Inactivation of the essential autophagy gene Atg5 results in selective accumulation of aggregation-prone proteins independently of substrate ubiquitination.
Abstract
Genetic ablation of autophagy in mice leads to liver and brain degeneration accompanied by the appearance of ubiquitin (Ub) inclusions, which has been considered to support the hypothesis that ubiquitination serves as a cis-acting signal for selective autophagy. We show that tissue-specific disruption of the essential autophagy genes Atg5 and Atg7 leads to the accumulation of all detectable Ub–Ub topologies, arguing against the hypothesis that any particular Ub linkage serves as a specific autophagy signal. The increase in Ub conjugates in Atg7−/− liver and brain is completely suppressed by simultaneous knockout of either p62 or Nrf2. We exploit a novel assay for selective autophagy in cell culture, which shows that inactivation of Atg5 leads to the selective accumulation of aggregation-prone proteins, and this does not correlate with an increase in substrate ubiquitination. We propose that protein oligomerization drives autophagic substrate selection and that the accumulation of poly-Ub chains in autophagy-deficient circumstances is an indirect consequence of activation of Nrf2-dependent stress response pathways.
Introduction
Protein aggregates are not normally observed in unstressed cells because of the cellular quality control systems that maintain cellular proteostasis by coordinating protein synthesis and degradation (Balch et al., 2008; Powers et al., 2009). Prolonged cellular stress increases the burden on these quality control mechanisms, resulting in the accumulation and subsequent aggregation of proteins linked to the pathogenesis of numerous neurodegenerative disorders (e.g., Alzheimer’s disease, Parkinson’s disease, Huntington’s disease [HD], and Lou Gehrig’s disease [amyotrophic lateral sclerosis] and prion encephalopathies; Ross and Poirier, 2004). Conditional mouse models of neurodegeneration such as HD (Yamamoto et al., 2000), SCA1 (spinocerebellar ataxia type 1; Zu et al., 2004), and tauopathy (Santacruz et al., 2005), in which expression of the mutant protein was turned off after the onset of pathogenesis, and inclusion body appearance established that there is a window of time during which neurons can recover from the proteotoxic stress induced by aggregation-prone proteins and that neurons are capable of disaggregation and/or elimination of inclusion bodies. It is unlikely that the proteasome contributes directly to the destruction of these aggregated proteins because substrates must be fully unfolded to be degraded by this protease (Finley, 2009). Indeed, the presence of intracellular protein aggregates in cell culture or in diseased brain tissue is associated with global disruption of ubiquitin (Ub) homeostasis and accumulation of undegraded proteasome substrates (Bence et al., 2001; Bennett et al., 2007). Macroautophagy (herein referred to as autophagy), the principal means by which cells target cytoplasmic constituents to the lysosome or vacuole, has been proposed to serve as an alternate pathway by which cells can eliminate protein aggregates (Kopito, 2000; Larsen and Sulzer, 2002). Because lysosomes are endowed with an acidic lumen and a battery of hydrolases, they are less dependent than proteasomes on unfolding of polypeptide substrates.
Morphological evidence of autophagosome proliferation in brains from patients and animal models of neurodegenerative diseases, together with biochemical and morphological studies in tissue culture cells overexpressing aggregation-prone proteins or exposed to proteasome inhibitors, suggests a tight linkage between pathogenic protein aggregation and induction of the autophagy pathway (for reviews see Nixon, 2006; Williams et al., 2006). The significance of these observations was initially controversial because it was not clear from those studies whether the induced autophagosomes were part of a cytoprotective response or autophagy-associated cell death (for review see Nixon, 2006). However, knockdown of the essential autophagy genes, Atg5 and LC3, leads to the accumulation of inclusion bodies and detergent-insoluble protein aggregates and retards the disappearance of aggregated huntingtin (htt) inclusions in cell culture models (Iwata et al., 2005; Yamamoto et al., 2006; Carra et al., 2009). Together with the finding that brain-specific genetic ablation of Atg5 or Atg7 in mice induces a neurodegenerative syndrome characterized by movement disorder, Ub pathology and accumulation of insoluble protein aggregates, it is clear that autophagy plays an essential, constitutive cytoprotective role in the brain, most likely by eliminating potentially toxic protein aggregates (Hara et al., 2006; Komatsu et al., 2006, 2007). A key challenge is to determine whether and how protein aggregates are selectively recognized and targeted to the autophagy machinery.
The invariant presence of Ub in inclusion bodies in neurodegenerative diseases and models thereof (Alves-Rodrigues et al., 1998), together with the finding of Ub-positive inclusions in the brains and livers of Atg5 or Atg7 knockout (KO) mice (Hara et al., 2006; Komatsu et al., 2006, 2007), has suggested the hypothesis that Ub might serve as a signal for selective autophagy in addition to its established role in proteasome targeting (Kirkin et al., 2009c). How Ub might serve to specify such different fates for proteins sharing the same cytoplasm is not well understood but has been proposed to be encoded in Ub linkage topology (Pickart and Fushman, 2004). Ub has seven lysines, each of which can be linked to other Ubs to form a chain. All possible Ub–Ub linkages (including linear, mixed, and branched chains) have been identified in cells, but the physiological significance of most of these is unclear (Peng et al., 2003; Kim et al., 2007; Ikeda and Dikic, 2008). Although K48-linked poly-Ub chains are considered to be canonical targeting signals for proteasomal degradation (Chau et al., 1989; Finley, 2009), proteasome inhibition leads to rapid accumulation of K48, K63, and K11 poly-Ub chains in mammalian cells (Bennett et al., 2007; Meierhofer et al., 2008; Xu et al., 2009) and all non-K63 linkages in yeast (Xu et al., 2006), suggesting that K48 may not be unique in its ability to target proteins for degradation by the proteasome (Finley, 2009). K63-linked chains, in addition to established functions in endocytosis and DNA repair (Chen and Sun, 2009), have been proposed to also serve as signals to direct misfolded or aggregated proteins to the autophagy pathway (Seibenhener et al., 2007; Olzmann and Chin, 2008; Tan et al., 2008; Kirkin et al., 2009c).
Another established signal for autophagic substrate selectivity is protein aggregation. For example, import of the vacuolar enzyme Ape1 (aminopeptidase 1) into the yeast vacuole is mediated by the cytoplasm to vacuole pathway, in which the substrate is recognized by a cytoplasmic receptor, Atg19, and encapsulated into autophagosomes that ultimately fuse with the vacuole (Scott et al., 2001). Formation of Ape1 oligomers, directed by an N-terminal presequence, is necessary and sufficient for recognition by Atg19 (Kim et al., 1997). Thus, at least in yeast, aggregation is the best-understood mechanism for selective cargo recognition by the autophagy pathway (Nair and Klionsky, 2005). Therefore, at least two modifications, ubiquitination and oligomer formation, have been proposed to serve as cis-acting signals to promote selective autophagy. The extent to which either or both of these mechanisms function in the clearance of aggregated disease-associated proteins like htt and the mechanisms by which they are recognized by the autophagy system are unknown.
