Abstract
Glucose and glutamine serve as the two primary carbon sources in proliferating cells, and uptake of both nutrients is directed by growth factor signaling. Although either glucose or glutamine can potentially support mitochondrial tricarboxylic acid (TCA) cycle integrity and ATP production, we found that glucose deprivation led to a marked reduction in glutamine uptake and progressive cellular atrophy in multiple mammalian cell types. Despite the continuous presence of growth factor and an abundant supply of extracellular glutamine, interleukin-3 (IL-3)-dependent cells were unable to maintain TCA cycle metabolite pools or receptor-dependent signal transduction when deprived of glucose. This was due at least in part to down-regulation of IL-3 receptor α (IL-3Rα) surface expression in the absence of glucose. Treatment of glucose-starved cells with N-acetylglucosamine (GlcNAc) to maintain hexosamine biosynthesis restored mitochondrial metabolism and cell growth by promoting IL-3-dependent glutamine uptake and metabolism. Thus, glucose metabolism through the hexosamine biosynthetic pathway is required to sustain sufficient growth factor signaling and glutamine uptake to support cell growth and survival.
Keywords: Metabolism, glucose, glutamine, hexosamine, growth factor signaling, glycosylation
Mammalian cells rely on growth factor signals to direct nutrient uptake from the extracellular environment to support survival, growth, and proliferation. Growth factor-dependent nutrient uptake is essential for both ATP production and cellular biosynthesis of the proteins, RNA, DNA, and lipid membranes required for production of a daughter cell. In contrast, cell-autonomous nutrient uptake and metabolism is one of the most consistent features displayed by cancer cells. In cancer cells, glucose and glutamine serve as primary sources of carbon for ATP production and biosynthesis. Glutamine also serves as an essential nitrogen donor for production of nucleotides, certain amino acids, and nicotinamide, in addition to its direct role in protein synthesis (DeBerardinis et al. 2008; Vander Heiden et al. 2009).
In recent years, much insight into the signaling mechanisms that control nutrient uptake has been gained. The phosphoinositide 3-kinase (PI3K)–Akt pathway is a critical mediator of glucose uptake through regulation of glucose transporters and glycolytic metabolism (DeBerardinis et al. 2008). Signaling control of amino acid metabolism is less well understood, although the mammalian target of rapamycin (mTOR) pathway can promote surface expression of the 4F2hc amino acid transporter (Slc3a2), and oncogenic Myc can stimulate enhanced consumption of glutamine (Edinger and Thompson 2002; Wise et al. 2008; Gao et al. 2009). Proper coordination of the metabolism of glucose and glutamine is likely to be important for the cell to maintain optimal bioenergetics while engaging in growth; yet it remains poorly understood how cells sense the availability of these two key nutrients and coordinate their uptake and metabolism.
Multiple mechanisms exist for the cell to detect its nutrient and bioenergetic resources. The LKB1–AMP-activated protein kinase (AMPK) pathway is activated in response to low cellular ATP levels and acts to inhibit both cell cycle progression and growth-promoting signaling pathways such as mTOR, as well as to stimulate energy production through increased fatty acid oxidation (Shaw et al. 2004; Jones et al. 2005; Gwinn et al. 2008; Shackelford and Shaw 2009). mTOR is responsive to essential amino acid availability and integrates this metabolic information with upstream growth factor signaling cues (Guertin and Sabatini 2007; Shaw 2008). Metabolite-derived protein modification can also be used as a direct link between metabolism and cell function (Wellen and Thompson 2010). For example, acetylation of certain substrates, including histones and a number of metabolic enzymes, has been shown recently to be regulated in a nutrient-responsive manner (Wellen et al. 2009; Wang et al. 2010; Zhao et al. 2010).
In yeast, glucose deprivation activates Snf1 kinase (the homolog to mammalian AMPK), which stimulates transcriptional activation of genes that enact a metabolic conversion to the use of alternate carbon sources, such as sucrose, galactose, or maltose (Zaman et al. 2008). In contrast, while AMPK also facilitates cellular response to glucose deprivation in mammalian cells, these adaptations do not include a switch to a different substrate for glycolysis, since mammalian cells lack an extracellular source of an alternative hexose. The major substrate available to mammalian cells to support cellular bioenergetics and biosynthesis in addition to glucose is glutamine. Glutamine is the most abundant amino acid in extracellular fluid, and has the potential to fully support the tricarboxylic acid (TCA) cycle in the absence of glucose and, for certain types of cells, to provide a gluconeogenic substrate. If amino acid uptake can be regulated by both signal transduction and compensating bioenergetic mechanisms, it stands to reason that glutamine uptake should be sustained or enhanced in response to glucose deprivation in the presence of growth factor.
In this study, we investigated the regulation of glutamine metabolism during glucose withdrawal in hematopoietic cells, uncovering a novel mechanism used to coordinate the metabolism of these two key nutrients. In the absence of glucose, glutamine consumption declines in interleukin-3 (IL-3)-dependent hematopoietic progenitors and IL-3-dependent cell lines due to reduced growth factor signaling, resulting from impaired surface expression of IL-3 receptor α (IL-3Rα). Specific restoration of the hexosamine branch of glucose metabolism, accomplished through treatment of cells with N-acetylglucosamine (GlcNAc), is sufficient to restore IL-3Rα surface expression and signaling, as well as IL-3-dependent growth, in the absence of glucose. GlcNAc-stimulated growth is supported in the absence of glucose through promotion of glutamine consumption in an IL-3-dependent manner. Increased glutamine uptake driven by activation of the hexosamine pathway enables growth by supplying substrate to the TCA cycle for energy production, by contributing carbon for lipid biosynthesis, and by promoting uptake of essential amino acids. This study demonstrates a novel mechanism used to coordinate glucose and glutamine metabolism through hexosamine pathway-dependent regulation of growth factor receptor surface expression and signaling.
Results
Glutamine consumption in hematopoietic cells is dependent on glucose availability
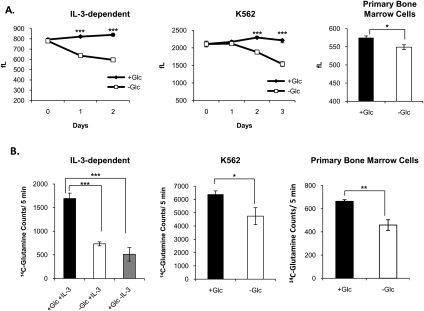
Glucose and glutamine are the two primary carbon sources used by proliferating cells, yet how or if cells coordinate the metabolism of these two key nutrients in order to fuel growth is poorly understood. Previous work has shown that growth factor-deprived cells experience a decline in the ability to take up both glucose and amino acids, suggesting that cells lack the intrinsic ability to take up sufficient nutrients to maintain bioenergetics and viability in the absence of growth factor-induced instruction (DeBerardinis et al. 2008). However, in growth factor-replete cells, we hypothesized that cells would compensate for limited availability of one nutrient by consuming increased amounts of the other. Thus, to understand the cellular response to glucose deprivation and the role of glutamine in compensating for the absence of glucose, we examined the response to glucose starvation in several types of hematopoietic cells capable of surviving for several days without glucose, including primary, immortalized, and leukemia cell types. Each of these cell types ceased to proliferate and atrophied over several days upon withdrawal of glucose (Fig. 1A; data not shown). Surprisingly, glutamine consumption was not increased in compensation for the absence of glucose, but rather was significantly reduced (Fig. 1B). In the case of IL-3-dependent cells, glutamine consumption after 2 d of glucose withdrawal in the presence of IL-3 was reduced to nearly the same levels as in the absence of growth factor (Fig. 1B).
Figure 1.
Glutamine consumption is reduced in the absence of glucose. (A) IL-3-dependent bax−/−bak−/− cells and K562 cells were cultured in the presence or absence of glucose for 2 or 3 d and cell size was measured in femtoliters (fL) each day (mean ± SD of triplicates). Primary bone marrow cells were cultured in the presence or absence of glucose for 4 d and size was measured in femtoliters (mean ± SD of triplicates). (B) Transport of 14C-glutamine over 5 min was measured (mean ± SD of triplicates) after the following treatments: IL-3-dependent cells were cultured in the presence or absence of glucose or IL-3 for 2 d, K562 cells were cultured in the presence or absence of glucose for 2 d, and primary bone marrow cells were cultured in the presence or absence of glucose for 4 d. For all indicated panels, P < 0.05 (*), P < 0.005 (**), and P < 0.0005 (***).
