitated complexes were analyzed by immunoblotting using α-AcK and Flag antibody (α-Flag).
To confirm the acetylation sites, either or both lysines were mutated to nonacetylatable arginine residues. The mutant AT hook proteins were expressed and purified from E. coli, and an in vitro acetylation assay was performed. Individually, K1493R and K1497R mutant proteins had an acetylation profile similar to the wild-type protein (Fig. 2B, lanes 2–4). The double K1493R and K1497R (RR) mutant, however, was not acetylated by the Ada2 complex, suggesting that both K1493 and K1497 are acetylated by Gcn5 (Fig. 2B, lane 5).
Next, we examined whether the lysine mutants could be acetylated in vivo. Mutant yeast strains expressing full-length K1493R, K1497R, or K1493R and K1497R (RR) Snf2 under the native promoter were established using PCR-mediated recombination methods. SWI/SNF was immunoprecipitated from wild type and mutants, and acetylation was detected with the AcK antibody. The levels of acetylation were slightly reduced in the K1493R mutant strain, while acetylation levels disappeared completely in the K1497R and RR mutant strains (Fig. 2C, lanes 3,4). These data indicate that K1493 and K1497 of Snf2 are acetylated in vivo.
Acetylation of Snf2 facilitates interaction between its bromodomain and the two acetyl lysines
The acetylated lysine residues are located between the AT hook domains of Snf2, which in human Snf2 have been demonstrated to have DNA-binding activity. Even though AT hook domains are not well conserved in other organisms (Supplemental Fig. S2), a short patch of the DNA-binding module is conserved (Bourachot et al. 1999). Indeed, we confirmed that the AT hook protein has DNA-binding activity (Supplemental Fig. S3A). Therefore, we reasoned that acetylation of Snf2 might alter its DNA-binding activity. To address this question, recombinant Snf2C was incubated with the Ada2 complex in the presence or absence of acetyl CoA in vitro, and then Snf2C proteins were repurified using glutathione sepharose (Supplemental Fig. S3B). To measure the efficiency of acetylation, the reaction was duplicated using 3H-acetyl CoA and incorporated 3H was quantified by scintillation counting. Based on counts per minute, ∼50% of Snf2C was acetylated (data not shown). EMSA was carried out using an increasing amount of Snf2C that was treated with or without acetyl CoA. Acetylated Snf2C and unmodified Snf2C bound to DNA with the same efficiency (Supplemental Fig. S3C); thus, acetylation of Snf2 did not affect DNA-binding activity per se.
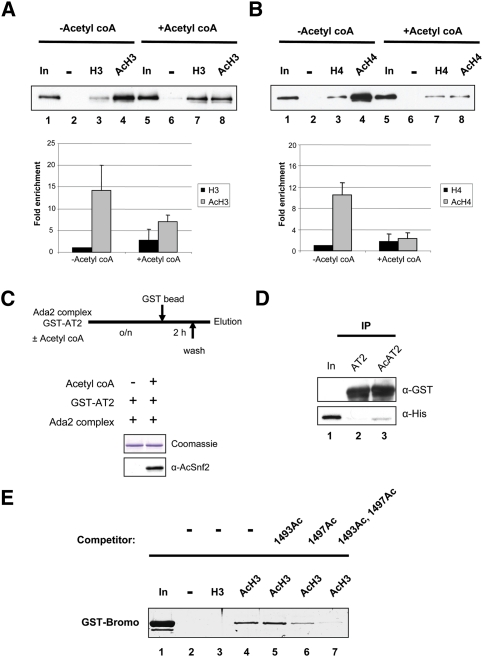
The bromodomain of Snf2 has been shown to facilitate association with hyperacetylated chromatin (Hassan et al. 2001; Chandy et al. 2006). The bromodomain is in proximity of the two identified acetylation sites in Snf2. Therefore, we reasoned the acetylation of Snf2 might affect its ability to bind acetylated histones. Thus, a peptide-binding assay was carried out using in vitro acetylated Snf2C or control Snf2C treated without acetyl CoA and either unmodified histone H3 or acetylated histone H3 peptides. Biotinylated histone peptides bound to Dynabeads were incubated with acetylated Snf2C or unmodified Snf2C. The bound proteins were analyzed by Western blot. Since the Snf2C fragment contains the bromodomain, unmodified Snf2C preferentially bound to the acetylated H3 peptide (about 14-fold over the unmodified peptide) (Fig. 3A, lanes 3,4). In contrast, acetylated Snf2C lost its preferential binding ability toward the acetylated histone H3 peptide (Fig. 3A, lanes 7,8). Acetylated H4 peptide yielded similar results, where acetylated Snf2C could not distinguish acetylated histone H4 peptide and unmodified histone H4 peptide (Fig. 3B, lanes 7,8), even though unmodified Snf2C bound to acetylated H4 peptide 10-fold more than unmodified H4 peptide (Fig. 3B, lanes 3,4).