A leading candidate for this recognition process is p62, a ubiquitous component of inclusion bodies which contains an LC3 recognition sequence (Ichimura et al., 2008), which binds directly to LC3, an essential lipid-anchored protein on the surface of preautophagosome and autophagosome membranes (Pankiv et al., 2007; Ichimura et al., 2008), and a Ub association (UBA) domain, which has been reported to bind selectively to K63-linked poly-Ub chains (Seibenhener et al., 2004; Tan et al., 2008). Thus, the presence of both the LC3 recognition sequence and UBA domains in p62 has suggested a model in which p62 serves as an adaptor that recruits ubiquitinated proteins to autophagosomes (Bjørkøy et al., 2005; Pankiv et al., 2007; Kirkin et al., 2009a; Lamark et al., 2009). Although the genetic demonstration that p62 is required for the formation of Ub-positive inclusions in Atg7-null mice (Komatsu et al., 2007) and in a Drosophila melanogaster model of neurodegeneration (Nezis et al., 2008) confirms that p62 lies at the interface between protein aggregation, autophagy and Ub pathology, the inability to detect either significant binding affinity of p62’s UBA toward free Ub (Long et al., 2008) or unanchored tetra-Ub or selectivity toward K63 chains (Raasi et al., 2005; Kirkin et al., 2009b) challenges the Ub adaptor model. Moreover, recent studies have established a critical role for p62 in stabilizing Nrf2 (nuclear factor E2-related factor 2), a transcription factor which serves as the master regulator of one of the major cellular defense pathways that protects cells against oxidative and electrophilic stress (Liu et al., 2007; Komatsu et al., 2010). Therefore, it is possible that the involvement of p62 in inclusion body pathology might reflect this role, as opposed to or perhaps in conjunction with its proposed function as an Ub adaptor.
In this study, we have used Ub absolute quantification (AQUA; Gerber et al., 2003; Kirkpatrick et al., 2005; Pan et al., 2009) mass spectrometry to analyze the Ub species that accumulate in brains and livers of mice harboring tissue-specific deletions of two genes that are essential for autophagy, Atg5 and Atg7. We find that autophagy-deficient mice accumulate all detectable poly-Ub linkages in roughly equal proportion and that this accumulation is abrogated when the autophagy KO is genetically combined with KO of p62 or Nrf2, thus revealing a novel connection between integration of stress signaling pathways, autophagy, and Ub homeostasis. These findings lead us to conclude that specific poly-Ub chains are unlikely to be the key determinants of interaction with p62 or of autophagic fate. To assess the role of substrate aggregation in autophagic targeting, we have exploited a novel flow cytometry–based assay, which shows that aggregation-prone proteins accumulate preferentially over diffuse cytosolic proteins after inactivation of Atg5 and that this accumulation does not correlate with increased poly-Ub–modified substrate. These data lead us to propose that substrate oligomerization is a common mechanism for selective autophagic cargo recognition and that in the absence of autophagy, aggregation-prone substrates accumulate, resulting in the activation of stress response pathways that regulate global Ub homeostasis.
Results
Accumulation of all poly-Ub species in brain- and liver-specific autophagy KO mouse models
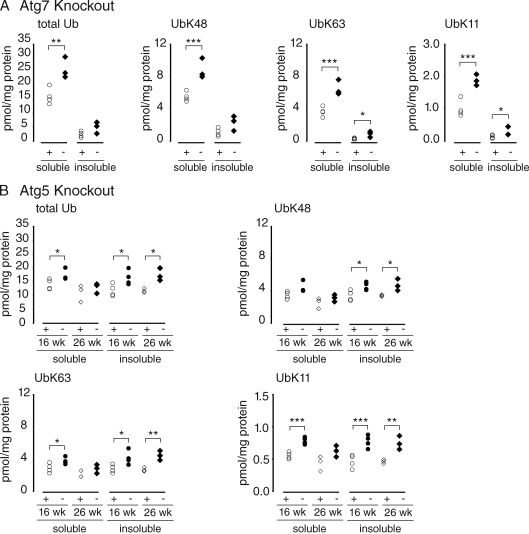
We reasoned that if specific poly-Ub linkages serve as selective signals for autophagy, those species should be enriched in tissues from mice in which the autophagy pathway has been disabled. To test this hypothesis, we used AQUA mass spectrometry to precisely quantify the abundance of tryptic peptides derived from total Ub and the major (K48, K63, and K11) Ub–Ub chain linkages in digests of poly-Ub–enriched brain homogenates from mice harboring brain-specific disruption of Atg5 or Atg7 (Fig. 1). Although other poly-Ub linkages (K6, K27, K29, and K33) can be detected in these tissues, together they constitute <1% of the total Ub isopeptides (unpublished data) and, unless they were found to be significantly changed, were not included in this analysis. Significant elevation of total brain Ub was observed in Atg7- and Atg5-null mice at as early as 6 wk of age (Fig. 1). Although the majority of brain Ub partitioned into a detergent-soluble fraction, the relative amount of Ub in the detergent-insoluble fraction increased with aging in all genotypes. By 26 wk, there was no further accumulation of soluble Ub species, suggesting that poly-Ub conjugates become less soluble with age. Importantly, Ub isopeptide species (K48, K63, and K11) increased in parallel in brain homogenates from both autophagy KO models. These findings, although consistent with a possible permissive role for poly-Ub chains in autophagy, fail to support the hypothesis that K63 (or any other particular poly-Ub linkage) serves as a signal to selectively target substrates to the autophagy pathway. Instead, these data point to a possible role for autophagy in regulating global Ub levels.
Figure 1.
Accumulation of Ub in autophagy-deficient brain. Ub profile from the 1% Triton X-100–soluble and –insoluble fraction of autophagy-deficient brain homogenates captured with P2UBA resin. (A) Age-matched control Atg7 littermates (Atg7flox/+;nestin-Cre:p62+/−; denoted +) at 6 wk (open circles; n = 4) or Atg7 KO (Atg7flox/flox;nestin-Cre:p62+/−; denoted −) at 6 wk (closed diamonds; n = 3) are shown. (B) Control Atg5 littermates (Atg5flox/+;nestin-Cre; denoted +) at 16 (open circles; n = 4) and 26 wk (open diamonds; n = 3) or Atg5 KO (Atg5flox/flox;nestin-Cre; denoted −) at 16 (closed circles; n = 4) and 26 wk (closed diamonds; n = 3) are shown. Each symbol represents one animal. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005.
