Abstract
Genomic DNA is packed in chromatin fibers organized in higher-order structures within the interphase nucleus. One level of organization involves the formation of chromatin loops that may provide a favorable environment to processes such as DNA replication, transcription, and repair. However, little is known about the mechanistic basis of this structuration. Here we demonstrate that cohesin participates in the spatial organization of DNA replication factories in human cells. Cohesin is enriched at replication origins and interacts with prereplication complex proteins. Down-regulation of cohesin slows down S-phase progression by limiting the number of active origins and increasing the length of chromatin loops that correspond with replicon units. These results give a new dimension to the role of cohesin in the architectural organization of interphase chromatin, by showing its participation in DNA replication.
Keywords: Cohesin, DNA replication, replication origin, MCM, chromatin loop
Chromosomal cis interactions underlie the basic conformation of chromatin in interphase and influence all aspects of DNA metabolism, including transcription and replication (for review, see Gause et al. 2008). In the G1 phase of the cell division cycle, origins of replication are “licensed” by the assembly of prereplicative complexes (pre-RCs) consisting of the origin recognition complex (ORC), Cdc6, Cdt1, and minichromosome maintenance (MCM) proteins. Later on, the activity of CDK and Dbf4–Cdc7 kinases promotes the loading of additional replication proteins and leads to DNA unwinding and the initiation of DNA synthesis (for review, see Mendez and Stillman 2003; Sclafani and Holzen 2007). Hundreds of “replication factories” are formed during S phase, each one containing one or several clusters of six to 10 origins that fire almost simultaneously (Jackson and Pombo 1998). It has been proposed that, in these factories, neighboring origins are located in physical proximity to each other and the interorigin DNA regions are looped out, forming rosette-like structures (Berezney et al. 2000). In support of this model, it has been determined that the size of DNA loops correlates with the length of replicons, the units of DNA duplicated from each origin (Buongiorno-Nardelli et al. 1982; Lemaitre et al. 2005). While this disposition facilitates the local concentration of the initiator proteins and kinases required to activate origins, the molecular mechanisms that mediate this type of architectural organization remain unknown. In this study, we describe a role for cohesin in the formation of chromatin loops and the determination of replicon size at replication factories.
Cohesin is a protein complex initially identified for its role in sister chromatid cohesion (Guacci et al. 1997; Michaelis et al. 1997; Losada et al. 1998). In recent years, however, cohesin has been shown to participate also in DNA double-strand break repair (for review, see Sjögren and Ström 2010) and the control of gene expression (Hadjur et al. 2009; Nativio et al. 2009; Hou et al. 2010; Kagey et al. 2010; Schmidt et al. 2010). All known functions of cohesin appear to involve its capacity to embrace DNA molecules within its ring-shaped structure (for review, see Nasmyth and Haering 2009). The complex is composed of two members of the structural maintenance of chromosomes (SMC) family of proteins—Smc1 and Smc3—and two additional subunits known as Rad21/Scc1 and SA/Scc3. At least three other proteins interact with cohesin and modulate its function: Pds5, Wapl, and Sororin (for review, see Peters et al. 2008).
Cohesin is loaded onto chromatin by a mechanism that depends on the Scc2–Scc4 heterodimer (Ciosk et al. 2000; Watrin et al. 2006). In Xenopus cell-free extracts, formation of pre-RCs at origins is a prerequisite for cohesin loading (Gillespie and Hirano 2004; Takahashi et al. 2004). In contrast, cohesin can associate with chromatin in the absence of pre-RCs in budding yeast and Drosophila (Uhlmann and Nasmyth 1998; MacAlpine et al. 2009). Once engaged with the DNA, cohesin rings are able to translocate and reach other genomic sites (Lengronne et al. 2004). Chromatin immunoprecipitation (ChIP) analyses in mammalian cells have shown that cohesin becomes preferentially enriched at discrete sites, many of which are also bound by the chromatin insulator CTCF (Parelho et al. 2008; Wendt et al. 2008). Cohesin cooperates with CTCF to promote the formation of loops at some loci and thereby regulate gene expression (Hadjur et al. 2009; Mishiro et al. 2009; Nativio et al. 2009; Hou et al. 2010). In addition, cohesin contributes to tissue-specific gene expression independently of CTCF by facilitating the interaction between regulatory elements such as enhancers and core promoters (Kagey et al. 2010; Schmidt et al. 2010).
Here we describe a novel role for cohesin in the process of DNA replication that involves its ability to stabilize chromatin loops. We report that cohesin is enriched at origins of replication and interacts with MCM proteins. Down-regulation of cohesin results in slow S-phase progression, caused by the formation of larger chromatin loops in G1 and a reduced frequency of origin firing during S phase. These changes make replication factories less efficient without affecting their total number. Our data indicate that cohesin exerts a fundamental architectural role in the interphase nucleus, and show for the first time its participation in the spatial organization of replication factories in human cells.
Results
Cohesin interacts with pre-RC proteins
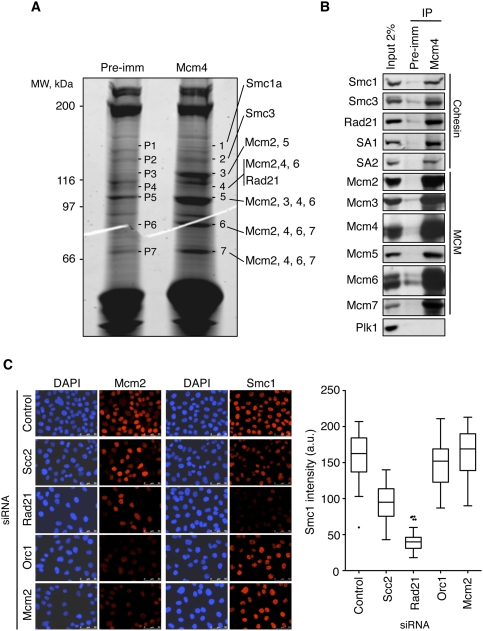
In order to gain insights into the regulation of human origins of replication, we conducted a proteomics search for proteins that interact with the MCM complex. After the immunoprecipitation of Mcm4 from a nuclear extract of cells synchronized in S phase, mass spectrometry analyses identified all six subunits of MCM (Mcm2–7) and three components of cohesin: Smc1, Smc3, and Rad21 (Fig. 1A; Supplemental Table S1), besides other factors that will be described elsewhere. The interaction between MCM and cohesin was confirmed by immunoprecipitation immunoblot assays, which also revealed the presence of both somatic versions of the SA subunit: SA1 and SA2 (Fig. 1B). Cohesin was recovered after immunoprecipitation assays with antibodies directed to other MCM subunits (Supplemental Fig. S1A) and, conversely, Mcm4 was present in cohesin immunoprecipitates (Supplemental Fig. S1B). The MCM–cohesin interaction was detected at all phases of the cell cycle (Supplemental Fig. S1C) and was not affected by ethidium bromide, suggesting that it is not mediated by bridging DNA molecules (Supplemental Fig. S1D).
