Neutrophil elastase escapes azurophilic granules, translocates to the nucleus, and degrades histones to promote chromatin decondensation necessary for NET formation.
Abstract
Neutrophils release decondensed chromatin termed neutrophil extracellular traps (NETs) to trap and kill pathogens extracellularly. Reactive oxygen species are required to initiate NET formation but the downstream molecular mechanism is unknown. We show that upon activation, neutrophil elastase (NE) escapes from azurophilic granules and translocates to the nucleus, where it partially degrades specific histones, promoting chromatin decondensation. Subsequently, myeloperoxidase synergizes with NE in driving chromatin decondensation independent of its enzymatic activity. Accordingly, NE knockout mice do not form NETs in a pulmonary model of Klebsiella pneumoniae infection, which suggests that this defect may contribute to the immune deficiency of these mice. This mechanism provides for a novel function for serine proteases and highly charged granular proteins in the regulation of chromatin density, and reveals that the oxidative burst induces a selective release of granular proteins into the cytoplasm through an unknown mechanism.
Introduction
Neutrophils are the first line of immune defense (Lekstrom-Himes and Gallin, 2000; Nathan, 2006), and they combat pathogens by phagocytosis, degranulation, and the release of neutrophil extracellular traps (NETs; Brinkmann et al., 2004; Nauseef, 2007; Papayannopoulos and Zychlinsky, 2009). NETs are composed of decondensed chromatin and antimicrobial factors, including neutrophil elastase (NE) and myeloperoxidase (MPO; Brinkmann et al., 2004; Urban et al., 2009), and capture and kill bacteria, fungi, and parasites (Urban et al., 2006; Guimarães-Costa et al., 2009; Ramos-Kichik et al. 2009). NETs are implicated in immune defense, sepsis, and autoimmunity (Clark et al., 2007; Kessenbrock et al., 2009; Papayannopoulos and Zychlinsky, 2009; Hakkim et al., 2010). Mast cells, eosinophils, and plant cells also release DNA, which suggests that this may be a common strategy in immunity (von Köckritz-Blickwede et al., 2008; Yousefi et al., 2008; Wen et al., 2009).
NE and MPO are stored in azurophilic granules of naive neutrophils (Borregaard and Cowland, 1997; Lominadze et al., 2005). NE is a neutrophil-specific serine protease that degrades virulence factors and kills bacteria (Lehrer and Ganz, 1990; Belaaouaj et al., 2000; Weinrauch et al., 2002). MPO catalyzes the oxidation of halides by hydrogen peroxide (Hazen et al., 1996; Eiserich et al., 1998; Nauseef, 2007). NE and MPO knockout mice are susceptible to bacterial and fungal infections (Belaaouaj et al., 1998; Aratani et al., 1999; Tkalcevic et al., 2000; Gaut et al., 2001; Belaaouaj, 2002). Interestingly, histones are the most abundant NET component and are potent antimicrobials (Hirsch, 1958; Kawasaki and Iwamuro, 2008; Urban et al., 2009).
Isolated human neutrophils release NETs 2–4 h after stimulation with microbes or activators of PKC such as PMA (Fuchs et al., 2007), but respond much faster when activated by platelet cells stimulated with LPS, a process thought to be relevant during sepsis (Clark et al., 2007).
NETs form via a novel form of cell death (Fuchs et al., 2007) that requires the production of reactive oxygen species (ROS). Neutrophils from chronic granulomatous disease patients with mutations in the NADPH oxidase that disrupt ROS production (Clark and Klebanoff, 1978) fail to form NETs (Fuchs et al., 2007; Bianchi et al., 2009). In neutrophils from healthy donors, ROS production is followed by the disassembly of the nuclear envelope. Chromatin decondenses in the cytoplasm and binds to granular and cytoplasmic antimicrobial proteins before NET release. Chromatin decondensation and the association with antimicrobial proteins are two essential steps during NET formation. The molecular mechanism linking ROS production to chromatin decondensation and binding to antimicrobial proteins is unknown.
Here we show that NE is essential to initiate NET formation and that it synergizes with MPO to drive chromatin decondensation. Our findings reveal a novel mechanism to drive massive chromatin decondensation, and provide evidence for a novel pathway that allows granular proteins to leak into the cytoplasm.
Results
Neutrophil extracts promote chromatin decondensation
To identify factors involved in NET formation, we developed a cell-free nuclear decondensation assay using intact nuclei and cytoplasmic extracts from neutrophils and other control cells. Only the neutrophil-derived low-speed supernatant (LSS), containing cytoplasm and granules, decondensed nuclei from neutrophils, peripheral blood mononuclear cells (PBMCs), human leukemia-60 (HL-60), and HeLa cells (Fig. 1, A and B), which indicates that neutrophil LSS contains specific factors that decondense nuclei. Further separation of the LSS into cytoplasmic (high-speed supernatant [HSS]) and membrane/granule (high-speed pellet [HSP]) fractions showed that the decondensation activity partitioned with the HSP (Fig. 1 C). Neutrophils contain azurophilic, specific, and gelatinase granules (Borregaard and Cowland, 1997). The decondensation activity fractionated with the azurophilic granules (Fig. 1 C, fraction 3; Kjeldsen et al., 1994), which suggests that granular factors are not only components of NETs but are also involved in their formation.
Figure 1.
NE cleaves histones and promotes nuclear decondensation in vitro. (A) Nuclei isolated from neutrophils were incubated in buffer or in neutrophil-derived LSS lysates for 120 min at 37°C and labeled with Sytox green. Bar, 10 µm. (B) Neutrophil extracts are sufficient to decondense nuclei from other cell types. Nuclear decondensation of LSS extracts from HL-60 cells differentiated with RA, HeLa cells, PBMCs, and neutrophils were tested with nuclei isolated from neutrophils (Neut). The nuclear area was quantified using ImageJ image processing software. Circles denote the median area and the bars indicate the range of the nuclear area values, calculated from the standard deviation of each dataset. (C) Decondensation activity is stored in azurophilic granules. Decondensation of nuclei treated with buffer, neutrophil LSS, neutrophil HSS, HSP, and the granular subfractions of gelatinase (1), specific (2), and azurophilic (3) granules were purified over a discontinuous Percoll gradient. Nuclei were also incubated with LSS or HSP in the presence or absence of NEi, SLPI, CGi, or ABAH. Inhibitors were used at the indicated concentrations. (D) The purity of gelatinase (1), specific (2), and azurophilic (3) granule fractions was tested by SDS-PAGE immunoblot analysis using antibodies against gelatinase, lactoferrin, MPO, and NE. NE fractionates exclusively with azurophilic granules (3). (E) NEi inhibits the degradation of nuclear H4 in vitro. After 120 min at 37°C, decondensation reactions were passed over a 0.65-µm filter to separate the nuclei from the extract. The total reaction (T), flowthrough (SF), or nuclear (NF) fractions were analyzed by immunoblotting. Top, nuclei in buffer; middle, nuclei in LSS; bottom, nuclei incubated with LSS and 5 µM NEi. (F) NE and MPO translocate to nuclei in vitro. LSS (L), nuclei alone (N), or LSS + nuclei (L+N), were incubated for the indicated time and separated over filters as in E. The levels of H1, H4, NE, and MPO in the nuclear fraction at different time points were analyzed by immunoblotting. (G) A plot of the corresponding nuclear decondensation, measured as in A, for the experiment in F. (H) Purified NE is sufficient to decondense nuclei in vitro. Nuclei were incubated with buffer (B) or 1 µM purified NE (NE) in the absence of MPO (no MPO, open circles) for the indicated duration (0–240 min). In parallel, 1 µM MPO was added to the same samples (+MPO, closed circles). Decondensation was measured by quantifying the nuclear area and was significantly enhanced in the presence of both NE and MPO (closed circles). (I) Purified NE cleaves nuclear histones in vitro. The samples from H along with NE incubated with purified neutrophil histones were resolved by SDS-PAGE and analyzed by immunoblotting. H4 was processively degraded but H2A, H2B, and H3 were only partially degraded by NE. Soluble purified histones were completely degraded. Notably, the addition of 1 µM MPO had no effect on histone degradation by NE (third column). ***, P < 0.0001.
NE is necessary and sufficient for chromatin decondensation
NE and MPO are two of the primary enzymes stored in azurophilic granules and are found in abundance in NETs. Unlike MPO, only NE sedimented exclusively to the azurophilic fraction (Fig. 1 D, lane 3; Lominadze et al., 2005). We tested whether the activity of NE and MPO is required for nuclear decondensation. Chromatin decondensation was blocked by two NE inhibitors, GW311616A (NEi) and serum leukocyte protease inhibitor (SLPI; Macdonald et al., 2001), but not by the MPO inhibitor 4-aminobenzoic acid hydrazide (ABAH; Fig. 1 C; Kettle et al., 1997).