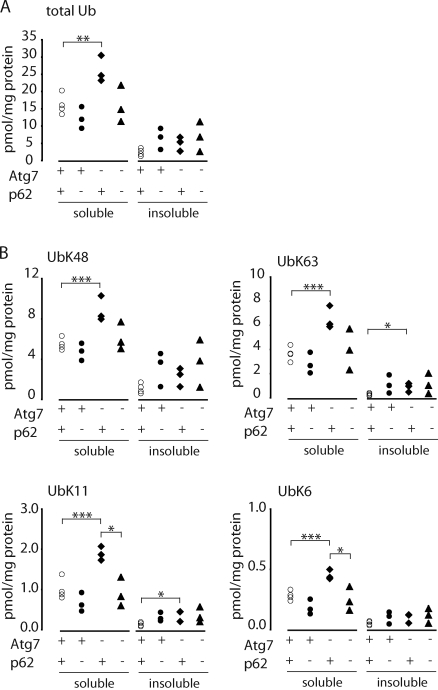
We used mass spectrometry to assess the role of Ub in the formation of p62 inclusion bodies. KO of p62 suppressed the elevated levels of total Ub (Fig. 2 A) and poly-Ub chains in brains from 6-wk-old Atg7−/− mice (Fig. 2 B), indicating that Ub recruited into inclusion bodies in a p62-dependent fashion is not enriched in K63 or any other specific poly-Ub linkage. We were also unable to observe any measurable binding of either free Ub or poly-Ub chains to immobilized recombinant p62 UBA in vitro (Fig. S1), which is consistent with the very weak affinity of this domain previously reported for mono-Ub (Long et al., 2008) or unanchored Ub chains (Raasi et al., 2005; Kirkin et al., 2009b). AQUA mass spectrometry analysis showed that >97% of p62 in Atg7−/− mouse brain is not associated with Ub (Fig. S1 D), suggesting that the vast majority of cellular p62 is not associated with polyubiquitinated proteins. Moreover, p62 levels did not increase in parallel with poly-Ub chains in detergent-insoluble extracts of livers (Fig. 3) or brains (not depicted) from autophagy-deficient mice. These data do not support a role for p62 in either the selective binding of proteins bearing poly-Ub chains or the recruitment of polyubiquitinated proteins to autophagic degradation.
Figure 2.
Suppressed accumulation of Ub in Atg7-/p62-deficient brains. (A and B) Quantitative mass spectrometry analysis of P2UBA captured total (A) and isopeptide-linked (B) Ub from the soluble and insoluble fractions of autophagy-deficient brains. Age-matched control Atg7 littermates (Atg7flox/+:nestin-Cre:p62+/−; denoted +) at 6 wk (open circles; n = 4), p62 KO (Atg7flox/flox:p62−/−; denoted −) at 6 wk (closed circles; n = 3), Atg7 KO (Atg7flox/flox:nestin-Cre:p62+/−; denoted −) at 6 wk (closed diamonds; n = 3), or Atg7-/p62-DKO (Atg7flox/flox:nestin-Cre:p62−/−) at 6 wk (closed triangles; n = 3) are shown. Each symbol represents one animal. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005.
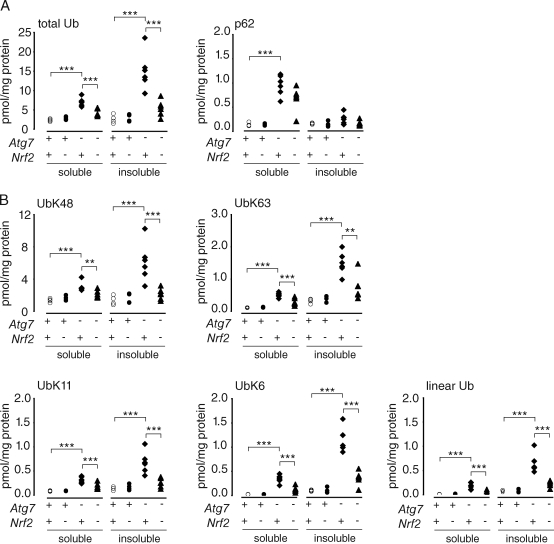
Figure 3.
Suppressed accumulation of Ub in Atg7-/Nrf2-deficient livers. (A and B) Quantitative mass spectrometry analysis of P2UBA captured p62 (A), total (A), and isopeptide-linked (B) Ub from the soluble and insoluble fraction of autophagy-deficient livers (28 d after pIpC injection). Control littermates (F/F or F/F:Nrf2+/−; denoted +; open circles; n = 4), Nrf2 KO (F/F:Nrf2−/−; denoted −; closed circles; n = 4), Atg7 KO (F/F:Mx1 or F/F:Mx1:Nrf2+/−; denoted −; closed diamonds; n = 6), or Atg7-/Nrf2-DKO (F/F:Mx1:Nrf2−/−; closed triangles; n = 6) are shown. Each symbol represents one animal. **, P ≤ 0.01; ***, P ≤ 0.005.
An alternative explanation for the observation that p62 ablation suppresses Ub accumulation and inclusion body formation in Atg7 KO mice (Komatsu et al., 2007) may be related to the recent demonstration that p62 plays a central role as an activator of the major cellular antioxidant stress response pathway mediated by the transcription factor Nrf2 (Komatsu et al., 2010). In unstressed cells, binding of cytoplasmic Nrf2 to Keap1 (kelch-like ECH-associated protein 1), a BTB (broad complex, tramtrack, and bric a brac) domain substrate adaptor, results in its constitutive ubiquitination by a Cul3-Rbx Ub ligase and degradation by the proteasome (Li and Kong, 2009; Nguyen et al., 2009), thereby maintaining low levels of Nrf2-activated gene expression. Oxidation of reactive thiols within Keap1 under conditions of oxidative stress reduces its interaction with Nrf2, allowing Nrf2 to translocate into the nucleus, where it induces the transcription of cytoprotective genes that contain antioxidant response elements (AREs; Rushmore et al., 1991; Venugopal and Jaiswal, 1996; Nioi et al., 2003; Hayes and McMahon, 2009). Recent data demonstrate that binding of p62 to Keap1 disrupts this interaction, leading to Nrf2 stabilization and increased translocation to the nucleus (Komatsu et al., 2010). Elevated levels of p62, resulting from autophagy deficiency in Atg7 KO mice, would thus be expected to promote Nrf2 stabilization and transcription of phase II antioxidant proteins. This hypothesis is supported by the finding that both the Nrf2 transcriptional response and hepatotoxicity, which are strongly activated in Atg7−/− liver, are attenuated in Atg7/Nrf2 double KO (DKO) liver (Komatsu et al., 2010).
Nrf2 regulates the accumulation of all poly-Ub species in autophagy KO liver
To determine whether the accumulation of poly-Ub species in Atg7−/− liver is dependent on Nrf2, we assessed poly-Ub linkages in livers from Atg7−/−, Nrf2−/−, and Atg7-/Nrf2-DKO mice (Fig. 3). Similar to the results from autophagy-deficient brain, total Ub and K48, K63, K11, and K6 chains were all significantly elevated in livers from Atg7-null mice, indicating that we did not identify a difference in the response to autophagy ablation between brain and liver (Fig. 3). Although the increased Ub species in liver was primarily insoluble (Fig. 3), age determined in what fraction the Ub accumulated, as demonstrated by our results comparing 6-, 16-, and 26-wk-old Atg5-null brain. At early times after Atg7 ablation, such as 8 d after polyinosinic acid–polycytidylic acid (pIpC) injection in liver, the increase in Ub was in the soluble fraction, whereas by 28 d after pIpC injection, the increase in Ub was in the insoluble fraction (Komatsu et al., 2007), as was also reflected by our mass spectrometry results of 28 d after pIpC-injected liver (Fig. 3). Although Nrf2 ablation alone did not influence total Ub or poly-Ub levels, the increase in these Ub species in Atg7−/− livers was abrogated in Atg7-/Nrf2-DKO (Fig. 3), indicating that accumulation of poly-Ub species in autophagy-deficient liver requires the transcription factor Nrf2. Together, these data fail to support the conclusion that specific poly-Ub chain topologies serve as signals to selectively target Ub-conjugated proteins to p62 or to the autophagy pathway. Instead, our data together with recent findings from Komatsu et al. (2010) support the conclusion that p62 may play an important and hitherto unappreciated regulatory role at the interface between oxidative stress sensing and control of protein degradation pathways.
