Abstract
Tumourigenesis is a multistep process that results from the sequential accumulation of mutations in key oncogene and tumour suppressor pathways. Personalized cancer therapy that is based on targeting these underlying genetic abnormalities presupposes that sustained inactivation of tumour suppressors and activation of oncogenes is essential in advanced cancers. Mutations in the p53 tumour-suppressor pathway are common in human cancer and significant efforts toward pharmaceutical reactivation of defective p53 pathways are underway1–3. Here we show that restoration of p53 in established murine lung tumours leads to significant but incomplete tumour cell loss specifically in malignant adenocarcinomas but not in adenomas. We define amplification of MAPK signaling as a critical determinant of malignant progression and also a stimulator of Arf tumour-suppressor expression. The response to p53 restoration in this context is critically dependent on the expression of Arf. We propose that p53 not only limits malignant progression by suppressing the acquisition of alterations that lead to tumour progression, but also, in the context of p53 restoration, responds to increased oncogenic signaling to mediate tumor regression. Our observations also underscore that the p53 pathway is not engaged by low levels of oncogene activity that are sufficient for early stages of lung tumour development. These data suggest that restoration of pathways important in tumour progression, as opposed to initiation, may lead to incomplete tumour regression due to the stage-heterogeneity of tumour cell populations.
Experimental restoration of p53 tumour-suppressor function has highlighted the potential for therapeutic intervention of this pathway to treat cancer and has uncovered a diversity of anti-tumour responses in different tumour types4–6. These studies imply that some types of cancer rely upon sustained inactivation of the p53 pathway as these tumours undergo rapid, and sometimes complete, regression once p53 is restored. To elucidate the role of p53 in lung cancer progression and to evaluate the therapeutic potential of p53 reactivation in this disease, we analyzed the effect of Trp53 restoration in a mouse model of KrasG12D driven lung cancer at different time points in tumor progression. Trp53 restoration was achieved by delivering tamoxifen to four-week old KrasLA2/+;Trp53LSL/LSL;Rosa26CreERT2 mice (Supplementary Fig. 1). We measured tumour volumes with micro-computed tomography (μCT) and tumour areas by histological analysis. Surprisingly, at this time point, Trp53 restoration did not significantly change tumour size or number (Fig. 1a, b). Efficient deletion of the `STOP' element from Trp53LSL alleles was evident in all tumours analyzed (Supplementary Fig. 2).
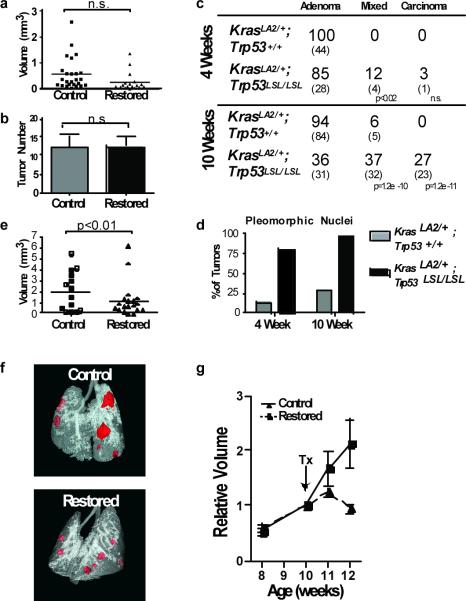
Figure 1. Lung adenomas are apathetic to Trp53 restoration, whereas adenocarcinomas are responsive.
a, Individual tumour volumes were determined by μCT two weeks after Trp53-restoration that began at four weeks of age. b, Histological sections were evaluated for tumor number in control (n=10) and restored (n=12) mice. c, Tumour number and grade at four and ten weeks of age. Individual tumours from KrasLA2/+;Trp53LSL/LSL (n=5) and KrasLA2/+;Trp53+/+ (n= 4) mice. The percentage and number (in parentheses) of tumours is indicated. d, Pleomorphic nuclei in tumour samples from c (see also Supplemental Fig. 5). e, Restoration of Trp53 in ten-week old animals results in significantly diminished tumour size. Tumour volumes determined by μCT two weeks after p53 restoration. f, Representative tomograms from μCT analysis at 12 weeks. g, Serial μCT analysis of individual tumours. Average relative size and standard error of tumours are plotted.
