Abstract
The cannabinoid receptor one (CB1) is a class A G-protein-coupled receptor thought to bind ligands primarily within its helical bundle. Evidence suggests, however, that the extracellular domain may also play a role. We have previously shown that the C-terminus of the extracellular loop 2 of CB1 is important in binding some compounds; receptors with mutations in this region (F268W, P269A, H270A, and I271A) bound some agonists with severely reduced affinity relative to the wild-type receptor. In the present work, we examine the impact of these mutations on binding a chemically diverse set of ligands. The receptors, F268W and I271A, exhibited a greater sensitivity to binding the inverse agonists/antagonists SLV319, AVE1625, NESS0327 relative to P269A and H270A, suggesting that the Pro and His are not involved in binding those compounds. In contrast, binding of the agonists, BAY593074 and WIN55212-2, was diminished in all four receptors, suggesting the conformational unit contributed by all four residues is important. A more marked loss in binding was observed for agonists of the nonclassical (CP55940) and classical (HU-210, JWH061, JWH179) cannabinoid classes and for a silent antagonist derivative (O-2050), pointing to the critical nature of this region for binding both the bicyclic/tricyclic core and the alkyl chain of these derivatives. However, moving the location of the alkyl chain on a series of pyrazole analogues shows it can be better accommodated in certain locations (O-1255) than others (O-1302, O-1690) and underscores the involvement of residues F268 and I271.
Keywords: G-protein-coupled receptor, cannabinoid receptor 1, extracellular loop 2, ligand binding
INTRODUCTION
G-protein-coupled receptors (GPCRs) such as the cannabinoid receptor 1 are of considerable importance, as they constitute a significant portion of the targets for FDA approved drugs [Overington et al., 2006]. Ever-increasing information implicates cannabinoid receptor modulation with appetite regulation, analgesia, intraocular pressure, and short-term memory [see Howlett et al., 2002]. Localized on presynaptic nerve terminals, cannabinoid receptors are thought to play a direct role in the inhibition of neurotransmitter release [Mackie and Hille, 1992]. Attempts to harness the therapeutic potential of the cannabinoid system have fallen short due to undesirable side effects [see Janero and Makriyannis, 2009; Pertwee, 2009]. However, recent advances suggest the possibility of selectively activating certain signaling pathways, a concept that may ultimately be used to assist in the development of novel drugs that achieve the desirable medicinal benefits while minimizing the undesirable side effects.
Like all GPCRs, the cannabinoid receptors have seven hydrophobic transmembrane domains linked by three extracellular and three intracellular loops. Two cannabinoid receptors subtypes have been identified to date: the cannabinoid receptor 1 (CB1), which is found predominantly in the central nervous system [Devane et al., 1988; Matsuda et al., 1990], and the cannabinoid receptor 2 (CB2), which is localized mainly to immune tissue [Munro et al., 1993]. The two receptors have 44% sequence identity, and both couple predominantly to Gi proteins, but CB1 has been shown to signal through Gs and Gq subunits under certain conditions [Abadji et al., 1999; Bonhaus et al., 1998; Glass and Felder, 1997; Lauckner et al., 2005].
Capable of binding Δ9-THC, the main psychoactive constituent of the plant Cannabis sativa, also known as marijuana, CB1 is also activated by endogenous lipid-derived eicosanoids such as anandamide and 2-arachidonoylglycerol, aminoalkylindole-based compounds such as WIN55212-2, and synthetic nonclassical cannabinoids such as CP55940, as well as the tricyclic classical cannabinoids, including Δ9-THC, HU-210, and JWH061. Antagonists and inverse agonists for the cannabinoid receptors based on a pyrazole scaffold have also been developed. These include SR141716A (rimonabant), AM251, and AM281. Modifications of the alkyl chain of cannabinoid receptor agonists can affect both compound efficacy and their specificity for the CB receptor subtypes [Huffman et al., 1999, 2003].
Relatively little is known about the orientation of CB1 ligands at the active site of the receptor. Aromatic residues, such as F201, W280, and W357 among the transmembrane domains of mouse CB1, are critical for SR141716A and WIN55212-2 binding; however, evidence suggests that these do not directly interact with nonclassical agonists, such as CP55940 [McAllister et al., 2003]. We [Chin et al., 1998] and others [Song and Bonner, 1996] have previously demonstrated the importance of K192 in CP55940 but not WIN55212-2 binding. A hydrogen bonding interaction between the K192 and the phenolic hydroxyl group of classical and nonclassical agonists [Chin et al., 1998; Song and Bonner, 1996] or the carboxamide oxygen of SR141716A [Hurst et al., 2002, 2006] appears critical for ligand binding. Other residues of the receptor implicated in ligand binding include S383 which is thought to induce a bend in TM7 critical for nonclassical agonist binding [Kapur et al., 2007] as well as C386, also located in TM7, whose side chain precludes SR141716A binding when labeled with MTS reagents [Fay et al., 2005].