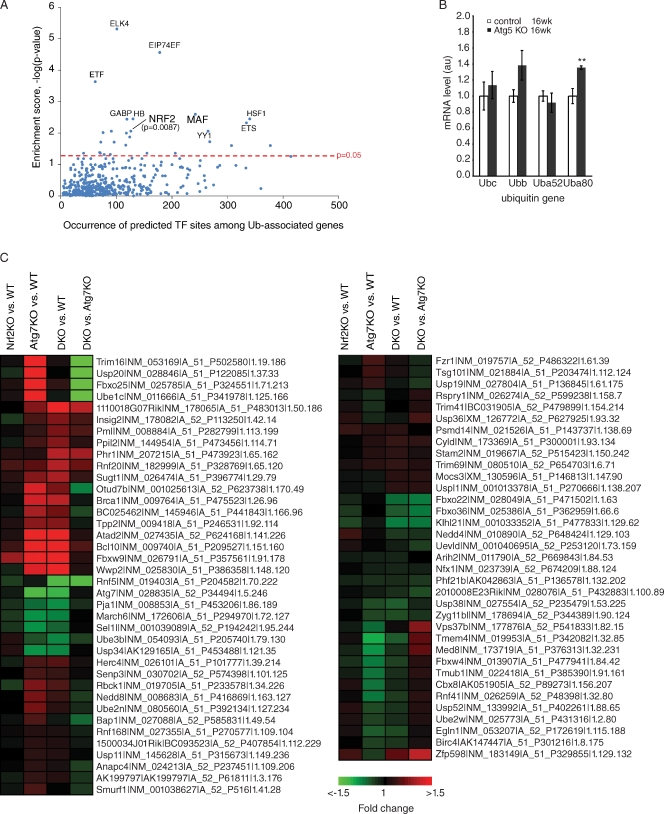
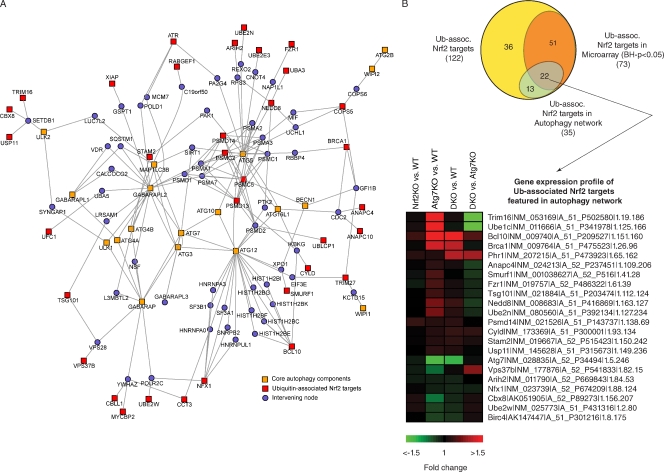
We used bioinformatics and functional genomics to assess whether Nrf2 might activate the Ub pathway or Ub-associated genes as part of a general stress response detoxification mechanism. To this end, we determined whether AREs were enriched in Ub or Ub-associated genes, defined by annotation information from the UniProt database (UniProt Consortium, 2008), Gene Ontology (Ashburner et al., 2000), and Panther classification (Thomas et al., 2003) systems. We found that Nrf2 (and its heterodimeric binding partner Maf, musculoaponeurotic fibrosarcoma oncogene) sites are enriched in Ub-associated genes compared with other transcription factor sites (P = 0.0087; Fig. 4 A) and that Ub-associated genes are significantly enriched among Nrf2 targets (not depicted). Previously, one Ub gene, UbC, was reported to contain an ARE consensus sequence (Rangasamy et al., 2004); however, our analysis, which incorporated stringent conservation information across four organisms, did not identify AREs within any of the Ub genes, although by RT-PCR we did detect a small increase in one Ub gene (Fig. 4 B). In support of the hypothesis that Nrf2 regulates expression of Ub-associated genes during autophagy deficiency, microarray analysis showed significant changes in numerous Ub-associated Nrf2 targets in Atg7−/− livers that returned to wild-type levels in the Atg7-/Nrf2-DKO livers (Fig. 4 C), of which we validated a few by quantitative real-time PCR (Fig. S2). Moreover, computational mapping of the second order network of core autophagy components revealed an enrichment of Ub-associated Nrf2 targets within the autophagy network compared with the genome (Fig. 5 A); several of these targets were changed in the Atg7−/− livers and returned to wild-type levels in the Atg7-/Nrf2-DKO livers (Fig. 5 B). These data suggest an unexpected degree of interconnectivity between Nrf2/p62 stress signaling, autophagy, and Ub homeostasis and provide a plausible alternative explanation for the observed p62 dependence of Ub accumulation in Atg7 KO mice.
Figure 4.
Nrf2 transcription factor–binding sites are enriched among Ub-associated genes. (A) Enrichment of predicted transcription factor (TF)–binding sites in the promoter region (−5,000 to 300) of Ub-associated genes versus its representation genome wide. The top 10 most enriched transcription factor–binding sites predicted among Ub-associated genes are labeled, with their enrichment scores (or −Log10–transformed Benjamini–Hochberg-adjusted p-values) plotted against its occurrence. (B) Elevated Ub mRNA levels from Atg5-deficient brains. Total RNA was extracted from control Atg5 littermates (Atg5flox/+;nestin-Cre) at 16 wk or Atg5 KO (Atg5flox/flox;nestin-Cre) at 16 wk (n = 4 for each genotype; **, P ≤ 0.01) and the amount of transcript from Ubc, Ubb, Uba52, and Uba80 genes was quantified using real-time RT-PCR and normalized to actin. Error bars indicate SEM. au, arbitrary unit. (C) Gene expression analysis of Ub-associated Nrf2 targets mapped to mouse orthologues. Probes detecting transcripts exhibiting differential expression (Benjamini–Hochberg-adjusted P < 0.05) in at least one of the four comparisons are shown. Heat map represents Log2 (fold changes) with respect to either wild type (WT) or Atg7 KO (as indicated). Labels show the symbol, RefSeq (accession numbers from the NCBI Nucleotide database), gene probe name, and the gene row and gene column (i.e., array coordinates).