Figure 3.
Acetylation of Snf2 facilitates intramolecular interactions between the bromodomain and the two acetyl lysine residues. (A) Acetylated or unacetylated Snf2C proteins were tested for binding to unmodified (H3) or acetylated histone H3 (AcH3) peptides. (B) Acetylated or unacetylated Snf2C proteins were tested for binding to unmodified (H4) or acetylated histone H4 (AcH4) peptides. (C) GST-AT2 (1466–1546 amino acids) purified from E. coli was acetylated using Ada2 complexes with or without acetyl CoA. AT2 protein that was treated with or without acetyl CoA was repurified using glutathione sepharose. Acetylation of AT2 proteins was confirmed by Western blot. (D) Coimmunoprecipitation assay was performed using His-tagged bromodomain of Snf2 and either unacetylated GST-AT2 (AT2) or acetylated GST-AT2 (AcAT2). (E) GST-bromodomain of Snf2 was incubated with biotinylated AcH3 peptide or biotinylated H3 peptide in the presence of excess Snf2 peptides acetylated at K1493 (1493Ac), K1497 (1497Ac), or K1493 and K1497 (1493Ac, 1497Ac), or the absence of Snf2 peptide. Bound proteins were visualized by Coomassie staining.
This finding raised the possibility that acetylation of Snf2 might promote an interaction between the two acetyl lysine residues and the bromodomain. To test this possibility, the protein fragment AT2, encompassing the two acetylation sites and the second AT hook domain, was purified and acetylated in vitro (Fig. 3C). Coimmunoprecipitation was performed using acetylated or unacetylated AT2, which contains an N-terminal GST tag, and a His-tagged bromodomain fragment of Snf2. Only acetylated GST-AT2 was able to coimmunoprecipitate with the bromodomain fragment (Fig. 3D, lane 3). These data indicate that acetylation of Snf2 facilitates an interaction between the bromodomain and the two acetyl lysine residues.
We tested whether the interaction between the Snf2 bromodomain and acetylated histones could be perturbed by the addition of excess acetylated Snf2 peptide. The GST-bromodomain fragment of Snf2 was incubated with biotintylated histone H3 or biotintylated and acetylated histone H3 peptide in the absence or presence of acetylated Snf2 peptides. The Snf2 bromodomain fragment favored the acetylated H3 peptide over the unmodified H3 peptide (Fig. 3E, lanes 3,4). Interestingly, excess 1497Ac peptide inhibited the interaction between the bromodomain protein fragment and the acetylated H3 peptide by about half (Fig. 3E, lanes 4,6). Moreover, in the presence of excess double-acetylated peptides (1493Ac and 1497Ac), the bromodomain no longer bound to the acetylated H3 peptide (Fig. 3E, lane 7), indicating that acetylation of both sites is necessary to bind to the bromodomain. This result is consistent with a recent study that bromodomains need two acetyl lysines for stable binding (Moriniere et al. 2009). Thus, interactions between the two acetyl lysine residues and the bromodomain of Snf2 facilitate dissociation of SWI/SNF from acetylated histones.