Azurophilic granules store two additional NE-related proteases, proteinase 3 (PR3) and cathepsin G (CG). Using protease-specific chromogenic peptides and histones as substrates, we show that NEi inhibits NE and PR3, but not CG (Fig. S1). SLPI is known to inhibit NE and CG but not PR3 (Rao et al., 1991). Considering the specificity of these inhibitors, these results indicate that PR3 is not sufficient to drive decondensation in vitro. In addition, CG inhibitor I (CGi), a CG-specific inhibitor, did not inhibit nuclear decondensation. Collectively, these data suggest that among these serine proteases, only NE is required for the decondensation of the nucleus.
NE degrades histones to promote nuclear decondensation
Because histones pack DNA, we examined whether histones H3 and H4 are degraded in nuclei treated with LSS. To distinguish between histone degradation and histone release, we separated the soluble unbound proteins (SF) from the nuclear fraction (NF) by filtration. Interestingly, H4, but not H3, was degraded in an NE-dependent manner (Fig. 1 E). We also monitored the kinetics of nuclear localization of NE and MPO in relation to histone degradation and nuclear decondensation. Both NE and MPO were detected in the nuclear fraction within the first 30 min. Notably, H1 was degraded early but decondensation coincided with H4 degradation at 150 min (Fig. 1 F), which suggests that nuclear decondensation is driven primarily by the degradation of core histones (Fig. 1 G). However, H1 may have to be degraded first to allow for the subsequent degradation of core histones.
Moreover, purified NE and PR3, but not CG, promoted nuclear decondensation in vitro (Fig. 1 H, Fig. S1 F, and not depicted). NE degraded H4 processively, whereas the other histones were only partially degraded (Fig. 1 I). Histone degradation was detectable by 30 min and coincided with nuclear decondensation (Fig. 1, H and I). As a control, we showed that NE completely degraded soluble histones purified from neutrophils (Fig. 1 I), which indicates that the pattern of histone degradation depends on chromatin structure and not on the intrinsic susceptibility of histones to this protease. Similarly, purified PR3 cleaved nuclear histones in vitro (Fig. S1 E). In contrast, and consistent with our previous observations, nuclear and purified histones were poor substrates for CG (unpublished data).
To examine whether the degradation of histones is causing chromatin decondensation, we preincubated permeabilized nuclei with antihistone or a control antibody against CD63, a transmembrane granular protein. Nuclear decondensation was reduced by 50% in nuclei incubated with antibodies against H4, and to a lesser extent with antibodies against other core histones (Fig. S2 A), but not by the control antibody. Accordingly, H4 degradation was reduced in nuclei treated with antihistone antibodies (Fig. S2 B).
Interestingly, in a fractionation experiment, we purified H1 as an inhibitor of NET formation (unpublished data). H1 promotes the condensed, closed state of chromatin, which restricts its accessibility (Roche et al., 1985; Bustin et al., 2005; Woodcock et al., 2006). Nuclei pretreated or coincubated with H1 were resistant to decondensation and histone degradation by LSS and HSP (Fig. S2, C–E). Moreover, MPO failed to partition with the NF in nuclei pretreated with H1, which indicates that chromatin is the primary binding site of MPO in vitro (Fig. S2 D). Therefore, NE and MPO require access to the core histones to degrade them and induce decondensation.
NE and MPO synergize to promote nuclear decondensation
Because MPO localizes to NETs and binds to nuclei in vitro (Fig. 1 F), we tested whether MPO promotes nuclear decondensation (Brinkmann et al., 2004). At 1 µM, MPO alone had little effect, but it dramatically enhanced chromatin decondensation in the presence of NE (Fig. 1 H). Interestingly, MPO did not affect histone degradation, which suggests that it does not up-regulate NE activity (Fig. 1 I). MPO promoted nuclear decondensation in a dose-dependent manner that did not require its substrate H2O2 and was not inhibited by ABAH (Fig. 2, A and B). Furthermore, horseradish peroxidase, used as a control for enzymatic activity, did not promote chromatin decondensation (unpublished data). We concluded that MPO enhances NET formation independent of its enzymatic activity.
Figure 2.
NE and MPO synergize. (A) Effect of MPO titration on nuclear decondensation in vitro. Nuclei were incubated with the indicated concentrations of MPO for 30 (open circles), 60 (gray circles), or 120 min (closed circles). Decondensation was concentration- but not time-dependent. (B) MPO promotes decondensation independent of its enzymatic activity. Nuclei were treated with 5 µM MPO alone or MPO in the presence of 100 µM ABAH (an MPO inhibitor), 100 µM H2O2 (MPO substrate), or both. (C) Nuclear decondensation was driven by titration of NE at different concentrations of MPO. MPO increases NE-mediated decondensation. (D) The effect of NE titration on nuclear decondensation. Nuclei were incubated with the indicated concentrations of NE in the absence (black line) or presence (red line) of 10 µM NEi for 120 min before measuring nuclear decondensation. (E) NE-mediated degradation of histones is dose-dependent during nuclear decondensation. Western immunoblot analysis of MPO, H4, and NE was performed. Nuclei mixed with increasing concentrations of NE and 0.3 µM MPO for 120 min were separated into nuclear (NF, left) and soluble (SF, right) fractions. Reactions were performed in the absence or presence of NEi. The samples where nuclei decondensed are indicated by positive signs below the NF blot. Asterisks mark the levels of NE bound to nuclei, in the presence or absence of NEi, at the highest concentration of NE. ***, P < 0.0001.
To better understand the synergy between NE and MPO, we measured its effect on nuclear decondensation driven by NE. Increasing amounts of MPO enhanced NE-mediated nuclear decondensation (Fig. 2 C). Surprisingly, high concentrations of NE inhibited nuclear decondensation (Fig. 2 D). This autoinhibition could be caused by NE autodegradation. Alternatively, at high concentrations, NE could inhibit histone degradation. Accordingly, DNA has been shown to inhibit NE (Belorgey and Bieth, 1995). We confirmed the inhibitory effect of DNA on NE and CG, and found that it was reversed by the addition of NaCl (Fig. S3, A and B). Intact nuclei also inhibited NE activity, but this effect was not reversed by the addition of NaCl. Therefore, after histone cleavage, NE activity may be down-regulated by binding to DNA. Autoinhibition was not observed for PR3, which completely dissolved the nuclei at high concentrations (Fig. S1, E and F).
To investigate whether high concentrations of NE decreased histone degradation, we incubated nuclei with increasing NE concentrations and separated the nuclear (NF) and soluble (SF) fractions over filters (Fig. 2 E). H4 was degraded under optimal, but not autoinhibitory, concentrations of NE. Notably, NE activity is not required for NE binding to nuclei, as it partitioned with nuclei in the presence of NEi. The lower levels of NE in the NF fraction in the absence of NEi suggest that NE degrades itself in the nucleus (Fig. 2 E, left, asterisks).
We asked whether NE and MPO interact inside the nucleus by incubating nuclei with NE and low concentrations of MPO. MPO nuclear localization was independent of NE activity, as significant amounts of MPO were found in the nuclear fraction in the presence of NEi (Fig. 2 E, right). Moreover, we observed NE-dependent MPO degradation in the nuclear, but not in the soluble fraction, which indicates that these proteins come into close proximity in the nucleus.
NE activity is required for NET formation in neutrophils
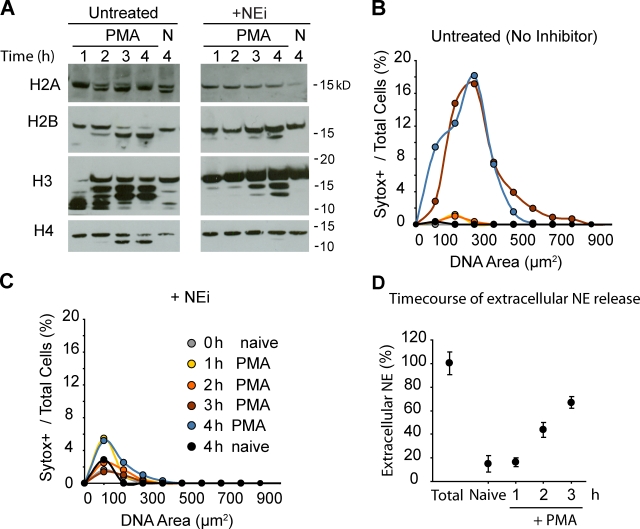
To investigate whether NE activity is required for NET formation, we pretreated neutrophils isolated from healthy donors with NEi or CGi, and induced NET formation with PMA in the presence of Sytox, a cell-impermeable DNA dye. Untreated neutrophils and cells treated with CGi formed NETs efficiently 2–4 h after activation. Notably, NEi-treated neutrophils failed to make NETs, and appeared necrotic as determined by the Sytox-positive condensed nuclei (Fig. 3 A). To quantitate NET formation, we measured the DNA area of each Sytox-positive neutrophil and plotted the percentage of Sytox-positive cells against their corresponding nuclear area (Fig. 3 B). Our quantitation confirmed that neutrophil death is dramatically reduced in the presence of NEi (11.2% vs. 63.7%), whereas the low nuclear areas of the dead cells indicate that cells did not make NETs but were necrotic.