Figure 5.
Ub-associated Nrf2 targets are enriched in the autophagy network. (A) A second-order network centered on the core autophagy components (orange nodes) identified via PPIs showed an enrichment (hypergeometric P = 0.01) of Ub-associated Nrf2 targets (red nodes) in the network. (B) Venn diagram showing the various counts and overlaps between the microarray and network analysis. Also shown are the Ub-associated Nrf2 targets featured in the autophagy network mapped to mouse orthologues. Probes detecting transcripts exhibiting differential expression (Benjamini–Hochberg [BH]-adjusted P < 0.05) in at least one of the four comparisons are shown. Heat map represents Log2 (fold changes) with respect to either wild type (WT) or Atg7 KO (as indicated). Labels show the symbol, RefSeq (accession numbers from the NCBI Nucleotide database), gene probe name, and the gene row and gene column (i.e., array coordinates).
Selective accumulation of mutant htt after loss of autophagy
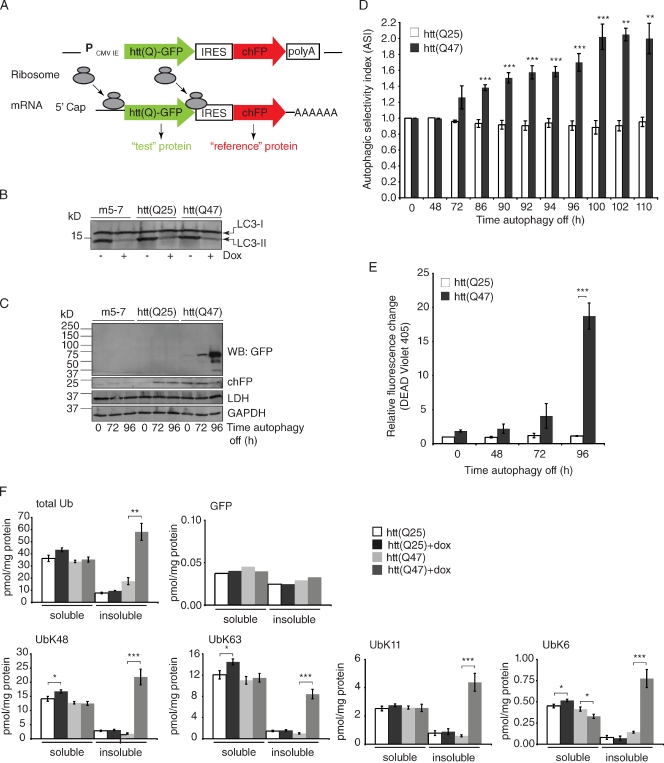
The accumulation of all Ub species in neural and liver-specific Atg5- and Atg7-deficient models and the dependence on Nrf2 led us to speculate that there must be additional features of a protein besides the presence of a poly-Ub chain that specify substrate targeting to autophagy. By analogy to the cytoplasm to vacuole pathway in yeast, in which oligomerization of AP-1 drives selective autophagy, we hypothesized that substrate oligomerization may constitute a signal for selective autophagy in mammals. To test this hypothesis, we developed a flow cytometry–based strategy to assess selective autophagy in mammalian cells. We constructed bicistronic reporters that express a 5′ green fluorescent test protein, translated in a cap-dependent manner, together with a 3′ reference protein, translated from an internal ribosome entry site (IRES; Fig. 6 A; Pestova et al., 2001; Ngoi et al., 2004; Santacruz et al., 2005). We used cherry fluorescent protein (chFP), which is very stable when expressed in mammalian cells (Shaner et al., 2005), as the reference protein. Because both fluorescent fusion proteins are simultaneously expressed under the control of the same cytomegalovirus promoter, their relative rates of synthesis should be equal, and thus changes in their relative steady-state abundance reflect different rates of degradation (Yen et al., 2008). To assess the role of autophagy in degrading the two fusion proteins, we stably expressed these constructs in m5-7 cells, an Atg5−/− mouse embryo fibroblast line harboring the Atg5 gene under the control of a doxycycline (dox)-repressible promoter (Hosokawa et al., 2006). The addition of dox to parental m5-7 cells or to lines expressing constructs composed of htt exon 1–GFP in the test position resulted in substantial depletion of phosphatidylethanolamine-modified LC3 (LC3-II; a marker of autophagosome formation; Fig. 6 B), as previously reported for m5-7 cells (Hosokawa et al., 2006), indicating that expression of these constructs does not affect the ability of Atg5 to be controlled by dox. After the addition of dox, there was no detectable change in the starvation-induced autophagy substrate, lactate dehydrogenase (LDH), or a substrate of chaperone-mediated autophagy, glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Fig. 6 C; Cuervo et al., 1994, 1997).
Figure 6.
Flow cytometry assay to quantify selective autophagy. (A) Diagram of the bicistronic reporter construct. Ribosomes are recruited to both the 5′ end of the bicistronic construct to translate the test fusion protein fused to GFP and at the IRES to translate the stable, nonaggregating reference protein chFP. The test protein is either htt exon 1 with a nonpathogenic poly-Gln repeat htt(Q25) fused to GFP or htt with an expanded poly-Gln tract, htt(Q47)-GFP, which promotes aggregation. (B) The addition of dox for 96 h decreased autophagy in the bicistronic stably expressing cell lines similar to the parental line, m5-7, as measured by the decreased levels of phosphatidylethanolamine-modified LC3 (LC3-II). (C) Western blot (WB) analysis of the accumulation of htt(Q)-GFP after autophagy shutoff. At steady-state (t = 0 h), the levels of both htt(Q25)-GFP and htt(Q47)-GFP were virtually undetectable. After autophagy shutoff, there was a robust increase in the level of htt(Q47)-GFP, as detected by GFP Western blot, whereas there was very little or no detectable increase in htt(Q25)-GFP, GAPDH, LDH, or chFP. (D) Quantitative flow cytometry time course analysis measuring selective autophagy in live cells. The ASI is the ratio of the relative increase in green and red fluorescence, as assessed by two-color flow cytometry, in response to dox. In control experiments, the ASI of two soluble, nonaggregating proteins, htt(Q25)-GFP and chFP, was 1. In contrast, the ASI of the aggregation-prone protein, htt(Q47)-GFP, compared with chFP after autophagy shutoff (+dox; 100 h) was 2. Data represent three independent experiments performed on different days. **, P ≤ 0.01; ***, P ≤ 0.005. (E) Flow cytometry time course analysis of cell death at different time points after autophagy shutoff (+dox). The degree of cell death correlated directly with increased htt(Q47)-GFP levels after autophagy shutoff and was significantly enhanced in cells expressing htt(Q47)-GFP compared with htt(Q25)-GFP (***, P ≤ 0.005). Data represent three independent experiments performed on different days. (F) AQUA using heavy isotope–labeled GFP standard in addition to the routine Ub standards after P2UBA pull-down of soluble and insoluble lysates from htt(Q)-GFP-IRES-chFP bicistronic expressing cells before (−dox) and after autophagy shutoff (+dox). The experimental GFP peptide was only detected in two of the four experiments, and the data shown are representative of these. In the other two experiments, the experimental GFP peptide was below the threshold of detection. (D–F) Error bars indicate SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005.