To establish the timing and stage of tumour progression when the effects of p53 mutation are evident, we compared KrasLA2/+;Trp53LSL/LSL tumours to tumours from age-matched KrasLA2/+;Trp53+/+ mice. At four weeks, most tumours were adenomatous hyperplasias or alveolar adenomas with monomorphous epithelial cells and uniform nuclei (Supplementary Fig. 3). In KrasLA2/+;p53LSL/LSL mice at this age, a small fraction of tumours had mixed histological features with focal areas of adenocarcinoma within lower grade lesions (Fig. 1c). Though most tumours in KrasLA2/+;Trp53+/+ and KrasLA2/+;Trp53LSL/LSL mice were of similar grade, the presence of pleomorphic nuclei was a prominent feature of KrasLA2/+;p53LSL/LSL tumours compared to KrasLA2/+;Trp53+/+ tumours (Fig. 1d). This observation may indicate that KrasLA2/+;p53LSL/LSL tumour cells have a higher degree of genetic instability that may facilitate tumour progression at a later time point.
At ten weeks of age, KrasLA2/+;Trp53LSL/LSL tumours were much more likely to exhibit nuclear pleomorphism and regions of advanced histopathology. Beyond these mixed grade tumors, frank adenocarcinomas were present exclusively in KrasLA2/+;Trp53LSL/LSL mice (Fig. 1c). Given that high-grade tumours were more prevalent in older mice, we restored Trp53 in ten-week old KrasLA2/+;Trp53LSL/LSL;Rosa26CreERT2 mice. Here, restoration of Trp53 resulted in significantly smaller tumours compared to controls after two weeks (Fig. 1e, f). To monitor the response of individual tumours, we performed serial μCT before and after Trp53 restoration. Although control tumours continued to grow during the two-week time course, we found that the average size of tumours after Trp53 restoration was static (Fig. 1g). However, the response of individual tumours was variable (Supplementary Fig. 4). Thus, we hypothesized that only a subset of tumours is sensitive to Trp53 restoration.
Upon Trp53 restoration, adenocarcinomas, but not adenomas, displayed multiple features of cellular senescence. Both p53 and the p53 target gene p21 were detectable in tumours as early as three days after restoration, but were specifically confined to tumours with adenocarcinomatous features (Fig. 2b and Supplementary Fig. 5). In addition to observing p21 induction in the adenocarcinomas of ten-week old mice (Fig. 2c), p21 was also induced in the few carcinomas found in the four-week old animals (data not shown). Moreover, Trp53 restoration caused reduced cellular proliferation (Fig. 2d). Of note, the percentage of tumours with adenocarcinomatous features was significantly diminished two weeks after Trp53 restoration with many tumours showing gaps between collections of low-grade tumor cells filled with foam macrophages (Fig. 2a, e). Despite this evidence of cell loss, we failed to find signs of apoptosis in lung tumours across an extensive time course after Trp53 restoration (Supplementary Fig. 6). Collectively, these data demonstrate that adenomas do not respond to p53 restoration, while adenocarcinomas are sensitive. Moreover, Trp53 mutation per se does not induce phenotypic changes in tumour grade but rather creates a permissive environment for the outgrowth of cells that have undergone additional steps in tumour progression. Finally, the alterations that drive tumour progression can also stimulate p53-dependent cellular responses.