Figure 1.
MCM complex interacts with cohesin. (A) Sypro-ruby staining of preimmune (pre-imm) and Mcm4 immunoprecipitates after SDS-PAGE fractionation. The indicated bands (1–7, and corresponding controls, P1–P7) were excised from the gel and analyzed by mass spectrometry. Mcm2–7 proteins and three components of the cohesin complex (Smc1a, Smc3, and Rad21) were identified (Supplemental Table S1). (B) Immunoprecipitations (IP) from HeLa nuclear extracts synchronized in S phase using Mcm4 or preimmune antibodies. Immunoprecipitates and input extract (2% of the amount used in the immunoprecipitation) were analyzed by Western blot with the indicated antibodies. Polo kinase 1 (Plk1) is shown as a negative control. (C) Immunostaining of chromatin-bound Mcm2, Smc1 proteins (red), and DNA (DAPI, blue) in control cells or after treatment with the indicated siRNAs. Bar, 50 μm. (Right panel) Box plot showing the quantification of Smc1 staining intensity in the different cell populations shown (n > 100 cells in each condition).
MCM and cohesin are loaded independently on chromatin
Both MCM and cohesin associate with chromatin at the exit of mitosis (Losada et al. 2000; Mendez and Stillman 2000). In order to examine whether they depend on each other for the process of chromatin loading, the chromatin association of cohesin and MCM was evaluated after RNAi-mediated silencing of a subunit of each complex (Rad21 and Mcm2) and a subunit of their known chromatin loaders (Scc2 and Orc1, respectively). Cohesin loading was impaired after Scc2 or Rad21 down-regulation, as expected, but not after Orc1 or MCM silencing (Fig. 1C). Conversely, MCM loading was prevented by Orc1 or Mcm2 down-regulation but not by the silencing of Rad21 or Scc2. These data strongly suggest that cohesin and MCM complexes are loaded onto chromatin by independent mechanisms, and pre-RC formation is not essential for the recruitment of cohesin to DNA in human cells.
Depletion of cohesin slows down S phase independently of cohesion and checkpoint responses
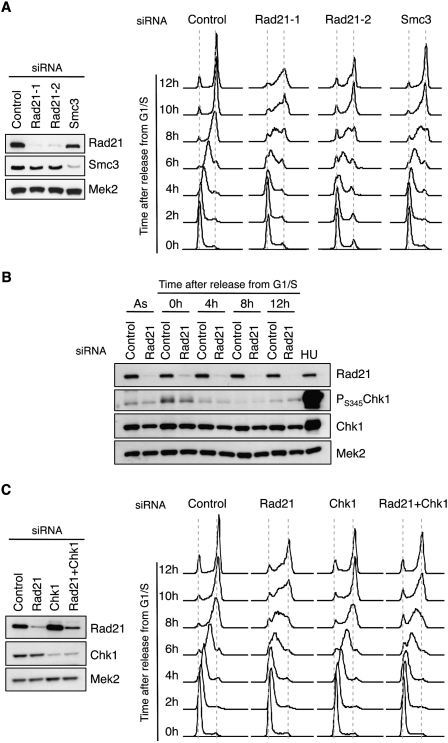
Given that cohesin interacts with pre-RC components, we next asked whether it played a role in DNA replication. Control and cohesin-depleted cells were synchronized at G1/S in order to monitor progression through S phase. Analyses of DNA content indicated that control cells completed S phase in ∼8 h upon release from the block, whereas cells depleted of cohesin required >12 h (Fig. 2A). This effect was observed with two different siRNA molecules directed to Rad21 and was confirmed by the down-regulation of another cohesin subunit: Smc3. Even if Smc3 silencing was slightly less efficient, it still caused a significant S-phase delay.
Figure 2.
Cohesin down-regulation impairs S-phase progression. (A, left) Immunoblots showing Rad21 and Smc3 levels in control cells or cells treated with three different siRNA oligonucleotides targeting cohesin (Rad21-1, Rad21-2, and Smc3). Mek2 levels are shown as loading control. (Right) DNA content analysis of the indicated siRNA-treated populations at different times after release from a G1/S block. (B) Detection of Rad21, PS345-Chk1, and total Chk1 levels after Rad21 down-regulation in asynchronous cells (As) or at the indicated times after release from a G1/S block. (HU) Cells treated with 2 mM hydroxyurea for 2 h, a control for checkpoint activation. The levels of Mek2 are shown as loading control. (C) S-phase progression, as in A, of cell populations treated with control, Rad21, Chk1, or Rad21 + Chk1 siRNA. Immunoblots on the left show the remaining levels of Rad21, Chk1, and Mek2 (loading control).
The participation of cohesin in DNA replication could depend on its ability to establish and/or maintain sister chromatid cohesion during S phase. To address this issue, we targeted sororin, a cohesin-interacting protein that is essential for cohesion but dispensable for cohesin loading (Schmitz et al. 2007). Sororin depletion did not delay S phase, indicating that the slower DNA replication caused by cohesin depletion is independent from the loss of sister chromatid cohesion (Supplemental Fig. S2).
We next checked whether the delay in S phase is mediated by a checkpoint response. Chk1 is the main effector kinase activated by ATR in response to DNA damage induced by replicative stress (for review, see Smits et al. 2010). The levels of activated Chk1 (pS345-Chk1) were similar in control and cohesin-depleted cells, either in asynchronous cultures or at different time points after release from a G1/S arrest (Fig. 2B). Besides, abrogation of Chk1 function by siRNA-mediated silencing did not rescue the strong defect in S-phase progression caused by cohesin loss (Fig. 2C). A slight effect was observed that is likely due to the lower efficiency of cohesin depletion after the double Rad21/Chk1 siRNA treatment (see immunoblots in Fig. 2C). This result was further confirmed by chemical inhibition of Chk1 or ATR/ATM with UCN-01 or caffeine, respectively (Supplemental Fig. S3A).
On the other hand, a modest activation of Chk2 was observed in cohesin-depleted cells, but almost exclusively in cells with 2C DNA content (Supplemental Fig. S3B,C). Chk2 kinase, a target of ATM, was probably activated by DNA breaks generated in mitosis by chromosome condensation or by the microtubule-pulling forces in the absence of proper cohesion. Indeed, cells that scored positive for γH2AX, a marker of double-strand breaks, were arrested mainly in mitosis and not in G1 or S phase (Supplemental Fig. S3D). Thus, even if the short-term down-regulation of cohesin activates ATM–Chk2 and results in a partial mitotic arrest, the checkpoint response is not responsible for the slow S-phase progression.