Recently, we have shown that the extracellular domains of CB1 can play an integral role in ligand binding [Ahn et al., 2009]. It had long been thought that class A GPCRs, including CB1, that bind small hydrophobic molecules, do so primarily in the transmembrane domain. However, we have shown that the C-terminal region of the extracellular loop 2 (EC2) differentiates agonist from inverse agonist binding. Point mutations at positions F268, P269, H270, and I271 of CB1 led to receptors that bound the agonists tested with substantially attenuated affinities but had no effect on inverse agonist binding affinity. Consistent with these findings, high concentrations of the nonclassical agonist CP55940 resulted in low EC50 and Emax values for agonist-induced G-protein coupling activity. Outstanding questions include whether the decrease in agonist binding was caused by the loss of a functional group that made direct contact with the ligand or if each of the mutations led to a conformational change that indirectly hindered agonist binding. Moreover, it is unclear what aspects of the chemical structure of the agonists tested precluded binding. This is an important issue because molecular modeling of the receptor suggests that the F268 in this region of the receptor could project into the core of the receptor bundle and ligand binding cavity [Ahn et al., 2009].
Molecular modeling studies of CB1 ligands by Thomas et al. [1998] and Salo et al. [2004] suggest the aryl rings of the SR141716A molecule may overlap with the alkyl side chain of the classical and nonclassical cannabinoid receptor agonists in the ligand binding pocket. SAR studies to determine the role of the various substituents of SR141716A determined that the aryl ring at the C-5 substituent or the N-1 substituent of SR141716A is important for antagonism while the C-3 substituent modulates receptor activation [Wiley et al., 2001]. Interestingly, the presence of an alkyl chain in place of the piperidine ring at the C-3 substituent of SR141716A imparts partial agonism at the mouse CB1.
Understanding the interactions of CB1 ligands and specific amino acid residues of the receptor is critical for the development of high-affinity selective therapeutic agents. In the present study, we examine the binding profile of a structurally diverse set of ligands to a series of CB1 receptors with mutations in the C-terminus of EC2. Consistent with our previous findings [Ahn et al., 2009], the C-terminus of EC2 plays an important role in differentiating agonist binding from inverse agonist/antagonist binding. However, compounds with the bicyclic/tricyclic core, such as is found in the classical and nonclassical cannabinoids, exhibit more severely reduced binding than some other agonists. Moreover, we find that mutation of the two residues, F268 and I271, among the four assessed in this region display more sensitivity to binding ligands including some inverse agonists/antagonists.
MATERIALS AND METHODS
Plasmid Preparation
Site-directed mutagenesis (QuickChange; Stratagene, La Jolla, CA) was performed on a template of the human CB1 cDNA cloned into pcDNA3.1 according to the manufacturer's protocol. DNA sequencing verified the mutations.
CB1 Membrane Preparation
CB1 receptor-containing membrane preparation was obtained as previously described [Abadji et al., 1999]. In brief, HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 3.5 mg/ml glucose at 37°C in 5% CO2. To transiently transfect HEK293T cells, 1 day before transfection, they were seeded at ~0.9 × 106 cells/100-mm dish. The calcium phosphate precipitation method [Chen and Okayama, 1987] was used to transfect the cells. Approximately 24 h after transfection, the cells were harvested, then washed twice with phosphate-buffered saline (PBS). The cells were resuspended in a mammalian protease inhibitor cocktail (4-2-aminoethyl)benzene-sulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin) (Sigma-Aldrich, St. Louis, MO) containing PBS solution and then lysed by nitrogen cavitation at 750 psi for 5 min, using a Parr cell disruption bomb. A 10-min spin at 4°C at 500 g was used to pellet cell debris and nuclei. The resulting supernatant was subsequently spun at 100,000 g for 45 min at 4°C. The membrane-containing pellet was then resuspended in TME buffer (25 mM Tris-HCl, 5 mM MgCl2, and 1 mM EDTA, pH 7.4) containing 7% sucrose (w/v). The Bradford assay [Bradford, 1976] was used to determine protein concentration. The membrane preparation was stored at 0.6 mg/ml at −70°C.
Ligand Binding
Ligand binding assays were performed as previously described [Abadji et al., 1999]. Briefly, for saturation binding assays, ~39 mg of membrane was incubated with increasing concentrations of [3H]SR141716A (43 Ci/mmol; GE Healthcare, Piscataway, NJ) for 90 min at 30°C in TME+0.1% fatty acid-free bovine serum albumin (BSA) in a final volume of 200 μl. Nonspecific binding was typically determined with a 5 μM concentration of unlabeled ligand. For competition binding assays, ~25–40 μg of membranes was incubated with 4 nM [3H]SR141716A and increasing concentrations of displacing ligand for 90 min at 30°C in TME+0.1% fatty acid-free BSA in a final volume of 200 μl. Nonspecific binding was typically determined with 5 μM of unlabeled ligand. The addition of 250 μl of TME+5% BSA was used to terminate the reaction before separating bound from unbound ligand by filtering with a 24-manifold cell harvester (Brandel, Gaithersburg, MD). Filters were washed with cold TME buffer. Scintillation counting was then used to determine the bound radioactivity.