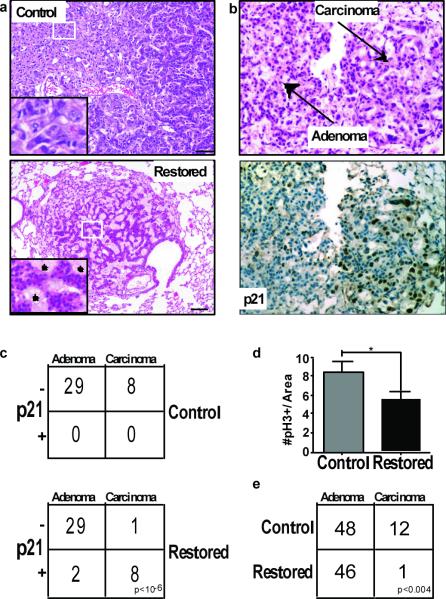
Figure 2. Adenocarcinoma cells are sensitive to Trp53 restoration and are specifically eliminated from lung tumours.
a, Histological sections from control and Trp53-restored tumours stained with hemotoxylin and eosin (H&E). Inset of Trp53-restored tumours show monomorphic nuclei and tumour fissures filled with foam macrophages (asterisks). Scale bars equal 50 microns. b, Tumour sections three days post Trp53 restoration. H&E stained tumour section (top) with adenoma and carcinoma areas indicated. Serial section stained with p21 antibody (bottom). c, Contingency tables showing coincidence of p21 and adenocarcinoma 3 days post Trp53 restoration. d, Average number and standard deviation of pH3 positive cells/μm2 for each tumor are shown. Trp53 restoration leads to significantly fewer mitoses. e, Tumour grades in control and Trp53 restored animals two weeks after initial treatment (Vehicle treated tumours n= 60, tamoxifen treated tumours n=47).
To interrogate the mechanisms that drive tumour progression, we performed IHC on tumours from KrasLA2/+;Trp53+/+ and KrasLA2/+;Trp53LSL/LSL animals to assess the activity of multiple relevant pathways during the adenoma-adenocarcinoma transition. We found that phosphorylation levels of Mek1/2 (p-Mek) and p42/p44 MAPK (p-Erk) was dramatically higher in adenocarcinomas compared to adenomas (Fig. 3a, b). This observation suggested that amplified MAPK signaling could drive progression of KrasG12D initiated lung cancer. High levels of MAPK signaling downstream of oncogenic Kras induces cell cycle arrest/senescence mediated by Arf and p53 in multiple cell types in vitro7–9. In contrast, physiological levels of oncogenic signaling elicited by a single oncogenic Kras allele do not activate the Arf/p53 tumour suppressor pathway and instead can drive proliferation and tumourigenesis in many cell types10–14. Some cancer models show that while mutation of Ras family members can initiate tumourigenesis, amplification of oncogenic Ras or loss of the wild-type Ras allele is associated with tumour progression15–17. Thus, we hypothesized that amplification of oncogenic signaling could simultaneously drive tumour progression and the outgrowth of cells that had inactivated the p53 pathway during the natural course of tumour evolution. Consistent with this hypothesis, FISH analysis revealed that a subset of adenocarcinomas with high p-Erk had copy number gains of the Kras locus. Conversely, copy number alterations of Kras were never seen in adenomas (Fig 3c). Together, these observations suggest that while the mechanisms that amplify oncogenic signaling are likely diverse, MAPK signal amplification typifies KrasLA2 lung adenocarcinomas and may drive progression toward malignancy.
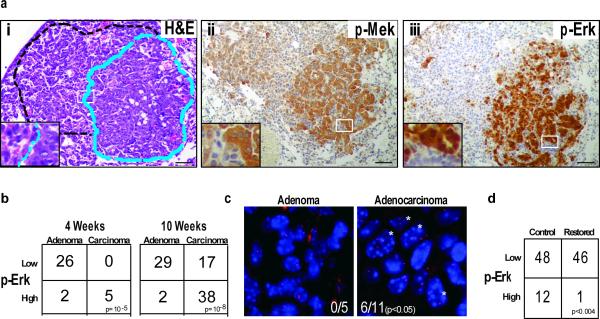
Figure 3. Adenocarcinomas are typified by amplified MAPK signaling.
a, Serial tumour sections of a mixed grade tumour stained with (i) H&E, (ii) anti-p-Mek, or (iii) anti-p-Erk. Dashed lines outline high-grade (blue) and low-grade (black) areas. b, Contingency tables representing number of tumours with high or low MAPK signaling, and the corresponding tumour grades in KrasLA2/+;Trp53+/+ and KrasLA2/+;Trp53LSL/LSL at the four and ten-week time points. c, A subset of tissue sections from a were stained with BAC probes surrounding the Kras locus on mouse chromosome 6. Kras signals (red) and DAPI/DNA (blue) counter stain are shown in an adenoma and a carcinoma. Asterisks indicate nuclei with greater than two Kras signals. d, Contingency plot of tumours with high p-Erk staining shows significantly fewer tumours with high p-Erk staining after Trp53 restoration.