Figure 3.
NE is required for NET formation. (A) NEi but not CGi, a CG inhibitor, blocks NET formation. Purified human neutrophils, untreated or pretreated with NEi or CGi, were either activated with PMA for 4 h or left unactivated (naive) in the presence of 10% FCS. Shown are fluorescence images of cells in the presence of the cell-impermeable DNA dye Sytox green (left), and phase contrast images (right). Bar, 50 µm. (B) Quantitation of chromatin decondensation in samples from A. A plot of the distribution of Sytox-positive neutrophils with respect to the chromatin area is shown. Naive cells at 4 h (orange), cells stimulated with PMA alone (black), or stimulated with PMA in the presence of NEi (yellow) or CGi (purple) were quantified. The overall percentage of Sytox-positive cells for each sample is shown in parentheses. Representative data out of six independent experiments are shown. (C) NEi blocks NET formation stimulated by C. albicans. Neutrophils were untreated or pretreated with 10 µM NEi in 10% human serum, and incubated with C. albicans for 3 h at MOI = 10 in the presence of Sytox. Extracellular DNA was visualized by Sytox fluorescence. PMA-stimulated and PMA + NEi control neutrophils from the same donor are shown as well. NET formation by C. albicans is less efficient than with PMA, and is blocked by NEi. (D) Quantitation of C. The distribution of Sytox-positive cells against their DNA area is shown. The right inset depicts the percentage of NETs as the number of cells whose DNA area exceeds 400 µm2. Representative data out of three independent experiments are shown. (E) NEi is not cytotoxic. Release of LDH, a cytoplasmic protein, is used to monitor cell lysis. LDH levels in the medium of naive neutrophils after a 5-h incubation in the presence of increasing levels of NEi (0, 1, 10, and 30 µM). LDH levels are normalized to the total LDH content of an equivalent number of neutrophils lysed with detergent (total). (F) 5 µM NEi and 5 µM CGi have no effect on the production of ROS in response to PMA. Neutrophils were either untreated or pretreated with NEi, CGi, or the NADPH oxidase inhibitor DPI as a control. Subsequently, cells were stimulated with PMA, and ROS were detected by monitoring luminol chemiluminescence in the presence of horseradish peroxidase. Mean values and the standard deviation from triplicate samples for each condition are presented (error bars). ***, P < 0.0001.
Candida albicans is a potent physiological activator of NET formation in isolated primary neutrophils. Neutrophils treated with NEi failed to make NETs efficiently when exposed to C. albicans (Fig. 3 C). The number of neutrophils that died without making NETs increased dramatically and was comparable to neutrophils treated with NEi and stimulated with PMA (Fig. 3 D). Interestingly, C. albicans induced NET formation with lower efficiency than PMA (Fig. 3 D), which indicates that when presented with microbes, neutrophils may respond in different ways, with some neutrophils consuming NE and MPO through phagocytosis or degranulation while others form NETs.
As controls, we show that NEi was not cytotoxic (Fig. 3 E) and did not inhibit ROS production, which was potently blocked by the NADPH oxidase inhibitor diphenyleneiodonium (DPI; Fig. 3 F; Cross and Jones, 1986).
NE cleaves histones during NET formation
Because PMA is a more potent inducer of NETs than C. albicans and other microbes, we used it to examine whether histones are degraded in an NE-dependent manner during NET formation. Consistent with our in vitro data, H2B and H4 were partially degraded during NET formation, whereas H3 shifted to a higher molecular weight species (Fig. 4 A). The presence of NEi inhibited the degradation of H4 and significantly reduced the degradation of H2B. NEi also prevented the formation of an H2A degradation product, which was observed in both naive and activated cells in the absence of the inhibitor. The higher molecular weight H3 species in both activated and naive neutrophils were independent of NE activity and may represent other posttranslational modifications. Histones were degraded 2–4 h after stimulation, coinciding with the peak of NET release (Fig. 4 B).
Figure 4.
NE partially degrades core histones during NET formation. (A) Histone cleavage during NET formation is inhibited by NEi. Western immunoblotting against histones in lysates of naive (N) and PMA-activated neutrophils (PMA) in the presence (+NEi) or in the absence of NEi (untreated). (B and C) Quantitation of chromatin decondensation for the samples shown in A. (B) Untreated neutrophils. (C) Neutrophils treated with NEi. Shown are naive neutrophils at 0 h (gray) or 4 h (black), activated with PMA for 1 (yellow), 2 (orange), 3 (red), or 4 h (blue). (D) NE is not significantly externalized before NET formation. The time course of the release of NE into the medium by neutrophils activated with PMA measured by ELISA is shown. MNase was added to solubilize NE bound to DNA. Samples were normalized to NE levels from plated naive neutrophils lysed with 0.1% Triton X-100 (Total).
Histone degradation was detected before cell death and peaked during NET formation (Fig. 4 B). NET release and neutrophil death occurred simultaneously between 2 h and 3 h after stimulation, as indicated by the large DNA area of all Sytox-positive neutrophils, which was blocked in the presence of NEi (Fig. 4 C). By monitoring the release of NE during stimulation, we confirmed that significant levels of the NE remained intracellular during the first 2 h (Fig. 4 D).
NE translocates to the nucleus during NET formation
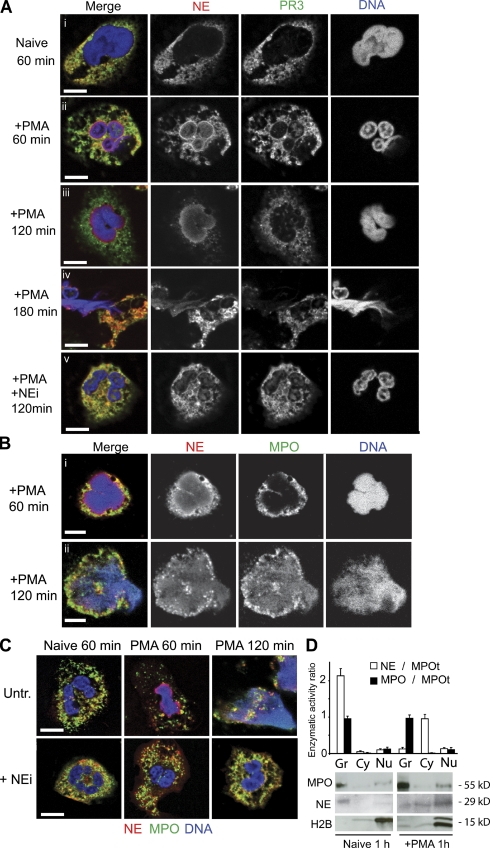
During NET formation, NE localized to the nucleus 60 min after stimulation, accompanied by reduced granular staining (Fig. 5 A, ii). In contrast, only low levels of PR3 were detected in the heterochromatin areas of the nucleus, arguing for a selective translocation of NE. After 120 min of stimulation, NE, but not PR3, was found predominantly in the decondensing nucleus in a diffuse gradient-like pattern (Fig. 5 A, iii). Interestingly, when neutrophils were treated with NEi, NE remained granular (Fig. 5 A, v), which suggests that NE activity is required for NE translocation. Because NE bound efficiently to isolated nuclei in the presence of NEi, NE activity may be required for granular release (Fig. 2 E).
Figure 5.
