Abstract
We have reported previously that reactive-site mutants of N-TIMP-3 [N-terminal inhibitory domain of TIMP-3 (tissue inhibitor of metalloproteinases 3)] modified at the N-terminus, selectively inhibited ADAM17 (a disintegrin and metalloproteinase 17) over the MMPs (matrix metalloproteinases). The primary aggrecanases ADAMTS (ADAM with thrombospondin motifs) -4 and -5 are ADAM17-related metalloproteinases which are similarly inhibited by TIMP-3, but are poorly inhibited by other TIMPs. Using a newly developed recombinant protein substrate based on the IGD (interglobular domain) of aggrecan, gst-IGD-flag, these reactive-site mutants were found to similarly inhibit ADAMTS-4 and ADAMTS-5. Further mutations of N-TIMP-3 indicated that up to two extra alanine residues can be attached to the N-terminus before the Ki (app) for ADAMTS-4 and ADAMTS-5 increased to over 100 nM. No other residues tested at the [−1] position produced inhibitors as potent as the alanine mutant. The mutants N-TIMP-3(T2G), [−1A]N-TIMP-3 and [−2A]N-TIMP-3 were effective inhibitors of aggrecan degradation, but not of collagen degradation in both IL-1α (interleukin-1α)-stimulated porcine articular cartilage explants and IL-1α with oncostatin M-stimulated human cartilage explants. Molecular modelling studies indicated that the [−1A]N-TIMP-3 mutant has additional stabilizing interactions with the catalytic domains of ADAM17, ADAMTS-4 and ADAMTS-5 that are absent from complexes with MMPs. These observations suggest that further mutation of the residues of N-TIMP-3 which make unique contacts with these metalloproteinases may allow discrimination between them.
Keywords: a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS), aggrecanase, collagenase, metalloproteinase, osteoarthritis, tissue inhibitor of metalloproteinases 3 (TIMP-3)
Abbreviations: ADAM, a disintegrin and metalloproteinase; ADAMTS, ADAM with thrombospondin motifs; CBB, Coomassie Brilliant Blue R-250; DMBA, dimethylaminobenzaldehyde; DMEM, Dulbecco's modified Eagle's medium; DMMB, Dimethylmethylene Blue; Dpa, N-3-(2,4-dinitrophenyl)-L-2,3-diaminopropionyl; GAG, glycosaminoglycan; GST, glutathione transferase; IGD, interglobular domain; IL-1α, interleukin-1α; Mca, (7-methoxycoumarin-4-yl)acetyl; MMP, matrix metalloproteinase; N-TIMP, N-terminal domain of tissue inhibitor of metalloproteinases; OA, osteoarthritis; OSM, oncostatin M; TIMP, tissue inhibitor of metalloproteinases
INTRODUCTION
The TIMPs (tissue inhibitors of metalloproteinases) are a family of four endogenous inhibitors that were originally characterized by their ability to inhibit the MMPs (matrix metalloproteinases). TIMPs have now been shown to also inhibit members of two other related metalloproteinase families, the ADAMs (a disintegrin and metalloproteinase) and the ADAMTSs (ADAM with thrombospondin motifs). Specifically, TIMP-1 has been shown to inhibit ADAM10 [1], whereas TIMP-2 has been shown to inhibit ADAM12 [2] and ADAMTS-1 [3]; TIMP-3 has the widest inhibitory activity among the TIMPs, since it has been shown to inhibit ADAM10 [1], ADAM12 [2], ADAM17 [4], ADAM28 [5], ADAM33 [6], ADAMTS-1 [3], ADAMTS-2 [7], ADAMTS-4 [8,9] and ADAMTS-5 [9]. Not all ADAMs and ADAMTSs are inhibited by the TIMPs; for example, none of the four human TIMPs inhibits ADAM8 or ADAM9 [10].
TIMPs comprise two domains: the N-terminal inhibitory domain and a C-terminal domain. The N-terminal inhibitory do-main is a wedge-like structure that inhibits the MMPs by insertion into the active site of the MMP. The crystal structure of the TIMP-1–MMP-3 catalytic domain complex [11] shows that the amino and carbonyl groups of Cys1 of TIMP-1 co-ordinate the catalytic Zn2+ ion and that the Thr2 and Val4 side chains fit into the S1′ and S3′ pockets of MMP-3 respectively. The importance of the N-terminus of the TIMP has also been highlighted by several mutagenesis [12–15] and chemical modification [16,17] studies. These studies indicated that the N-terminal cysteine residue is essential for MMP inhibitory activity. Mutation of this residue to serine or blocking the N-terminal α-amino group by the addition of an extra amino acid [15,17], acetylation [17] or carbamylation [16] severely reduces inhibitory activity against MMPs. The recent crystal structure of the ADAM17 catalytic domain–N-TIMP-3 (N-terminal domain of TIMP-3) complex [18] has confirmed that ADAM17 is also inhibited by TIMP-3 in a similar manner as the MMPs, suggesting that the mechanism by which TIMP-3 inhibits ADAMTSs is also similar.
We have reported previously, however, that two N-terminal reactive-site mutants of N-TIMP-3, N-TIMP-3(T2G) and [−1A]N-TIMP-3 (N-TIMP-3 with an extra alanine residue appended to Cys1) which, as predicted from other studies [15–17], are poor MMP inhibitors, still retain inhibitory activity against ADAM17 [19]. This inhibition indicates that there are subtle differences in TIMP inhibition of ADAMs compared with that of the MMPs that are not immediately obvious from the crystal structures. In the present paper, we report the testing of these N-terminal reactive-site mutants of N-TIMP-3 for their ability to inhibit the two primary aggrecanases ADAMTS-4 and ADAMTS-5 based on their similar TIMP inhibitory profiles to ADAM17 since they are potently inhibited only by TIMP-3. We have generated additional mutants with various lengths of alanine extensions and with other amino acids at the −1 position to determine whether these display further selectivity between the two aggrecanases.
Aggrecan degradation in IL-1α (interleukin 1α)-stimulated porcine articular cartilage explants, an ex vivo model of arthritis, is inhibited by N-TIMP-3, but not by TIMP-1 or TIMP-2 [20], suggesting that the ADAMTSs and not the MMPs are responsible for the degradation of cartilage aggrecan. Furthermore, aggrecan in the cartilage of ADAMTS-5-null mice was protected from degradation when challenged with arthritis induced by surgical joint destabilization [21], or with antigen-induced arthritis [22], suggesting that, in mice, the primary aggrecanase in this disease is ADAMTS-5. However, in cartilage from rheumatoid arthritis and OA (osteoarthritis) patients, there is evidence that aggrecan is degraded by both the ADAMTSs and MMPs; tissue sections stain positive for neo-epitopes produced by both MMP and ADAMTS activity [23]. Aggrecan fragments from both MMP- and ADAMTS-mediated degradation have also been found in OA joint cartilage and synovial fluid [24]. Degradation of the other cartilage structural macromolecule type II collagen is mediated primarily by the collagenases, which are members of the MMP family [25]. Consequently, the relative contribution of the ADAMTSs and the MMPs in the progression of human disease remains unclear. We therefore applied our reactive-site mutants of N-TIMP-3 to investigate the role of ADAMTS and MMPs in IL-1α-induced pig or IL-1α/OSM (oncostatin M)-induced human OA cartilage degradation.