To test whether cells with amplified MAPK signaling persist after Trp53 restoration, we analyzed KrasLA2/+;Trp53LSL/LSL;Rosa26CreERT2 animals across a time course after tamoxifen administration. Loss of high p-Erk staining cells was evident one week after Trp53 restoration (Supplementary Fig. 7). Compared to controls, tumours with Trp53 restored showed significantly fewer p-Erk positive tumours after two weeks (Fig 3d). This observation, in conjunction with the selective loss of histologically advanced tumour cells seen after Trp53 restoration, suggests that amplified MAPK signaling is incompatible with p53 expression.
Next, we micro-dissected tumours from KrasLA2/+;Trp53LSL/LSL mice and performed gene expression analysis to better characterize the molecular alterations that drive tumour progression. We developed an “adenocarcinoma signature” by comparing the gene expression profiles of adenocarcinomas and adenomas (Fig. 4a). Gene set enrichment analysis revealed that pathways associated with increased proliferation such as cell cycle processes, the Ras pathway, and c-myc target genes were enriched in adenocarcinomas compared to adenomas (Fig. 4b). Other gene sets associated with adenocarcinomas were chemokine regulation, inflammation, immune responses, as well as AP1 and PEA3 target genes. These data indicate that multiple pathways are enhanced upon progression to malignancy coincident with, or as a result of, amplified MAPK signaling.
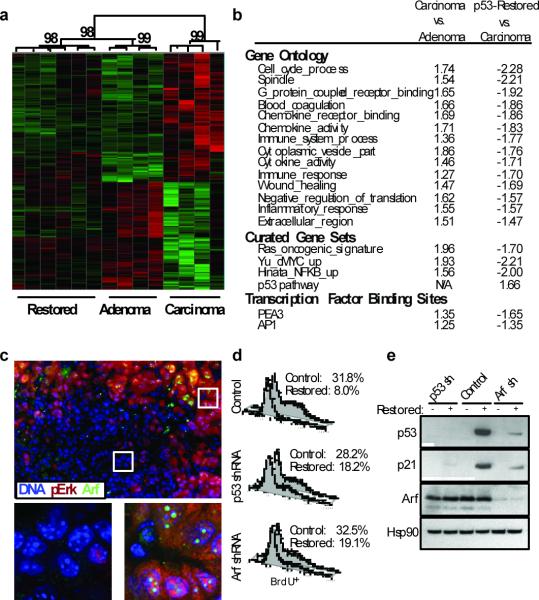
Figure 4. Arf is specifically expressed in adenocarcinomas and sensitizes lung cancer cells to Trp53 restoration.
a, Hierarchical clustering of samples based on the Adenocarcinoma Signature. Confidence values (percent) are indicated at the top of major clades. b, GSEA analysis. Notable gene sets are listed with normalized enrichment scores for each comparison. Positive and negative enrichment scores indicate correlation and anti-correlation respectively. c, Tissue sections from KrasLA2/+;Trp53LSL/LSL adenomas, mixed-grade tumors, and adenocarcinomas were co-labeled with antibodies to p-Erk (red) and Arf (green). Six adjacent fields of view of a mixed-grade tumour (top), and deconvoluted z-stack images (bottom) of dashed regions of an adenoma area (left) and adenocarcinoma area (right) are shown. d, Proliferation assay in adenocarcinoma-derived cell lines. Knockdown with retrovirally expressed shRNAs to Trp53 or Cdkn2a (Arf-specific transcript), or control cells 48-hours post Trp53 restoration. Percentage of BrdU+ cells is indicated. e, Adenocarcinoma cell lysates from d were subjected to immunoblot analysis for p53, p21, Arf, and Hsp90 (loading control).