Data Analysis
Ligand binding assays were performed in duplicate. The data are presented as the median with the corresponding 95% confidence limits from at least three independent experiments. IC50 values were calculated by nonlinear regression (fitted to a one-site competition model) using GraphPad Prism (GraphPad Software, San Diego, CA). The Cheng–Prusoff equation [Cheng and Prusoff, 1973] was used to calculate the Ki values using Kd values for the tracer obtained from saturation binding analyses. Analysis of variance (ANOVA) followed by Dunn's post hoc test was used to compare the wild-type Ki values with those of the mutant receptors. P-values of <0.05 were considered statistically significant.
RESULTS
The sequence of CB1 EC2 is shown in Figure 1. Using alanine scanning mutagenesis, we previously found mutations in the C-terminus of EC2 of CB1 that were sensitive to agonist but not to inverse agonist binding [Ahn et al., 2009]; P269A, H270A, and I271A receptors exhibited diminished binding for the agonists CP55940, methanandamide, and HU-210 but bound inverse agonists, SR141716A, AM251, and LY320135 with wild-type receptor affinity. The neighboring mutation, yielding the F268A receptor, displayed both impaired localization and ligand binding, while characterization of other amino acid substitutions at 268 identified a F268W receptor that was trafficked to the cell surface yet displayed the differential binding preference for inverse agonists comparable to the P269A, H270A, and I271A receptors. Therefore, we used this set of CB1 receptor mutants, F268W, P269A, H270A, and I271A, to reveal the molecular features for ligand binding within this region.
Fig. 1.
The EC2 loop of CB1. The dotted line represents the disulfide bridge between residues C257 and C264. Residues comprising the C-X-X-X-Ar motif (where Ar is any aromatic residue) are noted by the hatched marked circles. Mutating the residues in grey results in variants with attenuated ligand binding properties [Ahn et al., 2009] and were examined for SAFIR (structure-affinity relationship) here.
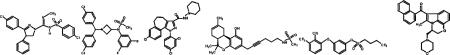
While our previous work suggested a clear demarcation in the binding affinity of inverse agonists and agonists to the EC2 mutant receptors, relative to wild-type, this may reflect commonalities in the structures tested as opposed to preferences for ligands with particular pharmacological effects. Therefore, we evaluated the binding affinity of a spectrum of agonists, antagonists, and inverse agonists with a range of structures to the CB1 mutant receptors. Since we had shown that the diarylpyrazole ligands, SR141716A and AM251, retained high-affinity binding to the C-terminal EC2 mutants, we tested the diarylpyrazoline SLV319, a CB1 inverse agonist that is more hydrophilic than SR141716A, the benzhydrylazetidine, AVE1625, and NESS0327 which has a 3-carbon linker between the monocholorinated aryl ring of SR141716A and the pyrazole core, and thus an antagonist with limited flexibility relative to the diarylpyrazole ligands. The Ki values from competition binding studies using [3H]SR141716A as tracer are shown in Table 1. For each of these compounds, the P269A and H270A receptors bound the ligand with an affinity comparable to the wild-type receptor (SLV319, Ki = 1.8 nM; AVE1625, Ki = 0.64 nM; NESS0327, Ki 5 4.6 nM) or at most 2-fold more weakly. The F268W and I271A receptors displayed greater differences; the F268W receptor showed a 3- and 5-fold loss in affinity for AVE1625 and NESS0327, respectively, and the I271A receptor a 5- and 3-fold loss for SLV319 and AVE1625, respectively.
TABLE 1.
Binding Affinity of Various Ligands to CB1 Receptors With Mutations in Positions 268–271 in EC2
 |
||||||
|---|---|---|---|---|---|---|
|
Ki (nM) |
||||||
| Receptor | SLV319 | AVE1625 | NESS0327 | O-2050 | BAY593074 | WIN55212-2 |
| WT | 1.8 (1.2–2.6) | 0.64 (0.37–1.1) | 4.6 (3.2–6.5) | 3.4 (2.6–4.6) | 249 (140–445) | 510 (395–658) |
| F268W | 2.0 (1.5–2.5) | 2.2 (1.7–2.7)* | 21.0 (18.3–24.2)* | 148 (87.4–251)* | 1857 (841–4101)* | 1829 (1296–2579)* |
| [WT:F268W]a | [1:1] | [1:3] | [1:5] | [1:44] | [1:7] | [1:4] |
| P269A | 2.1 (1.7–2.7) | 0.88 (0.70–1.1) | 3.0 (2.7–3.4) | 68.6 (48.5–97.1)* | 341 (275–422) | 2030 (1193–3455)* |
| [WT:P269A]a | [1:1] | [1:1] | [1:1] | [1:20] | [1:1] | [1:4] |
| H270A | 4.0 (2.8–5.8)* | 1.2 (0.89–1.7)* | 6.1 (5.2–7.3) | 40.2 (28.7–56.1)* | 412 (348–487) | 1999 (1609–2484)* |
| [WT:H270A]a | [1:2] | [1:2] | [1:1] | [1:12] | [1:2] | [1:4] |
| I271A | 8.6 (7.4–10.1)* | 1.8 (1.4–2.3)* | 11.2 (9.8–12.9)* | 166 (119–229)* | 2872 (1543–5348)* | 2825 (2516–3173)* |
| [WT:I271A]a | [1:5] | [1:3] | [1:2] | [1:49] | [1:12] | [1:6] |
Data are the median and corresponding 95% confidence limits of three independent experiments performed in duplicate. Ki values for each ligand were determined using 4 nM [3H]SR141716A.