To test the hypothesis that aggregation-prone proteins are selective autophagy substrates compared with stable, monomeric cytosolic proteins, we used htt exon 1, htt(Q47)-GFP, with an expanded poly-Gln tract that promotes aggregation as the test protein and chFP as the reference. As a control, we used the nonpathogenic htt(Q25)-GFP, which differs from htt(Q47)-GFP only in lacking the poly-Gln expansion and the propensity to aggregate (Busch et al., 2003). In the absence of dox, neither htt fusion protein was detectable by immunoblot (Fig. 6 C), and green fluorescence levels were similar to those of parental m5-7 cells (not depicted). The addition of dox led to a robust increase in htt(Q47)-GFP immunoreactivity (Fig. 6 C) and green fluorescence (Fig. 6 D), which was detectable after an initial lag of 48–72 h, correlating well with the time course of LC3-II disappearance. To confirm that dox addition does not affect the relative rates of cap-dependent and -independent translation, we monitored incorporation of [35S]Met into htt(Q47)-GFP and chFP (Fig. S3 A). Although cap-independent translation from an IRES can increase during cellular stress (Fernandez et al., 2002; Kozak, 2005), we did not measure a significant difference in the relative rates of incorporation of [35S]Met into htt(Q47)-GFP and chFP in cells treated with dox for 72 or 96 h (Fig. S3 A). Additionally, Northern blot analysis confirmed that the transcript was the expected size and failed to reveal increased levels of htt(Q47)-GFP-IRES-chFP mRNA in response to dox (Fig. S3 B). Together, these data indicate that m5-7 cells efficiently degrade htt(Q47)-GFP in a manner that depends on continued expression of Atg5. Addition of the proteasome inhibitor MG132, in the absence of dox, resulted in increased htt(Q47)-GFP and chFP levels, indicating that the proteasome contributes to the degradation of both proteins. However, addition of MG132 and dox resulted in an additive increase in htt(Q47)-GFP yet minimal increase in chFP (Fig. S3 C). Thus, chFP is primarily a proteasome substrate, whereas htt(Q47)-GFP can partition between the proteasome and autophagy pathways. To determine the autophagic substrate selectivity of htt(Q47)-GFP compared with chFP or htt(Q25)-GFP, we used two-color flow cytometry (Fig. 6 D). We used the ratio of relative increase in green and red fluorescence, assessed by two-color flow cytometry, in response to dox to generate the autophagic selectivity index (ASI). With htt(Q25)-GFP in the test position, the ASI at t = 100 h was close to 1, indicating the same amount of basal autophagy of the test and reference reporters (Fig. 6 D), and loss of autophagy did not result in increased cell death (Fig. 6 E). In contrast, with htt(Q47)-GFP as the test protein, the ASI at t = 100 h was 2 (Fig. 6 D) and corresponded with a substantial increase in cell death (Fig. 6 E). Overall, these data demonstrate that the aggregation-prone fragment containing an expanded polyQ tract selectively accumulates over monomeric, cytoplasmic proteins after autophagy inhibition and that autophagy is pivotal in reducing the toxicity of aggregation-prone proteins.
To determine whether accumulation of a selective autophagy substrate correlates with an increase in a specific poly-Ub species, which would be expected if a specific Ub conjugate served as a targeting signal, we used Ub AQUA analysis to assess total Ub and K48, K63, K11, and K6 isopeptides. The addition of dox to cells expressing htt(Q25)-GFP did not result in increased Ub species in either soluble or insoluble fractions, which is consistent with previous studies showing the absence of Ub-positive inclusions in unstressed Atg5 KO cells (Szeto et al., 2006; Kuma et al., 2007). Although expression of htt(Q47)-GFP alone did not lead to changes in the levels of Ub species, the addition of dox to cells expressing htt(Q47)-GFP led to accumulation of total Ub and all major Ub species in the insoluble fraction (Fig. 6 F). Moreover, the amount of Ub-associated htt(Q47)-GFP was <0.05 pmol/mg, representing <0.1% of the total Ub species that accumulated in htt(Q47)-GFP cells after autophagy shutoff (Fig. 6 F). These data suggest that a specific Ub conjugate does not target the aggregation-prone protein, htt(Q47)-GFP, for selective autophagy and that the increased Ub levels observed in autophagy-deficient cells are not caused by increased substrate ubiquitination.
Discussion
Accumulation of poly-Ub conjugates within intracellular inclusion bodies in response to loss or inactivation of the autophagy system is consistent with the hypothesis that polyubiquitination of proteins serves as a cis-acting signal for their selective recognition by and targeting to the autophagy machinery (Kirkin et al., 2009c). Alternatively, it is possible that loss of function or capacity in the autophagy pathway leads to activation of cellular stress response systems that increase the net balance between Ub conjugation and deconjugation. In this study, we observed that all detectable poly-Ub linkage topologies increase in parallel after loss of autophagy in three different mouse models of autophagy deficiency and in tissue culture cells after selective inactivation of the autophagy pathway. These results do not support the hypothesis that a unique Ub–Ub linkage topology is a cis-acting signal for selective substrate recognition by autophagy. Our inability to detect increased levels of Ub conjugation to autophagy substrates after inactivation of the core autophagy machinery, together with our observation of a tight correlation between aggregate formation and autophagic substrate selection, led us to propose that protein oligomerization drives autophagic substrate specificity. We propose that accumulation of poly-Ub chains in autophagy deficiency is an indirect consequence of p62-mediated activation of the Nrf2-dependent stress response pathway. Stabilization of Nrf2 and up-regulation of ARE-containing genes have been observed in several neurodegenerative disorders, although it remains to be determined whether sustained Nrf2 signaling plays a protective or causative role in these neurodegenerative disorders (de Vries et al., 2008; Johnson et al., 2008; van Roon-Mom et al., 2008). Interestingly, the accumulation of all Ub species in Atg7−/− and Atg5−/− mice is similar to the effect of expression of mutant htt in HD transgenic models and human HD patients (Bennett et al., 2007), suggesting that accumulation of Ub conjugates in all of these conditions may be the consequence of activation of related stress response pathways.