NE, PR3, and MPO localization during NET formation. (A–C) Naive and PMA-activated neutrophils in the presence or absence of NEi, fixed at the indicated time points and immunolabeled for NE (red) and PR3 (green; A), or NE (red) and MPO (green; B and C). DNA was stained with DRAQ5 (blue). (A) NE, and to a lesser extent PR3, translocate to the nucleus within 60 min after stimulation. (B) MPO associates with DNA before cell lysis but later than NE. (C) NEi prevents NE and MPO translocation to the nucleus, and chromatin decondensation. Bar, 5 µm. (D) NE is released from the granules during activation. Lysates from naive and activated neutrophils were prepared after 60 min of incubation and separated into cytoplasmic (HSS) and granule (HSP) fractions. The enzymatic activity (initial rate of change in absorbance) was normalized to the total amount of MPO (MPOt), which remains unchanged, and plotted as the fraction of NE activity over total MPO activity (open bars). The distribution of MPO activity in each sample over total MPO activity is also shown (shaded bars). Samples were also resolved by SDS-PAGE and analyzed by immunoblotting against MPO, NE, and histone H2B.
In contrast to NE, MPO remained granular until the later stages of NET formation (Fig. 5 B, i), where it colocalized with NE and DNA in neutrophils undergoing NET release (Fig. 5 B, ii). Consistently, NE translocated faster than MPO in vitro (Fig. 1 F). Furthermore, the translocation of MPO was also blocked by NEi (Fig. 5 C).
We detected high levels of NE in the nuclei of neutrophils undergoing NET formation by immunoblot analysis, whereas NE activity was selectively decreased in the granules compared with MPO (Fig. 5 D). We did not detect NE activity in the nuclear fraction of either sample, as expected from the inhibitory effect of DNA (Fig. S3 A; Belorgey and Bieth, 1995). Moreover, CD63, a transmembrane protein found in azurophilic granule membranes, was not perinuclear at any point during NET formation. This indicates that the granules do not fuse with the nuclear membrane. Instead, NE is released from the granules and then translocates to the nucleus, a hypothesis that remains to be confirmed (Fig. S4).
Although it is conceivable that in isolated neutrophils, NE is first externalized and then binds to the nucleus of necrotic neutrophils, our observations argue against this possibility. The translocation of NE to the nucleus is selective and occurs within 1 h after stimulation (Fig. 5 A, iii), whereas neutrophil death occurs only after 3 h of stimulation with PMA (Fig. 4 B). Moreover, NE is not significantly externalized during that time (Fig. 4 D). Finally, cell death is primarily caused by NET formation, as neutrophil death is strikingly reduced when NE activity is inhibited (Fig. 3 B). However, such a mechanism may represent an alternative route to NET formation during an infection where externalized NE may process nuclei derived from necrotic bystander cells.
NE is required for NET formation in vivo
We examined NET formation in the lungs of wild type (WT) and NE knockout mice infected intranasally with a sublethal dose of Klebsiella pneumoniae, a Gram-negative bacterium that causes pneumonia. Neutrophils were massively recruited to the lungs 48 h after infection in both groups of animals, confirming that the loss of NE does not interfere with neutrophil recruitment (Hirche et al., 2004). The lungs of WT mice contained decondensed web-like structures that stain with antibodies against MPO and an H2A-H2B-DNA complex (Fig. 6 A, i; Brinkmann et al., 2004). NETs have also been observed in a mouse pneumococcal pneumonia model (Beiter et al., 2006). In contrast, in the lungs of infected NE knockout animals, neutrophils exhibited condensed nuclei, and MPO did not colocalize with the DNA marker but remained granular (Fig. 6 A, ii).
Figure 6.
NE knockout mice fail to form NETs. (A) Representative fluorescence images of the lungs of WT (i) and NE knockout (ii) mice infected with K. pneumoniae, and stained with antibodies against MPO (green) and against a DNA/histone complex (red). The lungs of WT mice (i) contain decondensed web-like chromatin structures that stain for MPO (arrow). In contrast, in the lungs of NE knockout mice, all neutrophils appear naive, with condensed nuclei and granular MPO staining (asterisks). Bar, 20 µm. (B) NETs (arrows) trapping bacteria are detected in scanning electron micrographs of WT mouse lungs (i and ii) infected with K. pneumoniae. The lungs of NE knockout animals are devoid of NETs (iii and iv). Bars: (i and iii) 100 nm; (ii and iv) 1 µm. (C) Lysates from NE knockout mouse peritoneal neutrophils lack nuclear decondensation activity in vitro. Cell-free nuclear decondensation assays of mouse peritoneal nuclei treated for 2 h with LSS extracts from peritoneal neutrophils derived from WT and NE knockout mice are shown. ***, P < 0.0001.
We also analyzed these sections by scanning electron microscopy. NETs entrapping bacteria were present in abundance in the lungs of WT animals (Fig. 6 B, i and ii). In contrast, the lungs of NE knockout mice were devoid of structures resembling NETs on lung tissues and bacterial surfaces (Fig. 6 B, iii and iv). The absence of NETs in NE single knockout animals indicates that the extracellular DNA detected in the lungs of WT mice does not originate from bacterial biofilm, as such structures would be present in both groups of mice. Furthermore, these results suggest that PR3 cannot complement the loss of NE in NET formation.
In addition, we examined nuclear decondensation by extracts derived from mouse peritoneal neutrophils. LSS extracts from WT, but not from NE knockout mouse peritoneal neutrophils, decondensed nuclei in vitro (Fig. 6 C). Despite our efforts, peritoneal mouse neutrophils responded poorly to stimulation ex vivo, reflecting our current inability to potently stimulate NET formation in these isolated cells (Fig. S5). Collectively, these results indicate that NE is the major protease driving nuclear decondensation during NET formation in vivo.
Discussion
According to our model, ROS production leads to the release of NE and subsequently MPO from azurophilic granules. NE translocates first to the nucleus, where it digests nucleosomal histones and promotes extensive chromatin decondensation (Fig. 7). In contrast to this crude approach, posttranslational histone modifications are well-suited for fine and reversible spatiotemporal control. Our data also imply that the late binding of MPO to chromatin enhances decondensation independent of its enzymatic activity. This synergy may constitute an important trigger for timely cell rupture and NET release. MPO may drive chromatin decondensation by promoting a relaxed chromatin state, but the details of the molecular mechanism remain to be addressed.
Figure 7.
Model of NET formation. In resting neutrophils, NE and MPO are stored in the azurophilic granules. Upon activation and ROS production, NE escapes the granules and translocates to the nucleus. In the nucleus, NE cleaves histones and promotes chromatin decondensation. MPO binds to chromatin in the late stages of the process. MPO binding promotes further decondensation. NE and MPO cooperatively enhance chromatin decondensation, leading to cell rupture and NET release.
In addition to histone degradation, posttranslational modifications may also play a role during NET formation. We observed several shifts in the mobility of histone H3 that may represent such modifications (Fig. 4 A). In particular, histone citrullination has been observed in chromatin released by neutrophil-like human leukemia 60 (HL-60) cells and primary neutrophils (Neeli et al., 2008; Wang et al., 2009). The peptidylarginine deiminase 4 (PAD4) inhibitor Cl−-amidine was tested against HL-60 cells, where it was shown to block hypercitrullination and limit chromatin release in response to IL-8 and Shigella flexneri (Wang et al., 2009). However, Wang et al. (2009) did not test Cl−-amidine in primary neutrophils but showed that overexpression of active PAD4 is sufficient to induce NET formation in HL-60 cells, although the overall NET yield is low in these cells. Citrullination may account for some of the NE-independent shifts observed in histone H3 during NET formation only in conjunction with additional modifications because deimination has been reported to increase the mobility of these proteins (Wang et al., 2004). Alternatively, in the absence of NEi, NE may first cleave histone H3 to a lower molecular weight moiety. Over time, the cleaved histone H3 may accumulate additional modifications, which may account for the apparent higher molecular weight shifts. Whether PAD4 is required for NET formation remains to be tested in primary neutrophils and in vivo. Nevertheless, it is interesting to speculate that posttranslational histone modifications may regulate NET formation by modulating the ability of NE to cleave histones.
The intracellular release of NE underlies the existence of an as yet unknown mechanism that allows granular proteins to be selectively released into the cytoplasm. ROS could promote the release of NE directly by disrupting the association of NE with the proteoglycan matrix that is thought to down-regulate protease activity in resting cells (Serafin et al., 1986; Kolset and Gallagher, 1990; Reeves et al., 2002). Consistently, we find that NE activity is required for NE translocation. NE and MPO may be differentially released from the matrix because of differences in net surface charge and size. Alternatively, the release of these proteins may not involve such interactions because Niemann et al. (2004) did not detect serglycin in mature circulating neutrophils. In addition, ROS may drive lipid peroxidation, but the resulting increase in membrane permeability is thought to apply to ions and not larger molecules (Lemasters et al., 2002). Alternatively, the membrane of some granules may be more extensively disrupted through an unidentified mechanism. In support of a membrane breakdown mechanism, we have previously reported that the nuclear envelope and granular membranes disappear during NET formation (Fuchs et al., 2007). Understanding how NE and MPO translocate from the granules to the nucleus is a complex question that awaits further investigation.