EXPERIMENTAL
Materials
Restriction enzymes were from New England Biolabs. The plasmid pGEX-4T1, Pfu DNA polymerase, GFX Gel Band PCR Purification kit, GFX Microplasmid Purification kit and glutathione–Sepharose 4B column material were from GE Healthcare. Precision protein standards for SDS/PAGE were from Bio-Rad Laboratories. Proteinase inhibitor cocktail set II was purchased from Calbiochem. Chloramine T, DMBA (dimethylaminobenzaldehyde), DMMB (Dimethylmethylene Blue) and OSM were obtained from Sigma–Aldrich. ADAM17 and the Mca-PLAQAV-Dpa-RSSSR [where Mca is (7-methoxycoumarin-4-yl)acetyl and Dpa is N-3-(2,4-dinitrophenyl)-L-2,3-diaminopropionyl] fluorigenic substrate were from R&D Systems. The Mca-PLGL-Dpa-AR fluorigenic substrate was supplied by Bachem. TIMP-1, TIMP-2, N-TIMP-3, MMP-1ΔC (catalytic domain of MMP-1), MMP-2, MMP-3ΔC (catalytic domain of MMP-3), ADAMTS-4-2 and ADAMTS-5-4 were prepared as described previously [9,17,26–30]. Purified IL-1α was a gift from Professor J. Saklatvala (Kennedy Institute of Rheumatology, Imperial College London). Anti-ARGSV antibody was a gift from Professor B. Caterson (School of Biosciences, Cardiff University, Cardiff, U.K.). Porcine metacarpophalangeal joints were supplied by Fresh Tissue Supplies and dissected on-site within 24 h of slaughter. Human cartilage was obtained from consenting OA patients undergoing hip or knee joint replacement at Heatherwood and Wexham Park Hospital (Slough, Berkshire, U.K.).
Construction of N-TIMP-3 mutants
The plasmid pET-42b-N-TIMP-3-His was used as the template for mutagenesis by PCR as described in [19]. The common reverse primer 5′-AAAAGCGGCCGCGTTACAACCCAGGTGATA-3′ (NotI restriction site in bold italics) was used in all mutagenesis procedures. The forward primers used were: [−2A]N-TIMP-3, 5′-AAAACATATGGCAGCATGCACATGCTCGCCCAGC-3′; [−3A]N-TIMP-3, 5′-AAAACATATGGCAGCAGCATGCACATGCTCGCCCAGC-3′; [−5A]N-TIMP-3, 5′-AAAACATATGGCAGCAGCAGCAGCATGCACATGCTCGCCCAGC-3′; [−8A]N-TIMP-3, 5′-AAAACATATGGCAGCAGCAGCAGCAGCAGCAGCATGCACATGCTCGCCCAGC-3′; [−1G]NTIMP-3, 5′-AAAACATATGGGTTGCACATGCTCGCCCAGC-3′; [−1M]N-TIMP-3, 5′-AAAACATATGATGTGCACATGCTCGCCCAGC-3′; [−1E]N-TIMP-3, 5′-AAAACATATGGAGTGCACATGCTCGCCCAGC-3′ and [−1K]N-TIMP-3, 5′-AAAACATATGAAGTGCACATGCTCGCCCAGC-3′ (codons for the mutated amino acids are underlined and the NdeI restriction site in bold italics).
Expression and purification of N-TIMP-3 and mutants
N-TIMP-3 and its mutants were expressed in Escherichia coli BL-21(DE3) and purified by Ni2+-chelate chromatography as described previously [9]. Additionally, endotoxin was removed by including a wash step with 60% propan-2-ol and 6 M guanidinium chloride, during column chromatography [31]. The amount of endotoxin after purification was determined using the Limulus amoebocyte lysate assay (Cambrex) to be less than 1 pg/pmol of N-TIMP-3. The level of endotoxin that stimulated cartilage matrix degradation in porcine articular cartilage explants was >1 ng/ml.
MMP and ADAM17 inhibition kinetics
The ability of N-TIMP-3 mutants to inhibit the MMPs and ADAM17 was tested using the fluorigenic substrate Mca-PLG~L-Dpa-AR (~ indicates the scissile bond) [32] for the MMPs and Mca-PLA~QAV-Dpa-RSSSR for ADAM17. The N-TIMP-3 mutants were pre-incubated with MMP (1 nM MMP-1ΔC, 125 pM MMP-2, 500 pM MMP-3ΔC) or 250 pM ADAM17 ectodomain for 1 h at 37 °C before residual activity was measured by fluorescence increase (excitation 323 nm, emission 395 nm) using 2 μM substrate. The data were fitted to the tight binding inhibitor equation: v=[(E–I–k+[(E–I–k)2+4Ek]½)/(2E)], where v is the velocity of the reaction, E is the enzyme concentration, I is the initial inhibitor concentration, and k is the apparent inhibition constant, using Prism (GraphPad Software).
Construction, expression and purification of gst-IGD-flag
The expression plasmid pGEX-4T1 containing the GST (glutathione transferase) sequence fused with the IGD (interglobular domain) of aggrecan (Tyr330–Gly457), with a C-terminal FLAG (DYKDDDDK) sequence, cloned into the EcoRI and XhoI cloning sites (Figure 1A). The plasmid was transformed into E. coli BL-21(DE3) cells. Recombinant protein expression was induced with IPTG (isopropyl β-D-thiogalactopyranoside) (100 μM) overnight at 25 °C. Bacteria were harvested, washed and mechanically disrupted with a French Press (five times at 1500 psi, where 1 psi=6.9 kPa). After centrifugation at 1500 g for 15 min, the supernatant containing gst-IGD-flag was applied to a glutathione–Sepharose 4B column equilibrated with 50 mM Tris/HCl (pH 8.0). The column was washed with 50 mM Tris/HCl (pH 8.0) and 0.5 M NaCl and eluted with 10 mM reduced glutathione in 50 mM Tris/HCl (pH 8.0). The eluted protein substrate was dialysed three times against 10 vol. of 50 mM Tris/HCl (pH 8.0) and 150 mM NaCl for 4 h at 4 °C and concentrated to an A280 of >2.5 using poly(ethylene sulfate) membrane spin concentrators. The concentration of intact substrate was determined to be approx. 34 μM. The yield of substrate (approx. 20 mg of partially purified gst-IGD-flag per litre of bacterial culture) was sufficient for over 2000 assays.
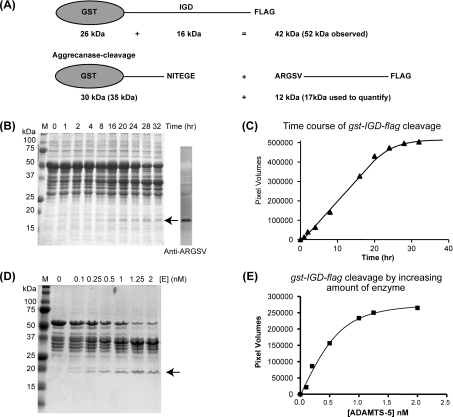
Figure 1. Characterization of the gst-IGD-flag substrate.