To access the response to p53 restoration in adenocarcinomas at the level of gene expression, we profiled micro-dissected tumours from KrasLA2/+;Trp53LSL/LSL;Rosa26CreERT2 mice seven days after Trp53 restoration. Each tumour was histologically analyzed for evidence consistent with a p53 response and the appearance of some adenocarcinoma features (Supplementary Fig. 7). We utilized the adenocarcinoma signature to perform hierarchical clustering on these p53-restored samples together with adenomas and adenocarcinomas. Interestingly, the p53-restored samples clustered with adenomas with high confidence. Additionally, adenocarcinoma specific gene sets were de-enriched in Trp53-restored tumours (Fig. 4a, b). These data are consistent with our observation that adenocarcinoma cells are specifically culled from these tumours leaving behind adenoma cells that are unresponsive to p53 restoration.
Hyperactive MAPK signaling has been shown to induce the transcription of Cdkn2a, leading to expression of Arf, an upstream activator of p539. Consistent with this mechanism of triggering the p53 pathway, we found that Cdkn2a expression was higher in adenocarcinomas compared to adenomas (Supplementary Fig. 8a). Moreover, in tumours from KrasLA2/+;Trp53LSL/LSL mice, p-Erk co-localizes with Arf staining by immunofluorescence and immunohistochemistry specifically in adenocarcinomas and the higher-grade areas of mixed-grade tumours (Fig 4c and Supplementary Fig. 8b, c). To test the functional requirement of Arf in the p53 response, we studied cell lines generated from KrasLA2/+;Trp53LSL/LSL;Rosa26CreERT2 lung adenocarcinomas. As we had observed in vivo, restoration of Trp53 in cell culture led to prolonged cell cycle arrest without appreciable cell death. Further, the gene expression program elicited by Trp53 restoration in these cell lines is similar to that observed in vivo, validating them as a relevant model with which to test Arf dependency for activation of p53 responses after Trp53 restoration (Supplementary Fig. 9). We expressed shRNAs targeted against either an Arf-specific transcript of Cdkn2a or Trp53 in cell lines prior to Trp53 restoration. As shown in Fig. 4d, inhibition of Arf or p53 allowed cells to continue to proliferate following addition of tamoxifen. Furthermore, Arf knockdown kept p53 levels low after Trp53 restoration and limited the induction of the p53 target gene p21 (Fig. 4e and Supplementary Fig. 9c). These data indicate that Arf is required for p53 restoration-induced cell cycle arrest in this system.
Our failure to detect apoptosis following Trp53 restoration stands in contrast to the report of Junttila et al18. The most likely explanation for this discrepancy is the different technologies used to control p53 function. Whereas our method restores a disrupted gene back to its wild-type state and allows otherwise normal p53 expression, Junttila et al. used a p53-ER fusion protein that might be expected to accumulate to high levels in advanced tumors that express Arf. Thus, in that setting, p53-ER may induce more robust responses that include apoptosis when activated by tamoxifen.
Given our results on the requirement of Arf in the p53 response in this setting, one might imagine that Arf and p53 mutations would have equivalent effects in lung cancer progression. However, unlike in KrasLA2/+;Trp53LSL/LSL mice, carcinomas rarely develop in KrasLA2 mice carrying Arf mutations19. Thus, the clear effects of p53 mutation on tumor progression likely reflect additional functions of p53, including its role in responding to DNA damage and other forms of genomic instability20.
The possibility that reactivation of defective p53 pathways will lead to variable and incomplete tumour regression is intriguing and may have important implications for anti-cancer therapies. As a tumour naturally progresses, multiple related, but genetically distinct clones co-evolve within the tumour. Conceptually akin to resistance mutations that are selected after targeted cancer therapies, pharmacological targeting of mutations that drive tumour progression, rather than initiation, may lead to significant pruning of some clones but leave behind others that still have significant potential to progress further in tumourigenesis. While it is generally accepted that the elimination of the most advanced tumour cells within a cancer will be a significant benefit in cancer therapy, strategies to eliminate, or prevent progression of, the less advanced residual tumour cells may be important to allow durable therapeutic responses.