Cohesin influences origin activity
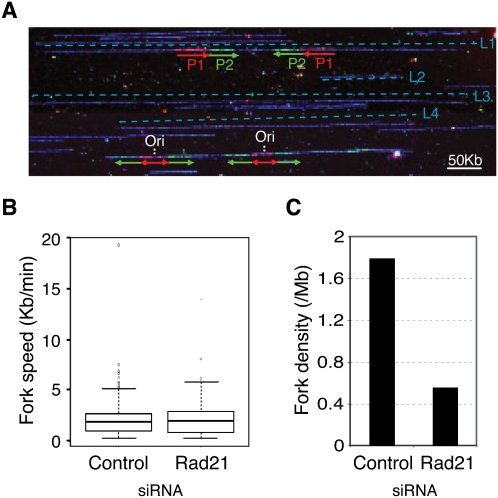
To gain insight into the molecular mechanism underlying the S-phase delay, we monitored DNA replication in single molecules by DNA combing after sequential pulse-labeling of cells with the nucleotide analogs IdU and CldU. The combing technique does not enrich for specific fractions of the genome and accurately reflects the global dynamics of DNA replication (Fig. 3A; Schwob et al. 2009). Fork velocity was similar in control and cohesin-depleted cells (∼2 kb/min) (Fig. 3; Supplemental Table S2). However, the density of forks (number of forks divided by the total length of DNA fibers, normalized by the percentage of cells in S phase) was reduced by threefold after cohesin down-regulation (Fig. 3C; Supplemental Table S2). These results were reproduced with cell cultures synchronized in early S phase (Supplemental Fig. S4; Supplemental Table S2). Because fork density is proportional to the number of active origins, the S-phase delay is likely caused by a reduced frequency of origin firing.
Figure 3.
Cohesin influences origin activity. (A) Representative image of combed DNA fibers after IdU + CldU double pulse-labeling. Immunodetection of IdU (red), CldU (green), or ssDNA (blue) are shown. Fork directionality and track length for the first (P1) and second (P2) pulses are shown by red and green arrows, respectively. The positions of two bicolor signals corresponding to moving forks and two “green–red–green” signals corresponding to replication origins (Ori) are indicated. Bar, 25 μm (50 kb). (B) Box plot showing fork progression rates in asynchronous populations of control or Rad21-depleted cells (control: n = 250 forks; Rad21: n = 175 forks). The horizontal line within the box represents the median. The box spans the interquartile range, and the vertical line spans the lower and upper quartiles. Outliers are shown as circles. (C) Global fork density in the same populations as in B, estimated by dividing the number of unambiguous forks by the total length of analyzed DNA and normalized to the number of cells in S phase (see the Materials and Methods; Supplemental Table S2).
As cohesin has been shown to participate in transcriptional regulation, its effect in DNA replication could also be caused by the altered expression of origin-activating genes. However, the levels of most initiator proteins—including ORC, MCM, Cdc45, and GINS subunits—did not change after cohesin down-regulation with three different siRNA molecules (Supplemental Fig. S5A). An intriguing exception was Cdc6, whose levels were partially reduced after cohesin depletion. This effect correlated with reduced transcription of CDC6 after cohesin or CTCF down-regulation (Supplemental Fig. S5B). Cdc6 protein promotes the loading of MCM complexes onto chromatin (for review, see Borlado and Mendez 2008). Importantly, the amount of Cdc6 present in cohesin-depleted cells was sufficient to ensure normal association of MCM proteins with the chromatin (Supplemental Fig. S5C). As an additional control, we checked that an even stronger reduction in Cdc6 levels by siRNA in HeLa cells did not significantly compromise S-phase progression (Supplemental Fig. S5D). As far as we can determine, the effects of cohesin depletion in DNA replication are not caused by changes in the expression of initiator proteins.
Genome-wide enrichment of cohesin at replication origins
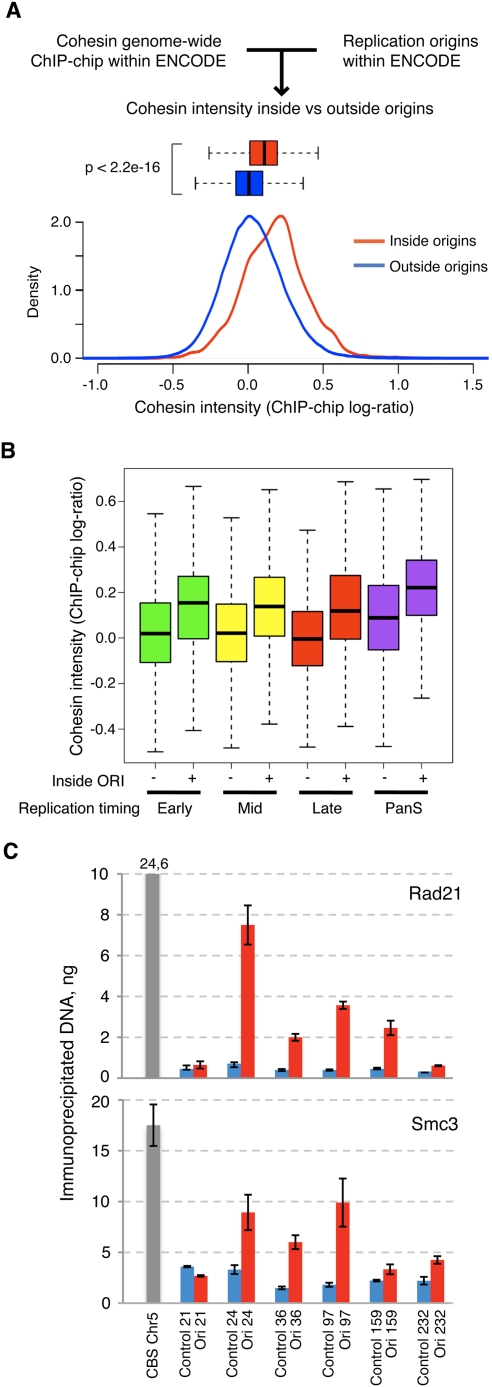
A recent “ChIP–chip” study has mapped the distribution of cohesin across the human genome in HeLa cells. While the strongest cohesin-binding sites (CBSs) are coincidental with the binding sites of the insulator protein CTCF, cohesin also associates with many other positions with lower affinity (Wendt et al. 2008). We therefore checked whether cohesin was enriched in human replication origins, taking advantage of the recent identification of 283 origins within the ENCODE representation of the genome, also in HeLa cells (The ENCODE Project Consortium 2007; Cadoret et al. 2008). To this aim, the entire data set of cohesin-binding positions defined by ChIP–chip was filtered to select those located within ENCODE. The average intensity of cohesin signals at the 9489 genomic intervals located at origins was significantly higher than at regions not overlapping with origins (P < 2.2 × 10−16) (Fig. 4A). As a control, the intensity of cohesin signals in groups of 9489 genomic intervals selected at random was compared with the rest of ENCODE sequences to assess the likelihood of finding such an enrichment by chance. This exercise was repeated 10,000 times, obtaining an average P-value of 0.5 (Supplemental Fig. S6), confirming the statistical significance of the enrichment of cohesin at origins. When the timing of replication for each ENCODE region was considered (Karnani et al. 2007), the enrichment of cohesin was detected in early-, mid-, and late-S-phase origins, as well as those origins without defined replication timing (“pan-S”) (Fig. 4B).