(A) Schematic representation of the gst-IGD-flag substrate and its products upon aggrecanase activity. The observed molecular masses are indicated in parentheses. (B) Time course of cleavage of gst-IGD-flag by ADAMTS-5-4. gst-IGD-flag (17 μM) was incubated with recombinant ADAMTS-5-4 (500 pM) at 37 °C. At different time points (0–32 h), reactions were stopped and analysed by SDS/PAGE (10% acrylamide). The 17 kDa product band (arrow) also stained positive for the aggrecanase generated neo-epitope ARGSV. (C) The product was analysed by densitometry to calculate the extent of cleavage, expressed as pixel volumes. (D and E) Cleavage of gst-IGD-flag by different amounts of enzyme ([E]). gst-IGD-flag (16 μM) substrate was incubated for 16 h at 37 °C with increasing amounts of ADAMTS-5-4 (0–2 nM). M, molecular mass markers (size in kDa).
gst-IGD-flag aggrecanase assays
Equal volumes (5 μl) of 17 μM gst-IGD-flag and 1 nM ADAMTS-4 which lacks the C-terminal spacer domain (ADAMTS-4-2 [29]) or 0.5 nM ADAMTS-5 which lacks the C-terminal cysteine-rich spacer and thrombospondin domains (ADAMTS-5-4 [30]) were incubated at 37 °C for the indicated period of time. Inhibitor, if used, was pre-incubated with the ADAMTS enzyme (total volume 5 μl) at 37 °C for 1 h before addition of 5 μl of the substrate for 1 h. The reactions were stopped by addition of an equal volume (10 μl) of 2× SDS/PAGE sample buffer containing 10 mM EDTA. SDS/PAGE (10% acrylamide) was run under reducing conditions using a modification of the ammediol/glycine/HCl buffer system described by Wyckoff et al. [33]. Following staining with CBB (Coomassie Brilliant Blue R-250), the amount of product was determined by densitometric quantification of the 17 kDa band using the GS-710 scanning densitometer (Bio-Rad Laboratories) and analysed using the 1D Phoretix Software (Nonlinear Dynamics). Under conditions where substrate cleavage was linear, the amount of substrate cleaved is directly related to the velocity of the reaction. For inhibition kinetics, the data were fitted to the same tight binding equation as in the MMP inhibition kinetics.
Cartilage explant cultures
Porcine articular cartilage explants were prepared as described previously [20]. Explanted human cartilage pieces (approx. 5 mm×5 mm×2.5 mm) from either the femoral head or the knee were rested overnight in DMEM (Dulbecco's modified Eagle's medium) with 20% (v/v) fetal bovine serum before being washed twice with DMEM without serum. Each cartilage piece was placed in one well of a round-bottomed 96-well plate with 200 μl of serum-free DMEM with or without IL-1α (10 ng/ml) and OSM (50 ng/ml), together with various concentrations of each TIMP. Each treatment was prepared in triplicate. After 3 days, the conditioned media were harvested and stored at −20 °C until use. For longer-term culture of porcine articular cartilage, the media were replaced every 4 days and the harvested media were stored as above.
Analysis of cartilage degradation
GAG (glycosaminoglycan) release into the culture medium was determined as described previously [20]. Hydroxyproline released into the culture medium was determined using a modification of the assay as described by Bergman and Loxley [34].
Molecular modelling with AutoDock 4.2
Molecular docking of N-TIMP-3 and the mutants on to the three-dimensional X-ray structure of the catalytic domain of MMP-1 and ADAMTS-5 was carried out using the AutoDock software package (version 4.2) as implemented through the graphical user interface AutoDockTools (ADT 1.5.4) [35]. The structures used were from the N-TIMP-1–MMP-1 complex (PDB code 2J0T [36]), the N-TIMP-3-ADAM17 complex (PDB code 3CKI [18]), ADAM17 (PDB code 3G42 [37]), ADAMTS-4 (PDB code 2RJP [38]) and ADAMTS-5 (PDB code 2RJQ [38]). To create the initial co-ordinates for docking studies, the water molecules were removed from the crystal structures and excluded from the calculations. The non-catalytic ZnCl2 found in the ADAMTS-5 structure was also removed. Hydrogen atoms were added and Gasteiger charges computed. For the catalytic zinc, the formal charge of +2 and van der Waals radius of 0.87 Å (1 Å=0.1 nm) were used, as suggested by Hu and Shelver [39]. To minimize computation time, the only bonds allowed to rotate were in mutated residues of the TIMP. The docking area was defined by a box, centred on the TIMP-binding position above the metalloproteinase. Grids points of 100×100×100 with 0.5 Å spacing were calculated around the docking area for all the ligand atom types using AutoGrid4. A total of 200 separate docking calcu-lations were performed for each N-TIMP. Each docking calculation consisted of 2500000 energy evaluations using the Lamarckian genetic algorithm local search over 27000 generations. A mutation rate of 0.02 and a crossover rate of 0.8 were used to generate new docking trials for subsequent generations. The results of the docking experiments were visualized using PyMOL (DeLano Scientific; http://www.pymol.org/).
RESULTS
Aggrecanase assay using the gst-IGD-flag substrate
The gst-IGD-flag substrate purified from the soluble extract of the E. coli contained a considerable amount of lower-molecular-mass products after purification (Figure 1B, lane 1). Either the GST tag or the FLAG tag was associated with these bands, indicating degradation (results not shown). The degradation was not prevented by the addition of protease inhibitors (results not shown). Further purification using ion-exchange and size-exclusion chromatography did not separate the degradation products from the intact substrate under native conditions (results not shown). However, the intact substrate accounted for over 70% of the CBB-stainable protein bands (Figure 1B, lane 2), and the expected product after ADAMTS cleavage (Figure 1B, arrow) was distinguishable from the degradation products. The apparent molecular masses of the intact substrate and cleavage products were 52, 35 and 17 kDa respectively. These were higher than the calculated molecular masses (Figure 1A). The 17 kDa band reacted with the neo-epitope antibody which recognizes the newly generated N-terminal A374RGS377 of the aggrecan core protein, indicating that it is an authentic C-terminal fragment generated by aggrecanase activity (Figure 1B).
The linear range of gst-IGD-flag cleavage was determined by incubating 17 μM gst-IGD-flag with 0.5 nM ADAMTS-5-4 for up to 32 h at 37 °C (Figure 1B). Densitometric analysis of the products after SDS/PAGE separation and CBB-staining indicated that cleavage was linear up to 400000 pixel volumes, equivalent to incubation with 0.5 nM ADAMTS-5-4 for 20 h (Figure 1C). The enzyme concentration-dependence of substrate cleavage was determined by incubating gst-IGD-flag with increasing amounts of ADAMTS-5-4 (0–2.5 nM) for 20 h at 37 °C before visualization by CBB staining (Figure 1D). Cleavage was linear up to 0.5 nM enzyme (Figure 1E).
Inhibition of aggrecanases by reactive-site mutants of N-TIMP-3
Using the gst-IGD-flag assay, we tested the ability of N-TIMP-3(T2G) and [−1A]N-TIMP-3 to inhibit the aggrecanases. The remaining activity after inhibition was plotted as percentage activity against inhibitor concentration in Figures 2(A) and 2(B) for ADAMTS-4-2 and ADAMTS-5-2 respectively, and the data were fitted to the tight binding inhibition equation to obtain the Ki (app) values shown in Table 1, with the S.E.M. values representing the goodness of fit. The mutants retained inhibitory activity for ADAMTS, with Ki (app) values between 18 and 30 nM for ADAMTS-4-2 and approx. 1.5 nM for ADAMTS-5-4. N-TIMP-3 shows co-operativity in its inhibition of ADAM17 with a Hill coefficient, h, for N-TIMP-3 inhibition of ADAM17 of 3.59 [19], but the inhibition curves reported in the present paper fitted the equation for tight binding non-co-operative inhibition, reflecting an h value of 1. All of the N-terminal reactive-site mutants tested show selective inhibition for the ADAMTS enzymes over the MMPs.