Metabolite pools are depleted in the absence of glucose
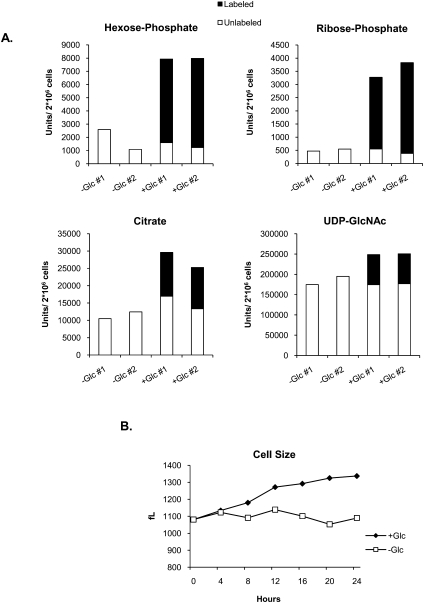
It is possible that cells may obtain energy from a source other than glutamine in the absence of glucose. To address this issue, total metabolite pools in the presence or absence of glucose were analyzed. We starved IL-3-dependent hematopoietic cells of glucose for 24 h and then added back [U-13C6]glucose (labeled at all six carbons). After 1 h, metabolites were extracted and analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). Most of the metabolites from key pathways supported by glucose—including glycolysis, the pentose phosphate pathway, the TCA cycle, and the hexosamine biosynthetic pathway—were depressed in glucose-deprived cells and increased within 1 h of glucose addition (Fig. 2A; Supplemental Table S1). Glycolytic and pentose phosphate pathway metabolites, such as hexose-phosphate and ribose-phosphate, were markedly higher in glucose-treated as compared with untreated cells, and the cellular pools of these metabolites were ∼80% labeled by 1 h after 13C-glucose readdition, indicative of the strict glucose dependence of these metabolite pools (Fig. 2A; Supplemental Tables S1, S2). TCA cycle metabolites such as citrate, which could be generated from glucose and/or nonglucose sources, were also higher after glucose treatment, with 40%–50% of the cellular citrate pools containing label after 1 h, suggesting that, while cells may sustain some TCA cycle activity in the absence of glucose by using alternate carbon sources, alternate substrates do not fully compensate (Fig. 2A; Supplemental Tables S1, S2). Thus, glucose is required to sustain metabolite pools in multiple pathways, including the TCA cycle, despite extracellular availability of glutamine. In the presence of IL-3, readdition of glucose to cells rapidly restores metabolite pools (Fig. 2A), enabling cells to resume growing within hours (Fig. 2B).
Figure 2.
Metabolite pools are depleted in the absence of glucose. (A) IL-3-dependent cells were withdrawn from glucose for 24 h, after which 15 mM 13C6-glucose was added into the culture medium. Cells were harvested and LC-MS/MS metabolite analysis was performed on duplicate samples 1 h after glucose addition. Select metabolites in glycolysis (hexose-phosphate), the pentose phosphate pathway (ribose-phosphate), the TCA cycle (citrate), and the hexosamine pathway (UDP-GlcNAc), with background subtracted, are shown. Data represent total (labeled [▪] + unlabeled [□]) metabolite pools (for additional metabolites, see Supplemental Tables S1, S2). (B) IL-3-dependent cells were withdrawn from glucose for 24 h, after which 15 mM glucose was added into the culture medium. Cell size was measured over the next 24 h. Result is representative of two independent experiments.
IL-3Rα surface expression is dependent on glucose availability
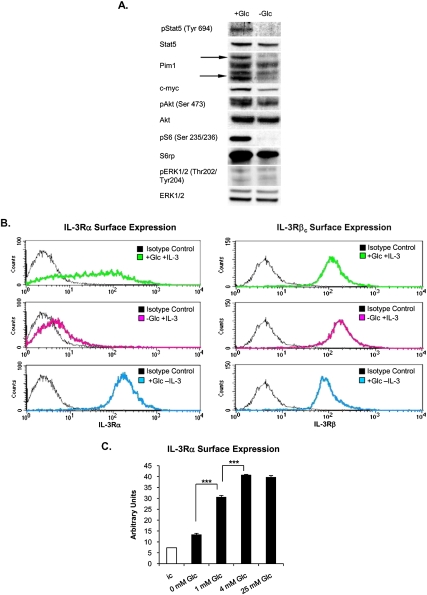
The above results demonstrate that, despite availability of growth factor, cellular uptake of glutamine or other energy sources declines in the absence of glucose. Since glutamine consumption is growth factor-dependent (Fig. 1B), we investigated whether growth factor signaling was impaired in the absence of glucose. While signal transduction in most cell types is a composite of the activity of multiple growth factors present in serum, IL-3-dependent hematopoietic cells depend primarily on IL-3 for initiation of signaling through the Jak–Stat, PI3K–Akt–mTOR, and RAS–MEK–ERK pathways (Steelman et al. 2004; Baker et al. 2007). IL-3-dependent phosphorylation of Stat5 and levels of its transcriptional product, Pim1, were severely reduced in glucose-deprived cells (Fig. 3A). In addition, c-myc levels and Akt Ser 473 phosphorylation were reduced, and ribosomal protein S6 (S6rp) phosphorylation was essentially absent (Fig. 3A). ERK phosphorylation levels were low in the presence or absence of glucose at the time point shown (Fig. 3A). Since multiple IL-3-dependent signaling pathways were suppressed in the absence of glucose, we investigated whether the receptor itself might be impacted. The IL-3R consists of α subunits, which are specific to IL-3, and β subunits, which are common to IL-3, IL-5, and GM-CSF, and on which signaling components dock in order to initiate signaling (Baker et al. 2007). A dodecameric complex consisting of equal parts IL-3Rα, IL-3Rβc, and IL-3 is thought to form upon IL-3 binding (Dey et al. 2009). In replete medium containing IL-3, a wide range of IL-3Rα surface levels is observed among cells (Fig. 3B). In the absence of IL-3, IL-3Rα surface expression is markedly up-regulated (Fig. 3B; Lum et al. 2005). Strikingly, however, IL-3Rα is nearly absent from the cell surface when cells are cultured in IL-3-replete medium lacking glucose (Fig. 3B). Glucose dose-dependent regulation of IL-3Rα is observed, with increasing levels of surface expression at 0, 1, and 4 mM glucose (Fig. 3C). No further increase in surface receptor levels was observed above 4 mM (Fig. 3C). Surface expression of IL-3Rβc, in contrast, was minimally affected by either growth factor or glucose availability (Fig. 3B).
Figure 3.
IL-3Rα surface expression is dependent on glucose availability. (A) Western blot analysis from IL-3-dependent cells cultured in the presence or absence of glucose for 48 h. (B) Cells were cultured for 48 h in replete media (with both glucose and IL-3) or in media lacking either glucose or IL-3. Surface expression of IL-3Rα and IL-3Rβc was examined by FACS analysis. (C) Cells were cultured for 48 h in glucose-free media supplemented with 0, 1, 4, or 25 mM glucose, and IL-3Rα surface expression was analyzed by FACS. (ic) Isotype control. Data represent mean ± SD of triplicates. (***) P < 0.0005.
The hexosamine biosynthetic branch of glucose metabolism regulates IL-3Rα surface expression
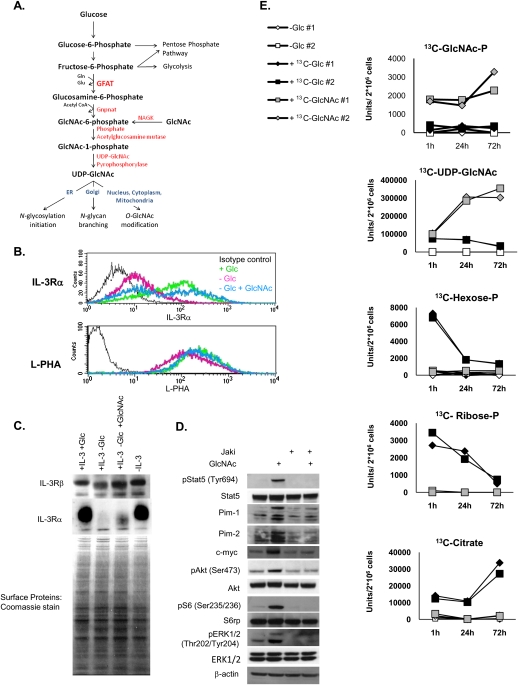
Since IL-3Rα is a glycoprotein, we asked whether its surface expression might be sensitive to glucose availability for glycosylation. N-linked glycosylation of membrane proteins requires production of UDP-GlcNAc by the hexosamine pathway for both initiation of N-glycosylation in the endoplasmic reticulum (ER) and N-glycan branching in the Golgi apparatus (Fig. 4A; Kornfeld and Kornfeld 1985; Dennis et al. 2009). To assess whether glucose flux through the hexosamine pathway is involved in regulation of IL-3Rα surface expression, we substituted GlcNAc for glucose in the culture medium. GlcNAc comes into the cell through bulk endocytosis and is phosphorylated by the salvage pathway enzyme N-acetylglucosamine kinase (NAGK) in order to enter the hexosamine pathway at GlcNAc-6-P (Fig. 4A; Lau and Dennis 2008). Since GlcNAc can enter the hexosamine pathway without contributing significantly to glycolysis (Dennis et al. 2009), this approach allowed us to examine whether metabolite flux through the hexosamine pathway is sufficient to promote IL-3Rα surface expression in the absence of other metabolic fates of glucose. Treatment of cells with 15 mM GlcNAc in the absence of glucose significantly rescued IL-3Rα surface expression in the absence of glucose (Fig. 4B). In addition, GlcNAc treatment of glucose-deprived cells maintained the binding of the lectin phytohemagglutinin-L (L-PHA)—which recognizes GlcNAc-branched N-glycans generated by a medial Golgi remodeling pathway that is limited by UDP-GlcNAc concentrations—to levels comparable with those observed in the presence of glucose (Fig. 4B). FACS results were confirmed by Western blot analysis of cell surface proteins, which indicated a dramatic reduction in surface IL-3Rα in the absence of glucose and partial rescue by GlcNAc (Fig. 4C). Reduced surface expression in the absence of glucose was not reflective of reduced gene expression, since IL-3Rα mRNA levels were actually somewhat higher in glucose-withdrawn cells (Supplemental Fig. S1A). IL-3Rβc remained at the cell surface in all conditions (Fig. 4C). Western blot analysis also pointed to a heterogeneous size distribution of the receptor, consistent with Golgi processing of its associated N-glycans; treatment of membrane protein fractions with the deglycosylating enzyme PNGase F removed this heterogeneity and generated a band consistent with the 41-kDa size of the unglycosylated IL-3Rα (Supplemental Fig. S1B). Signaling downstream from the IL-3R was rescued by addition of GlcNAc, paralleling surface expression of the α subunit (Fig. 4D). Additionally, Jak inhibition completely blocked the ability of GlcNAc to promote signaling (Fig. 4D). Jak activation at the IL-3R occurs upon IL-3 binding and is necessary for signaling to Stat5 and PI3K (Baker et al. 2007), consistent with a coordinate regulation of these pathways through regulation of the IL-3R itself.