Figure 4.
Cohesin is enriched at origins of replication regardless of their timing of replication. (A) Signal distribution of cohesin abundance at genomic intervals located inside (red) and outside (blue) origins. Signal distributions are also represented in box plots (boxes contain the second and third data quartiles, and whiskers cover the two extreme quartiles). (B) Box plots showing the distribution of cohesin signal inside or outside origins in genomic regions that replicate in early-, mid-, or late-S phase or at any given time during S phase (panS). (C) ChIP analysis showing the relative abundance of Rad21 and Smc3 at six genomic regions containing replication origins (red bars), and six adjacent, control regions (blue bars) (Supplemental Table S4). The amount of immunoprecipitated DNA and standard error in a triplicate experiment are represented. A known CBS located at chromosome 5 was used as positive control (gray bar).
In order to validate the bioinformatics approach, ChIP assays were conducted with antibodies against two cohesin subunits—Rad21 and Smc3—to evaluate their presence at several origins within ENCODE. A CBS from chromosome 5 was used as positive control (Wendt et al. 2008). A significant enrichment of cohesin was observed with both antibodies in five out of six origins tested, relative to the background levels of cohesin at adjacent nonorigin sequences in each case (Fig. 4C). These complementary approaches indicate that cohesin is present at replication origins in human cells, regardless of their timing of replication, and likely contributes to their activation.
Cohesin depletion reduces the intensity but not the number of DNA replication foci
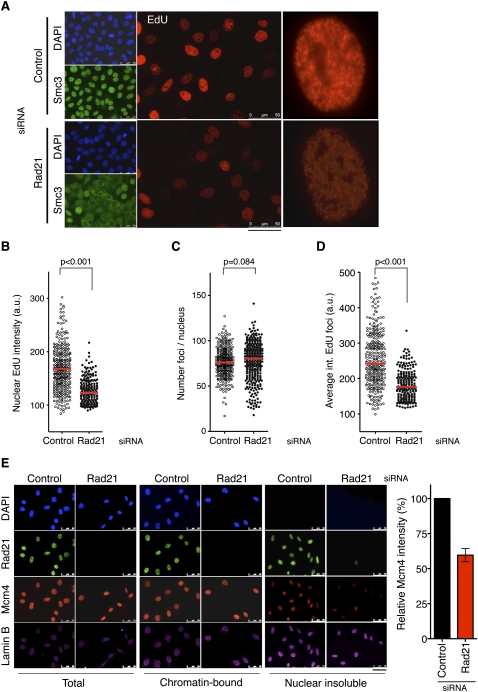
The effect of cohesin on DNA replication was further analyzed by the visualization of DNA synthesis at replication foci using the nucleotide analog 5-ethynyl 2′-deoxyuridine (EdU). After a short pulse, cohesin-depleted nuclei showed a global reduction in EdU incorporation, consistent with less-efficient replication (Fig. 5A,B). This effect was confirmed by the staining of PCNA foci (data not shown). Interestingly, the average number of detected foci per nucleus remained approximately constant (Fig. 5C), while the intensity of individual foci was reduced (Fig. 5D). Together with the DNA-combing data, these results suggest that fewer origins are activated within each replication factory when cohesin levels are reduced.
Figure 5.
Cohesin down-regulation affects DNA replication foci and impairs MCM localization to the nucleoskeleton. (A) Visualization of replication foci by EdU incorporation (red) in control or Rad21 silenced cells. Smc3 staining (green) and DNA staining with DAPI (blue) are shown. Bar, 50 μm. A single nucleus from each population is shown at higher magnification. (B) Box plot showing the automatic, unbiased quantification of nuclear EdU intensity in control or Rad21-depleted cells (n > 200 in each condition). (C) Quantification, as in B, of the number of detected foci per nucleus. (D) Quantification, as in B, of the average intensity of individual foci per nucleus. (E) Control cells or cells treated with Rad21 siRNA were synchronized in G1/S and submitted to serial in situ extractions to access the chromatin-bound fraction and the insoluble fraction reflecting the nucleoskeleton. DNA was stained with DAPI (blue), and Rad21 (green), Mcm4 (red), and Lamin B (magenta) were detected by immunofluorescence. Bar, 50 μm. For image acquisition, the same exposure time was used for each fluorophore in samples subjected to the same treatment. The histogram shows the average intensity of Mcm4 staining in the nuclear insoluble fraction (n > 40 cells for each condition in each of three independent experiments).
Replication foci contain the large macromolecular structures responsible for DNA synthesis. Even if newly synthesized DNA can be detected by the incorporation of nucleotide analogs, certain components of the replisome, such as the MCM complex, have never been visualized at foci, probably because of limited antibody accessibility (for review, see Takahashi et al. 2005). To further investigate how the association of cohesin to origins and MCM proteins could influence the operation of replication factories, we considered a classic hypothesis that postulates that these factories are assembled at a nuclear scaffolding structure, formed by lamins and other proteins, frequently referred to as the nucleoskeleton or “nuclear matrix” (Hozak et al. 1993). The existence of this nuclear network in vivo is still a matter of debate, but multiple studies support its biological relevance in transcription, replication, and repair (for review, see Misteli 2007; Elcock and Bridger 2008). Here, we use the term nucleoskeleton to refer to the structure that remains in the nuclei after treatments with detergent to remove cytosolic and nucleosoluble proteins, and nuclease digestion to solubilize and remove chromatin fragments. Cohesin has been detected at the nucleoskeleton (Sadano et al. 2000; Gregson et al. 2001), and we speculated that it could contribute to the tethering of replication proteins to this structure. In support of this notion, cohesin down-regulation significantly reduced the amount of MCM proteins at the nucleoskeleton in cells synchronized in G1/S, while their total concentration or chromatin-bound levels were virtually unaffected (Fig. 5E). In contrast, MCM down-regulation did not affect the presence of cohesin in the nucleoskeleton (Supplemental Fig. S7). These results suggest that cohesin tethers MCM complexes to the nuclear compartment in which replication factories are assembled.