In addition, we examined the sedimentation of acetylated Snf2C by glycerol gradient centrifugation because an acetylation-mediated intramolecular interaction would affect the sedimentation of Snf2C. For this purpose, Snf2C was acetylated by the Ada2 complex, and the N-terminal GST tag was cut off. Note that there was ∼50% of unacetylated Snf2C as well. Snf2C was expected to be 35.5 KDa, but it ran 47 KDa in SDS-PAGE (Supplemental Fig. S4A). All eluted fractions were analyzed by immunoblotting using an Snf2 antibody that recognizes all forms of Snf2C, and an AcSnf2 antibody that only recognizes the acetylated form. As a result, immunoblotting against Snf2 showed a peak in fraction number 9–10, a little smaller than ovalbumin (45 kDa) (Supplemental Fig. S4B). However, when the same fractions were probed with AcSnf2 antibody, peaks were in fraction number 10–12 (Supplemental Fig. S4B), suggesting that acetylation of Snf2 induces a conformational change. It also indicated that fast migration of acetylated Snf2 was not due to dimerization, since peaks were even smaller than bovine serum albumin (BSA) (66 kDa). These data strengthen the idea that acetylation of Snf2 facilitates intramolecular interaction between acetyl lysines and the bromodomain.
Acetylation site mutant SWI/SNF is enriched at gene promoters
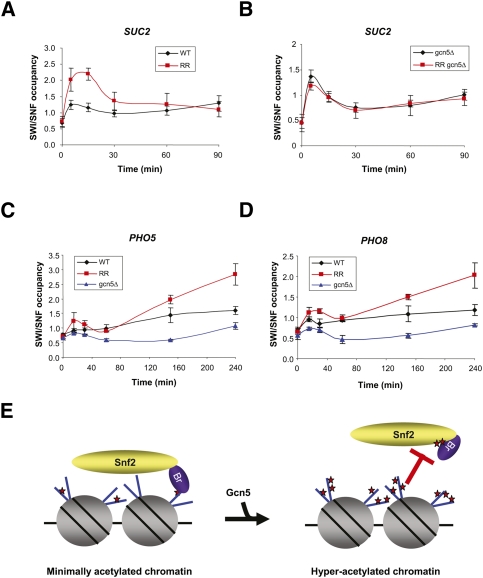
Since the acetylation of Snf2 inhibits the preferential interaction between the bromodomain and acetylated histones, we decided to examine the occupancy of the wild-type and mutant SWI/SNF complex at the SUC2 gene locus. SUC2 encodes an invertase that is transcriptionally repressed in high-glucose media and induced in low-glucose media. Both SWI/SNF and Gcn5 are required for maximal SUC2 gene expression (Hirschhorn et al. 1992; Wu and Winston 1997; Geng and Laurent 2004). Wild-type and RR mutant Snf2 were tagged with a C-terminal double Flag tag, and expression levels of both tagged proteins were found to be similar (data not shown). We monitored by ChIP assay SWI/SNF occupancy in both the wild-type and RR mutant over time, following shifting the cells from YPD (2% dextrose) to low-dextrose medium (0.05%), which induces SUC2 expression. Wild-type Snf2 was enriched by ∼1.5-fold at the SUC2 promoter at 5 min after induction with respect to time 0 and then dropped back to basal levels (Fig. 4A, black line). In contrast, occupancy of the RR mutant peaked broadly at ∼5–15 min after induction. Compared with the wild-type Snf2, the peak in occupancy for the RR mutant was about twofold at 5 min and 15 min post-induction.(Fig. 4A, red line). As a control, we examined occupancies of wild-type and RR mutant Snf2 in the gcn5Δ background. Both wild-type and mutant Snf2 behaved about the same in the gcn5Δ background (Fig. 4B), and their patterns were similar to the wild type (Supplemental Fig. S5). This finding excludes a possibility that introduced mutations (K to R) on Snf2 simply result in increased SWI/SNF occupancy in Figure 4A. Moreover, this result illustrates that enhanced occupancy of the RR mutant at the SUC2 promoter required Gcn5, consistent with the possibility that it requires acetylated histones at the promoter.