Figure 2. The Ki (app) of N-TIMP-3 mutants for ADAMTS-4-2 and ADAMTS-5-4.
N-TIMP-3 or the mutants (0–1 μM) were incubated with ADAMTS-4-2 [1 nM (A and C)] or ADAMTS-5-4 [500 pM, (B and D)] for 1 h at 37 °C, before the addition of gst-IGD-flag (17 μM). After 16 h at 37 °C, SDS/PAGE and densitometric analysis of the 17 kDa product band was used to determine the percentage of residual activity. Data were fitted to the tight binding equation to obtain the Ki (app).
Table 1. Ki (app) of N-terminal reactive-site mutants for MMP-1ΔC, MMP-2, MMP-3ΔC, ADAMTS-4-2 and ADAMTS-5-4.
| Ki (app) (nM) | ||||||
|---|---|---|---|---|---|---|
| Inhibitor | MMP-1ΔC | MMP-2 | MMP-3ΔC | ADAM17 | ADAMTS-4-2 | ADAMTS-5-4 |
| N-TIMP-3 | 1.7±0.2 | 2.7±0.3 | 53.6±5.4 | 13.7±0.2 | 1.8±0.2 | 0.5±0.1 |
| N-TIMP-3(T2G) | >1000 | >1000 | >1000 | 35.6±1.9 | 18.3±0.1 | 1.5±0.1 |
| [−1A]N-TIMP-3 | 800 | 970 | >1000 | 33.9±2.8 | 22.2±0.1 | 1.7±0.1 |
Effect of extending the number of alanine residues appended to the N-terminus on the inhibition of ADAM17 and the aggrecanases
The alanine-extension mutants [−2A]N-TIMP-3, [−3A]N-TIMP-3, [−5A]N-TIMP-3 and [−8A]N-TIMP-3 with two, three, five and eight additional alanine residues at the N-terminus respectively were tested for their ability to inhibit ADAMTS-4 and ADAMTS-5 (Figures 2C and 2D respectively). These mutants were also tested against MMP-1ΔC, MMP-2, MMP-3ΔC and ADAM17. The results are summarized in Table 2. All of the alanine-extension mutants were poor MMP inhibitors, with less than 50% inhibition observed at the highest concentration tested (1 μM). The [−2A]N-TIMP-3 remained a potent inhibitor for ADAMTS-5 [Ki (app)=1.8 nM], but was weaker for ADAMTS-4 [Ki (app)=35.9 nM] and ADAM17 [Ki (app)=65.3 nM]. With three additional alanine residues, the apparent inhibition constants for ADAM17, ADAMTS-4 and ADAMTS-5 were approx. 100-fold worse compared with that of the wild-type inhibitor. The Ki (app) for ADAM17 was 239 nM, similar to that for ADAMTS-4. The inhibitory ability of the [−5A]N-TIMP-3 and the [−8A]N-TIMP-3 mutants decreased further, where the [−8A]N-TIMP-3 showed no significant inhibition at the highest concentration tested (1 μM). N-terminal sequence analysis indicated that all of these mutants started with the expected alanine residue.
Table 2. Ki (app) of N-terminal alanine-extension mutants for MMP-1ΔC, MMP-2, MMP-3ΔC, ADAMTS-4-2 and ADAMTS-5-4.
| Ki (app) (nM) | ||||||
|---|---|---|---|---|---|---|
| Inhibitor | MMP-1ΔC | MMP-2 | MMP-3ΔC | ADAM17 | ADAMTS-4-2 | ADAMTS-5-4 |
| [−1A]N-TIMP-3 | 800 | 970 | >1000 | 33.9±2.8 | 22.2±0.1 | 1.7±0.1 |
| [−2A]N-TIMP-3 | >1000 | 790 | >1000 | 65.3±5.2 | 35.9±8.4 | 1.8±0.9 |
| [−3A]N-TIMP-3 | >1000 | >1000 | >1000 | 239 | 173.7 | 87.6 |
| [−5A]N-TIMP-3 | >1000 | >1000 | >1000 | 843 | 385 | 593 |
| [−8A]N-TIMP-3 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 |
Effect of changing the amino acid at position [−1] on the inhibition of ADAMTS-4 and ADAMTS-5
To investigate the properties of the enzyme pocket that accommodates the N-terminus, the mutants [−1G]N-TIMP-3, [−1E]N-TIMP-3, [−1M]N-TIMP-3 and [−1K]N-TIMP-3 were generated and their ability to inhibit MMPs, ADAM17, ADAMTS-4 and ADAMTS-5 were tested. N-terminal sequencing indicated that the N-termini of these mutants were differently processed by E. coli. The N-terminal residues of [−1E]N-TIMP-3, [−1M]N-TIMP-3 and [−1K]N-TIMP-3 were methionine, making them [−ME]N-TIMP-3, [−MM]N-TIMP-3 and [−MK]N-TIMP-3. The mutant [−1G]N-TIMP-3 did not return a sequence, suggesting that the N-terminus was chemically modified, probably by N-acetylation (henceforth [−xG]N-TIMP-3). As the [−2A]N-TIMP-3 mutant was still active against the ADAMTSs, the Ki (app) values of these mutants for the various metalloproteinases were determined.
As summarized in Table 3, all of the mutants were poor MMP inhibitors, with less than 50% inhibition at the highest concentration tested (1 μM), apart from [−MK]N-TIMP-3, which inhibited MMP-1 and MMP-2 with Ki (app) values of 0.5 and 0.75 μM respectively. On the other hand, [−MM]N-TIMP-3 inhibited ADAMTS-4, ADAMTS-5 and ADAM17 with Ki (app) values of 75, 69 and 153 nM respectively. The other [−1] mutants were weaker inhibitors of these enzymes.
Table 3. Ki (app) of position [−1] mutants for MMP-1ΔC, MMP-2, MMP-3ΔC, ADAMTS-4-2 and ADAMTS-5-4.
| Ki (app) (nM) | ||||||
|---|---|---|---|---|---|---|
| Inhibitor | MMP-1ΔC | MMP-2 | MMP-3ΔC | ADAM17 | ADAMTS-4-2 | ADAMTS-5-4 |
| [−1A]N-TIMP-3 | 800 | 970 | >1000 | 33.9±2.8 | 22.2±0.1 | 1.7±0.1 |
| [−xG]N-TIMP-3 | >1000 | >1000 | >1000 | 465 | 107 | 118 |
| [−ME]N-TIMP-3 | >1000 | >1000 | >1000 | 529 | 326 | 327 |
| [−MM]N-TIMP-3 | >1000 | >1000 | >1000 | 153 | 75.7 | 69.4 |
| [−MK]N-TIMP-3 | 500 | 750 | >1000 | 325 | 280 | 217 |
Inhibition of porcine cartilage degradation by reactive-site mutants of N-TIMP-3
The aggrecanase-selective mutants N-TIMP-3(T2G), [−1A]N-TIMP-3 and [−2A]N-TIMP-3 were tested in the IL-1α-stimulated porcine articular cartilage explant model of cartilage degradation. As reported previously [20], upon stimulation with IL-1α, there was a 5-fold increase in GAG release from explanted porcine articular cartilage, which was not inhibited by TIMP-1 or TIMP-2, but was dose-dependently inhibited by N-TIMP-3. Additionally, all three of the N-TIMP-3 mutants tested were able to inhibit this release dose-dependently, although they were slightly weaker than N-TIMP-3 (Figure 3).