The presence of a UBA domain in p62 and its consistent enrichment in Ub-positive inclusion bodies (Zatloukal et al., 2002; Nagaoka et al., 2004) led to the proposal that it serves as an adaptor that targets proteins for degradation by autophagy (Bjørkøy et al., 2005). This model is not consistent with our data showing that p62 levels do not increase in parallel with Ub chains in detergent-insoluble extracts from autophagy-deficient mice and that Ub-associated p62 does not constitute a significant fraction of total p62. Consistent with our work showing that p62 regulates global Ub metabolism through Nrf2 rather than through direct binding, previous studies have failed to detect significant affinity between p62’s isolated UBA domain and unanchored Ub chains (Raasi et al., 2005; Kirkin et al., 2009b) and demonstrated that the p62 UBA domain interacts with Ub with ∼0.5 mM or weaker affinity (Long et al., 2008). However, Ub binding by p62 likely requires p62 oligomerization (Kirkin et al., 2009b). Data from Kirkin et al. (2009b) showed that in vitro translated p62 binds to GST-Ub (or GST-4xUb) and is in part dependent on an intact p62 PB1 (Phox and Bem1p) domain that mediates p62 oligomerization. We attempted to mimic the avidity-based binding made possible by oligomerization by covalently conjugating p62’s UBA domain to beads and still failed to observe Ub binding, perhaps suggesting that PB1-mediated p62 oligomerization may array UBA domains in a geometry conducive to binding Ub that is not recapitulated on beads. We cannot completely rule out the direct involvement of p62’s UBA domain in Ub-dependent autophagic substrate degradation; however, our Ub measurements in the context of Atg7 and p62 or Nrf2 DKO tissues are in agreement with recent data implicating p62 as an activator of the Nrf2-mediated stress response pathway (Liu et al., 2007; Komatsu et al., 2010) together with data demonstrating Nrf2 (Ishii et al., 2000) and Maf (Jain et al., 2010) regulation of p62. Although, we were unable to identify with confidence any Nrf2 ARE sites for p62, even if we extended the search to 10 kb upstream, Jain et al. (2010) reported the existence of a Maf motif in p62’s promoter and showed that Nrf2 itself does not bind to this Maf element but that the Nrf2–MafG complex does. Together, these data suggest the existence of an Nrf2:p62 feedback loop. We propose that increased soluble p62, resulting from Atg7 depletion, leads to inhibition of Keap1 binding, stabilization of Nrf2, and, consequently, increased transcription of Nrf2-dependent genes, including those directly affecting Ub metabolism.
Our observation that aggregation rather than ubiquitination status strongly correlates with selective accumulation of mutant htt after Atg5 inactivation supports the hypothesis that aggregation or oligomerization is a key signal for substrate capture by the autophagosome. Substrate oligomerization as a cis-acting selective autophagy feature is not unique to misfolded aggregation-prone proteins because the autophagy substrates P granules (Zhang et al., 2009) and midbody rings (Pohl and Jentsch, 2009) all oligomerize and form inclusions after inactivation of autophagy in the absence of evidence of their modification with poly-Ub. Moreover, although p62 accumulates in Atg7−/− mice, it was not highly enriched on Ub-associated affinity beads, suggesting that degradation of this selective autophagy substrate also correlates poorly with ubiquitination status. Although our data do not support the hypothesis that covalent poly-Ub conjugation to substrates is the targeting signal for selective autophagy, they do not exclude the possibility that ubiquitination, either of substrates or components of the autophagy pathway, could play a permissive role in autophagy. The presence of a functional poly-Ub–binding UBA domain on the LC3-binding protein NBR1 (Kirkin et al., 2009b), together with immunocytochemical evidence of Ub within lysosomes (Schwartz et al., 1988; Doherty et al., 1989), suggests that proteins other than p62 may mediate an intimate relationship between the autophagy and Ub systems. Ub is a versatile signaling molecule that plays very different roles in a variety of cellular processes ranging from membrane trafficking to DNA repair (Chen and Sun, 2009; Finley, 2009). Further studies will be needed to more precisely elucidate its role in autophagy.
Materials and methods
Antibodies and reagents
Anti-LC3B polyclonal antibody was generated against the N-terminal 15 residues from human LC3B (Iwata et al., 2005). Anti-Atg5 (SO4) and anti-Atg12 (NM2) polyclonal antibodies were provided by N. Mizushima (Mizushima et al., 2001). Anti-GFP monoclonal antibody was purchased from Roche. To detect chFP, we used Living Colors DsRed polyclonal antibody (Takara Bio Inc.). Anti-Ub monoclonal antibody (FK2) was obtained from Enzo Life Sciences, Inc. Anti-p62 polyclonal antibody was raised against the C terminus of p62 (residues 256–439) and was a gift from J. Shin (School of Medicine, Sungkyunkwan University, Suwon, South Korea). Rabbit Anti-LDH (H-160) was purchased from Santa Cruz Biotechnology, Inc., and monoclonal anti-GAPDH (clone 6C5) was purchased from Millipore. Dox (Sigma-Aldrich) was prepared in water, Geneticin (G418) was obtained from Invitrogen, and proteasome inhibitor (MG132) was obtained from Enzo Life Sciences, Inc. FuGENE 6 transfection reagent was purchased from Roche.
Plasmids
pcDNA3.1-chFP–α-tubulin was a gift from R. Tsien (University of California, San Diego, La Jolla, CA; Shaner et al., 2004, 2005) and was used to create pcDNA3.1-chFP. All of the bicistronic vectors used were generated from pIRES2-DsRed-Express (Takara Bio Inc.). The DsRed was removed, and constructs of interest were cloned in the multiple cloning site or the DsRed position (for the majority of constructs this was chFP). pcDNA3.1–htt(Q)-GFP constructs have been described previously (Kazantsev et al., 1999; Bence et al., 2001), and the expression level of htt(Q)-GFP in the bicistronic reporter was comparable with commonly used pcDNA3.1–htt(Q)-GFP constructs, as visualized by immunoblotting (Fig. S4 A), and by fluorescence microscopy, htt(Q47)-GFP formed characteristic inclusion bodies, whereas htt(Q25)-GFP and chFP were diffuse (Fig. S4 B). We also generated bicistronic constructs with htt(Q)-GFP expressed downstream of the IRES (Fig. S4 C); however, because expression of the gene downstream of the IRES is less robust than expression of the upstream gene (Fig. S4 C), htt(Q47)-GFP levels were below the threshold to form inclusion bodies visible by fluorescence microscopy (Fig. S4 D).
Cell culture and transfection
Atg5 Tet-Off cells were provided by from N. Mizushima and N. Hosokawa (Tokyo Medical and Dental University, Bunkyo-ku, Tokyo, Japan), and generation of these cells has been previously described (Hosokawa et al., 2006). Clone m5-7 was further transfected with the aforementioned bicistronic constructs, pcDNA3.1-chFP or pcDNA3.1–htt(Q97)-GFP, and stable transformants were selected and maintained in the presence of 1 mg/ml G418 (Invitrogen). The cells expressing the bicistronic reporters were sorted by gating on both GFP and chFP fluorescence. To further enrich the population of cells expressing the construct of interest, the cells were expanded and resorted an additional two times for a total of three sorts. The final sorted population was then used to sort single cells into each well of a 96-well plate. 10–20 single clones of each cell line were analyzed for GFP, chFP, Atg5, and LC3-II levels and the responsiveness to dox to ensure our results were not caused by a clonal phenomenon. All m5-7–derived cell lines were maintained according to the original described conditions with the exception that cells were not starved before any experiment (Hosokawa et al., 2006). In brief, cells were maintained in DME with 4.5 g/liter glucose and l-Gln without Na pyruvate (Mediatech) containing 10% FBS with or without 10 ng/ml dox (Sigma-Aldrich) at 37°C in 5% CO2. Plasmid transfection was performed using Fugene 6 (Roche) according to the manufacturer’s protocol. For experiments with proteasome inhibitor (MG132), the final concentration was 1 µM.