Figure 4.
Acetylation site mutant SWI/SNF is enriched at the gene promoters. (A) Wild-type or RR mutant cells grown in dextrose (2%) media were shifted to low-dextrose media (0.05%). Cells were harvested at indicated time points. Cross-linking of wild-type (Snf2-Flag; black line) and mutant (RR Snf2-Flag; red line) Snf2 to the SUC2 promoter was analyzed by ChIP using anti-Flag antibody. The level of Snf2 occupancy at the SUC2 promoter was normalized to the level of Snf2 signal at the control TELVIR locus. (B) Occupancy of wild-type Snf2 (black line) and RR mutant Snf2 (red line) at the SUC2 promoter was analyzed by ChIP assay in the gcn5Δ background. (C,D) Wild type (black line), RR mutant (red line), and gcn5Δ (blue line) grown in synthetic dextrose (SD) media were shifted to SD media without phosphate. Cells were harvested at indicated time points. Occupancy of SWI/SNF at PHO5 and PHO8 promoters was analyzed by ChIP using Flag antibody. (E) The model illustrating an in vivo role for Snf2 acetylation by Gcn5.
It is also possible that increased SWI/SNF occupancy may result in increased gene expression. Therefore, we compared levels of SUC2 mRNA in wild-type and RR mutant and gcn5Δ and RR gcn5Δ strains. As a result, there were not significant differences between wild-type Snf2 and the RR mutant in SUC2 expression (Supplemental Fig. S6).
Next, we investigated two other promoters, PHO5 and PHO8, which are regulated by Gcn5 and SWI/SNF. These genes are activated by switching cells to media without phosphate. Upon phosphate depletion, SWI/SNF occupancy slowly increased over time in wild-type cells. However, in RR mutant cells, SWI/SNF was enriched at the PHO5 and PHO8 promoters after 2 h (Fig. 4C,D, respectively), which was later than in the case of SUC2. This could be because PHO genes are more slowly induced, compared with SUC2 genes (Adkins et al. 2004). In gcn5Δ, wild-type SWI/SNF occupancy was reduced to preinduction levels at all times (Fig. 4C,D, blue lines), indicating that histone acetylation is a prerequisite for duration of SWI/SNF at the promoter nucleosomes. The difference of wild-type SWI/SNF occupancy at SUC2 and PHO promoters in gcn5Δ could be due to the difference in expression kinetics. SWI/SNF is known to be targeted by sequence-specific transcription factors upon induction. Since SUC2 is rapidly expressed, in gcn5Δ, SWI/SNF recruitment to SUC2 might be driven by transcription factors and is transient such that any effect of loss of histone acetylation on SWI/SNF occupancy is not readily apparent. However, as noted above, enhanced occupancy of the RR mutant was gcn5-dependent at SUC2. In contrast, PHO genes are slowly induced and, in this situation, interaction of SWI/SNF with acetylated histones may be more important to maintain its occupancy. Hence, the effects of loss of histone acetylation in gcn5Δ are more apparent at PHO gene promoters. Thus, SWI/SNF occupancy in gcn5Δ was reduced. Taken together, at both SUC2 and PHO genes, the Snf2 acetylation mutant appeared to occupy the promoter to a greater extent and for a longer period than the wild-type Snf2 protein. This supports in vitro findings that the acetylation of Snf2 facilitates dissociation of SWI/SNF from acetylated histones.