Statistically significant differences from wild-type (P<0.05) using analysis of variance followed by Dunn's post hoc test.
Values represent the Ki ratio for WT to EC2 mutant receptors for the specified compound.
To probe whether the cannabinoid agonists CP55940 and HU-210 previously found to bind receptors F268W, P269A, H270A, and I271A, with attenuated affinity [Ahn et al., 2009] did so as a result of a shift in the receptor toward the inactive state, we determined the affinity of the “silent” antagonist O-2050 for these EC2 mutant receptors (Table 1). The structure of O-2050 is based on a tricyclic ring structure more similar to a classical CB receptor agonist than a diapyrazole inverse agonist such as SR141716A. However, it is considered devoid of any agonism at the CB1 receptor, likely attributable to the introduction of a triple bond in its alkyl side chain reducing the chain flexibility potentially needed for CB1 activation by classical and nonclassical agonists [Griffin et al., 1999; Reggio, 2003]. O-2050, which should bind the inactive and active forms of the CB1 receptor with comparable affinity, was found to bind to F268W, P269A, H270A, and I271A receptors with substantially reduced affinity (44-, 20-, 12-, and 49-fold, respectively) relative to the wild-type receptor (Ki = 3.4 nM). This suggests that binding the tricyclic ring structure and/or the bulky alkyl chain require the conformation and/or contacts of the wild-type EC2 C-terminus and that the mutants in this region are not simply shifted to the inactive state.
To examine the affinity of the EC2 mutant receptors for other structurally diverse agonists, BAY593074 (wild-type receptor Ki = 249 nM), and WIN55212-2, an aminoalkylindole (wild-type receptor Ki 5 510 nM), were tested. The binding affinity was diminished relative to that of the wild-type receptor (up to 12-fold for BAY593074 and up to 6-fold for WIN55212-2) but not as substantially as with O-2050. Thus, binding the classical and nonclassical agonists (and their analogues), relative to these agonists, seems more sensitive to alterations in the composition of CB1 EC2. Interestingly, both WIN55212-2 and SR141716A are thought to form aromatic stacking interactions critical for high-affinity ligand binding [McAllister et al., 2003] and mutations that inhibit the affinity of one ligand typically also decrease the other's affinity for the receptor [McAllister et al., 2003]. Here, the C-terminal EC2 mutations do not affect SR141716A binding affinity and although they decrease the affinity of WIN55212-2 to the receptor, the attenuation is not as severe as to the classical and nonclassical agonists.
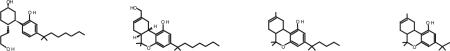
To gain insight on whether the length of the alkyl side chain of CB receptor agonists was a factor in the attenuation of binding affinity, we tested the effect of shortening the alkyl side chain of the classical cannabinoid receptor agonist structure. The 5-carbon-long dimethylpentyl Δ8-THC (JWH061) chain is shorter than the 7-carbon-long dimethylheptyl side chain common to the nonclassical compound CP55940 and on the classical compound HU-210. This is reduced by 3 carbons to yield dimethyl Δ8-THC (JWH179) containing a 2-carbon-long chain [Huffman et al., 2003]. Shortening the alkyl side chain did not attenuate the loss of binding affinity for any of the EC2 mutants and the fold-loss was found to be comparable to the reduction in binding observed for CP55940 and HU-210 to the F268W, P269A, H270A, and I271A receptors relative to the wild-type CB1 receptor (Table 2). These data suggest the length of the alkyl chain as configured on the Δ8-THC series was not the sole determinant of the diminished agonist binding affinity and underscores the need for the C-terminus EC2 residues in accommodating the bicyclic/tricyclic core.
TABLE 2.