Cohesin regulates the size of chromatin loops in interphase
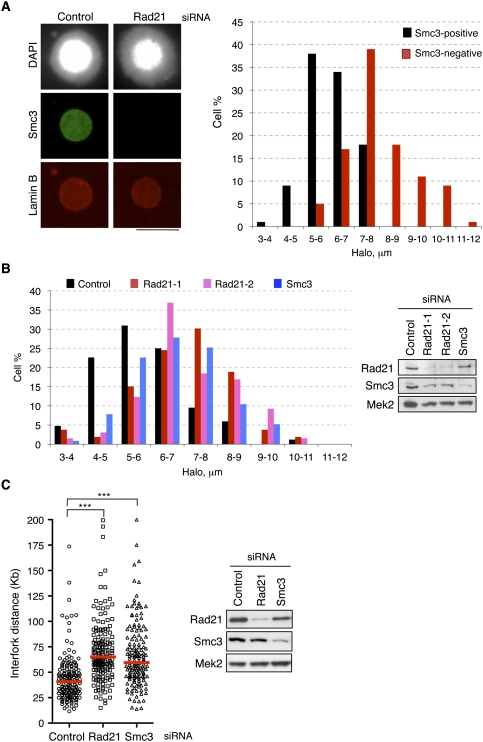
As mentioned above, cohesin regulates gene expression by stabilizing long-range interactions between distant chromatin sites, thereby forming loops. In an analogous manner, cohesin might contribute to the higher-order organization of replication factories by bringing together a group of neighboring origins and looping out the intervening DNA. To test this idea, we took advantage of the “fluorescent DNA halo” technique to estimate the average length of DNA loops in interphase nuclei. When cells are permeabilized with detergent and depleted of soluble proteins by extraction with high-salt buffers, supercoiled DNA loops unwind and form a halo around an insoluble scaffold that can be visualized by fluorescence staining (Vogelstein et al. 1980). This technique has been instrumental in establishing the correspondence between chromatin loops and replicon size (Buongiorno-Nardelli et al. 1982), in defining replicon remodeling events in Xenopus (Lemaitre et al. 2005), and in demonstrating that origins located near the base of DNA loops are activated preferentially (Courbet et al. 2008). If cohesin participates in the formation or stabilization of chromatin loops at replication factories, its down-regulation should result in fewer, longer loops, leading to an increase in the average halo radius. Control cells and cells treated with Rad21 siRNA were synchronized at G1/S to avoid variability in loop size during cell cycle progression. Both populations were mixed and treated as described above to generate DNA halos. When cells positive and negative for cohesin staining were compared, it became apparent that loss of cohesin induced a striking increase in the halo radius (Fig. 6A), indicative of larger DNA loops. Down-regulation of cohesin with additional siRNA molecules gave similar results (Fig. 6B). The presence of larger loops would correlate with longer replicon units, in agreement with the limited origin usage observed. This idea was confirmed by measuring interfork distances in extended DNA fibers after pulse-labeling of cells with IdU and CldU. Interfork distances increased upon Rad21 and Smc3 down-regulation (Fig. 6C). In contrast, down-regulation of CTCF did not significantly affect either halo size or interfork distance, and S-phase progression was essentially normal (Supplemental Fig. S8). These experiments indicate that cohesin determines the size of interphase chromatin loops that can be visualized by the DNA halo technique independently of CTCF. Combined with the functional effects on DNA replication described above, we conclude that cohesin participates in the higher-order organization of replication factories and modulates the size of chromatin loops that likely correspond to replicon units.
Figure 6.
Cohesin regulates the length of chromatin loops. (A) Nuclei from control and Rad21-depleted cells synchronized in G1/S were mixed, attached to the same coverslip, and subjected to the treatment to generate DNA halos. (Left) Immunofluorescence of these nuclei showing DNA (grayscale image), Smc3 (green), and Lamin B (red) stainings. Bar, 25 μm. Smc3-positive and Smc3-negative halo radii were measured (n = 100 for each condition). (Right) Histogram showing radii measurements grouped in nine intervals (a–b: value ≥a and <b). (B) Distribution of DNA halo size after the down-regulation of Rad21 or Smc3 proteins with the indicated siRNA oligonucleotides. Immunoblots show the efficiency of RNAi-mediated silencing. (C) Interfork distance determined by single-molecule analysis of replication forks after down-regulation of cohesin subunits Rad21 and Smc3 (n = 150 for each condition). Three asterisks indicate P-value < 0.001.
Discussion
Cohesin loading and pre-RC formation
In the first part of this study, we describe a physical interaction between cohesin and the MCM complex in human cells that is consistent with a previous report of an interaction between Smc1 and Mcm7 (Ryu et al. 2006). Whether the association of cohesin with chromatin depends on the previous formation of pre-RCs at origins has been a matter of discussion. Here we show that cohesin associates normally with chromatin after the down-regulation of ORC or MCM, arguing that cohesin loading is independent of pre-RC formation in human cells, as it happens in yeast or Drosophila cells (Uhlmann and Nasmyth 1998; MacAlpine et al. 2009). Therefore, the requirement of pre-RCs for cohesin loading that has been reported in Xenopus extracts (Gillespie and Hirano 2004; Takahashi et al. 2004) could be a particularity of this system. Xenopus extracts recapitulate the early embryonic cycles, a quick succession of chromosome duplication and segregation events with no active transcription. In this context, the genomic positions where pre-RCs are assembled may constitute the only “entry points” for cohesin. In addition, considering the results of our study, the loading of cohesin at pre-RC sites in Xenopus would ensure its physical presence around origins, where it would contribute to the dynamics of DNA replication.
Cohesin is present at replication origins and modulates their activity
Cohesin can be detected at thousands of sites along the genome (Wendt et al. 2008; Kagey et al. 2010). While a complete genome-wide correlation between CBSs and replication origins cannot be established because of the lack of a comprehensive map of the latter, using a bioinformatics approach we indeed identified an enrichment of cohesin at the origins located within the ENCODE representation of the genome. When data from the cohesin ChIP–chip assay (Wendt et al. 2008) were compared with the genomic positions of origins mapped within ENCODE by nascent strand analyses in the same cell line (Cadoret et al. 2008), it became clear that origins are preferential sites for cohesin binding. This observation, further validated by cohesin ChIP assays, seems a conserved feature through evolution because it has also been reported in yeast (Glynn et al. 2004; Lengronne et al. 2004), Drosophila (MacAlpine et al. 2009), and even Bacillus subtilis (Gruber and Errington 2009), and suggests a role for cohesin in origin activity. Actually, we found that cohesin down-regulation slows down S-phase progression by a mechanism that is independent of sister chromatid cohesion, regulation of gene expression, and checkpoint responses. Instead, single-molecule analyses revealed that cohesin down-regulation reduced the number of active origins and increased the average interfork distance, without affecting fork speed. These results imply that the presence of cohesin at origins modulates their activity, providing a novel link between the DNA replication and cohesion machineries, which is independent from the reported effect of cohesin acetylation on fork progression (Terret et al. 2009).