Figure 4.
Metabolite flux through the hexosamine pathway restores IL-3Rα surface expression and signaling in the absence of glucose. (A) The hexosamine biosynthetic pathway is gated by the rate-limiting enzyme GFAT (glutamine fructose-6-phosphate amidotransferase), which transfers an amide group from glutamine to fructose-6-phosphate to produce glucosamine-6-phosphate. Glucosamine-6-P is acetylated by Gnpnat (glucosamine phosphate N-acetyltransferase) to generate GlcNAc-6-P. The final product of the pathway is UDP-GlcNAc, which is used for both N-linked glycosylation in the ER and Golgi and for O-GlcNAc protein modification in the nucleus and cytoplasm. The metabolite GlcNAc can enter the hexosamine pathway after phosphorylation by the salvage pathway enzyme NAGK. (B) IL-3-dependent cells were cultured in IL-3-containing medium in the presence or absence of glucose or in glucose-free medium supplemented with 15 mM GlcNAc for 48 h. IL-3 was present in all conditions. FACS analysis of surface expression of IL-3Rα or surface binding of L-PHA was performed. Results are representative of at least three independent experiments. (C) Cells were treated for 2 d in the presence or absence of glucose, GlcNAc, and IL-3, as indicated. Cell surface proteins were biotinylated, isolated over NeutrAvidin column, and analyzed by Western blot and Coomassie stain. (D) Cells were withdrawn from glucose for 24 h and then treated with GlcNAc in the presence or absence of Jak inhibitor for an additional 24 h. IL-3 was present in all conditions. Cells were harvested and signaling was analyzed by Western blot. (E) IL-3-dependent cells were starved of glucose for 24 h (in the presence of IL-3), and then 15 mM 13C6-glucose, 15 mM 13C6-GlcNAc, or equal volume water control was added to cells. Cells were harvested at 1, 24, and 72 h after nutrient addition. LC-MS/MS analysis of labeled metabolites was performed. The experiment was done with duplicates for each condition. Representative labeled pools of metabolites from the hexosamine pathway (GlcNAc-P and UDP-GlcNAc), glycolysis (Hexose-P), pentose phosphate pathway (Ribose-P), and the TCA cycle (citrate) are shown at each time point. For additional metabolites in each pathway, see Supplemental Table S2.
We next sought to rule out the possibility that GlcNAc was also being used as a fuel source in addition to its role in the hexosamine pathway. This is a critical control, since even a minor amount of GlcNAc entering glycolysis would obscure the interpretation of these experiments. Thus, to examine the metabolic fate of GlcNAc and compare it with that of glucose, we traced the fate of N-acetyl-D-[U-13C6]glucosamine (13C6-GlcNAc) and [U-13C6]glucose after a 24-h glucose starvation period. An additional set of glucose-starved cells was treated with an equal volume of water, as an unlabeled control. Metabolites were extracted at 1, 24, and 72 h after nutrient addition. As expected, metabolites in the hexosamine pathway were rapidly labeled by either glucose or GlcNAc, although higher levels of labeled hexosamine pathway metabolites were observed following GlcNAc as compared with glucose treatment (Fig. 4E; Supplemental Table S2). Examination of metabolites in other metabolic pathways—including glycolysis, the pentose phosphate pathway, and the TCA cycle—revealed that, while glucose carbon readily entered each of these pathways, GlcNAc carbon was not detectable above background in any of these pathways (Fig. 4E; Supplemental Table S2). Thus, while glucose can be directly used to support energy production and cellular biosynthetic reactions, GlcNAc cannot.
Restoration of hexosamine pathway flux enables growth in the absence of glucose
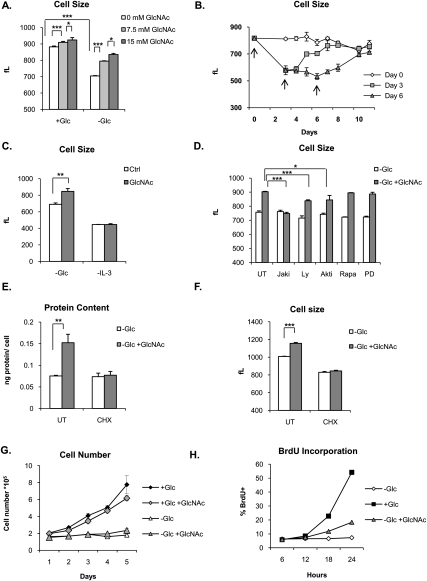
Since rescue of the hexosamine pathway in the absence of glucose was sufficient to promote IL-3Rα surface expression and downstream signaling, we next examined whether it could also promote IL-3-dependent growth or proliferation. Cells were treated with GlcNAc in the presence or absence of glucose. After 4 d, glucose-withdrawn cells were significantly smaller than cells cultured in the presence of glucose. Treatment with GlcNAc was able to rescue cell size in a dose-dependent manner, despite the absence of glucose (Fig. 5A). GlcNAc also significantly, but modestly, increased cell size in the presence of glucose (Fig. 5A). GlcNAc treatment not only enabled cells to maintain their size in the absence of glucose, but could actually promote growth. In cells that had been withdrawn from glucose for up to 6 d, addition of GlcNAc stimulated cells to grow back to near their original size over several days (Fig. 5B). Similar effects on growth were observed in other IL-3-dependent hematopoietic cells that we tested, including primary murine bone marrow cells and M-NFS-60 murine myeloblastic cells (Supplemental Fig. S2). GlcNAc regulation of cell size occurred in a growth factor-dependent manner; in the absence of IL-3, GlcNAc treatment had no effect (Fig. 5C). Inhibition of Jak signaling also blocked GlcNAc-stimulated growth, whereas inhibitors of other pathways had either moderate (PI3K or Akt inhibition) or minimal (mTORC1 or MAPK inhibition) effects, reflecting the involvement of multiple pathways in IL-3-dependent growth (Fig. 5D). Resistance to rapamycin was likely due to increased expression of Pim kinases (Fig. 4D), which regulate growth independently of mTOR in hematopoietic cells (Fox et al. 2005; Hammerman et al. 2005). We next confirmed that hexosamine-dependent changes in cell size were in fact reflective of increased biomass; indeed, protein content was increased in the presence of GlcNAc, and cycloheximide treatment blocked the ability of GlcNAc to promote cell growth (Fig. 5E,F). Despite the ability of GlcNAc to rescue cell growth, GlcNAc-treated cells failed to proliferate in the absence of glucose (Fig. 5G). GlcNAc mildly stimulated BrdU incorporation, but the effect was much more modest than produced by glucose (Fig. 5H). The BrdU incorporation in cells treated with GlcNAc in the absence of glucose was associated with accumulation of a portion of cells that had initiated S phase, although few if any of those cells were able to complete the cell cycle and return to G1 (data not shown). Thus, restoration of the hexosamine branch of glucose metabolism is sufficient for growth but not proliferation in these cells.
Figure 5.
Activation of the hexosamine pathway with GlcNAc is sufficient to promote growth in the absence of glucose. (A) IL-3-dependent cells were cultured in the presence or absence of glucose plus 0, 7.5, or 15 mM GlcNAc, as indicated. IL-3 was present in all conditions. Cell size after 4 d was measured in femtoliters (fL) (mean ± SD of triplicates). (B) Cells were starved of glucose for 11 d, with 15 mM GlcNAc added at day 0, day 3, or day 6, indicated by the arrows. Media were changed (including fresh IL-3) every 3 d. Cell size was measured on indicated days (mean ± SD of triplicates). (C) Cells were starved of glucose or IL-3 ±15 mM GlcNAc for 4 d, and cell size was measured by Coulter Counter (mean ± SD of triplicates). (D) Cells were withdrawn from glucose for 24 h, and then GlcNAc was added to cells for an additional 48 h in the presence of 0.5 μM Jaki (JAK inhibitor I), 10 μM Ly (LY294002, PI-3K inhibitor), 10 μM Akti (Akt inhibitor VIII), 100 nM Rapa (Rapamycin, mTOR inhibitor), and 30 μM PD (PD 98059; MAPKK inhibitor). (UT) Untreated. IL-3 was present in all conditions. Cell size was measured in femtoliters (fL) (mean ± SD of triplicate wells). (E) Cells were starved of glucose in the presence or absence of 15 mM GlcNAc and 1 μg/mL cycloheximide for 5 d. (UT) Untreated. IL-3 was present in all conditions. Protein content was measured by BCA protein assay and normalized to total cell number (mean ± SD of triplicate wells). Result is representative of two independent experiments. (F) Cell size in glucose-starved cells was measured (mean ± SD of triplicates) after 3 d of treatment in the presence or absence of GlcNAc and 2 μg/mL cycloheximide. (UT) Untreated. IL-3 was present in all conditions. Results are representative of three independent experiments. (G) Cells were cultured ±glucose (11 mM) and ±GlcNAc (15 mM) for 5 d. IL-3 was present in all conditions. Cell number was assessed by Coulter counter (mean ± SD of triplicates). (H) Cells were starved of glucose for 24 h, after which either 15 mM glucose or 15 mM GlcNAc was added to cells in the presence of 10 μM BrdU. BrdU incorporation was measured by FACS every 6 h for 24 h. Result is representative of two independent experiments. For all indicated panels, P < 0.05 (*), P < 0.005 (**), and P < 0.0005 (***).