Gcn5 uses a novel target to inactivate SWI/SNF
In this study, we identified two lysine residues on the Snf2 protein that are acetylated by Gcn5 in vivo and in vitro (Figs. 1, 2). Acetylation of Snf2 facilitates an interaction between the two acetyl lysine residues and the Snf2 bromodomain, which promotes dissociation of SWI/SNF from acetylated histones (Fig. 3). Indeed, when these two lysine residues are mutated to arginine (double R mutant), occupancy of the Snf2 protein is enriched at the SUC2, PHO5, and PHO8 promoters (Fig. 4). The two identified acetylation sites in Snf2 are not conserved in other species (Supplemental Fig. S2). However, it has been reported recently that a human homolog of Snf2, SMARCA4, is acetylated at three lysine residues (K188, K455, and K1033) by mass spectrometry analysis (Choudhary et al. 2009). Among them, K1033, which is located right after the DEXDc domain, is conserved from yeast to humans, excluding Saccharomyces cerevisiae Snf2, of which the equivalent amino acid is arginine (Supplemental Fig. S7A). Interestingly, amino acid sequences surrounding K1033 in SMARCA4 resemble that of K1497 in scSnf2, with consensus sequences being GKXXXKacS/T (Supplemental Fig. S7B). RSC complex binding to acetylated histones has also been shown to be inactivated by Gcn5. Strikingly, the subunit inactivated by Gcn5 within the RSC complex is not the Snf2 homolog subunit Sth1, but instead a different subunit: Rsc4. When Rsc4 is acetylated at Lys 25 by Gcn5, an acetylation-mediated intramolecular interaction inhibits binding of its bromodomain to actylated histone H3 Lys 14 (VanDemark et al. 2007). Thus, while the SWI/SNF and RSC complexes are highly homologous, the crucial subunit for regulation of their interaction with acetylated histone differs. Such differences may play an important role in the specialization of each complex.
A model for the function of Snf2 acetylation
Why is acetylation of Snf2 important in vivo? One possible explanation is that this acetylation allows cells to recycle the SWI/SNF complex. This idea is supported by the observation that SWI/SNF is required for transcription of ∼5% of all yeast genes, although it is relatively rare (100–500 copies per yeast cells) (Peterson and Workman 2000). The other possibility is that Snf2 acetylation fine-tunes gene expression. It may prevent prolonged retention of SWI/SNF on the promoter, and therefore stop unnecessary transcription.
Based on our observations, we proposed a model for SWI/SNF regulation by Snf2 acetylation. In this model (Fig. 4E), upon gene activation, SWI/SNF is targeted to a gene by a sequence-specific transcription factor. It then recognizes basal levels of histone acetylation and alters chromatin structure by means of removing histones or sliding nucleosomes. Meanwhile, Gcn5-containing complexes acetylate both histones and the Snf2 subunit, which promotes the dissociation of SWI/SNF by an acetylation-mediated intramolecular interaction. Alternatively, SWI/SNF might act on the newly Gcn5-acetylated nucleosomes until it becomes acetylated itself and dissociates. Thus, Snf2 acetylation by Gcn5 could be a molecular switch for the interaction between SWI/SNF and chromatin. Our findings show that there is an important interplay between chromatin-modifiying complexes that helps to regulate both gene expression and complex activity.
Materials and methods
Additional information is provided in the Supplemental Material.
Strains and antibodies
All strains used in the study are listed in Supplemental Table S1.
Anti-acetylated lysine (EMD; ST1027), anti-GST (Santa Cruz Biotechnology; Z-5), anti-His (Qiagen), and anti-Flag antibodies (Sigma; M2) were purchased. Snf2 antibody was kindly provided by J. Reese. Anti-acetylated Snf2 antibody was raised by Covance using synthesized peptide CDIPKPRTAGKacTSVKacSARTSTRGR.
ChIP assay
ChIPs were performed as described previously (Li et al. 2007). Quantitative PCR (qPCR) was carried out using PerfeCTa SYBR Green FastMix (Quanta) with an ABI 7900HT real-time PCR machine.
The following primer sets were used: SUC2 UAS (5′-GACGTTTCTTTTCGAGGAATGCTTAA-3′, 5′-CAGGAGGAAGGATTATAGGATCGAAATC-3′), PHO8 UAS (5′-AACGCTCTCGGAAGAGCAGATTGA-3′, 5′-TGTACAAAGAGGACATCCGCTGCT-3′), PHO5 UAS (5′-ACATTGGCATTAGCTAGGAGGGCA-3′, 5′-TGTAGCGAGCTACTTTCGGTCAGT-3′), TELVIR (5′-CTGAGTTTAACGGTGATTATTAGGTGGATT-3′, 5′-TTTTAGTTTACGCGTTATGACAATTTTATGTAG-3′).