An architectural role for cohesin at DNA replication factories
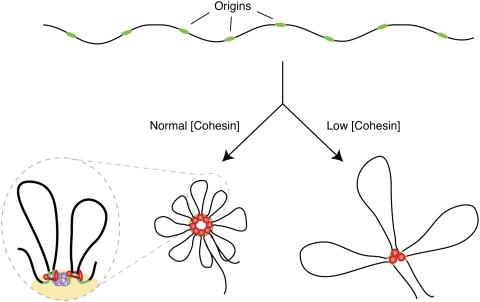
The assembly of DNA replication factories in human cells entails the physical association of a cluster of origins and the formation of chromatin loops (for review, see Cayrou et al. 2010). In this study, we show that cohesin down-regulation led to a significant increase in the length of DNA loops in which chromatin is organized. This result, combined with the negative impact of cohesin loss on DNA replication, leads us to propose that cohesin is required for the formation and/or stabilization of loops at replication foci (Fig. 7). In this model, cohesin would mediate the long-range intrachromosomal interactions necessary to bring together a cluster of replication origins. Loop formation would occur at late mitosis and during G1, at the time of origin selection and licensing (Dimitrova and Gilbert 1999; Mendez and Stillman 2000). In the resultant structures, origins would be located at the bases of the loops, where they are more prone to fire (Courbet et al. 2008). Upon cohesin down-regulation, replication foci would be structured in a different manner, with fewer origins, longer loops, and, therefore, larger replicon units (Fig. 7). This alternative arrangement explains the S-phase phenotypes and the fact that cohesin down-regulation reduces the average intensity of each replication factory without reducing the total number of replication foci.
Figure 7.
Architectural role of cohesin at replication foci: a model. Potential replication origins (green) within a DNA region (black) are grouped in rosette-like structures by the action of cohesin (red dots). Loss of cohesin may destabilize this structural arrangement resulting in fewer, longer loops. The magnified illustration shows cohesin stabilization of loops and its interaction with MCM at a replication factory (yellow). See the text for details.
Interestingly, down-regulation of CTCF neither delayed DNA replication nor affected halo size. The latter observation may seem surprising, but it could be explained because the “DNA halo” technique allows the visualization of chromatin loops anchored to insoluble nuclear structures, such as those in replication factories (Hozak et al. 1993), rather than DNA loops that are formed transiently to regulate transcription. In any case, it is possible that other proteins cooperate with cohesin to organize loops at replication factories, much as CTCF, the mediator complex, or tissue-specific transcription factors cooperate with cohesin to regulate gene expression in different contexts (Phillips and Corces 2009; Kagey et al. 2010; Schmidt et al. 2010).
Materials and methods
Preparation of nuclear extracts for immunoprecipitation
HeLa cells (2 × 107) were resuspended in 1 mL of osmotic buffer (10 mM HEPES at pH7.9, 0.2 M potassium acetate, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol [DTT], 1 mM NaVO4, 5 mM β-glycero-phosphate, 0.1 mM phenyl methane sulphonyl fluoride [PMSF], 0.5 mM NaF, protease inhibitor cocktail [Roche]). Triton X-100 was added at 0.1% and cells were incubated for 5 min on ice. After centrifugation (600g for 5 min), the nuclei-enriched pellet was resuspended in 1 mL of hypotonic buffer (10 mM HEPES at pH 7.9, 1 mM DTT, 1 mM NaVO4, 5 mM β-glycero-phosphate, 0.1 mM PMSF, 0.5 mM NaF, protease inhibitors as above) and incubated for 5 min on ice. KOAc (0.25 M) and CaCl2 (1.5 mM) were added and the samples were digested with 7.5 U of micrococcal nuclease (Sigma) for 25 min at 24°C to shear the chromatin into fragments shorter than 2 kb. Digestion was stopped with 2 mM ethylene glycol tetraacetic acid (EGTA). The insoluble fraction was removed by centrifugation (16,000g for 20 min) and supernatants were used for immunoprecipitation.
Protein analysis by mass spectrometry
Nuclear extracts prepared from HeLa cells synchronized in S phase were used in immunoprecipitation reactions with preimmune and Mcm4 sera (∼8 mg of extract per reaction), and the precipitated proteins were fractionated by SDS-PAGE. Bands from both lanes of this gel were excised, reduced, and alkylated before digestion with sequencing-grade trypsin (Promega). Supernatants were dried under vacuum and resuspended in 0.1% formic acid and 5% MS-grade acetonitrile (Lab-Scan). Tryptic peptides were analyzed by reverse-phase chromatography coupled to tandem mass spectrometry (HPLC-MS/MS) in an Ultimate 3000 (Dionex) coupled to a LTQ linear ion trap (Thermo Scientific) as described (Casado-Vela et al. 2009). All fragmentation spectra were searched against Swiss-Prot version 57.4 database containing 470,369 entries using the SEQUEST search engine implemented in Proteome Discoverer version 1.1. A tolerance of ±1.50 Da for precursor ions and ±0.5 Da for fragment ions was allowed. Carboxyamidomethylated cysteine was selected as fixed modification and oxidation on methionine was set as variable modification. Only those peptides with ≤5% false discovery rate (FDR) against the decoy database were accepted as true positives. The hits obtained in the analyses of the immunoprecipitates from preimmune sera were subtracted from those found in the Mcm4 immunoprecipitates.
Cell manipulations, EdU incorporation, and protein immunodetection
HeLa cell culture, siRNA transfections (a list of the targeted RNA sequences is provided in Supplemental Table S3), and subsequent analyses by immunoblot or flow cytometry were carried out as described (Ibarra et al. 2008). Synchronization of cell cultures in G1/S was achieved by incubation in medium supplemented with 2 mM thymidine for 18 h. For checkpoint abrogation, HeLa cells were treated with UCN-01 or caffeine as described (Ibarra et al. 2008). For the visualization of DNA replication foci, 10 μM EdU was added to the culture media for 15 min. Cells were washed with phosphate-buffered saline (PBS), fixed with 2% paraformaldehyde for 15 min, and permeabilized with 0.5% Triton for 5 min, and EdU was stained as described (Salic and Mitchison 2008). EdU foci intensity was analyzed with Acapella Image Analysis software (Perkin-Elmer). For protein immunofluorescence detection, cells were grown on glass coverslips or μCLEAR-bottom 96-well dishes (Greiner Bio-One), overlaid with the indicated primary antibodies for 1 h at room temperature (RT), washed three times with PBS, and overlaid with the corresponding Texas Red-, Alexa 647-, or FITC-conjugated secondary antibodies. Nuclear DNA was stained with 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI). To access chromatin-bound proteins, cells were treated with 0.5% Triton X-100 for 5 min prior to fixation. To visualize nuclear scaffold-associated proteins, cells were treated as described (Gregson et al. 2001). Images were acquired on a Leica CTR6000 microscope or the OPERA LX system (Perkin-Elmer). A list of the antibodies used in this study is provided in Supplemental Table S4.