This release mechanism may represent a new concept in cell biology associated with the oxidative burst that may not be restricted to neutrophils. However, the release of NE is reminiscent of the lysosomal membrane permeabilization observed during cell death (Boya and Kroemer, 2008). As in the case of NET formation, lysosomal lysis implicates ROS and requires protease activation. In such a case, neutrophils may have adapted the same concept for NET formation.
An important role of NETs may be the sequestration of the neutrophil’s toxic antimicrobials to minimize tissue damage (Henson and Johnston, 1987; Weiss, 1989; Xu et al., 2009). Using antimicrobials as key factors in NET formation ensures that these toxic proteins are sequestered by chromatin. A two-step mechanism provides the necessary time for these proteins to bind to chromatin before neutrophil rupture. Allowing NE to operate early protects other NET proteins from being degraded by NE while it is still active inside the nucleus. Moreover, the inhibitory effect of DNA on NE activity may slow down the degradation of DNA-bound proteins, preserving their antimicrobial capacity. Interestingly, although histone degradation fragments are more potent antimicrobials than the intact proteins (Kawasaki and Iwamuro, 2008), the degradation of histones reduces cytotoxicity against the host during sepsis (Xu et al., 2009).
The requirement of neutrophil-specific factors explains why thus far extracellular trap formation is restricted to granulocytes, which also produce the highest levels of ROS (Segal, 2005). Mast cells express mast cell protease II, and eosinophil elastase is found in eosinophils (Woodbury et al., 1978; Lungarella et al., 1992). However, eosinophils may not require proteases, as they release their mitochondrial DNA, which lacks histones (Yousefi et al., 2008). Moreover, the contribution of other cell-specific peroxidases (Henderson and Kaliner, 1979; Ten et al., 1989) in ET formation may be closely associated with their ability to interact with DNA.
The immunodeficiency of NE and MPO knockout mice may be due in part to a defect in NET formation. Our findings may help explain the nonredundancy of NE with other related serine proteases. Mutations in NE are associated with neutropenias that are treatable with granulocyte colony-stimulating factor (G-CSF; Ancliff et al., 2001; Aprikyan and Dale, 2001). Patients treated with G-CSF display neutrophils devoid of granular proteins (Horwitz et al., 2007). In spite of having near normal neutrophil counts, death from sepsis still poses a high risk in patients treated with G-CSF. With G-CSF treatment, mortality caused by sepsis has decreased from 6% to 0.9% per year (Rosenberg et al., 2006). The remaining risk in neutropenic patients with mutations in NE may be due in part to a defect in NET formation. Because the molecules involved in NET formation play important roles in other neutrophil antimicrobial mechanisms, it is difficult to dissect the contribution of NETs in immune defense. Notably, microbes have evolved virulence factors to degrade NETs (Beiter et al., 2006; Buchanan et al., 2006; Walker et al., 2007; Lauth et al., 2009). Therefore, it could be informative to document the types of microbes that infect patients with different immunodeficiencies, and associate them to their susceptibility to NETs.
The advances presented here uncover a novel mechanism for NET formation that may help to better understand and treat human immunodeficiency, sepsis, and autoimmune disease.
Materials and methods
Cell isolation and NET formation
Blood was drawn from healthy volunteers, and neutrophils were isolated over a Histopaque 1119 bed and a discontinuous Percoll gradient as described previously (Fuchs et al., 2007). Cells were stored in HBSS(−) (without calcium or magnesium) before experiments. PBMCs were isolated from the same preparation.
Preparation of extracts, nuclei, and granular fractionation
Extracts were prepared from 5 × 107 neutrophils/ml, which were lysed by douncing in 20 mM Hepes, pH 7.4, 100 mM KCl, 100 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, protease inhibitor cocktail pellets (Roche), and 0.1 mM PMSF. LSS was prepared by removal of nuclei by centrifugation at 300 g for 10 min. HSS was prepared by centrifugation of LSS at 100,000 g for 1 h. For granule preparations, cells were lysed by nitrogen cavitation followed by light douncing. The granule fractionation was performed by centrifugation (37,000 g, 20 min) of LSS prepared in 20 mM Hepes, pH 7.4, 100 mM KCl, 100 mM sucrose, 3 mM NaCl, 3 mM MgCl2, and 1 mM EGTA, over a discontinuous (1.050, 1.090, and 1.120 g/ml) Percoll gradient as described previously (Kjeldsen et al., 1994; Lominadze et al., 2005). Gradient fractions were isolated and centrifuged at 100,000 g for 1 h to remove the residual Percoll. Granule fractions were resuspended into the original volume of LSS. HL-60 lysates and nuclei were prepared from HL-60 cells differentiated with 5 µM retinoic acid for 96 h. Intact nuclei were isolated as described by Celis (1998).
Cell-free nuclear decondensation assay
Reactions of 10 µl of LSS extract at ∼10 mg/ml total protein (derived from 3–5 × 107 neutrophils/ml), the equivalent amount of HSP, or isolated granules, were mixed with 104 nuclei and Sytox. Before incubation, 3-µl aliquots were transferred onto 12-well, 5-mm diagnostic slides (Menzel-Glaser) and covered with 20 × 50 mm coverslips. Reactions were performed in a humidified chamber at 37°C for 120 min (unless otherwise specified). Sytox-labeled nuclei were analyzed by fluorescence microscopy.
Cell-free nuclear decondensation assay over filters
100-µl reactions containing 105 nuclei and 90 µl LSS extract, buffer, or granules were incubated for 2 h at 37°C. The reactions were then centrifuged through Ultrafree-MC Amicon polyvinylidene fluoride (PVDF) filters with a 0.65-µm pore size (Millipore) to separate the soluble flowthrough, containing the cytoplasm and the granules, from the nuclear material and bound proteins, which are retained by the filter. The filters were washed three times in cell lysis buffer, and the nuclear material was eluted with 120 µl of 1× SDS sample buffer. 20 µl of 6× SDS sample buffer was added to the 100 µl FT. Before the initiation of the reactions, a 3-µl aliquot was removed and placed on diagnostic slides, and 0.3 µl of 10× Sytox was added to measure nuclear decondensation by microscopy at the indicated time points.
Quantitation of chromatin decondensation and NET formation
Sytox images of unfixed neutrophils or nuclei were analyzed using ImageJ image processing software. The area of Sytox signal for 300–500 cells per sample was individually measured. For quantitation of nuclear decondensation, we plotted the mean DNA area derived from each nucleus (circles) and the standard deviation of the values (bars) denoting the range of areas for each condition. For NET formation, the distribution of the number of cells across the range of nuclear area was obtained using the frequency function in Excel (Microsoft). The data were converted to a percentage of Sytox-positive cells by dividing the Sytox-positive counts by the total number of cells as determined from corresponding phase-contrast images, and plotted as the percentage of all cells that were positive for Sytox for each DNA area range.
NET formation
We seeded 5 × 104 neutrophils per well in 24-well plates, in HBSS(+) (including calcium and magnesium), supplemented with 10% FCS. Cells were allowed to settle onto uncoated plates for 1 h before stimulation with 100 nM PMA. Sytox green (1:15,000) was added and NETs were visualized by fluorescence microscopy. Wherever indicated, cells were pretreated with 5 µM NEi (GW311616A; Sigma-Aldrich) or CGi (219372; EMD) for 1 h before stimulation. Stimulation with C. albicans was performed in HBSS(+) medium supplemented with 10% human serum at an MOI of 10. C. albicans were grown overnight at 30°C in YPD media and subcultured to reach an exponential phase. Sytox images were taken 3 h after infection.
Histone degradation during NET formation
At each time point, the medium was removed and 5 × 105 neutrophils were resuspended in 300 µl of 1× SDS loading buffer. Samples were resolved by SDS-PAGE electrophoresis, transferred to PVDF, blocked in 5% BSA, and labeled with primary antibodies and secondary horseradish peroxidase–conjugated antibodies (see Antibodies for Western immunoblotting).
Extracellular NE was measured by removing half the volume of media. NE levels were quantitated by ELISA. MNase (2 U/ml) was present throughout the time course to solubilize any extracellular NE bound to DNA.