Methods
Mice
Animal studies were approved by the Committee for Animal Care, and conducted in compliance with the Animal Welfare Act Regulations and other federal statutes relating to animals and experiments involving animals and adheres to the principles set forth in the Guide for the Care and Use of Laboratory Animals, National Research Council, 1996 (institutional animal welfare assurance no. A-3125–01). All animals were maintained on a mixed C57BL/6J × 129SvJ strain. KrasLA2, Trp53LSL and Rosa26CreERT2 mice have been described6,14. Tamoxifen (Sigma) was dissolved in corn oil (Sigma) and administered i.p. twice weekly at a 200μg/g total body weight. Lung tumour volumes were acquired from isoflurane-anesthetized mice using eXplore Locus MicroCT imager. Image acquisition was performed using eXplore Scan Control software, using a 45 μm voxel size program, and 3D reconstruction was performed using eXplore Reconstruction Utility software. High-resolution tomograms were generated and individual tumour volumes were measured and calculated using MicroView software (all from GE Healthcare). For serial μCT analysis, the relative size of each tumour was compared to the tumor volume at the ten-week time point.
Statistics
p-values indicated in contingency tables and FISH analysis were established by the Fisher's Exact test. Significance values from tumor volume studies and, proliferation (pH3) analyses were evaluated by Student's two-tailed t-test. Microarray statistics are described below.
Histology
All tissues were fixed in 10% formalin overnight, transferred to 70% ethanol, then embedded in paraffin. Four-micron sections were cut.
Tumor Grading
All histological analyses were assisted by a board certified veterinary pathologist (R.B.). A tumour was scored positive for pleomorphic nuclei if it contained three or more such cells.
Immunohistochemistry
Slides were stained with hemotoxylin and eosin or with antibodies to p53 (Novocastra NCL-p53-CM5p), phosphorylated Mek1/2 (p-Mek), phosphorylated p42/p44 MAPK (p-Erk), cleaved caspase 3 (CC3), phosphorylated histone H3 (pH3) (Cell Signaling Technology), p19/Arf, or p21 (F5) (Santa Cruz Biotech). Antigen retrieval was performed in 10mM sodium citrate pH 6.0 by heating in a pressure cooker for 10 minutes.
Immunofluorescence
Images were captured with a 60X, 1.3 NA objective on a Delta Vision deconvolution microscope (Applied Precision Inc.). Image Z-stacks were deconvoluted with SoftWorx software package, and maximum projections saved as tiff files. TUNEL staining was performed via manufacturers instructions (In Situ Cell Death Kit, Roche).
FISH
BAC clone RP23-188E5 was labeled with Texas Red dye. Hybridizations and probe labeling was performed at the Memorial Sloan Kettering Cancer Center, cytogenetics core facility.
Cell line derivation, retrovirus production, and shRNA cloning
Tumours were isolated from KrasLA2/+;Trp53LSL/LSL;Rosa26CreERT2 animals, dissociated with collagenase and trypsin for one hour at 37°C, and then grown in DME plus 10% fetal bovine serum. Retroviruses were produced by transfection of Phoenix cells (Lipofectamine 2000) and shRNAs were designed and cloned as previously described22. The 97nt oligo used to generate the shRNA targeting the Cdkn2a locus was: 5'-TGC TGT TGA CAG TGA GCG ACG CTC TGG CTT TCG TGA ACA TTA GTG AAG CCA CAG ATG TAA TGT TCA CGA AAG CCA GAG CGC TGC CTA CTG CCT CGG A-3'. Cell cycle analysis was performed according to the APC BrdU Flow Kit (BD Parmingen). Data is representative of 5 cell lines derived from independent tumours. Cell lysates were prepared with RIPA buffer before separation on 4–12% BisTRIS NuPAGE gels (Invitrogen). Mouse p53 protein was detected using NCL-p53-505 (Novocastra). Hsp90 served as a loading control and was detected using mAb (BD Transduction Laboratories). Other antibodies were as for IHC. Data is representative of two independent cell lines sorted by GFP expression (expressed from within the shRNA retrovirus22) for purity.