Figure 3. Dose-dependent inhibition of GAG release upon IL-1α stimulation in porcine articular cartilage by N-TIMP-3 reactive-site mutants.
Porcine articular cartilage explants were stimulated with IL-1α (10 ng/ml) for 3 days with the various TIMPs at the concentrations indicated (0.01, 0.1 and 1 μM). GAG released in the medium was measured by DMMB. n=3 for each treatment.
Collagen degradation of IL-1α-stimulated cartilage was not inhibited by aggrecanase-selective TIMP-3 mutants
Upon stimulation with IL-1, collagen degradation in porcine cartilage mediated by MMPs occurs after 21 days [40]. The cartilage-stimulation experiments were therefore extended for a longer time period at a single dose of 100 nM of the various TIMPs to determine the effects of the mutants on aggrecan and collagen catabolism.
As in the previous experiment, TIMP-1 and TIMP-2 did not affect the amount of GAG release from the cartilage upon IL-1α stimulation (Figure 4A). There was no significant difference in GAG release between stimulated cartilage explants in the presence of N-TIMP-3 and the unstimulated cartilage explants, indicating that TIMP-3 is an effective inhibitor of aggrecanases. However, there was a gradual loss of aggrecan from the tissue even without IL-1α stimulation, and by the end of 38 days, this equated to the loss of approx. 45% of the total GAG content. The N-TIMP-3 mutants significantly reduced the release of GAG at the early time points, compared with TIMP-1 and TIMP-2. However, there was a slow gradual loss of aggrecan above that observed for N-TIMP-3. This equated to approx. 85% of the total GAG content lost after 38 days.
Figure 4. Effects of the N-TIMP-3 mutants on aggrecan and collagen degradation in porcine articular cartilage under long-term stimulation with IL-1α.
Porcine articular cartilage explants were stimulated with IL-1α (10 ng/ml) with the various TIMPs at 0.1 μM for 38 days. The amount of GAG and hydroxyproline (Hypro) released into the medium was measured using the DMMB and DMBA assay respectively. The cumulative amounts of GAG (A) and hydroxyproline (B) loss against time were plotted. n=3 for each treatment.
Collagen degradation as measured by hydroxyproline release into the medium started at day 21 and rapidly progressed thereafter: after 35–38 days, the cartilage tissue was almost completely dissolved. TIMP-1, TIMP-2 and N-TIMP-3 inhibited collagen degradation, whereas the aggrecanase-selective N-TIMP-3 mutants were unable to inhibit collagen degradation (Figure 4B).
Inhibition of aggrecan degradation but not collagen degradation in human cartilage explants stimulated by IL-1α and OSM by N-TIMP-3 mutants
Stimulation of cartilage from human OA patients by IL-1α alone was insufficient to cause aggrecan degradation in 3 days (results not shown), but, in combination with OSM, there was a 2–3-fold increase in GAG release (Figure 5A), which represented approx. 3% of total GAG in the tissue. Cartilage from one knee joint was used to test the effects of the different doses of inhibitor. TIMP-1 unexpectedly showed dose-dependent inhibition of GAG release in this tissue, in contrast with porcine cartilage. TIMP-2, however, showed no inhibition of GAG release, making it unlikely that an MMP was involved. N-TIMP-3 was the most potent inhibitor of stimulated GAG release, with 0.1 μM completely inhibiting it. The three reactive-site mutants N-TIMP-3(T2G), [−1A]N-TIMP-3 and [−2A]N-TIMP-3 inhibited GAG release dose-dependently, although they were not as potent as N-TIMP-3. Within 3 days of culture, there was no stimulatory increase in collagen degradation, even when the tissue was treated with IL-1α and OSM (Figure 5B). The collagen degraded in 3 days of culture represented approx. 1.5% of the total collagen in the tissue. This suggests that the collagenolytic MMPs are already activated in OA cartilage. The hydroxyproline release from the OA cartilage was dose-dependently inhibited by TIMP-1, TIMP-2 and N-TIMP-3, but the aggrecanase-selective N-TIMP-3 mutants had no effect on the collagen degradation (Figure 5B).
Figure 5. Inhibition of GAG release but not hydroxyproline release of explanted human articular cartilage stimulated with IL-1α.
Cartilage from the knee of an OA patient was stimulated with IL-1α (10 ng/ml)+OSM (50 ng/ml) and the various TIMPs (0.01, 0.1 and 1 μM concentrations) for 3 days. GAG (A) and hydroxyproline (Hypro) (B) released into the medium were taken as measures of cartilage degradation. n=3 for each treatment. Cartilage samples from the hips of five OA patients (the previous and four additional) were treated with IL-1α (10 ng/ml) and the various TIMPs at a concentration of 100 nM. Data from the different patients were normalized by taking the amount of (C) GAG released upon stimulation as 100 and (D) hydroxyproline (Hypro) released without stimulation as 100, shown as means±S.D., n=3 for each patient; *P<0.03.
The experiments were repeated on cartilage obtained from the hips of four other patients using a single concentration of 0.1 μM of the respective TIMPs (n=5, including the previous patient). The GAG and hydroxyproline released upon stimulation without TIMP was normalized to a value of 100 in order to compare the different patients (Figure 5C and 5D). At a concentration of 0.1 μM, TIMP-1 showed a 50% inhibition of GAG release, whereas N-TIMP-3 inhibited GAG release to below that of control levels. TIMP-2 had no effect on GAG release. The three N-TIMP-3 mutants inhibited GAG release to control levels, but did not have any effect on collagen degradation, whereas the three wild-type TIMPs inhibited collagen degradation approx. 50% at a concentration of 0.1 μM.
Docking of N-TIMP-3 and modelled mutants to MMP-1, ADAM17, ADAMTS-4 and ADAMTS-5
In the absence of a crystal structure, we carried out molecular docking experiments to attempt to explain the inhibitory capabilities of the various mutants of TIMP-3. Docking of N-TIMP-3 with the ADAM17 component from the N-TIMP-3–ADAM17 complex (PDB code 3CKI) was first performed to validate the docking method. All 200 docking runs returned structures that fitted in the active-site cleft of the enzyme. The same N-TIMP-3 structure docked with a different ADAM17 structure (PDB code 3G42) returned 52 active-site fits. The positions of residues Lys315 and Met345, which are located on the active-site cleft of ADAM17, differ between the two ADAM17 structures. In the ADAM17 (PDB code 3G42) structure, they create a more restricted active-site cleft, which may explain the fewer active-site fits in this docking experiment. The same N-TIMP-3 structure docked into MMP-1, ADAMTS-4 and ADAMTS-5 returned 69, 2 and 27 active-site fits respectively. The modelled complex of N-TIMP-3 with the MMP-1 model was similar to the experimentally determined structure of the N-TIMP-1–MMP-1 complex [36]. The fit of N-TIMP-3 into the active site of ADAMTS-4 and ADAMTS-5, however, differed from those with MMP-1 and ADAM17: the catalytic Zn2+ ion was still chelated by the N-terminal cysteine residue of the TIMP, but the TIMP was tilted towards the prime side of the active site so that Phe34 of the TIMP that normally interacts with Tyr351Val352 of ADAM17 was not in contact with either ADAMTS, creating a water-accessible gap between the TIMP-3 and the ADAMTS components. The interaction of the bulky Arg84 of TIMP-3 with the loop G323VST326 in ADAMTS-4 or G372HHS375 of ADAMTS-5 probably prevented TIMP-3 binding in the same way as in previously characterized TIMP–metalloproteinase complexes. Mutation of all of these residues on both molecules to alanine in silico generated complexes that are more similar to the canonical TIMP–metalloproteinase interaction, supporting the above possibility.