Detection of cohesin presence at origins of replication within ENCODE
The cohesin ChIP–chip data (Wendt et al. 2008; NCBI Gene Expression Omnibus, accession GSE9613) were kindly provided by K. Shirahige, J.M. Peters, and G. Legube. Raw CEL files corresponding to sample GSM243190 (Scc1) were processed with Tiling Analysis software (TAS, Affymetrix). The Scc1 ChIP signal was normalized on the input (whole-cell extract), and ChIP–chip log-ratios were normalized to 37-base-pair (bp) genomic intervals. Then, normalized data was filtered to use only the intervals that map inside pilot ENCODE regions (1% of the genome). After filtering, a total of 468,098 intervals were used. Replication timing segments were downloaded from University of California at Santa Cruz (UCSC) browser (hg18), track University of Virginia DNA Replication Temporal Segmentation (UVa DNA Rep Seg at http://genome.ucsc.edu) using the UCSC Table Browser (Karolchik et al. 2004). The four subtracks are the replication timing segments “early,” “mid,” “late,” and “panS,” as defined by Karnani et al. (2007). All genomic information was referred to hg18 (NCBI human build 36). The genomic coordinates for origins of replication located within ENCODE (Cadoret et al. 2008) were converted from hg17 to hg18 (NCBI build 36) using UCSC liftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver).
The 468,098 genomic intervals from the cohesin ChIP–chip arrays were mapped into the origins of replication and the replication timing segments. A total of 9489 intervals mapped into origins (2.07% of the ENCODE regions). The number of intervals in each of the replication timing segments was 101,760 early, 124,135 mid, 128,484 late, and 93,915 panS. In addition, 19,804 intervals did not map in any of the four segments. Genomic interval overlaps, statistical tests, and density distributions were calculated using R functions (http://cran.r-project.org). Densities were obtained with the default parameters of the density function.
Preparation of fluorescent DNA halos
Cells (2 × 106 per milliliter) were treated with nuclei buffer (10 mM Tris at pH 8, 3 mM MgCl2, 0.1 M NaCl, 0.3 M sucrose, protease inhibitors) plus 0.5% Nonidet P40 for 10 min on ice. Cells were attached to coverslips using cytospin (1800 rpm for 5 min); stained with 2 μg/mL DAPI for 4 min; and immersed in a buffer containing 25 mM Tris (pH 8), 0.5 M NaCl, 0.2 mM MgCl2, 1 mM PMSF, and protease inhibitors for 1 min, then in Halo Buffer (10 mM Tris at pH 8, 2 M NaCl, 10 mM ethylene diamine tetraacetic acid [EDTA], 1 mM DTT, protease inhibitors) for 4 min. Next, cells were washed in a buffer containing 25 mM Tris (pH 8), 0.2 M NaCl, and 0.2 mM MgCl2 for 1 min, and in the same buffer omitting the NaCl for 1 min. Finally, cells were fixed in 2% formaldehyde for 10 min and processed for immunofluorescence. The halo radius (R) of each nucleus was determined by measuring the total area of the nucleus (At) and the central area, highly stained with DAPI, of the nuclear scaffold (As) using MetaMorph software (Molecular Devices) and applying the formula R = √(At/π) − √(As/π).
ChIP and real-time PCR
For ChIP assays, HeLa cells growing on plates were treated with 1% formaldehyde for 15 min. The cross-linking reaction was stopped by the addition of 0.125 M glycine. After 5 min, cells were washed with ice-cold PBS, scrapped from plates, and harvested by centrifugation. Cells were resuspended in lysis buffer (50 mM Tris-HCl at pH 8.1, 10 mM EDTA, 1% SDS, 1 mM PMSF, protease inhibitors) for 10 min on ice. Chromatin was sheared in a bath sonicator (Diagenode Bioruptor) to an average length of 0.2–0.8 kb. Samples were centrifuged (10,000g for10 min) and the supernatants containing fragmented chromatin were collected. Aliquots of each sample were kept at −80°C to serve as “input” samples. The remaining sample was diluted 10-fold in dilution buffer (20 mM Tris-HCl at pH 8.0, 2 mM EDTA at pH 8.0, 150 mM NaCl, 1% Triton X-100) and precleared for 90 min at 4°C with protein A agarose beads (Sigma) preincubated with 100 μg/mL bovine serum albumin (BSA) and 1 μg/mL salmon sperm DNA. Eight micrograms of antibody was added to the precleared samples, followed by overnight incubation on a rotating platform. Protein A agarose beads were added and incubation proceeded for 2 h at 4°C. Beads were then sequentially washed in “low-salt” wash buffer (20 mM Tris-HCl at pH 8.1, 2 mM EDTA at pH 8.0, 0.1% SDS, 1% Triton X-100, 150 mM NaCl), “high-salt” wash buffer (same as before, except 0.5 M NaCl), LiCl wash buffer (10 mM Tris-HCl at pH 8.1, 1 mM EDTA, 0.25 M LiCl, 1% NP-40, 1% deoxycholate), and TE (Tris-HCl at pH 8, 1 mM EDTA). Beads were incubated in 250 μL of elution buffer (1% SDS, 0.1 M NaHCO3). The supernatants from two consecutive elutions were pooled and cross-linking was reversed by the addition of 0.2 M NaCl and incubation for 6 h at 65°C. Forty millimolar Tris-HCl (pH 6.5), 2 mM EDTA, and 200 μg/mL RNase A were added and samples were incubated for 30 min at 37°C. Proteinase K was added at 100 μg/mL and incubation proceeded for 2 h at 45°C. DNA was recovered by phenol/chloroform extraction and ethanol precipitation. For quantitative analysis, real-time PCR was performed using the primers listed in Supplemental Table S3, SybrGreen Master Mix, and the standard program of 7900HT fast real-time PCR (Applied Biosystems) (2 min at 50°C, 10 min at 95°C; 40× [15 sec at 95°C, 1 min at 60°C]), with triplicates of each sample. The quantity of immunoprecipitated DNA was calculated after normalization of PCR efficiency with different amounts of input DNA. The binding to nonspecific rabbit IgG was subtracted.