Antibodies for Western immunoblotting
1:1,000 rabbit anti-H2A (2578, recognizes the C terminus; Cell Signaling Technology), 1:5,000 anti-H2B (07-371, recognizes aa 118–126; Millipore), 1:10,000 anti-H3 (07-690, recognizes the C terminus; Millipore), 1:5,000 anti-H4 (04-858, epitope mapped to aa 25–28; Millipore), and the pan-histone antibody MAB052 (1:500; Millipore) for analysis of H1. Rabbit anti-NE (ab21595; Abcam) was used at 1:200, or rabbit anti-NE (EMD) was used at 1:500. 1:10,000 anti-MPO (A0398; Dako), 1:500 anti-gelatinase (Dako), and 1:500 anti-lactoferrin (Sigma-Aldrich) were also used. Secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc.) were used at a 1:20,000 dilution.
Immunostaining and microscopy
Cells were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, blocked with 3% BSA, and stained with the following primary antibodies: 1:500 mouse anti-NE (in house), 1:200 rabbit anti-NE (EMD), 1:200 rabbit anti-MPO (Dako), 1:200 EPC rabbit anti-PR3, 1:200 mouse anti-CD63 (CBL 553; Millipore),and DRAQ5 (Biostatus Limited). Secondary antibodies conjugated with Cy2 and Cy3 (Invitrogen) were used. Confocal images were obtained using a confocal fluorescence microscope (TCS-SP; Leica) and a Fluotar 100× objective lens; images were captured using Leica software. Epifluorescence images were obtained using an epifluorescence microscope (Axioplan; Carl Zeiss, Inc.) and 10×, 20×, or 40× objective lenses. Images were captured using a ProgRes camera and software (Jenoptik).
Luminol assay
For ROS production, 106 neutrophils, either untreated or treated with 5 µM NEi, 5 µM CGi, or 10 µM of the NADPH inhibitor DPI, were activated with 100 nM PMA. ROS was measured by monitoring luminol fluorescence (50 µM) in the presence of 1.2 U/ml horseradish peroxidase.
Cytotoxicity assay
Cells were either untreated or treated with NEi at the indicated concentrations. After 5 h of incubation, the medium was collected, and LDH release was measured using the LDH cytotoxicity detection kit (Takara Bio, Inc.) and normalized to the total LDH of an equivalent number of lysed neutrophils.
Mouse lung infections
Five WT 129S2 mice and five NE (129S2 NE KO) knockout mice were infected with 2 × 103 Klebsiella pneumonia (KP52145; Benghezal et al., 2007). The bacteria were picked from an overnight plate and diluted into PBS. Mice were anesthetized and infected intranasally with 50 µl of PBS solution containing bacteria. 48 h later, mice were sacrificed, and lungs were removed and fixed with 2% PFA overnight. Lungs were embedded into paraffin and sectioned into 5-µm sections. Paraffin was removed with Neo Clear (Merck) and samples were rehydrated in ethanol. Samples were incubated in PBS containing 0.075% Tween 20 for 30 min at 50°C to expose the antigens, blocked with 5% FCS/5% donkey serum, and stained with rabbit anti-MPO (1:50; Dako) and a mouse Fabs against a H2A–H2B–DNA complex (a gift from Marc Monestier, Department of Microbiology and Immunology, School of Medicine, Temple University, Philadelphia, Pennsylvania; Losman et al., 1992) fused to ATT0 550. Lung inflammation and neutrophil recruitment were evaluated by hematoxylin and eosin stains. The experiment was repeated twice with five and seven mice per group, respectively.
For scanning electron microscopy, paraffin-embedded samples were rehydrated, postfixed with glutaraldehyde, contrasted using repeated changes of 0.5% OsO4 and 0.05% tannic acid, dehydrated in a graded ethanol series, critical-point dried, and coated with 5 nm platinum/carbon. Samples were obtained with a field emission scanning electron microscope (Leo 1550; Carl Zeiss, Inc.), and images were analyzed with SmartSEM software (Carl Zeiss, Inc.).
Animal experiments are in compliance with the German animal protection law and have been officially approved by the Landesamt fur Gesundheit und Soziales, Berlin.
Mouse peritoneal cells
Mouse peritoneal cells were collected 5 h after injection of 1 ml of thioglycolate into the peritoneal cavity of 10-wk-old mice. The peritoneum was lavaged with 10 ml of PBS to collect the neutrophils. Neutrophils were washed in PBS and plated as described in the procedure for human neutrophils. NET formation was monitored by microscopy for 24 h after stimulation. Quantitation of NET formation was performed by measuring the area of DNA for each cell by staining with Sytox, as described in the procedures for human neutrophils.
Cell-free nuclear decondensation in the presence of anti-histone antibodies or H1
Reactions were performed as described in the “Cell-free nuclear decondensation assay” section. However, before incubating with 1 µM NE, nuclei were treated with 1 µg/ml anti-histone antibodies in a 10-µl reaction volume on ice. Subsequently, reactions were brought to 100 µl and NE was added. For samples pretreated with H1, 1 µl of 104 nuclei were treated with 5 µl containing the indicated concentration of recombinant histone H1.1 (New England Biolabs, Inc.) for 60 min at 25°C. The nuclei were then tested for nuclear decondensation by mixing 1 µl of treated nuclei with 9 µl containing buffer, LSS, or HSP.
Enzymatic assays
Protease activity measurements were performed by incubating samples with 300 µM of the chromogenic peptides, elastase substrate I and CG substrate I (EMD), at 25°C, while monitoring absorbance at 410 nm using a SpectraMax 190 plate reader (MDS Analytical Technologies). MPO activity assays were performed by monitoring the absorbance at 450 nm of 0.1 mg/ml O-phenylenediamine (Sigma-Aldrich) at 25°C in the presence of 1 mM H2O2. Where indicated (Fig. S3), 0.3 mg/ml of plasmid DNA or 2 × 104/ml nuclei derived from RA-differentiated HL-60 cells were added to protease activity reactions. NE ELISA was performed using the Human Elastase ELISA test kit (Hycult biotechnology).
Statistical analysis
Raw measurements were analyzed in Graph Pad Prism 5 software using Kruskal-Wallis analysis of variance and Dunn’s multiple comparisons test for pairwise comparisons (***, P < 0.0001). All experiments have been repeated at least three times unless indicated.
Online supplemental material
Fig. S1 shows the specificity of the NE inhibitor. Fig. S2 shows that histone degradation mediates NE-mediated nuclear decondensation. Fig. S3 shows that DNA and nuclear binding inhibits NE activity. Fig. S4 shows that azurophilic granules do not fuse with the nuclear membrane during NET formation. Fig. S5 shows that mouse peritoneal neutrophils do not form NETs in response to PMA. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006052/DC1.
Acknowledgments
We thank Volker Brinkmann for obtaining scanning electron micrograph images of mouse lungs, Kaaweh Molawi and Juana de Diego for useful comments on the manuscript, and Marc Monestier for his generous gift of the antibody against the histone-DNA complex. V. Papayannopoulos designed the project and all the experiments. V. Papayannopoulos performed all experiments with the exception of experiments performed by K.D. Metzler and where assisted by A. Hakkim. V. Papayannopoulos wrote the manuscript. K.D. Metzler performed all human neutrophil immunofluorescence stains and imaging (Fig. 4), the nuclear digestion with purified NE (Fig. 1, H and I), the blots of granule fractions (Fig. 1 D), and the NE ELISA (Fig. 4 B). A. Hakkim developed the mouse infection model and performed the mouse lung infections and lung isolations. A. Hakkim and V. Papayannopoulos adjusted and optimized the sublethal dose of K. pneumoniae for knockout animals. V. Papayannopoulos performed lung processing, staining, and image analysis of lung sections. A. Zychlinsky advised and coordinated the project. A. Zychlinksy supervised the writing of the manuscript.
This work was funded by the Max Planck Society. V. Papayannopoulos was supported by an EMBO Long term fellowship. None of the authors have any commercial or other conflict of interest.