Effects of Truncating the Alkyl Side Chain of Dimethylpentyl Δ8-THC on Ligand Binding Affinity to Cannabinoid Receptors With Mutations in Positions 268–271 in EC2
 |
||||
|---|---|---|---|---|
|
Ki (nM) |
||||
| Receptor | CP55940a | HU-210a | JWH061 | JWH179 |
| WT | 24.8 (14.6–42.3) | 3.7 (2.1–6.5) | 29.9 (18.5–48.3) | 7.3 (5.4–9.8) |
| F268W | 8132 (5793–11421)* | 1672 (757.9–3689)* | 3459 (2619–4568)* | 1112 (922–1342)* |
| [WT:F268W]b | [1:328] | [1:452] | [1:116] | [1:152] |
| P269A | 2081 (1256–3446)* | 22.6 (10.3–49.5)* | 1728 (1414–2111)* | 798 (620–1027)* |
| WT:P269Ab | [1:84] | [1:6] | [1:58] | [1:109] |
| H270A | 188 (152–232)* | 13.8 (9.4–20.1)* | 373 (226–618)* | 90.0 (79.3–102)* |
| WT:H270Ab | [1:8] | [1:4] | [1:12] | [1:12] |
| I271A | 3306 (1422–7686)* | 310 (117–822)* | 773 (623–958)* | 194 (158–240)* |
| WT:I271Ab | [1:133] | [1:84] | [1:26] | [1:27] |
Data are the median and corresponding 95% confidence limits of three independent experiments performed in duplicate. Ki values for each ligand were determined using 4 nM [3H]SR141716A.
Statistically significant differences from wild-type (P<0.05) using analysis of variance followed by Dunn's post hoc test.
Ki values from Ahn et al. [2009], except for H270A, which was obtained here.
Values represent the Ki ratio for WT to EC2 mutant receptors for the specified compound.
To further probe the impact of the alkyl side chain on binding affinity to the EC2 receptor mutants, a series of SR141716A derivatives with an alkyl chain linked to different constituents was examined (Table 3). Thus, the bulk of these compounds resembles SR141716A that bound the mutant receptors with wild-type like affinity (Ki = 5.2 nM). All three compounds (O-1302, O-1690, and O-1255) were bound by the mutant receptors (P269A and H270A) like the wild-type (O-1302, Ki = 1.7 nM; O-1690, Ki = 1.3 nM; O-1255, Ki = 83.4 nM). In contrast, the F268W and I271A receptors displayed significantly diminished affinity for O-1302 and O-1690, though by only 4- and 6-fold, respectively, for O-1255. This suggests that the position of the alkyl chain relative to the cyclic core of the molecule impacts binding affinity and thus contributes to the loss of binding observed for CP55940 and HU-210 especially by the F268W and I271A receptors.
TABLE 3.
Ki Values for Binding SR141716A Analogues to the CB1 Receptors With Mutations in Positions 268–271 in EC2
 |
||||
|---|---|---|---|---|
|
Ki (nM) |
||||
| Receptor | SR141716Aa | O-1302 | O-1690 | O-1255 |
| WT | 5.2 (3.4–8.1) | 1.7 (1.2–2.3) | 1.3 (0.8–2.0) | 83.4 (49.7–139.9) |
| F268W | 7.3 (6.4–8.3) | 26.2 (20.2–34.1)* | 23.3 (16.9–32.0)* | 310.2 (242.8–396.2)* |
| [WT:F268W]b | [1:1] | [1:15] | [1:18] | [1:4] |
| P269A | 5.3 (3.8–7.3) | 1.1 (0.82–1.5) | 0.96 (0.71–1.3) | 74.7 (28.5–195.8) |
| [WT:P269A]b | [1:1] | [1:1] | [1:1] | [1:1] |
| H270A | 3.5 (2.5–5.0) | 2.6 (1.6–4.2) | 2.0 (1.4–2.8) | 113 (87.0–147) |
| [WT:H270A]b | [1:1] | [1:1] | [1:1] | [1:1] |
| I271A | 2.4 (1.9–2.9) | 38.4 (33.2–44.6)* | 77.3 (72.0–82.9)* | 460 (361–587)* |
| [WT:I271A]b | [2:1] | [1:23] | [1:59] | [1:6] |
Data are the median and corresponding 95% confidence limits of three independent experiments performed in duplicate. Ki values for each ligand were determined using 4 nM [3H]SR141716A.
Statistically significant differences from wild-type (P<0.05) using analysis of variance followed by Dunn's post hoc test.
Ki values from Ahn et al. [2009], except for H270A, which was obtained here.
Values represent the Ki ratio for WT to EC2 mutant receptors for the specified compound.
DISCUSSION
Taken together, these data suggest the C-terminus of CB1 EC2 participates in ligand binding and is particularly important for accommodating both the bicyclic/tricyclic core and alkyl side chain of the classical and nonclassical cannabinoids. Data from binding isotherms to the O-1255 series further suggest that the composition and location of both components are problematic as the alkyl chain in a different orientation on O-1255 was bound by the receptors without the substantial loss in affinity observed for CP55940 and HU-210. Moreover, the JWH061 and JWH179 series, with shorter alkyl side chains, displayed binding loss comparable to that for CP55940 and HU-210.
We find that mutation of F268 and I271 yield receptors that are especially sensitive to ligand binding, while mutation of the two residues internal to these, P269 and H270, yielded receptors that retained wild-type receptor like affinity for many compounds tested. This is consistent with the notion that the F268 and I271 residues project more deeply into the ligand binding pocket and/or form a hydrophobic lid on the ligand [Ahn et al., 2009].