Fork progression rate, global fork density, and interfork distance estimation
For DNA combing, cells were pulsed sequentially with 25 μM IdU for 20 min and 200 μM CldU for 20 min. After harvesting, cells were trypsinized, resuspended in PBS, and embedded in 0.5% low-melting-point agarose. We included 2 × 104 to 3 × 104 cells per agarose plug. Plugs were treated twice for 12 h in PK buffer (10 mM Tris HCl at pH 7.5, 100 mM EDTA, 20 mM NaCl, 0.5% SDS, 0.2 mg/mL Proteinase K); washed five times for 1 h in 10 mM Tris-HCl (pH 7.5), 50 mM EDTA, and 20 mM NaCl; and equilibrated in 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.7). Plugs were melted in preheated MES-EDTA at 65°C and treated overnight with 2 U of β-agarase (Biolabs) at 42°C. After heating for 10 min at 65°C, the DNA solution was applied to silanized coverslips using a DNA-combing apparatus (Pasteur Instruments). DNA-covered coverslips were incubated for 1 h at 65°C, glued on microscope slides, and denatured (0.5 N NaOH for 25 min), blocked (1% BSA, 0.2% Tween in PBS at pH 7.5), and incubated with antibodies to detect IdU, CldU, and ssDNA. Slides were mounted with ProlongGold Anti-fade Reagent (Molecular Probes). Image acquisition was made on a Leica microscope with MetaMorph software. For fork speed measurements, only the unambiguous, uninterrupted, bicolor, and single tracks were considered. Track length in kilobases was divided by pulse time to derive fork speed. Fork density was calculated relative to total DNA, normalized by the number of replicating cells. Tricolor tracks corresponding to origins and termination events were scored as two forks. Image analysis was performed with IdeFIx software developed in E. Schwob's laboratory (IGMM), and statistical analysis was done with R 2.9.1 software.
Extended DNA fibers for interfork distance measurements after sequential labeling with IdU and CldU were prepared as described (Terret et al. 2009).
Acknowledgments
We thank D. Megías, the CNIO Confocal Microscopy Unit, L. Martínez, A. García, and the CNIO Flow Cytometry Unit for excellent technical assistance; M. Drac and T. Gostan of the Montpellier DNA-combing facility for surface preparation and automated image analysis; A. Cuadrado for the RT–PCR analyses of CDC6 expression levels; K. Shirahige, J.M. Peters, and G. Legube for providing the genomic coordinates of CBSs; M.E. Terret for the protocol for DNA fiber preparation; R. Freire for the gift of Chk1 siRNA; members of the Chromosome Dynamics and DNA Replication groups for many discussions; and O. Fernández-Capetillo, M. Lemaire, and M. Serrano for comments on the manuscript. This work was supported by the Spanish Ministry of Science and Innovation (J.M., A.L., and I.C.), Consolider Ingenio 2010 and Fundación Caja Madrid (J.M. and A.L.), Comunidad de Madrid (I.C.), and Institut National du Cancer and Association pour la Recherche sur le Cancer (E.S.). E.G. and V.C. were supported by the Fondation pour la Recherche Médicale and Institut National du Cancer, respectively. A.I. was supported by a fellowship from the Basque Government.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.608210.
Supplemental material is available for this article.
References
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Borlado LR, Mendez J 2008. CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29: 237–243 [DOI] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M, Micheli G, Carri MT, Marilley M 1982. A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature 298: 100–102 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casado-Vela J, Martinez-Torrecuadrada JL, Casal JI 2009. Differential phosphorylation patterns between the Cyclin-A2/CDK2 complex and their monomers. Protein Expr Purif 66: 15–21 [DOI] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Mechali M 2010. Programming DNA replication origins and chromosome organization. Chromosome Res 18: 137–145 [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993 [DOI] [PubMed] [Google Scholar]
- Elcock LS, Bridger JM 2008. Exploring the effects of a dysfunctional nuclear matrix. Biochem Soc Trans 36: 1378–1383 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Schaaf CA, Dorsett D 2008. Cohesin and CTCF: Cooperating to control chromosome conformation? Bioessays 30: 715–718 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T 2004. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol 14: 1598–1603 [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2: E259 doi: 10.1371/journal.pbio.0020259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson HC, Schmiesing JA, Kim JS, Kobayashi T, Zhou S, Yokomori K 2001. A potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem 276: 47575–47582 [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137: 685–696 [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A 2010. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci 107: 3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozak P, Hassan AB, Jackson DA, Cook PR 1993. Visualization of replication factories attached to nucleoskeleton. Cell 73: 361–373 [DOI] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A 1998. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A 2007. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res 17: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496 doi: 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M 2005. Mitotic remodeling of the replicon and chromosome structure. Cell 123: 787–801 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM 2009. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B 2003. Perpetuating the double helix: Molecular machines at eukaryotic DNA replication origins. Bioessays 25: 1158–1167 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M 2009. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J 28: 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T 2007. Beyond the sequence: Cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5: e1000739 doi: 10.1371/journal.pgen.1000739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MJ, Kim BJ, Lee JW, Lee MW, Choi HK, Kim ST 2006. Direct interaction between cohesin complex and DNA replication machinery. Biochem Biophys Res Commun 341: 770–775 [DOI] [PubMed] [Google Scholar]
- Sadano H, Sugimoto H, Sakai F, Nomura N, Osumi T 2000. NXP-1, a human protein related to Rad21/Scc1/Mcd1, is a component of the nuclear matrix. Biochem Biophys Res Commun 267: 418–422 [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci 105: 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Watrin E, Lenart P, Mechtler K, Peters JM 2007. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol 17: 630–636 [DOI] [PubMed] [Google Scholar]
- Schwob E, de Renty C, Coulon V, Gostan T, Boyer C, Camet-Gabut L, Amato C 2009. Use of DNA combing for studying DNA replication in vivo in yeast and mammalian cells. Methods Mol Biol 521: 673–687 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren C, Ström L 2010. S-phase and DNA damage activated establishment of sister chromatid cohesion: Importance for DNA repair. Exp Cell Res 316: 1445–1453 [DOI] [PubMed] [Google Scholar]
- Smits VA, Warmerdam DO, Martin Y, Freire R 2010. Mechanisms of ATR-mediated checkpoint signalling. Front Biosci 15: 840–853 [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC 2004. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol 6: 991–996 [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Wigley DB, Walter JC 2005. Pumps, paradoxes and ploughshares: Mechanism of the MCM2–7 DNA helicase. Trends Biochem Sci 30: 437–444 [DOI] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV 2009. Cohesin acetylation speeds the replication fork. Nature 462: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K 1998. Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Pardoll DM, Coffey DS 1980. Supercoiled loops and eucaryotic DNA replication. Cell 22: 79–85 [DOI] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM 2006. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16: 863–874 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]