Gene Expression Analysis
Microarray Analysis
RNA was extracted from micro-dissected adenocarcinomas, adenomas, and Trp53-restored tumours from KrasLA2/+;Trp53LSL/LSL mice using Trizol (Invitrogen). RNA was processed, labeled, and hybridized to Affymetrix 430A_v2 chips according to manufacturer's instructions. Affymetrix data analysis was done using statistical tools provided by the r/Bioconductor projects (http://cran.r-project.org/; http://www.bioconductor.org/). Data import and quality control assessment was done using the packages Affy and AffyPLM23. Data was summarized and normalized using gcRMA24. Differential expression analysis was carried out using limma25. All the R commands, input data and environment-specific details required to duplicate these analyses are located here: http://luria.mit.edu/caw_web/Feldser_supplemental/EtOH TAM/ http://luria.mit.edu/caw_web/Feldser_supplemental/tumours/
Hierarchical Clustering with p-values
The ‘Adenocarcinoma Signature’ consists of 71 genes up in adenocarcinomas and 57 genes up in adenomas (p< 0.05 and Fold Change >1.75). Expression values were row normalized using z-scores and the resulting data were subjected to hierarchical clustering (Ward's method) and a sample dendrogram was produced using the R package pvclust26. The heatmap was generated using Spotfire DecisionSite 9.1.1. The required R commands and the input data are located here: http://luria.mit.edu/caw web/Feldser supplemental/pvclust/
Gene Set Enrichment Analysis (GSEA)
GSEA (http://www.broad.mit.edu/gsea/) was used to examine the distribution of the curated gene sets from the Broad Institute's MsigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp) in lists of genes ordered according to differential expression between various conditions. Positive and negative enrichment scores indicate correlation and anti-correlation respectively. The resulting data and the files required to duplicate these analyses are available here: http://luria.mit.edu/caw_web/Feldser_supplemental/EtOH_TAM_GSEA/ http://luria.mit.edu/caw_web/Feldser_supplemental/tumours_GSEA/
Supplementary Material
Acknowledgements
We would like to thank M. DuPage and C. Kim for critical reading of the manuscript. We are indebted to D. Crowley, E. Vasile, and M. Perry in the Koch Institute Core facilities (supported by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute). We are grateful to M. Luo for microarray support. We thank M. Leversha at MSKCC for FISH. D.M.F. has been supported by the American Cancer Society (New England Area Fellow), and is a current Fellow of the Leukemia and Lymphoma Society. K.K.K. is supported by the John Reed Fund of the MIT undergraduate research program. M.M.W. was a Merck Fellow of the Damon Runyon Cancer Research Foundation and a Genentech postdoctoral fellow. T.J. is the David H. Koch Professor of Biology and a Daniel K. Ludwig Scholar. The Howard Hughes Medical Institute supported this work.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Selivanova G, Wiman KG. Reactivation of mutant p53: molecular mechanisms and therapeutic potential. Oncogene. 2007;26:2243–2254. doi: 10.1038/sj.onc.1210295. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, El-Deiry WS. Restoration of p53 to limit tumor growth. Curr Opin Oncol. 2008;20:90–96. doi: 10.1097/CCO.0b013e3282f31d6f. [DOI] [PubMed] [Google Scholar]
- 4.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007 doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 7.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 8.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 9.Lin AW, et al. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 11.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkisian CJ, et al. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 13.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 15.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 16.Bremner R, Balmain A. Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell. 1990;61:407–417. doi: 10.1016/0092-8674(90)90523-h. [DOI] [PubMed] [Google Scholar]
- 17.Liu ML, et al. Amplification of Ki-ras and elevation of MAP kinase activity during mammary tumor progression in C3(1)/SV40 Tag transgenic mice. Oncogene. 1998;17:2403–2411. doi: 10.1038/sj.onc.1202456. [DOI] [PubMed] [Google Scholar]
- 18.Junttila. Nature. 2010 in press. [Google Scholar]
- 19.Young NP, Jacks T. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci U S A. 2010;107:10184–10189. doi: 10.1073/pnas.1004796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 21.Meylan E, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.