In addition to the classical and nonclassical cannabinoids, we have previously shown that mutations in the C-terminus of EC2 display substantially reduced binding affinity for methanandamide, the C-2 methylated anandamide analogue. Interestingly, McAllister et al. [2003] showed the alanine substitution at F190 (corresponding to F189 in human CB1) in mouse CB1 extracellular loop 1 (EC1) resulted in a 6-fold loss in anandamide binding. Together with computational modeling, the results suggest that F189 is a key residue for anandamide binding as a CH-π interacts with the C5–C6 double bond of anandamide. Based on our previous molecular modeling, we have proposed an intramolecular interaction between F268 and two residues from EC1, F189, and H178. It is possible that the mutations in the C-terminus of EC2 influence the interaction with EC1, resulting in impaired methanandamide binding. However, we cannot rule out the possibility that the C-terminus of EC2 plays a role in agonist-specific receptor activation. In this study, we also demonstrate that the C-terminal EC2 mutant receptors exhibited reduction in binding affinity for the aminoalkylindole agonist, WIN55212-2 though not as markedly as for other classes of agonists. It is believed that the binding pocket for WIN55212-2 overlaps that of SR141716A. High affinity binding for both ligands appears to require a network of aromatic residues for receptor stabilization and/or aromatic stacking interaction with ligands.
The Cys-X-X-X-Ar (where Ar is aromatic) motif of EC2 that is conserved among GPCRs that bind biogenic amines and peptides is also found in CB1 with F268 serving as the aromatic residue. In other receptors, the aromatic residue is thought to contribute indirectly [Cherezov et al., 2007] or directly [Lim et al., 2008; Palczewski et al., 2000] to ligand binding. For example, in the β2-adrenergic receptor, the analogous residue phenylalanine 194 participates in hydrophobic interactions thought critical for stabilizing phenylalanine 193 for its interaction with ligand [Cherezov et al., 2007; Rosenbaum et al., 2007]. A more direct role for this residue is observed in bovine rhodopsin in which the corresponding residue, tyrosine 191, is part of the β4 strand that contributes to the chromophore binding pocket [Palczewski et al., 2000]. In addition, F209, the corresponding residue in the V1a vasopressin receptor is thought to project into the binding crevice and mutation of this residue impairs ligand binding and intracellular signaling [Conner et al., 2007]. A recent reporter cysteine accessibility mapping (RCAM) study suggests the EC2 residues, especially among the C-X-X-X-Ar motif, are responsible for a conformational change upon the agonist binding [Unal et al., 2010].
Although the CB1 receptor belongs to the class A GPCR family, the CB1 EC2 loop is somewhat unusual relative to other receptors in this class. It is short (18 residues), whereas the EC2 loops of the adrenergic receptors (about 25 residues) and rhodopsin (about 26 residues) are significantly longer. CB1 has an EC2 intraloop disulfide bond and no disulfide linkage between EC2 and TM3. Moreover, receptor modeling [Ahn et al., 2009] suggests that the CB1 EC2 adopts a small helical segment, while other receptors such as rhodopsin exhibit an antiparallel β-hairpin structure. Whereas the binding pocket for most ligands is thought to reside within the TM domain, it appears that CB1 EC2, especially F268 and I271, is highly organized to assist in forming the ligand binding site. Any therapeutic strategy targeting CB1 will require consideration of the conformation of this region.
ACKNOWLEDGMENTS
The authors thank Dr. John Huffman for providing JWH061 and JWH179 (grant DA-03590 to J.W.H.), and Drs. Jenny Wiley and Anu Mahadevan (Organix) for the Organix (O-series) compounds (grants DA-03672 to J.L.W. and DA-05488 to A.M.). We also thank Dr. Jenny Wiley for her careful reading of the manuscript.
Grant sponsor: National Institutes of Health; Grant numbers: DA-020763; DA-03590; DA-03672; DA-05488.