We also compared the effects of GlcNAc on cell growth to those of another metabolite that can promote hexosamine pathway activation: glucosamine. Glucosamine enters the cell through glucose transporters and is phosphorylated by hexokinase to generate glucosamine-6-phosphate (Brown 1951; Oguchi et al. 1975; Uldry et al. 2002). We found that either glucosamine or GlcNAc was able to stimulate cell growth in the absence of glucose, but that glucosamine treatment also led to an increase in cell number (Supplemental Fig. S3A,B). While glucosamine can enter cells more readily than GlcNAc, it can also be deaminated by glucosamine-6-phosphate deaminase (GNPDA) to produce ammonia and the glycolytic metabolite fructose-6-phosphate, potentially enabling glycolytic in addition to hexosamine pathway flux (Wolosker et al. 1998; Zhang et al. 2003). Glucosamine treatment indeed led to increased production of ammonia (Supplemental Fig. S3C). Considering the known action of GNPDA and the increased production of ammonia observed in our cells upon glucosamine treatment, the data suggest that glucosamine is able to be deaminated in order to enter the glycolytic pathway in addition to contributing to the hexosamine pathway.
N-linked glycosylation is required for GlcNAc-regulated IL-3Rα surface expression and growth
The murine IL-3Rα subunit contains five potential asparagine residues that might be modified by N-linked glycosylation (Hara and Miyajima 1992). To confirm that N-linked glycosylation was required for hexosamine pathway regulation of IL-3Rα and cell growth, cells were treated with the N-glycosylation inhibitors tunicamycin (tun), castanospermine (CSN), and deoxynojirimycin (DJ). Tun is an inhibitor of GlcNAc-phosphotransferase (GPT), the enzyme that catalyzes the first step in N-linked glycosylation, in which GlcNAc-1-phosphate is transferred from UDP-GlcNAc to dolichol phosphate. CSN and DJ are inhibitors of ER glucosidases that block remodeling of N-glycans in the Golgi (Elbein 1987). Indeed, upon inhibition of N-linked glycosylation with tun, CSN, or DJ, GlcNAc failed to regulate surface expression of IL-3Rα or its downstream signaling (Supplemental Fig. S4A,B). Correspondingly, the effects of GlcNAc on cell size were markedly blunted upon inhibition of N-linked glycosylation (Supplemental Fig. S4C). We also examined eIF2α phosphorylation, which occurs in response to ER stress to inhibit translation. Glucose deprivation led to increased eIF2α phosphorylation, as expected, but GlcNAc had minimal ability to reverse it, suggesting that its effects cannot be ascribed to resolution of glucose withdrawal-induced ER stress (Supplemental Fig. S4B). Conversely, we asked whether increased use of UDP-GlcNAc for N-glycan branching might promote increased IL-3Rα surface expression. Four N-acetylglucosaminyltransferases (Mgat1, Mgat2, Mgat4, and Mgat5) sequentially regulate N-glycan branching in the medial Golgi. Metabolic regulation of growth factor receptors, such as the epidermal growth factor receptor (EGFR), has been shown to be regulated through Mgat5 due to its activity in generating tetra-antennary N-glycans that are further extended with N-acetyllactosamine. These glycans interact with galectins at the cell surface, which can serve multiple functions, including suppression of ligand-independent activation and prevention of endocytosis (Dennis et al. 2009). Thus, we asked whether Mgat5 could regulate IL-3Rα surface expression by overexpressing either wild-type Mgat5 or a mutant Mgat5 (L188R, a point mutation blocking its Golgi localization) (Partridge et al. 2004). Indeed, IL-3Rα surface levels were modestly but significantly higher in cells overexpressing wild-type Mgat5 than in those overexpressing mutant Mgat5 (Supplemental Fig. S4D). This is expected because mono-, di-, tri-, and tetrabranched N-glycans display a gradation of affinities for galectins, and the surface presence of these N-glycans is dependent on the levels of Mgat1, Mgat2, Mgat4, and Mgat5, as well as UDP-GlcNAc. Together, these results suggest that UDP-GlcNAc use indeed regulates the surface expression of IL-3Rα.
Activation of the hexosamine pathway fuels growth by stimulating glutamine uptake
We next sought to understand how activation of the hexosamine pathway could enable growth in the absence of glucose as a fuel supply. While GlcNAc can enable surface presentation of and signaling from the IL-3R, it cannot act as a fuel source (Fig. 4E), and, in the absence of glucose, another carbon source must be obtained to support growth.
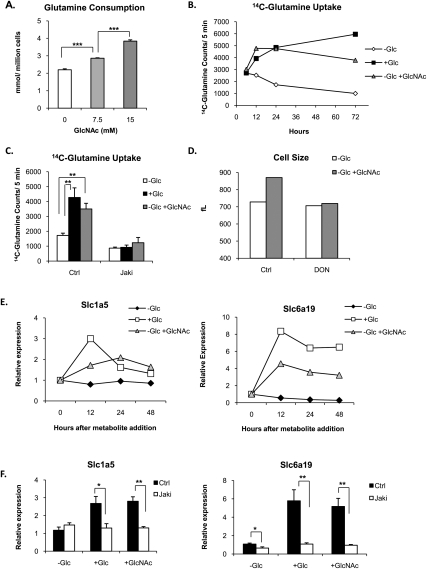
In cells using aerobic glycolysis, glutamine plays a key role in supplying the TCA cycle with substrate (DeBerardinis et al. 2007). Thus, we asked whether increased consumption of glutamine from the medium might account for replenishment of the TCA cycle and cell growth upon GlcNAc treatment. Indeed, in the absence of glucose, GlcNAc treatment increased glutamine consumption in a dose-dependent manner (Fig. 6A). To gain a sense for the timing by which GlcNAc treatment induces glutamine uptake, we measured the cells' capacity to transport 14C-glutamine at multiple time points after GlcNAc treatment of glucose-starved cells, finding that, by 12 h, glutamine uptake capacity was already potently induced. The ability of glucose or GlcNAc to induce glutamine uptake was comparable and followed a similar time course (Fig. 6B). GlcNAc-induced glutamine uptake was IL-3-dependent, since Jak inhibition blocked its uptake (Fig. 6C). Treatment with the glutamine analog 6-diazo-5-oxo-L-norleucine (DON) was able to largely block the effects of GlcNAc on cell growth, suggesting that increased glutamine uptake and metabolism is required for hexosamine-stimulated cell growth (Fig. 6D).
Figure 6.
GlcNAc treatment promotes increased glutamine consumption. (A) IL-3-dependent cells were withdrawn from glucose in the presence of 0, 7.5, or 15 mM GlcNAc for 3 d. IL-3 was present in all conditions. Glutamine consumption from the medium was measured by BioProfile Flex automated metabolite analyzer (mean ± SD of triplicates). (B) After 24 h of glucose starvation, 15 mM glucose or 15 mM GlcNAc was added to cells. IL-3 was present throughout the experiment. At 6, 12, 24, and 72 h after metabolite addition, a portion of the cells was harvested and the glutamine transport capacity of the cells was determined by measuring glutamine uptake over 5 min, as described in the Materials and Methods. (C) Cells were starved of glucose for 24 h, and then either 15 mM glucose or 15 mM GlcNAc was added to cells in the presence or absence of Jaki. After an additional 24 h, cells were harvested and 14C-glutamine uptake over 5 min was measured (mean ± SD of triplicates). IL-3 was present in all conditions. (D) Cells were starved of glucose for 24 h, and then treated in the presence or absence of 15 mM GlcNAc and 60 μM DON. After 48 h, cell size was measured. IL-3 was present in all conditions. (E) Cells were starved of glucose for 24 h, then 15 mM glucose or 15 mM GlcNAc was added to cells. IL-3 was present throughout the experiment. RNA was isolated at 0, 12, 24, and 48 h after metabolite addition, and gene expression was analyzed by quantitative RT–PCR. Data were normalized to 18S rRNA. Results are representative of two independent experiments. (F) Cells were starved of glucose for 24 h, and then glucose or GlcNAc was added for an additional 24 h, in the presence or absence of Jaki. IL-3 was present in all conditions. RNA was isolated from triplicate wells and gene expression was determined by quantitative RT–PCR, normalized to 18S rRNA (mean ± SD). For all indicated panels, P < 0.05 (*), P < 0.005 (**), and P < 0.0005 (***).