Footnotes
Abbreviations used in this paper:
- ABAH
- 4-aminobenzoic acid hydrazide
- CG
- cathepsin G
- CGi
- CG inhibitor I
- DPI
- diphenyleneiodonium
- G-CSF
- granulocyte colony-stimulating factor
- HSP
- high-speed pellet
- HSS
- high-speed supernatant
- LSS
- low-speed supernatant
- MPO
- myeloperoxidase
- NE
- neutrophil elastase
- NET
- neutrophil extracellular trap
- PAD4
- peptidylarginine deiminase 4
- PBMC
- peripheral blood mononuclear cell
- PR3
- proteinase 3
- PVDF
- polyvinylidene fluoride
- ROS
- reactive oxygen species
- SLPI
- serum leukocyte protease inhibitor
References
- Ancliff P.J., Gale R.E., Liesner R., Hann I.M., Linch D.C. 2001. Mutations in the ELA2 gene encoding neutrophil elastase are present in most patients with sporadic severe congenital neutropenia but only in some patients with the familial form of the disease. Blood. 98:2645–2650 10.1182/blood.V98.9.2645 [DOI] [PubMed] [Google Scholar]
- Aprikyan A.A., Dale D.C. 2001. Mutations in the neutrophil elastase gene in cyclic and congenital neutropenia. Curr. Opin. Immunol. 13:535–538 10.1016/S0952-7915(00)00254-5 [DOI] [PubMed] [Google Scholar]
- Aratani Y., Koyama H., Nyui S., Suzuki K., Kura F., Maeda N. 1999. Severe impairment in early host defense against Candida albicans in mice deficient in myeloperoxidase. Infect. Immun. 67:1828–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiter K., Wartha F., Albiger B., Normark S., Zychlinsky A., Henriques-Normark B. 2006. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16:401–407 10.1016/j.cub.2006.01.056 [DOI] [PubMed] [Google Scholar]
- Belaaouaj A. 2002. Neutrophil elastase-mediated killing of bacteria: lessons from targeted mutagenesis. Microbes Infect. 4:1259–1264 10.1016/S1286-4579(02)01654-4 [DOI] [PubMed] [Google Scholar]
- Belaaouaj A., McCarthy R., Baumann M., Gao Z., Ley T.J., Abraham S.N., Shapiro S.D. 1998. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat. Med. 4:615–618 10.1038/nm0598-615 [DOI] [PubMed] [Google Scholar]
- Belaaouaj A., Kim K.S., Shapiro S.D. 2000. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 289:1185–1188 10.1126/science.289.5482.1185 [DOI] [PubMed] [Google Scholar]
- Belorgey D., Bieth J.G. 1995. DNA binds neutrophil elastase and mucus proteinase inhibitor and impairs their functional activity. FEBS Lett. 361:265–268 10.1016/0014-5793(95)00173-7 [DOI] [PubMed] [Google Scholar]
- Benghezal M., Adam E., Lucas A., Burn C., Orchard M.G., Deuschel C., Valentino E., Braillard S., Paccaud J.P., Cosson P. 2007. Inhibitors of bacterial virulence identified in a surrogate host model. Cell. Microbiol. 9:1336–1342 10.1111/j.1462-5822.2006.00877.x [DOI] [PubMed] [Google Scholar]
- Bianchi M., Hakkim A., Brinkmann V., Siler U., Seger R.A., Zychlinsky A., Reichenbach J. 2009. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 114:2619–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard N., Cowland J.B. 1997. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 89:3503–3521 [PubMed] [Google Scholar]
- Boya P., Kroemer G. 2008. Lysosomal membrane permeabilization in cell death. Oncogene. 27:6434–6451 10.1038/onc.2008.310 [DOI] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Buchanan J.T., Simpson A.J., Aziz R.K., Liu G.Y., Kristian S.A., Kotb M., Feramisco J., Nizet V. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16:396–400 10.1016/j.cub.2005.12.039 [DOI] [PubMed] [Google Scholar]
- Bustin M., Catez F., Lim J.H. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell. 17:617–620 10.1016/j.molcel.2005.02.019 [DOI] [PubMed] [Google Scholar]
- Celis J.E. 1998. Cell biology: a laboratory handbook. Vol. 2 Academic Press, San Diego: 945 pp [Google Scholar]
- Clark F.A., Klebanoff S.J. 1978. Chronic granulomatous disease: studies of a family with impaired neutrophil chemotactic, metabolic and bactericidal function. Am. J. Med. 65:941–948 10.1016/0002-9343(78)90745-3 [DOI] [PubMed] [Google Scholar]
- Clark S.R., Ma A.C., Tavener S.A., McDonald B., Goodarzi Z., Kelly M.M., Patel K.D., Chakrabarti S., McAvoy E., Sinclair G.D., et al. 2007. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 13:463–469 10.1038/nm1565 [DOI] [PubMed] [Google Scholar]
- Cross A.R., Jones O.T. 1986. The effect of the inhibitor diphenylene iodonium on the superoxide-generating system of neutrophils. Specific labelling of a component polypeptide of the oxidase. Biochem. J. 237:111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiserich J.P., Hristova M., Cross C.E., Jones A.D., Freeman B.A., Halliwell B., van der Vliet A. 1998. Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature. 391:393–397 10.1038/34923 [DOI] [PubMed] [Google Scholar]
- Fuchs T.A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V., Weinrauch Y., Brinkmann V., Zychlinsky A. 2007. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176:231–241 10.1083/jcb.200606027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut J.P., Yeh G.C., Tran H.D., Byun J., Henderson J.P., Richter G.M., Brennan M.L., Lusis A.J., Belaaouaj A., Hotchkiss R.S., Heinecke J.W. 2001. Neutrophils employ the myeloperoxidase system to generate antimicrobial brominating and chlorinating oxidants during sepsis. Proc. Natl. Acad. Sci. USA. 98:11961–11966 10.1073/pnas.211190298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães-Costa A.B., Nascimento M.T., Froment G.S., Soares R.P., Morgado F.N., Conceição-Silva F., Saraiva E.M. 2009. Leishmania amazonensis promastigotes induce and are killed by neutrophil extracellular traps. Proc. Natl. Acad. Sci. USA. 106:6748–6753 10.1073/pnas.0900226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkim A., Fürnrohr B.G., Amann K., Laube B., Abed U.A., Brinkmann V., Herrmann M., Voll R.E., Zychlinsky A. 2010. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. USA. 107:9813–9818 10.1073/pnas.0909927107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S.L., Hsu F.F., Duffin K., Heinecke J.W. 1996. Molecular chlorine generated by the myeloperoxidase-hydrogen peroxide-chloride system of phagocytes converts low density lipoprotein cholesterol into a family of chlorinated sterols. J. Biol. Chem. 271:23080–23088 10.1074/jbc.271.38.23080 [DOI] [PubMed] [Google Scholar]
- Henderson W.R., Kaliner M. 1979. Mast cell granule peroxidase: location, secretion, and SRS-A inactivation. J. Immunol. 122:1322–1328 [PubMed] [Google Scholar]
- Henson P.M., Johnston R.B., Jr 1987. Tissue injury in inflammation. Oxidants, proteinases, and cationic proteins. J. Clin. Invest. 79:669–674 10.1172/JCI112869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirche T.O., Atkinson J.J., Bahr S., Belaaouaj A. 2004. Deficiency in neutrophil elastase does not impair neutrophil recruitment to inflamed sites. Am. J. Respir. Cell Mol. Biol. 30:576–584 10.1165/rcmb.2003-0253OC [DOI] [PubMed] [Google Scholar]
- Hirsch J.G. 1958. Bactericidal action of histone. J. Exp. Med. 108:925–944 10.1084/jem.108.6.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M.S., Duan Z., Korkmaz B., Lee H.H., Mealiffe M.E., Salipante S.J. 2007. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 109:1817–1824 10.1182/blood-2006-08-019166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H., Iwamuro S. 2008. Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug Targets. 8:195–205 [DOI] [PubMed] [Google Scholar]
- Kessenbrock K., Krumbholz M., Schönermarck U., Back W., Gross W.L., Werb Z., Gröne H.J., Brinkmann V., Jenne D.E. 2009. Netting neutrophils in autoimmune small-vessel vasculitis. Nat. Med. 15:623–625 10.1038/nm.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettle A.J., Gedye C.A., Winterbourn C.C. 1997. Mechanism of inactivation of myeloperoxidase by 4-aminobenzoic acid hydrazide. Biochem. J. 321:503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeldsen L., Sengeløv H., Lollike K., Nielsen M.H., Borregaard N. 1994. Isolation and characterization of gelatinase granules from human neutrophils. Blood. 83:1640–1649 [PubMed] [Google Scholar]
- Kolset S.O., Gallagher J.T. 1990. Proteoglycans in haemopoietic cells. Biochim. Biophys. Acta. 1032:191–211 [DOI] [PubMed] [Google Scholar]