Abbreviations used
- AVE1625
N-{1-[bis(4-chloro-phenyl)-methyl]azetidin-3-yl}-N-(3,5-difluorophenyl)methanesulfonamide
- BAY593074
3-[2-cyano-3-(trifluoromethyl)phenoxy]phenyl-4,4,4-trifluoro-1-butanesulfonate
- CB1
cannabinoid receptor 1
- CP55940
(1R,3R,4R)-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-4-(3-hydroxypropyl)cyclohexan-1-ol
- EC2
extracellular loop 2
- GPCR
G-protein-coupled receptor
- HEK293T
human embryonic kidney cell
- HU-210
(−)-11-hydroxydimethylheptyl-Δ8-tetrahydrocannabinol
- JWH061
3-(1′,1′-dimethylpentyl)-Δ;8-tetrahydrocannabinol
- JWH179
3-(1′,1′-dimethylethyl)-Δ8-tetrahydrocannabinol
- NESS0327
N-piperidinyl-[8-chloro-1-(2,4-dichlorophenyl)-1, 4,5,6 - xtetrahydrobenzo [6,7]cyclohepta[1, 2 - c]pyrazole - 3 -carboxamide
- O - 1255
[I] N [/I] - pentylphenyl - 5 - (4-chloro-phenyl)-3- ([I]N[/I]-piperidin-1-yl)carboxamide-4-methyl-pyrazole
- O-1302
[I]N[/I]-(piperidin-1-yl)-1-(2,4-dichlorophenyl)-4-methyl-5-(4-pentylphenyl)-1H-pyrazole-3-carboxamide
- O-1690
N-(N-piperdyl)-3-(2,4-dichlrophenyl)-4-(4′-(1′-methylpentyl)phenyl)-5-methylpyrazole O-2050, (6aR,10aR)-3-(1-methanesulfonylamino-4-hexyn-6-yl)-6a,7, 10,10a-tetrahydro-6,6,9-trimethyl-6H-dibenzo[b,d]pyran
- PBS
phosphate-buffered saline
- SLV319
4S-(−)-3-(4-chloroenyl) - N - methyl - N′ - [ (4 - chlorophenyl) sulfonyl] -4 - phenyl - 4, 5-dihydro-1H-pyrazole-1-caboxamidine
- SR141716A
N-(piperidin-1-yl)-5-(4- chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3- carboxamide
- THC
tetrahydrocannabinol
- TME
Tris/Mg2+/EDTA
- WIN 55212-2
(R)-(+)-[2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de)-1,4-benzoxazin-6-yl]-1-napthalenylmethanone
REFERENCES
- Abadji V, Lucas-Lenard JM, Chin C, Kendall DA. Involvement of the carboxyl terminus of the third intracellular loop of the cannabinoid CB1 receptor in constitutive activation of Gs. J Neurochem. 1999;72:2032–2038. doi: 10.1046/j.1471-4159.1999.0722032.x. [DOI] [PubMed] [Google Scholar]
- Ahn KH, Bertalovitz AC, Mierke DF, Kendall DA. Dual role of the second extracellular loop of the cannabinoid receptor 1: ligand binding and receptor localization. Mol Pharmacol. 2009;76:833–842. doi: 10.1124/mol.109.057356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhaus DW, Chang LK, Kwan J, Martin GR. Dual activation and inhibition of adenylyl cyclase by cannabinoid receptor agonists: evidence for agonist-specific trafficking of intracellular responses. J Pharmacol Exp Ther. 1998;287:884–888. [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CN, Lucas-Lenard J, Abadji V, Kendall DA. Ligand binding and modulation of cyclic AMP levels depend on the chemical nature of residue 192 of the human cannabinoid receptor 1. J Neurochem. 1998;70:366–373. doi: 10.1046/j.1471-4159.1998.70010366.x. [DOI] [PubMed] [Google Scholar]
- Conner M, Hawtin SR, Simms J, Wootten D, Lawson Z, Conner AC, Parslow RA, Wheatley M. Systematic analysis of the entire second extracellular loop of the V(1a) vasopressin receptor: key residues, conserved throughout a G-protein-coupled receptor family, identified. J Biol Chem. 2007;282:17405–17412. doi: 10.1074/jbc.M702151200. [DOI] [PubMed] [Google Scholar]
- Devane WA, Dysarz FA, III, Johnson MR, Melvin LS, Howlett AC. Determination and characterization of a cannabinoid receptor in rat brain. Mol Pharmacol. 1988;34:605–613. [PubMed] [Google Scholar]
- Fay JF, Dunham TD, Farrens DL. Cysteine residues in the human cannabinoid receptor: only C257 and C264 are required for a functional receptor, and steric bulk at C386 impairs antagonist SR141716A binding. Biochemistry. 2005;44:8757–8769. doi: 10.1021/bi0472651. [DOI] [PubMed] [Google Scholar]
- Glass M, Felder CC. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: evidence for a Gs linkage to the CB1 receptor. J Neurosci. 1997;17:5327–5333. doi: 10.1523/JNEUROSCI.17-14-05327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin G, Wray EJ, Rorrer WK, Crocker PJ, Ryan WJ, Saha B, Razdan RK, Martin BR, Abood ME. An investigation into the structural determinants of cannabinoid receptor ligand efficacy. Br J Pharmacol. 1999;126:1575–1584. doi: 10.1038/sj.bjp.0702469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- Huffman JW, Liddle J, Yu S, Aung MM, Abood ME, Wiley JL, Martin BR. 3-(1′,1′-Dimethylbutyl)-1-deoxy-delta8-THC and related compounds: synthesis of selective ligands for the CB2 receptor. Bioorg Med Chem. 1999;7:2905–2914. doi: 10.1016/s0968-0896(99)00219-9. [DOI] [PubMed] [Google Scholar]