To investigate how glutamine uptake might be up-regulated, we examined expression of amino acid transporters in response to GlcNAc treatment. After 24 h of glucose starvation, addition of either glucose or GlcNAc resulted in an increase in mRNA levels of two transporters (Slc6a19 [B0AT1] and Slc1a5 [ASCT2]) within 12 h after addback (Fig. 6E), consistent with the time course observed for stimulation of glutamine uptake. Slc1a5 is up-regulated in multiple types of cancer and has been implicated in mediating net glutamine uptake in cancer cells (Fuchs and Bode 2005). Glutamine uptake into the cell through Slc1a5 has been shown to be essential for regulating mTOR activity (Fuchs et al. 2007; Nicklin et al. 2009) by making it available for coupling glutamine export to leucine import into the cell through Slc7a5 (Nicklin et al. 2009). Slc6a19 preferentially transports neutral amino acids, including leucine, isoleucine, and glutamine. Mutations in SLC6a19 are responsible for Hartnup disorder (Broer et al. 2004; Kleta et al. 2004; Bohmer et al. 2005). IL-3R signaling was required for induction of these transporters, since the Jak inhibitor completely blocked induction of these genes (Fig. 6F).
Glutamine uptake promotes growth through multiple mechanisms
We next investigated how hexosamine-stimulated glutamine uptake supports cell growth. In order to grow, cells must be able to produce ATP and engage in biosynthesis. Glutamine serves as an important anapleurotic substrate, enabling production of ATP and biosynthetic precursors (DeBerardinis et al. 2007, 2008). In addition, glutamine can be exchanged for essential amino acids to allow mTORC1 signaling and protein synthesis (Nicklin et al. 2009). We therefore tested the ability of glutamine to be used as a TCA cycle substrate for ATP production, to contribute carbon for lipid biosynthesis, and to promote uptake of essential amino acids.
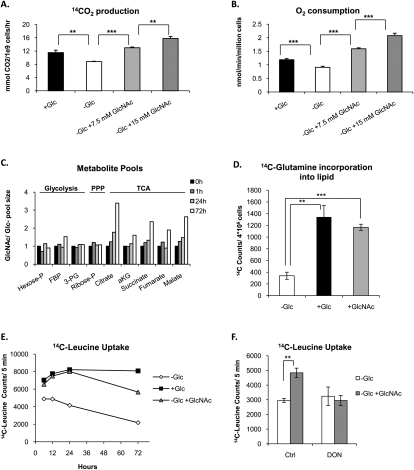
Glutamine enters the TCA cycle after being converted to α-ketoglutarate (αKG). αKG is then subject to oxidative decarboxylation by αKG dehydrogenase, generating CO2. Thus, to determine whether glutamine was indeed being oxidized in the TCA cycle, we treated cells with [U-14C5]glutamine to monitor production of CO2 from glutamine. Indeed, 14CO2 production decreased in the absence of glucose and increased in a dose-dependent manner upon GlcNAc treatment (Fig. 7A). Also consistent with increased TCA cycle activity, GlcNAc treatment resulted in a dose-dependent rescue of oxygen consumption (Fig. 7B). LC-MS/MS analysis further showed that, in comparison with glucose-withdrawn cells, GlcNAc treatment selectively led to increased pool sizes of TCA cycle metabolites, but not glycolytic or pentose phosphate pathway metabolites (Fig. 7C; Supplemental Table S1). Carbon from glucose or glutamine is converted into lipid through export of the TCA cycle metabolite citrate to the cytoplasm, where it is cleaved by ATP-citrate lyase to generate acetyl-CoA for de novo fatty acid synthesis. While the majority of acetyl-CoA comes from glucose under standard conditions, glutamine carbon should be able to fully support the TCA cycle in the absence of glucose, contributing both the acetyl-CoA and oxaloacetate carbon necessary to produce citrate (see Supplemental Fig. S5; DeBerardinis et al. 2007). To assess whether glutamine carbon is used to support lipid synthesis upon GlcNAc treatment, we measured incorporation of carbon from [U-14C5]glutamine into lipids. Indeed, either glucose or GlcNAc stimulated use of glutamine carbon for lipid synthesis in these cells (Fig. 7D). These results indicate that glutamine-derived carbon can contribute to the acetyl-CoA used for lipid synthesis in the presence or absence of glucose. Levels of glutamine carbon incorporation into lipid were not significantly different between glucose- and GlcNAc-treated cells. However, total lipid synthesis in glucose-replete cells must be substantially higher than in GlcNAc-treated cells, since glucose-replete cells were undergoing proliferative expansion. While these results indicate that glutamine carbon can be used to support lipid biosynthesis, they do not rule out the possibility that other carbon sources, such as other amino acids or serum acetate, might also contribute. With this in mind, we also tested whether treatment with GlcNAc could promote uptake of the essential amino acid leucine. Leucine uptake was stimulated after treatment of glucose-starved cells with either glucose or GlcNAc and could be inhibited by treatment with DON, indicating its glutamine dependence (Fig. 7E,F). Thus, GlcNAc treatment promotes metabolism of glutamine in the TCA cycle, use of glutamine carbon for biosynthesis, and glutamine-dependent uptake of leucine.
Figure 7.
Increased glutamine consumption promotes growth through multiple mechanisms. (A) IL-3-dependent cells were withdrawn from glucose in the presence or absence of 7.5 or 15 mM GlcNAc for 3 d. IL-3 was present in all conditions. Cells were then incubated with 14C5-glutamine and production of 14CO2 was measured after 8 h (mean ± SD of triplicates). Results are representative of two independent experiments. (B) Oxygen consumption was measured (mean ± SD of quadruplicate samples) after 2 d of glucose withdrawal in the presence of IL-3 ±7.5 or 15 mM GlcNAc. Results are representative of three independent experiments. (C) IL-3-dependent cells were starved of glucose for 24 h, and then 15 mM 13C6-glucose, 15 mM 13C6-GlcNAc, or equal volume water control was added to cells. Cells were harvested at 1, 24, and 72 h after nutrient addition. LC-MS/MS analysis of labeled and unlabeled metabolites was performed. The experiment was done with duplicates for each condition. The ratio of unlabeled metabolite pools in GlcNAc-treated versus glucose-withdrawn control cells was determined at each time point for metabolites in glycolytic and pentose phosphate pathways and the TCA cycle. Pools of TCA cycle metabolites, but not glycolytic or pentose phosphate metabolites, increased in response to GlcNAc treatment. See also Supplemental Tables S1 and S2. (D) Cells were starved of glucose overnight and then glucose or GlcNAc was added to cells, along with 14C5-glutamine, for 24 h. Cells were harvested, lipids were extracted, and incorporation of 14C into lipids was measured by scintillation counting (mean ± SD of triplicates). Results are representative of two independent experiments. (E) Cells were starved of glucose for 24 h, and then either 15 mM glucose or 15 mM GlcNAc was added to cells. Uptake of 14C-leucine was measured at indicated time points (mean ± SD of triplicate wells). (F) Cells were withdrawn from glucose for 24 h in IL-3-containing medium ±60 μM DON. GlcNAc was added to cells in each condition for an additional 24 h and uptake of 14C-leucine was assessed (mean ±SD of triplicates). For all indicated panels, P < 0.005 (**) and P < 0.0005 (***).
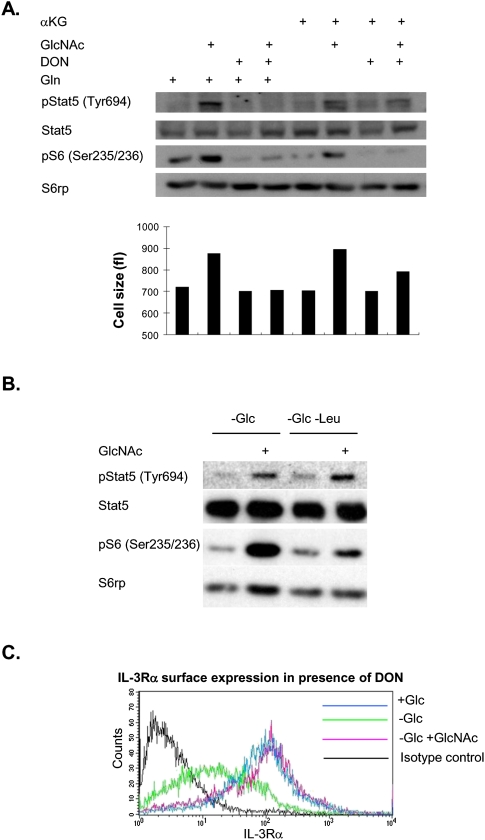
Since glutamine is known to regulate mTOR signaling through stimulation of leucine uptake (Nicklin et al. 2009), we asked whether increased glutamine uptake was required for the effects of GlcNAc on signaling. Inhibition of glutamine metabolism with DON indeed blocked the effects of GlcNAc on phosphorylation of S6rp, a target downstream from mTOR (Fig. 8A). As expected, leucine was required for increased S6rp phosphorylation in the presence of GlcNAc (Fig. 8B).
Figure 8.
Glutamine is required for continued signaling downstream from the IL-3R. (A) Cells were starved of glucose for 3 d in medium containing either 4 mM glutamine or 1 mM dimethyl-αKG ±15 mM GlcNAc and ±60 μM DON. Signaling was analyzed by Western blot. Cell size in femtoliters (fL) was assessed prior to lysis of cells. Results are representative of three independent experiments. (B) Cells were withdrawn from glucose ±0.4 mM leucine and ±15 mM GlcNAc for 2 d as indicated, and signaling was assessed by Western blot. Results are representative of two independent experiments. (C) Surface expression of the IL-3Rα was determined by flow cytometry in cells treated with 60 μM DON.