The model of [−1A]N-TIMP-3 was created by the addition of an alanine residue at the N-terminus of the N-TIMP-3 crystal structure, with the alanine residue being allowed to rotate in the docking experiments. When docked with MMP-1, 29 structures in the active site were obtained containing TIMP-3 docked in the MMP-1 active site. However, to accommodate the extra alanine residue, the TIMP was tilted in such a manner that Phe34 was not in contact with the enzyme, and the TIMP leant to one side of the active-site cleft (Figure 6A). When [−1A]N-TIMP-3 was docked with ADAM17 (PDB code 3CKI), 108 structures were returned containing TIMP-3 in the active-site cleft. The TIMP molecule was tilted towards the prime side of the active site, such that Phe34 of the TIMP was no longer in contact with the enzyme, as in the models of N-TIMP-3 docked into ADAMTS-4 and ADAMTS-5 (Figure 6B). In this model, however, the access of the catalytic Zn2+ ion to water is blocked by the side chain of the alanine residue. Docking [−1A]N-TIMP-3 with ADAMTS-4 and ADAMTS-5 produced 4 and 28 models in which TIMP-3 is in contact with the active site respectively. These were very similar to the corresponding complexes generated with N-TIMP-3, except that access of the catalytic Zn2+ ion to water was blocked by the NH2 group of the alanine (Figures 6C and 6D). The models of [−1A]N-TIMP-3 had additional interactions with the deeper active-site clefts of ADAM17, ADAMTS-4 and ADAMTS-5 that were not present in the MMP-1 complexes. These were with Met345 in ADAM17, the loop G323VST326 in ADAMTS-4 and G372HHS375 in ADAMTS-5 (highlighted in Figure 6).
Figure 6. In silico models of N-TIMP-3 and [−1A]N-TIMP-3 with ADAM17, MMP-1, ADAMTS-4 and ADAMTS-5.
Computer-simulated lowest free energy ranked models of complexes of N-TIMP-3 (salmon ribbon) or [−1A]N-TIMP-3 (blue ribbon) with MMP-1 (A), ADAM17 (B), ADAMTS-4 (C) or ADAMTS-5 (D) are shown. The catalytic Zn2+ ion is shown in magenta, water-accessible enzyme surfaces are grey and regions which interact only with N-TIMP-3, [−1A]N-TIMP-3 or with both are in salmon, blue and green respectively. Phe34 and the N-terminal extra alanine residues are shown with sticks. In the [−1A]N-TIMP-3 complexes with MMP-1 and ADAM17, the TIMP molecule is tilted and the interaction of Phe34 of the inhibitor with the enzymes are lost. Both N-TIMP-3 and [−1A]N-TIMP-3 are bound to ADAMTS-4 and ADAMTS-5 in a similar tilted manner. Residues which make the deeper active-site clefts of ADAM17 (Met345), ADAMTS-4 (G322VST325) and ADAMTS-5 (G372HHS375) that interact with the TIMP and are not present in MMP-1 are highlighted in red.
DISCUSSION
Using the newly developed gst-IGD-flag substrate, we demonstrate that the N-terminal mutants of N-TIMP-3 that have essentially lost their MMP-inhibitory activities, N-TIMP-3(T2G) and [−1A]N-TIMP-3, retain their ability to inhibit ADAMTS-4 and ADAMTS-5. Furthermore, the [−2A]N-TIMP-3 mutant also retained strong affinity with ADAMTS-5, but has a lower affinity for ADAMTS-4 and ADAM17.
The ability of these three mutants to dose-dependently inhibit aggrecan degradation in the IL-1α-stimulated model of porcine cartilage degradation reinforces our previous observations that the enzymes involved in the degradation of aggrecan in this system are from the ADAMTS family and are not MMPs [20]. There was no inhibition of collagen degradation when the N-TIMP-3 mutants were tested in the long-term IL-1α-stimulated porcine articular cartilage. As theTIMP-3 mutants used in the present study lack the ability to inhibit the MMPs, they were not expected to exhibit a direct effect on MMP-mediated collagen degradation, confirming that the selectivity of these N-TIMP-3 mutants was maintained ex vivo. Pratta et al. [41] reported that, in IL-1α-induced bovine nasal cartilage degradation, the blocking of aggrecan degradation by an ADAMTS-selective chemical inhibitor resulted in inhibition of collagen degradation. The discrepancy between the two studies may be due to different levels of aggrecan protection: in the present study, a slower persistent loss of aggrecan was observed in the explant cultures when the mutants were used at a concentration of 0.1 μM above the amount observed in the control and N-TIMP-3-treated explants, whereas aggrecan degradation was almost completely blocked in the study by Pratta et al. [41]. The amount of intact aggrecan left in the explants therefore may not have been sufficient to influence collagen degradation. Interestingly, mice deficient in MMP-13 are also protected in an animal model of OA [42], indicating that inhibition of MMP-13 is also an effective cartilage protectant. The inhibition of both aggrecanases and collagenases, as shown with 0.1 μM wild-type TIMP-3, may be an effective way to protect cartilage from degradation during the progression of OA.
The N-TIMP-3 reactive-site mutants are able to inhibit IL-1α with OSM-stimulated GAG release from human cartilage explants in culture. This inhibition, however, had no effect on the amount of collagen degradation detected, indicating that, by the stage when joint-replacement surgery was required, protection of aggrecan degradation had little effect on the outcome of collagen degradation. Furthermore, hydroxyproline release was fully activated in human OA cartilage without IL-1α and OSM treatment and it was dose-dependently inhibited by TIMP-1, TIMP-2 and TIMP-3, suggesting that the collagenolytic MMPs were fully active at this stage. Further studies using our ADAMTS-selective mutants in combination with TIMP-2 and TIMP-3 will enable us to dissect the relative contributions of the ADAMTSs and the MMPs during the progression of joint cartilage degradation and the benefits of targeting either protease at different stages of the disease.
On the basis of the crystal structure of ADAM17–N-TIMP-3 complex, it was considered that there would not be sufficient space in the active site of the enzyme to accommodate TIMPs with one or two additional amino acids at the N-terminus. However, our enzyme-inhibition studies described in the present paper have shown that up to two alanine residues can be accommodated. The in silico docking experiments performed indicate that the [−1A]N-TIMP-3 mutant is most likely to be tilted so that Phe34 of the TIMP is no longer in contact with the enzyme, while still blocking the access of the catalytic Zn2+ ion to the water molecule required for peptide hydrolysis. Our in vitro inhibition data suggest that, with MMP-1, this leads to a large loss of binding energy which renders the TIMP inactive, but this is not the case in ADAM17, ADAMTS-4 and ADAMTS-5. Additional interactions with Met345 in ADAM17, G322VST325 in ADAMTS-4 and G372HHS375 in ADAMTS-5 may compensate for the loss of binding energy associated with the loss of interactions involving Phe34. The amino acids within the TIMP molecule that are likely to be in contact with these extended loops include the residues around Leu94 and Arg84, which were identified by Lee et al. [43,44] as being important in ADAM17 inhibition. In addition, comparison of the crystal structures of ADAMTS-4 in the presence and absence of a synthetic inhibitor, and those of ADAMTS-5, showed that the loops G322VST325 and G372HHS375, designated the S2′ loop by Mosyak et al. [38], are able to adopt at least two different conformations. Similarly, ADAM17 has been shown to have a more flexible active site than MMP-2 upon binding to a non-competitive inhibitor [45]. The flexible nature of these active sites may allow TIMP-3 with one or two extra alanine residues at the N-terminus to still form tight enzyme–inhibitor complexes. Further structural studies of [−1A]N-TIMP-3 and the [−2A]N-TIMP-3 in complex with these enzymes will be required to validate these docking predictions, whereas the Leu94 and Arg84 residues may provide a further avenue in TIMP engineering to obtain TIMPs capable of differentiating between the two aggrecanases.