- Huffman JW, Miller JR, Liddle J, Yu S, Thomas BF, Wiley JL, Martin BR. Structure-activity relationships for 1′,1′-dimethylalkyl-Delta8-tetrahydrocannabinols. Bioorg Med Chem. 2003;11:1397–1410. doi: 10.1016/s0968-0896(02)00649-1. [DOI] [PubMed] [Google Scholar]
- Hurst D, Umejiego U, Lynch D, Seltzman H, Hyatt S, Roche M, McAllister S, Fleischer D, Kapur A, Abood M, et al. Biarylpyrazole inverse agonists at the cannabinoid CB1 receptor: importance of the C-3 carboxamide oxygen/lysine3.28(192) interaction. J Med Chem. 2006;49:5969–5987. doi: 10.1021/jm060446b. [DOI] [PubMed] [Google Scholar]
- Hurst DP, Lynch DL, Barnett-Norris J, Hyatt SM, Seltzman HH, Zhong M, Song ZH, Nie J, Lewis D, Reggio PH. N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-p yrazole-3-carboxamide (SR141716A) interaction with LYS 3.28(192) is crucial for its inverse agonism at the cannabinoid CB1 receptor. Mol Pharmacol. 2002;62:1274–1287. doi: 10.1124/mol.62.6.1274. [DOI] [PubMed] [Google Scholar]
- Janero DR, Makriyannis A. Cannabinoid receptor antagonists: pharmacological opportunities, clinical experience, and translational prognosis. Expert Opin Emerg Drugs. 2009;14:43–65. doi: 10.1517/14728210902736568. [DOI] [PubMed] [Google Scholar]
- Kapur A, Hurst DP, Fleischer D, Whitnell R, Thakur GA, Makriyannis A, Reggio PH, Abood ME. Mutation studies of Ser7.39 and Ser2.60 in the human CB1 cannabinoid receptor: evidence for a serine-induced bend in CB1 transmembrane helix 7. Mol Pharmacol. 2007;71:1512–1524. doi: 10.1124/mol.107.034645. [DOI] [PubMed] [Google Scholar]
- Lauckner JE, Hille B, Mackie K. The cannabinoid agonist WIN55,212-2 increases intracellular calcium via CB1 receptor coupling to Gq/11 G proteins. Proc Natl Acad Sci USA. 2005;102:19144–19149. doi: 10.1073/pnas.0509588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HD, Jongejan A, Bakker RA, Haaksma E, de Esch IJ, Leurs R. Phenylalanine 169 in the second extracellular loop of the human histamine H4 receptor is responsible for the difference in agonist binding between human and mouse H4 receptors. J Pharmacol Exp Ther. 2008;327:88–96. doi: 10.1124/jpet.108.140343. [DOI] [PubMed] [Google Scholar]
- Mackie K, Hille B. Cannabinoids inhibit N-type calcium channels in neuroblastoma-glioma cells. Proc Natl Acad Sci USA. 1992;89:3825–3829. doi: 10.1073/pnas.89.9.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- McAllister SD, Rizvi G, Anavi-Goffer S, Hurst DP, Barnett-Norris J, Lynch DL, Reggio PH, Abood ME. An aromatic microdomain at the cannabinoid CB1 receptor constitutes an agonist/inverse agonist binding region. J Med Chem. 2003;46:5139–5152. doi: 10.1021/jm0302647. [DOI] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. Emerging strategies for exploiting cannabinoid receptor agonists as medicines. Br J Pharmacol. 2009;156:397–411. doi: 10.1111/j.1476-5381.2008.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggio PH. Pharmacophores for ligand recognition and activation/inactivation of the cannabinoid receptors. Curr Pharm Des. 2003;9:1607–1633. doi: 10.2174/1381612033454577. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Salo OM, Lahtela-Kakkonen M, Gynther J, Jarvinen T, Poso A. Development of a 3D model for the human cannabinoid CB1 receptor. J Med Chem. 2004;47:3048–3057. doi: 10.1021/jm031052c. [DOI] [PubMed] [Google Scholar]
- Song ZH, Bonner TI. A lysine residue of the cannabinoid receptor is critical for receptor recognition by several agonists but not WIN55212-2. Mol Pharmacol. 1996;49:891–896. [PubMed] [Google Scholar]
- Thomas BF, Gilliam AF, Burch DF, Roche MJ, Seltzman HH. Comparative receptor binding analyses of cannabinoid agonists and antagonists. J Pharmacol Exp Ther. 1998;285:285–292. [PubMed] [Google Scholar]
- Unal H, Jagannathan R, Bhat MB, Karnik SS. Ligand-specific conformation of extracellular loop-2 in the angiotensin II type 1 receptor. J Biol Chem. 2010;285:16341–16350. doi: 10.1074/jbc.M109.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley JL, Jefferson RG, Grier MC, Mahadevan A, Razdan RK, Martin BR. Novel pyrazole cannabinoids: insights into CB1 receptor recognition and activation. J Pharmacol Exp Ther. 2001;296:1013–1022. [PubMed] [Google Scholar]