Unexpectedly, inhibition of glutamine uptake with DON also blocked the sustained IL-3-dependent phosphorylation of Stat5 (Fig. 8A), despite having no effect on GlcNAc induction of IL-3Rα surface expression (Fig. 8C). Unlike S6rp phosphorylation, GlcNAc-dependent regulation of Stat5 phosphorylation occurred in a leucine-independent manner (Fig. 8B). To investigate whether mitochondrial metabolism of glutamine was required for sustained Stat5 phosphorylation, we substituted cell-permeable dimethyl-αKG for glutamine in the culture medium. After αKG substitution, GlcNAc had only modest effects on phosphorylation of S6rp, and even this slight promotion of S6 phosphorylation was inhibited by DON, consistent with S6rp phosphorylation being dependent on glutamine–leucine exchange (Fig. 8A). It is likely that the slight rescue of S6rp phosphorylation by αKG is due to synthesis of glutamine from αKG. Since DON is capable of inhibiting transport of glutamine (Taylor et al. 1992), glutamine–leucine exchange may also be inhibited. In contrast, GlcNAc was able to potently promote phosphorylation of Stat5 when αKG was substituted for glutamine, and this effect was not blocked by DON, suggesting that mitochondrial metabolism of glutamine is required for Stat5 phosphorylation (Fig. 8A). Prior studies have shown that Stat5 phosphorylation can be promoted under conditions in which reactive oxygen species (ROS) production is elevated (Sattler et al. 1999; Iiyama et al. 2006). Consistent with the increased mitochondrial glutaminolysis and oxygen consumption that we observe during GlcNAc treatment, and that is expected to increase mitochondrial ROS production, we find that antioxidant treatment inhibits GlcNAc-stimulated Stat5 phosphorylation (Supplemental Fig. S6). Cell size correlated well with signaling, with DON blocking GlcNAc-stimulated growth in the presence of glutamine and exerting partial effects in the presence of αKG (Fig. 8A). These data suggest that IL-3-dependent glutamine consumption feeds back positively on signaling pathways, regulating mTOR-dependent signaling through promotion of essential amino acid uptake and Stat5 phosphorylation in a manner dependent on mitochondrial metabolism of glutamine.
Discussion
In this study, we identify a novel mechanism through which glucose and glutamine metabolism is coordinated. The data suggest that glucose availability to the hexosamine pathway can regulate growth factor receptor surface expression in a manner dependent on N-linked glycosylation. Hence, in the absence of glucose, reduced surface expression of IL-3Rα limits the ability of IL-3 to stimulate growth and proliferation. Surprisingly, restoration of hexosamine pathway flux in the absence of glucose through treatment with GlcNAc not only enables IL-3Rα surface expression and signaling to occur, but also IL-3-dependent growth. Since GlcNAc itself can only serve as a fuel signal, not a fuel source, another carbon source must be used by the cell under these conditions. We found that a critical component for GlcNAc-mediated cell growth in the absence of glucose is growth factor-dependent stimulation of glutamine uptake, which enables carbon flux into the TCA cycle, even when glucose carbon is unavailable. Increased glutamine consumption can fuel growth through mitochondrial metabolism of glutamine for production of ATP and biosynthetic precursors, and through promotion of uptake of essential amino acids. Stimulation of glutamine uptake and mitochondrial metabolism is also expected to feed back positively on the hexosamine pathway; both glutamine and acetyl-CoA are critical for continued activity of the hexosamine pathway (Fig. 4A). The inability of GlcNAc to promote proliferation in the absence of glucose is not surprising, since glucose is required for metabolic pathways such as the pentose phosphate pathway, which is important for nucleotide biosynthesis and NADPH production (Tong et al. 2009). Thus, the data suggest a model in which glucose-dependent flux through the hexosamine pathway regulates surface expression of IL-3Rα to entrain glutamine and essential amino acid metabolism to available levels of both growth factor and glucose (Supplemental Fig. S5).
The above data demonstrate that cells do not simply switch to glutamine metabolism when depleted of extracellular or intracellular glucose. Instead, cells must have an adequate supply of glucose to maintain the hexosamine pathway in order to support growth factor-induced uptake of glutamine. Furthermore, the requirement for glutamine to take up leucine suggests that a two-tiered checkpoint for amino acid consumption exists. Such a system makes sense bioenergetically; if a cell does not have glucose, biosyntheses that require intermediates derived from glycolysis or pathways that branch off from glycolysis cannot be conducted. Since proliferation cannot occur in the absence of glucose, cells do not waste organismal resources by acquiring and metabolizing glutamine. The second checkpoint—that glutamine is required for uptake of leucine—suggests that cells are only permitted to access the organism's common supply of essential amino acids when replete in the ability to maintain bioenergetics and support nonessential amino acid, lipid, and nucleotide synthesis through access to both glucose and glutamine.
Together, these findings define a novel mechanism through which cellular metabolism regulates signal transduction through metabolic regulation of glycosylation. Precise mechanistic details through which the hexosamine pathway regulates IL-3Rα surface expression, growth, and glutamine consumption will require further investigation. The data are consistent with a model in which new synthesis of IL-3Rα is required for continued surface expression in growth factor-replete conditions. In the absence of IL-3, high levels of IL-3Rα surface expression are likely due to inhibiting receptor endocytosis and/or degradation. Further investigation of IL-3Rα trafficking will be required to clarify these details. The generality of our findings will require investigation of additional growth factor receptors. UDP-GlcNAc use in the Golgi for N-glycan branching has been shown to regulate surface expression and signaling of other growth factor receptors, including EGFR, IGFR, and PDGFR (Partridge et al. 2004; Lau et al. 2007; Lau and Dennis 2008). The TGFβ receptor surface expression has been shown to be regulated in a glucose-dependent manner in mouse embryonic fibroblasts (MEFs) and endothelial cells (Wu and Derynck 2009); it will be of interest to determine whether the hexosamine pathway is also important for its regulation. Since mice lacking the Golgi branching enzyme Mgat5 display a metabolic phenotype that is consistent with our findings during glucose deprivation—including reduced anabolic signaling, reduced lipid synthesis and storage, and reduced oxygen consumption (Cheung et al. 2007)—it is likely that the metabolic regulation that we observe for IL-3Rα will be similar for other growth factor receptors. Golgi modification of N-glycans can also impact tumorigenesis (Lau and Dennis 2008). Surface GlcNAc-branched N-glycans are elevated upon cell transformation, and Mgat5−/− mice exhibit reduced tumor growth and metastasis (Yamashita et al. 1985; Pierce and Arango 1986; Granovsky et al. 2000).
It is also possible that other targets in addition to the IL-3R may mediate some of the effects of GlcNAc on growth. Other membrane proteins, such as nutrient transporters, could potentially be regulated by GlcNAc availability in the ER and Golgi as well. Increased gene expression of transporters and receptors in growth factor-stimulated cells may be supported by metabolic input to the hexosamine/Golgi pathways. For example, activation of T cells stimulates CTLA-4 receptor expression that is largely intracellular and reaches threshold levels at the cell surface with metabolic input to UDP-GlcNAc (Lau et al. 2007). In addition, UDP-GlcNAc production in the hexosamine pathway is used for O-GlcNAc modification of intracellular proteins in order to regulate cellular activities such as signaling and transcription (Love and Hanover 2005; Zachara and Hart 2006). O-GlcNAcylation is undoubtedly impacted in this system by glucose withdrawal and GlcNAc treatment, and thus may also participate in the effects on growth and glutamine consumption that we observed.
In summary, we report a novel mechanism used by hematopoietic cells to link glucose and glutamine metabolism to fuel growth through hexosamine pathway-dependent regulation of growth factor receptor surface expression. Like glucose, glutamine uptake is not cell-autonomous, but instead is directly controlled by growth factor signaling. Furthermore, the dependence of glutamine uptake on glucose flux through the hexosamine pathway provides a satisfying explanation for why glucose remains the essential bioenergetic substrate for mammalian cells, despite an abundant extracellular supply of oxidizable amino acids.