AUTHOR CONTRIBUTION
Ngee Lim designed, performed and analysed the experiments reported, prepared the Figures and wrote the paper. Masahide Kashiwagi designed the new substrate and made the initial observations of the TIMP-3 mutants. Robert Visse designed and analysed the molecular docking experiments. Jonathan Jones procured the human tissue. Jan Enghild was involved in the N-terminal sequencing of the N-TIMP-3 mutants. Keith Brew interpreted the data and edited the paper before submission. Hideaki Nagase designed the experiments, interpreted the data and wrote the paper.
FUNDING
The work was supported by the Wellcome Trust [grant number 075473], the Arthritis Research Campaign and National Institutes of Health [grant number AR40994].
References
- 1.Amour A., Knight C. G., Webster A., Slocombe P. M., Stephens P. E., Knäuper V., Docherty A. J., Murphy G. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 2000;473:275–279. doi: 10.1016/s0014-5793(00)01528-3. [DOI] [PubMed] [Google Scholar]
- 2.Jacobsen J., Visse R., Sørensen H. P., Enghild J. J., Brew K., Wewer U. M., Nagase H. Catalytic properties of ADAM12 and its domain deletion mutants. Biochemistry. 2008;47:537–547. doi: 10.1021/bi701629c. [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez-Manzaneque J. C., Westling J., Thai S. N., Luque A., Knäuper V., Murphy G., Sandy J. D., Iruela-Arispe M. L. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2002;293:501–508. doi: 10.1016/S0006-291X(02)00254-1. [DOI] [PubMed] [Google Scholar]
- 4.Amour A., Slocombe P. M., Webster A., Butler M., Knight C. G., Smith B. J., Stephens P. E., Shelley C., Hutton M., Knäuper V., et al. TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998;435:39–44. doi: 10.1016/s0014-5793(98)01031-x. [DOI] [PubMed] [Google Scholar]
- 5.Mochizuki S., Shimoda M., Shiomi T., Fujii Y., Okada Y. ADAM28 is activated by MMP-7 (matrilysin-1) and cleaves insulin-like growth factor binding protein-3. Biochem. Biophys. Res. Commun. 2004;315:79–84. doi: 10.1016/j.bbrc.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Zou J., Zhu F., Liu J., Wang W., Zhang R., Garlisi C. G., Liu Y. H., Wang S., Shah H., Wan Y., Umland S. P. Catalytic activity of human ADAM33. J. Biol. Chem. 2004;279:9818–9830. doi: 10.1074/jbc.M309696200. [DOI] [PubMed] [Google Scholar]
- 7.Wang W. M., Ge G., Lim N. H., Nagase H., Greenspan D. S. TIMP-3 inhibits the procollagen N-proteinase ADAMTS-2. Biochem. J. 2006;398:515–519. doi: 10.1042/BJ20060630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto G., Aoki T., Nakamura H., Tanzawa K., Okada Y. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4) FEBS Lett. 2001;494:192–195. doi: 10.1016/s0014-5793(01)02323-7. [DOI] [PubMed] [Google Scholar]
- 9.Kashiwagi M., Tortorella M., Nagase H., Brew K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5) J. Biol. Chem. 2001;276:12501–12504. doi: 10.1074/jbc.C000848200. [DOI] [PubMed] [Google Scholar]
- 10.Amour A., Knight C. G., English W. R., Webster A., Slocombe P. M., Knäuper V., Docherty A. J., Becherer J. D., Blobel C. P., Murphy G. The enzymatic activity of ADAM8 and ADAM9 is not regulated by TIMPs. FEBS Lett. 2002;524:154–158. doi: 10.1016/s0014-5793(02)03047-8. [DOI] [PubMed] [Google Scholar]
- 11.Gomis-Rüth F. X., Maskos K., Betz M., Bergner A., Huber R., Suzuki K., Yoshida N., Nagase H., Brew K., Bourenkov G. P., et al. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature. 1997;389:77–81. doi: 10.1038/37995. [DOI] [PubMed] [Google Scholar]
- 12.Huang W., Meng Q., Suzuki K., Nagase H., Brew K. Mutational study of the amino-terminal domain of human tissue inhibitor of metalloproteinases 1 (TIMP-1) locates an inhibitory region for matrix metalloproteinases. J. Biol. Chem. 1997;272:22086–22091. doi: 10.1074/jbc.272.35.22086. [DOI] [PubMed] [Google Scholar]
- 13.Meng Q., Malinovskii V., Huang W., Hu Y., Chung L., Nagase H., Bode W., Maskos K., Brew K. Residue 2 of TIMP-1 is a major determinant of affinity and specificity for matrix metalloproteinases but effects of substitutions do not correlate with those of the corresponding P1′ residue of substrate. J. Biol. Chem. 1999;274:10184–10189. doi: 10.1074/jbc.274.15.10184. [DOI] [PubMed] [Google Scholar]
- 14.Wei S., Chen Y., Chung L., Nagase H., Brew K. Protein engineering of the tissue inhibitor of metalloproteinase 1 (TIMP-1) inhibitory domain: in search of selective matrix metalloproteinase inhibitors. J. Biol. Chem. 2003;278:9831–9834. doi: 10.1074/jbc.M211793200. [DOI] [PubMed] [Google Scholar]
- 15.Wingfield P. T., Sax J. K., Stahl S. J., Kaufman J., Palmer I., Chung V., Corcoran M. L., Kleiner D. E., Stetler-Stevenson W. G. Biophysical and functional characterization of full-length, recombinant human tissue inhibitor of metalloproteinases-2 (TIMP-2) produced in Escherichia coli: comparison of wild type and amino-terminal alanine appended variant with implications for the mechanism of TIMP functions. J. Biol. Chem. 1999;274:21362–21368. doi: 10.1074/jbc.274.30.21362. [DOI] [PubMed] [Google Scholar]
- 16.Higashi S., Miyazaki K. Reactive site-modified tissue inhibitor of metalloproteinases-2 inhibits the cell-mediated activation of progelatinase A. J. Biol. Chem. 1999;274:10497–10504. doi: 10.1074/jbc.274.15.10497. [DOI] [PubMed] [Google Scholar]
- 17.Troeberg L., Tanaka M., Wait R., Shi Y. E., Brew K., Nagase H. E. coli expression of TIMP-4 and comparative kinetic studies with TIMP-1 and TIMP-2: insights into the interactions of TIMPs and matrix metalloproteinase 2 (gelatinase A) Biochemistry. 2002;41:15025–15035. doi: 10.1021/bi026454l. [DOI] [PubMed] [Google Scholar]
- 18.Wisniewska M., Goettig P., Maskos K., Belouski E., Winters D., Hecht R., Black R., Bode W. Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE–N-TIMP-3 complex. J. Mol. Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 19.Wei S., Kashiwagi M., Kota S., Xie Z., Nagase H., Brew K. Reactive site mutations in tissue inhibitor of metalloproteinase-3 disrupt inhibition of matrix metalloproteinases but not tumor necrosis factor-α-converting enzyme. J. Biol. Chem. 2005;280:32877–32882. doi: 10.1074/jbc.C500220200. [DOI] [PubMed] [Google Scholar]
- 20.Gendron C., Kashiwagi M., Hughes C., Caterson B., Nagase H. TIMP-3 inhibits aggrecanase-mediated glycosaminoglycan release from cartilage explants stimulated by catabolic factors. FEBS Lett. 2003;555:431–436. doi: 10.1016/s0014-5793(03)01295-x. [DOI] [PubMed] [Google Scholar]