- Lauth X., von Köckritz-Blickwede M., McNamara C.W., Myskowski S., Zinkernagel A.S., Beall B., Ghosh P., Gallo R.L., Nizet V. 2009. M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 1:202–214 10.1159/000203645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer R.I., Ganz T. 1990. Antimicrobial polypeptides of human neutrophils. Blood. 76:2169–2181 [PubMed] [Google Scholar]
- Lekstrom-Himes J.A., Gallin J.I. 2000. Immunodeficiency diseases caused by defects in phagocytes. N. Engl. J. Med. 343:1703–1714 10.1056/NEJM200012073432307 [DOI] [PubMed] [Google Scholar]
- Lemasters J.J., Qian T., He L., Kim J.S., Elmore S.P., Cascio W.E., Brenner D.A. 2002. Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Antioxid. Redox Signal. 4:769–781 10.1089/152308602760598918 [DOI] [PubMed] [Google Scholar]
- Lominadze G., Powell D.W., Luerman G.C., Link A.J., Ward R.A., McLeish K.R. 2005. Proteomic analysis of human neutrophil granules. Mol. Cell. Proteomics. 4:1503–1521 10.1074/mcp.M500143-MCP200 [DOI] [PubMed] [Google Scholar]
- Losman M.J., Fasy T.M., Novick K.E., Monestier M. 1992. Monoclonal autoantibodies to subnucleosomes from a MRL/Mp(-)+/+ mouse. Oligoclonality of the antibody response and recognition of a determinant composed of histones H2A, H2B, and DNA. J. Immunol. 148:1561–1569 [PubMed] [Google Scholar]
- Lungarella G., Menegazzi R., Gardi C., Spessotto P., de Santi M.M., Bertoncin P., Patriarca P., Calzoni P., Zabucchi G. 1992. Identification of elastase in human eosinophils: immunolocalization, isolation, and partial characterization. Arch. Biochem. Biophys. 292:128–135 10.1016/0003-9861(92)90060-A [DOI] [PubMed] [Google Scholar]
- Macdonald S.J., Dowle M.D., Harrison L.A., Shah P., Johnson M.R., Inglis G.G., Clarke G.D., Smith R.A., Humphreys D., Molloy C.R., et al. 2001. The discovery of a potent, intracellular, orally bioavailable, long duration inhibitor of human neutrophil elastase—GW311616A a development candidate. Bioorg. Med. Chem. Lett. 11:895–898 10.1016/S0960-894X(01)00078-6 [DOI] [PubMed] [Google Scholar]
- Nathan C. 2006. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 6:173–182 10.1038/nri1785 [DOI] [PubMed] [Google Scholar]
- Nauseef W.M. 2007. How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 219:88–102 10.1111/j.1600-065X.2007.00550.x [DOI] [PubMed] [Google Scholar]
- Neeli I., Khan S.N., Radic M. 2008. Histone deimination as a response to inflammatory stimuli in neutrophils. J. Immunol. 180:1895–1902 [DOI] [PubMed] [Google Scholar]
- Niemann C.U., Cowland J.B., Klausen P., Askaa J., Calafat J., Borregaard N. 2004. Localization of serglycin in human neutrophil granulocytes and their precursors. J. Leukoc. Biol. 76:406–415 10.1189/jlb.1003502 [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V., Zychlinsky A. 2009. NETs: a new strategy for using old weapons. Trends Immunol. 30:513–521 10.1016/j.it.2009.07.011 [DOI] [PubMed] [Google Scholar]
- Ramos-Kichik V., Mondragón-Flores R., Mondragón-Castelán M., Gonzalez-Pozos S., Muñiz-Hernandez S., Rojas-Espinosa O., Chacón-Salinas R., Estrada-Parra S., Estrada-García I. 2009. Neutrophil extracellular traps are induced by Mycobacterium tuberculosis. Tuberculosis (Edinb.). 89:29–37 10.1016/j.tube.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Rao N.V., Wehner N.G., Marshall B.C., Gray W.R., Gray B.H., Hoidal J.R. 1991. Characterization of proteinase-3 (PR-3), a neutrophil serine proteinase. Structural and functional properties. J. Biol. Chem. 266:9540–9548 [PubMed] [Google Scholar]
- Reeves E.P., Lu H., Jacobs H.L., Messina C.G., Bolsover S., Gabella G., Potma E.O., Warley A., Roes J., Segal A.W. 2002. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 416:291–297 10.1038/416291a [DOI] [PubMed] [Google Scholar]
- Roche J., Girardet J.L., Gorka C., Lawrence J.J. 1985. The involvement of histone H1[0] in chromatin structure. Nucleic Acids Res. 13:2843–2853 10.1093/nar/13.8.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg P.S., Alter B.P., Bolyard A.A., Bonilla M.A., Boxer L.A., Cham B., Fier C., Freedman M., Kannourakis G., Kinsey S., et al. 2006. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 107:4628–4635 10.1182/blood-2005-11-4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal A.W. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23:197–223 10.1146/annurev.immunol.23.021704.115653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafin W.E., Katz H.R., Austen K.F., Stevens R.L. 1986. Complexes of heparin proteoglycans, chondroitin sulfate E proteoglycans, and [3H]diisopropyl fluorophosphate-binding proteins are exocytosed from activated mouse bone marrow-derived mast cells. J. Biol. Chem. 261:15017–15021 [PubMed] [Google Scholar]
- Ten R.M., Pease L.R., McKean D.J., Bell M.P., Gleich G.J. 1989. Molecular cloning of the human eosinophil peroxidase. Evidence for the existence of a peroxidase multigene family. J. Exp. Med. 169:1757–1769 10.1084/jem.169.5.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkalcevic J., Novelli M., Phylactides M., Iredale J.P., Segal A.W., Roes J. 2000. Impaired immunity and enhanced resistance to endotoxin in the absence of neutrophil elastase and cathepsin G. Immunity. 12:201–210 10.1016/S1074-7613(00)80173-9 [DOI] [PubMed] [Google Scholar]
- Urban C.F., Reichard U., Brinkmann V., Zychlinsky A. 2006. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 8:668–676 10.1111/j.1462-5822.2005.00659.x [DOI] [PubMed] [Google Scholar]
- Urban C.F., Ermert D., Schmid M., Abu-Abed U., Goosmann C., Nacken W., Brinkmann V., Jungblut P.R., Zychlinsky A. 2009. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5:e1000639 10.1371/journal.ppat.1000639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Köckritz-Blickwede M., Goldmann O., Thulin P., Heinemann K., Norrby-Teglund A., Rohde M., Medina E. 2008. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 111:3070–3080 10.1182/blood-2007-07-104018 [DOI] [PubMed] [Google Scholar]
- Walker M.J., Hollands A., Sanderson-Smith M.L., Cole J.N., Kirk J.K., Henningham A., McArthur J.D., Dinkla K., Aziz R.K., Kansal R.G., et al. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 13:981–985 10.1038/nm1612 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wysocka J., Sayegh J., Lee Y.H., Perlin J.R., Leonelli L., Sonbuchner L.S., McDonald C.H., Cook R.G., Dou Y., et al. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 306:279–283 10.1126/science.1101400 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li M., Stadler S., Correll S., Li P., Wang D., Hayama R., Leonelli L., Han H., Grigoryev S.A., et al. 2009. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 184:205–213 10.1083/jcb.200806072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Drujan D., Shapiro S.D., Weiss J., Zychlinsky A. 2002. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 417:91–94 10.1038/417091a [DOI] [PubMed] [Google Scholar]
- Weiss S.J. 1989. Tissue destruction by neutrophils. N. Engl. J. Med. 320:365–376 10.1056/NEJM198902093200606 [DOI] [PubMed] [Google Scholar]
- Wen F., White G.J., VanEtten H.D., Xiong Z., Hawes M.C. 2009. Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol. 151:820–829 10.1104/pp.109.142067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury R.G., Everitt M., Sanada Y., Katunuma N., Lagunoff D., Neurath H. 1978. A major serine protease in rat skeletal muscle: evidence for its mast cell origin. Proc. Natl. Acad. Sci. USA. 75:5311–5313 10.1073/pnas.75.11.5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C.L., Skoultchi A.I., Fan Y. 2006. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14:17–25 10.1007/s10577-005-1024-3 [DOI] [PubMed] [Google Scholar]
- Xu J., Zhang X., Pelayo R., Monestier M., Ammollo C.T., Semeraro F., Taylor F.B., Esmon N.L., Lupu F., Esmon C.T. 2009. Extracellular histones are major mediators of death in sepsis. Nat. Med. 15:1318–1321 10.1038/nm.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S., Gold J.A., Andina N., Lee J.J., Kelly A.M., Kozlowski E., Schmid I., Straumann A., Reichenbach J., Gleich G.J., Simon H.U. 2008. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 14:949–953 10.1038/nm.1855 [DOI] [PubMed] [Google Scholar]