Materials and methods
Cell culture
IL-3-dependent bax−/−bak−/− hematopoietic cells (hereafter referred to as IL-3-dependent cells) have been described previously and were used to avoid confounding features of apoptotic cell death in these experiments (Lum et al. 2005). Cells were cultured in suspension in RPMI 1640, 10% FCS, penicillin/streptomycin, 2 mM L-glutamine, 10 mM HEPES, 50 μM β-mercaptoethanol, and 3 ng/mL recombinant IL-3 (BD Biosciences). Unless otherwise indicated, experiments were set up at densities of 2 × 105 to 2.5 × 105 cells per milliliter. To generate IL-3-dependent bax−/−bak−/− cells stably expressing wild-type and mutant Mgat5, pMX-PIE vectors expressing Mgat5 or Mgat5(L188R) (a point mutation that blocks localization of the enzyme to the Golgi), described previously in Partridge et al. (2004), were electroporated into the cells using an Amaxa Nucleofector I. pMX-PIE contains an internal ribosome entry site (IRES) between the multiple cloning site and enhanced green fluorescent protein (EGFP) sequence, and stable transfectants were selected by sorting for EGFP+ cells. Continued expression was verified by FACS analysis of EGFP expression and MGAT5 enzyme assays. Both wild-type and mutant protein were overexpressed, although cells maintained higher levels of the mutant (data not shown). M-NFS-60 cells were obtained from American Type Culture Collection (ATCC) and were cultured in the same medium as the IL-3-dependent bax−/−bak−/− cells. IL-3-dependent primary bone marrow cells were isolated from wild-type mice. Bone marrow was obtained from the tibia and femurs of two mice. White blood cells were isolated by Histopaque 1083 centrifugation. Cells were washed in PBS and plated into 10 mL of medium (80% DMEM, 20% FCS, 2 mM L-glutamine, penicillin/streptomycin, 10 mM HEPES, 50 μM β-mercaptoethanol, 3 ng/mL IL-3). Every 2 d, suspension cells were collected and replated in fresh medium until cells began proliferating after ∼4 wk. Cells were expanded and used for experiments. For all experiments performed in the absence of glucose, media were prepared as above, except that glucose-free RPMI as well as dialyzed FCS were used. Chemicals were obtained from the following sources: GlcNAc, glucosamine, tun, DON, PD 98059, LY294002, rapamycin, NAC, and BHA were from Sigma; CSN and DJ were from Alexis Biochemicals; JAK inhibitor I was from EMD Biosciences.
Lectin binding
To assess cell surface binding of L-PHA, a total of either 0.5 million or 1 million cells per condition were used. Cells were spun down at 1200 rpm for 3 min, washed in FACS buffer (PBS + 0.5% FCS), and then incubated in 10 μg/mL L-PHA-fluorescein (Vector Laboratories) in FACS buffer for 20 min. Cells were washed, filtered, and analyzed for surface binding using a Becton Dickinson LSR flow cytometer.
Mgat5 activity assay
Enzyme activity was determined as transfer of 6-3H-UDP-GlcNAc to the synthetic acceptors. Cell pellets from confluent plates of IL-3-dependent cells stably expressing the wild-type or mutant form of Mgat5 were lysed in 0.9% NaCl, 1% Triton X-100, and protease inhibitor on ice. The reaction contained 10 μL of cell lysate, 1 μCi of [63H]-UDP-GlcNAc (44,000 dpm/nmol), 50 mM MES (pH 6.5), 0.5 M GlcNAc, 1 mM UDP-GlcNAc, 1 μCi of 3H-UDP-GlcNAc, and 25 mM AMP in a final volume of 20 μL, with synthetic acceptors 1 mM βGlcNAc(1,2)αMan(1,6)βGlc-O(CH2)7CH3 for Mgat5, and 1 mM αMan(1,6)βGlc-O(CH2)7CH3, plus 2 mM MnCl2 for Mgat1. Reactions were incubated for 180 min at 37°C for Mgat5 and 45 min at 37°C for Mgat1 assays. Endogenous activity was measured in the absence of acceptor, and was subtracted from values determined in the presence of added acceptor. Reactions were stopped with 1 mL of H2O, and enzyme products were separated from radioactive substrates by binding them to 50-mg C18 cartridges (Alltech) preconditioned with methanol rinsing and water washes. Reactions were loaded and the columns were washed three times with water. Radiolabeled products were eluted directly into scintillation vials with two separately applied 250-μL aliquots of methanol, and the radioactivity was determined by liquid scintillation counting.
Western blotting
For signaling blots, cells were lysed in RIPA buffer and briefly sonicated. Lysates were quantitated using BCA Protein Assay (Pierce and Arango) and equal protein concentrations were loaded onto gels. For cell surface proteins, Cell Surface Protein Isolation kit (Pierce and Arango) was used according to the manufacturer's instructions. Briefly, surface proteins were biotinylated using Sulfo-NHS-SS-biotin, cells were lysed, and biotinylated proteins were isolated using a NeutrAvidin agarose column. For total membrane protein preparation, the CelLytic MEM Protein Extraction kit (Sigma) was used as per the manufacturer's instructions to isolate hydrophobic proteins. Digestion with PNGase F (Sigma) was performed on a hydrophobic fraction.
After running gels, proteins were transferred onto nitrocellulose membranes. Membranes were blocked in 5% milk and primary antibody incubations were performed in 3% BSA (overnight at 4°C). The antibodies used were Tubulin (Sigma); phospho-Stat5 (Upstate Biotechnologies); Pim1, Pim2,c-myc, IL-3Rα, and IL-3Rβ (Santa Cruz Biotechnology); and phospho-Akt-ser473, Akt, phospho-S6rp, S6rp, phospho-eif2α, eif2α, and Stat5 (Cell Signaling Technology). Blots were developed using ECL or ECL Plus Reagent (Amersham) and were exposed to Hyperfilm ECL (Amersham). Coomassie staining was performed using SimplyBlue Safe Stain (Invitrogen).
Glutamine consumption and ammonia production
Glutamine consumption and ammonia production were measured from media using a BioProfile FLEX automated metabolite analyzer (Nova Biomedical). Cells were spun down, supernatant was collected, and the concentration of metabolite in the supernatant was determined; the amount consumed or produced by cells was determined by subtracting the concentration in the supernatant from the concentration in medium-only control that had been incubated for the same amount of time as cells at 37°C.
14C-glutamine and 14C-leucine uptake assays
For glutamine uptake, cells were incubated with 5 μL of [U-14C5]glutamine (labeled at all five carbons) (GE Healthcare) in 0.2 mL of glutamine-deficient medium for 5 min at 37°C. After incubation, cells were spun down and washed three times with ice-cold PBS, after which cells were lysed with 200 μL of a 0.2% SDS/0.2 N sodium hydroxide (NaOH) solution, incubated for 1 h, neutralized with 20 μL of 1 N hydrochloric acid (HCl), and analyzed with a β scintillation counter (PerkinElmer Life Sciences). The protocol was similar for leucine uptake except for incubation medium: 5 μL of [U-13C6]leucine (PerkinElmer Life Sciences) in 0.2 mL of leucine-deficient medium.
Metabolite profiling by mass spectrometry
IL-3-dependent cells were starved of glucose overnight (12 million cells for each of three conditions). After 24 h of glucose starvation, cells were treated with 15 mM [U-13C6]glucose, 15 mM N-acetyl-D-[U-13C6]glucosamine (labeled at glucose carbons), or equal volume of water control. At 1 h, 24 h, and 72 h after metabolite addition, duplicate samples (2 million cells each) were taken from each condition. Cells were spun down at 1500 rpm for 5 min at room temperature, media were removed, and cells were extracted in 300 μL of 40:40:20 methanol:acetonitrile:water for 15 min at −20°C. Extracts were spun down at 2500 rpm and supernatant was transferred into a fresh tube. Pellet was resuspended in 100 μL of extraction solvent and spun down again, and two supernatants were combined. Positive ion mass spectrometry was performed as described previously on the eluate of an amino propyl column via a Finnigan TXQ Quantum Ultra triple-quadrupole mass spectrometer equipped with an electrospray ionization source (Thermo Fisher Scientific, Inc.) (Bajad et al. 2006). A TSQ Quantum Discovery MAX mass spectrometer, also equipped with an electrospray ionization source, was used to collect data on negative-mode ions after separation on a 25-cm C18 column coupled with a tributylamine ion-pairing agent to aid in the retention of polar compounds, as described previously (Lu et al. 2008). To quantify metabolites, peak heights were initially assigned using XCalibur software (Thermo Fisher Scientific, Inc.) and then evaluated manually.
14C-glutamine incorporation into lipid
Incorporation of [U-14C5]glutamine into lipid was performed as described previously, with minor modifications (Wise et al. 2008). Briefly, IL-3-dependent cells were starved of glucose overnight. Fifteen millimolar glucose, 15 mM GlcNAc, or equal volume water was added to cells in the presence of 0.1 μCi/mL 14C5-glutamine for an additional 24 h. Four-million cells per condition in triplicate were harvested. Cells were washed three times in PBS, harvested in 0.4 mL of 0.5% Triton X-100, and transferred to a glass tube. Organic extraction was performed by adding each of the following in succession, vortexing after each addition: 2 mL of methanol, 1 mL of chloroform, 1 mL of chloroform, 1 mL of water. After centrifugation at 2000 rpm for 10 min, the lower organic phase was removed and transferred to a new glass tube. A second extraction was performed on aqueous phase. Organic phases were combined and evaporated to dryness under a nitrogen stream. Lipids were resuspended in chloroform and transferred to scintillation vials, and 14C counts were measured.
Quantitative RT–PCR
RNA was isolated using Trizol (Invitrogen) and cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was performed on a 7900HT Sequence Detection System (Applied Biosystems) using TaqMan Gene Expression Assays (Applied Biosystems). Gene expression data were normalized to 18S rRNA.
Analysis of receptor surface expression by flow cytometry
A total of either 0.5 million or 1 million cells per condition were spun down. Cells were washed in FACS buffer and then incubated for 15 min in primary antibody (1:200 biotinylated rat anti-mouse CD123 [IL-3Rα] [BD Biosciences] or 1:200 PE-conjugated rat anti-mouse CD131 [IL-3Rβ]). Cells were washed three times in FACS buffer. For IL-3Rα, cells were then incubated in 1:200 streptavidin-PE secondary antibody (BD Biosciences) for 15 min. Cells were washed three times in FACS buffer and then surface expression was analyzed on a Becton Dickinson LSR Flow Cytometer.