All raw data are presented in Supplemental Table S2.
Peptide pull-down assay
GST-Snf2C (0.5 μg) was acetylated by the Ada2 complex with or without acetyl-CoA. Acetylated GST-Snf2 was repurified using glutathione sepharose (GE Healthcare). Repurified Snf2C was incubated with 0.5 μg of biotinylated H3 peptide (Millipore); biotinylated and acetylated K9/K14 H3 peptide (Millipore); biotinylated H4 peptide (Millipore); or biotinylated and pan-acetyl H4 peptide (Millipore). Added histone peptides were immobilized with streptavidin Dynabeads M-280 (Invitrogen), and bound proteins were analyzed by immunoblotting. Band intensity was measured using UN-SCAN-IT software. All experiments were performed at least three times.
Peptide competition assay
GST-bromodomain (1 μg) was incubated with either acetylated H3 peptide (0.5 μg) or unmodified H3 peptide (0.5 μg). After 2 h of incubation, either monoacetylated or double-acetylated Snf2 peptide (20 μg: CDIPKPRTAGK[ac]TSVK[ac]SARTSTRGR) was added to the mixture for competition. H3 peptides were immobilized using streptavidin Dynabead M-280 (Invitrogen). After washing, bound proteins were analyzed by SDS-PAGE and Coomassie staining.
Acknowledgments
We thank J. Reese for Snf2 antibody, all members of the Workman laboratory for helpful discussions and suggestions, and S. Pattenden for critical reading of the manuscript. This work was supported with funding from NIGMS grant 5R37GM47867-19 and the Stowers Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1979710.
Supplemental material is available for this article.
References
- Adkins MW, Howar SR, Tyler JK 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 14: 657–666 [DOI] [PubMed] [Google Scholar]
- Agalioti T, Chen G, Thanos D 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392 [DOI] [PubMed] [Google Scholar]
- Bourachot B, Yaniv M, Muchardt C 1999. The activity of mammalian brm/SNF2α is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol Cell Biol 19: 3931–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD 1996. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851 [DOI] [PubMed] [Google Scholar]
- Chandy M, Gutierrez JL, Prochasson P, Workman JL 2006. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot Cell 5: 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299–311 [DOI] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60 [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496 [DOI] [PubMed] [Google Scholar]
- Geng F, Laurent BC 2004. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J 23: 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Awad S, Prochasson P 2006. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J Biol Chem 281: 18126–18134 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6: 2288–2298 [DOI] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol 304: 355–370 [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL 2007. Histone acetyltransferase complexes: One size doesn't fit all. Nat Rev Mol Cell Biol 8: 284–295 [DOI] [PubMed] [Google Scholar]
- Lee KK, Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL 2004. Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem Soc Trans 32: 899–903 [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P 2006. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3: 429–438 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ 2006. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol Cell Biol 26: 4095–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. 2009. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461: 664–668 [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, et al. 2007. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130: 499–511 [DOI] [PubMed] [Google Scholar]
- Ornaghi P, Ballario P, Lena AM, Gonzalez A, Filetici P 1999. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J Mol Biol 287: 1–7 [DOI] [PubMed] [Google Scholar]
- Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M 2009. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat Struct Mol Biol 16: 412–420 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev 10: 187–192 [DOI] [PubMed] [Google Scholar]
- Prochasson P, Neely KE, Hassan AH, Li B, Workman JL 2003. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol Cell 12: 983–990 [DOI] [PubMed] [Google Scholar]
- Reinke H, Gregory PD, Horz W 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol Cell 7: 529–538 [DOI] [PubMed] [Google Scholar]
- Roberts SM, Winston F 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR 2007. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell 27: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Winston F 1997. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res 25: 4230–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]