- 21.Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 22.Stanton H., Rogerson F. M., East C. J., Golub S. B., Lawlor K. E., Meeker C. T., Little C. B., Last K., Farmer P. J., Campbell I. K., et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 23.Lark M. W., Bayne E. K., Flanagan J., Harper C. F., Hoerrner L. A., Hutchinson N. I., Singer I. I., Donatelli S. A., Weidner J. R., Williams H. R., et al. Aggrecan degradation in human cartilage: evidence for both matrix metalloproteinase and aggrecanase activity in normal, osteoarthritic, and rheumatoid joints. J. Clin. Invest. 1997;100:93–106. doi: 10.1172/JCI119526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struglics A., Larsson S., Pratta M. A., Kumar S., Lark M. W., Lohmander L. S. Human osteoarthritis synovial fluid and joint cartilage contain both aggrecanase- and matrix metalloproteinase-generated aggrecan fragments. Osteoarthritis Cartilage. 2006;14:101–113. doi: 10.1016/j.joca.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Murphy G., Nagase H. Progress in matrix metalloproteinase research. Mol. Aspects Med. 2008;29:290–308. doi: 10.1016/j.mam.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung L., Shimokawa K., Dinakarpandian D., Grams F., Fields G. B., Nagase H. Identification of the 183RWTNNFREY191 region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J. Biol. Chem. 2000;275:29610–29617. doi: 10.1074/jbc.M004039200. [DOI] [PubMed] [Google Scholar]
- 27.Troeberg L., Fushimi K., Scilabra S. D., Nakamura H., Dive V., Thogersen I. B., Enghild J. J., Nagase H. The C-terminal domains of ADAMTS-4 and ADAMTS-5 promote association with N-TIMP-3. Matrix Biol. 2009;28:463–469. doi: 10.1016/j.matbio.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki K., Kan C. C., Hung W., Gehring M. R., Brew K., Nagase H. Expression of human pro-matrix metalloproteinase 3 that lacks the N-terminal 34 residues in Escherichia coli: autoactivation and interaction with tissue inhibitor of metalloproteinase 1 (TIMP-1) Biol. Chem. 1998;379:185–191. doi: 10.1515/bchm.1998.379.2.185. [DOI] [PubMed] [Google Scholar]
- 29.Kashiwagi M., Enghild J. J., Gendron C., Hughes C., Caterson B., Itoh Y., Nagase H. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J. Biol. Chem. 2004;279:10109–10119. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- 30.Gendron C., Kashiwagi M., Lim N. H., Enghild J. J., Thøgersen I. B., Hughes C., Caterson B., Nagase H. Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J. Biol. Chem. 2007;282:18294–18306. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- 31.Franken K. L., Hiemstra H. S., van Meijgaarden K. E., Subronto Y., den Hartigh J., Ottenhoff T. H., Drijfhout J. W. Purification of his-tagged proteins by immobilized chelate affinity chromatography: the benefits from the use of organic solvent. Protein Expression Purif. 2000;18:95–99. doi: 10.1006/prep.1999.1162. [DOI] [PubMed] [Google Scholar]
- 32.Knight C. G., Willenbrock F., Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 33.Wyckoff M., Rodbard D., Chrambach A. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate-containing buffers using multiphasic buffer systems: properties of the stack, valid Rf- measurement, and optimized procedure. Anal. Biochem. 1977;78:459–482. doi: 10.1016/0003-2697(77)90107-5. [DOI] [PubMed] [Google Scholar]
- 34.Bergman I., Loxley R. Lung tissue hydrolysates: studies of the optimum conditions for the spectrophotometric determination of hydroxyproline. Analyst. 1969;94:575–584. doi: 10.1039/an9699400575. [DOI] [PubMed] [Google Scholar]
- 35.Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer S., Wei S., Brew K., Acharya K. R. Crystal structure of the catalytic domain of matrix metalloproteinase-1 in complex with the inhibitory domain of tissue inhibitor of metalloproteinase-1. J. Biol. Chem. 2007;282:364–371. doi: 10.1074/jbc.M607625200. [DOI] [PubMed] [Google Scholar]
- 37.Park K., Gopalsamy A., Aplasca A., Ellingboe J. W., Xu W., Zhang Y., Levin J. I. Synthesis and activity of tryptophan sulfonamide derivatives as novel non-hydroxamate TNF-α converting enzyme (TACE) inhibitors. Bioorg. Med. Chem. 2009;17:3857–3865. doi: 10.1016/j.bmc.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 38.Mosyak L., Georgiadis K., Shane T., Svenson K., Hebert T., McDonagh T., Mackie S., Olland S., Lin L., Zhong X., et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu X., Shelver W. H. Docking studies of matrix metalloproteinase inhibitors: zinc parameter optimization to improve the binding free energy prediction. J. Mol. Graphics Modell. 2003;22:115–126. doi: 10.1016/S1093-3263(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 40.Cawston T., Plumpton T., Curry V., Ellis A., Powell L. Role of TIMP and MMP inhibition in preventing connective tissue breakdown. Ann. N.Y. Acad. Sci. 1994;732:75–83. doi: 10.1111/j.1749-6632.1994.tb24726.x. [DOI] [PubMed] [Google Scholar]
- 41.Pratta M. A., Yao W., Decicco C., Tortorella M. D., Liu R. Q., Copeland R. A., Magolda R., Newton R. C., Trzaskos J. M., Arner E. C. Aggrecan protects cartilage collagen from proteolytic cleavage. J. Biol. Chem. 2003;278:45539–45545. doi: 10.1074/jbc.M303737200. [DOI] [PubMed] [Google Scholar]
- 42.Little C. B., Barai A., Burkhardt D., Smith S. M., Fosang A. J., Werb Z., Shah M., Thompson E. W. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 2009;60:3723–3733. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee M. H., Rapti M., Knäuper V., Murphy G. Threonine 98, the pivotal residue of tissue inhibitor of metalloproteinases (TIMP)-1 in metalloproteinase recognition. J. Biol. Chem. 2004;279:17562–17569. doi: 10.1074/jbc.M312589200. [DOI] [PubMed] [Google Scholar]
- 44.Lee M. H., Maskos K., Knäuper V., Dodds P., Murphy G. Mapping and characterization of the functional epitopes of tissue inhibitor of metalloproteinases (TIMP)-3 using TIMP-1 as the scaffold: a new frontier in TIMP engineering. Protein Sci. 2002;11:2493–2503. doi: 10.1110/ps.0216202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solomon A., Rosenblum G., Gonzales P. E., Leonard J. D., Mobashery S., Milla M. E., Sagi I. Pronounced diversity in electronic and chemical properties between the catalytic zinc sites of tumor necrosis factor-α-converting enzyme and matrix metalloproteinases despite their high structural similarity. J. Biol. Chem. 2004;279:31646–31654. doi: 10.1074/jbc.M401310200. [DOI] [PubMed] [Google Scholar]