14CO2 production from glutamine
IL-3-dependent cells were treated for 3 d in IL-3-containing medium in the presence or absence of glucose or GlcNAc. After 3 d, cells were counted and resuspended in RPMI without bicarbonate or glucose, with additions of 10 mM glucose or 7.5 or 15 mM GlcNAc, at 1 million cells per milliliter. [U-14C5]glutamine (0.05 μCi/μL) was added to cells at 5 μL/mL. One-milliliter amounts of cells were pipetted into 20-mL glass vials. Each tube was sealed using a rubber stopper with a suspended central well containing filter paper saturated with freshly prepared 5% potassium hydroxide (KOH). Cells were incubated for 8 h in a shaking 37°C water bath. At the end of incubation, 1 mL of 0.6 N acetic acid was added by inserting a needle through the rubber stopper, and vials were left overnight at room temperature to allow CO2 to react with KOH in filter paper (generates potassium bicarbonate [KHCO3]). Filter papers were removed, and 14C counts were determined by scintillation counter.
Oxygen consumption
Oxygen consumption in whole cells was determined as described previously (Schumacker et al. 1993). Briefly, IL-3-dependent cells were treated in IL-3-containing medium ±glucose and ±7.5 or 15 mM GlcNAc for 48 h. Ten-million cells per condition were spun down, resuspended in 1 mL of media, and transferred into a water-jacketed, airtight 3-mL chamber with a Clarke oxygen electrode (Hansatech Instruments). Oxygen consumption was recorded using Oxygraph software and rate of consumption was determined during minutes 4–8 of recording for each sample, when consumption rates were steady.
Acknowledgments
We thank members of the Thompson laboratory for helpful feedback and discussions. We thank Dr. Tullia Lindsten for expert help in isolating primary bone marrow cells. This work was supported by grants from the NIH and NCI to C.B.T., and by a Damon Runyon Post-doctoral Fellowship from the Damon Runyon Cancer Research Foundation (DRG-1955-07) to K.E.W.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1985910.
Supplemental material is available for this article.
References
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD 2006. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A 1125: 76–88 [DOI] [PubMed] [Google Scholar]
- Baker SJ, Rane SG, Reddy EP 2007. Hematopoietic cytokine receptor signaling. Oncogene 26: 6724–6737 [DOI] [PubMed] [Google Scholar]
- Bohmer C, Broer A, Munzinger M, Kowalczuk S, Rasko JE, Lang F, Broer S 2005. Characterization of mouse amino acid transporter B0AT1 (slc6a19). Biochem J 389: 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer A, Klingel K, Kowalczuk S, Rasko JE, Cavanaugh J, Broer S 2004. Molecular cloning of mouse amino acid transport system B0, a neutral amino acid transporter related to Hartnup disorder. J Biol Chem 279: 24467–24476 [DOI] [PubMed] [Google Scholar]
- Brown DH 1951. The phosphorylation of D (+) glucosamine by crystalline yeast hexokinase. Biochim Biophys Acta 7: 487–493 [DOI] [PubMed] [Google Scholar]
- Cheung P, Pawling J, Partridge EA, Sukhu B, Grynpas M, Dennis JW 2007. Metabolic homeostasis and tissue renewal are dependent on β1,6GlcNAc-branched N-glycans. Glycobiology 17: 828–837 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB 2007. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci 104: 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB 2008. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20 [DOI] [PubMed] [Google Scholar]
- Dennis JW, Nabi IR, Demetriou M 2009. Metabolism, cell surface organization, and disease. Cell 139: 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R, Ji K, Liu Z, Chen L 2009. A cytokine–cytokine interaction in the assembly of higher-order structure and activation of the interleukine-3:receptor complex. PLoS ONE 4: e5188 doi: 10.1371/journal.pone.0005188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB 2002. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell 13: 2276–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD 1987. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem 56: 497–534 [DOI] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB 2005. The Pim kinases control rapamycin-resistant T cell survival and activation. J Exp Med 201: 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BC, Bode BP 2005. Amino acid transporters ASCT2 and LAT1 in cancer: Partners in crime? Semin Cancer Biol 15: 254–266 [DOI] [PubMed] [Google Scholar]
- Fuchs BC, Finger RE, Onan MC, Bode BP 2007. ASCT2 silencing regulates mammalian target-of-rapamycin growth and survival signaling in human hepatoma cells. Am J Physiol Cell Physiol 293: C55–C63 doi: 10.1152/ajpcell.00330.2006 [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW 2000. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med 6: 306–312 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM 2007. Defining the role of mTOR in cancer. Cancer Cell 12: 9–22 [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman PS, Fox CJ, Birnbaum MJ, Thompson CB 2005. Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood 105: 4477–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Miyajima A 1992. Two distinct functional high affinity receptors for mouse interleukin-3 (IL-3). EMBO J 11: 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiyama M, Kakihana K, Kurosu T, Miura O 2006. Reactive oxygen species generated by hematopoietic cytokines play roles in activation of receptor-mediated signaling and in cell cycle progression. Cell Signal 18: 174–182 [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293 [DOI] [PubMed] [Google Scholar]
- Kleta R, Romeo E, Ristic Z, Ohura T, Stuart C, Arcos-Burgos M, Dave MH, Wagner CA, Camargo SR, Inoue S, et al. 2004. Mutations in SLC6A19, encoding B0AT1, cause Hartnup disorder. Nat Genet 36: 999–1002 [DOI] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S 1985. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem 54: 631–664 [DOI] [PubMed] [Google Scholar]
- Lau KS, Dennis JW 2008. N-Glycans in cancer progression. Glycobiology 18: 750–760 [DOI] [PubMed] [Google Scholar]
- Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW 2007. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129: 123–134 [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA 2005. The hexosamine signaling pathway: Deciphering the ‘O-GlcNAc code.’ Sci STKE 2005: re13 doi: 10.1126/stke.3122005re13 [DOI] [PubMed] [Google Scholar]
- Lu W, Bennett BD, Rabinowitz JD 2008. Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci 871: 236–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120: 237–248 [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi M, Miyatake Y, Ayabe J, Akamatsu N 1975. Phosphorylation of D-glucosamine by rat liver glucokinase. J Biochem 77: 1117–1121 [DOI] [PubMed] [Google Scholar]
- Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW 2004. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science 306: 120–124 [DOI] [PubMed] [Google Scholar]
- Pierce M, Arango J 1986. Rous sarcoma virus-transformed baby hamster kidney cells express higher levels of asparagine-linked tri- and tetraantennary glycopeptides containing [GlcNAc-β (1,6)Man-α (1,6)Man] and poly-N-acetyllactosamine sequences than baby hamster kidney cells. J Biol Chem 261: 10772–10777 [PubMed] [Google Scholar]
- Sattler M, Winkler T, Verma S, Byrne CH, Shrikhande G, Salgia R, Griffin JD 1999. Hematopoietic growth factors signal through the formation of reactive oxygen species. Blood 93: 2928–2935 [PubMed] [Google Scholar]
- Schumacker PT, Chandel N, Agusti AG 1993. Oxygen conformance of cellular respiration in hepatocytes. Am J Physiol 265: L395–L402 [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ 2009. The LKB1–AMPK pathway: Metabolism and growth control in tumour suppression. Nat Rev Cancer 9: 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ 2008. mTOR signaling: RAG GTPases transmit the amino acid signal. Trends Biochem Sci 33: 565–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6: 91–99 [DOI] [PubMed] [Google Scholar]
- Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA 2004. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR–ABL in cell cycle progression and leukemogenesis. Leukemia 18: 189–218 [DOI] [PubMed] [Google Scholar]
- Taylor PM, Mackenzie B, Hundal HS, Robertson E, Rennie MJ 1992. Transport and membrane binding of the glutamine analogue 6-diazo-5-oxo-L-norleucine (DON) in Xenopus laevis oocytes. J Membr Biol 128: 181–191 [DOI] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB 2009. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev 19: 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Ibberson M, Hosokawa M, Thorens B 2002. GLUT2 is a high affinity glucosamine transporter. FEBS Lett 524: 199–203 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327: 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB 2010. Cellular metabolic stress: Considering how cells respond to nutrient excess. Mol Cell 40: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosker H, Kline D, Bian Y, Blackshaw S, Cameron AM, Fralich TJ, Schnaar RL, Snyder SH 1998. Molecularly cloned mammalian glucosamine-6-phosphate deaminase localizes to transporting epithelium and lacks oscillin activity. FASEB J 12: 91–99 [DOI] [PubMed] [Google Scholar]
- Wu L, Derynck R 2009. Essential role of TGF-β signaling in glucose-induced cell hypertrophy. Dev Cell 17: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Tachibana Y, Ohkura T, Kobata A 1985. Enzymatic basis for the structural changes of asparagine-linked sugar chains of membrane glycoproteins of baby hamster kidney cells induced by polyoma transformation. J Biol Chem 260: 3963–3969 [PubMed] [Google Scholar]
- Zachara NE, Hart GW 2006. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta 1761: 599–617 [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR 2008. How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang W, Zou D, Chen G, Wan T, Li N, Cao X 2003. Cloning and functional characterization of GNPI2, a novel human homolog of glucosamine-6-phosphate isomerase/oscillin. J Cell Biochem 88: 932–940 [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]