Cell lysis and immunoblot analysis
Cells were lysed in 50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, and 0.1% SDS containing 1× protease inhibitors (Roche). Extracts were sonicated and centrifuged at 14,000 rpm in a microcentrifuge (Eppendorf 5417c; F45-30-11 rotor at 14,000 rpm) for 10 min at 4°C, and supernatants were quantified using the bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific). Protein was electrophoresed through Tris-Gly gels and transferred to polyvinylidene fluoride (Bio-Rad Laboratories). Membranes were probed overnight at 4°C with antibody. Protein was visualized with ECL plus (GE Healthcare) and quantified using the Typhoon Imager (GE Healthcare) and ImageQuant software (Molecular Diagnostics).
Flow cytometry
We used the FlasherII Diva Digital equipped with a violet-enhanced krypton laser (407 nm), a dye laser (598 nm; em.625/40 nm), and a 488-nm laser (525/50 nm; Stanford University FACS facility). There was effectively no detectable bleed through between the GFP and chFP (and where applicable violet 405) signals; however, the samples were compensated offline with FlowJo (Tree Star, Inc.) using cells expressing only single colors as controls. Cell death was measured using LIVE/DEAD Violet 405 (Invitrogen) according to the manufacturer’s protocol.
Poly-Ub affinity capture
Samples were prepared essentially as described previously (Bennett et al., 2007). Brain hemispheres were homogenized for 90 s (pulsing) using a Teflon micro-rotator in 600 µl of lysis buffer (1% Triton X-100, 150 mM NaCl, 20 mM Tris, pH 8.0, 10% glycerol, 10 mM N-ethylmaleimide [NEM], and complete protease inhibitor tablets, no EDTA [Roche]), followed by sonication (3×) for 10–15 s. Lysates were then centrifuged (2×) for 10 min at 14,000 rpm in a microcentrifuge at 4°C followed by 1× for 5 min at 14,000 rpm in a microcentrifuge at 4°C. The insoluble fraction was resuspended in 300 µl of 2% SDS followed by sonication (2×) for 15–30 s. Both the soluble and insoluble fractions were then spun for an additional 5 min at 14,000 rpm in a microcentrifuge (4°C for soluble and RT for insoluble) and then quantified using BCA (Thermo Fisher Scientific). 1 mg of protein (soluble) was diluted to a final volume of 300 µl of the aforementioned lysis buffer (no NEM or protease inhibitors), to which 75 µl human P2UBA resin (UBA domain from human ubiquilin 2 [UBQLN2] also known as PLIC-2; 1:1 slurry) was added and incubated overnight at 4°C with constant rotation. 1 mg of protein (insoluble) was diluted into 14 ml of lysis buffer (no NEM or protease inhibitors; final SDS concentration was 0.015%), to which 75 µl human P2UBA resin (1:1 slurry) was added and incubated overnight at 4°C with constant rotation.
Livers were homogenized in 0.25 M sucrose, 10 mM Hepes, pH 7.5, and 1 mM DTT. To generate Triton X-100–soluble and –insoluble fractions, Triton X-100 (final concentration 1%) and NaCl (final concentration 150 mM) were added, and the samples were vortexed and lysed on ice for 15 min, followed by sonication (3×; 15 s). Lysates were then centrifuged for 10 min at 14,000 rpm in a microcentrifuge at 4°C. The resulting supernatant was used as the Triton X-100–soluble fraction, and the insoluble fraction was resuspended in 2% SDS followed by sonication (3×; 15 s). Both the soluble and insoluble fractions were then spun for an additional 5 min at 14,000 rpm in a microcentrifuge (4°C for soluble and RT for insoluble) and then quantified using BCA. 2 mg of protein (soluble) was diluted to a final volume of 500 µl in 20 mM Tris, pH 7.6, 150 mM NaCl, and 1% Triton X-100, to which 60 µl human P2UBA resin (1:1 slurry) was added and incubated overnight at 4°C with constant rotation. 250 mg of protein (insoluble) was diluted into 14 ml of buffer (20 mM Tris, pH 7.6, 150 mM NaCl, and 1% Triton X-100), to which 60 µl human P2UBA resin (1:1 slurry) was added and incubated overnight at 4°C with constant rotation.
Mass spectrometry
Samples were prepared as described previously (Bennett et al., 2007). Mass spectrometry was also previously described (Bennett et al., 2007) with four additional ions that were measured by the AQUA method: human p62 (423–437; NYDIGAALDTIQYSK; unlabeled, m/z = 836.41; and heavy isotope labeled, m/z = 839.92), UbK6 (MQIFVK-[GG]-TLTGK; unlabeled, m/z = 460.60; and heavy isotope labeled, m/z = 462.93), linear Ub fusion (GGMQIFVK; unlabeled, m/z = 440.24; and heavy isotope labeled, m/z = 443.23), and GFP (FEGDTLVNR; unlabeled, m/z = 525.76; and heavy isotope labeled, m/z = 528.76). The GFP peptide is unique to GFP and is not found in chFP.
For Orbitrap analysis, the samples were analyzed by liquid chromatography tandem mass spectrometry using a previously described chromatography system and conditions (Bennett et al., 2007). The outlet of the chromatography column was directly coupled to a mass spectrometer (LTQ-Orbitrap XL; Thermo Fisher Scientific). Throughout the chromatography run, three tandem mass spectrometry scans were performed in the linear ion-trap during each parent-ion Orbitrap scan. Orbitrap scans were acquired at a nominal resolution setting of 60,000. The relative intensities of the isotope-labeled standards and analytes were obtained by measuring the intensities of the monoisotopic molecular ions after averaging ∼10 scans across the elution profile of the standard.
Quantitative real-time RT-PCR analysis
Transcript levels of the Ub genes were measured as previously described (Ryu et al., 2008) with one change: the transcript levels were normalized to actin. For RT-PCR of the Ub-associated Nrf2 targets and Nqo1, the SuperScript First-Strand Synthesis system for RT-PCR (Invitrogen) was used, and cDNA was synthesized from 2 µg of total RNA. Quantitative PCR was performed with qPCR Mastermix or qPCR Mastermix Plus for SYBR Green I (Eurogentec S.A.) in an ABI 7700 system (Applied Biosystems). Ribosomal RNA was used as an internal control. The sequences of the primers used were as follows: Nqo1 forward, 5′-AGCTGGAAGCTGCAGACCTG-3′; Nqo1 reverse, 5′-CCTTTCAGAATGGCTGGCA-3′; Nqo1 probe, 5′-ATTTCAGTTCCCATTGCAGTGGTTTGGG-3′; ribosomal RNA forward, 5′-CGGCTACCACATCCAAGGAA-3′; ribosomal RNA reverse, 5′-GCTGGAATTACCGCGGCT-3′; ribosomal RNA probe, 5′-TGCTGGCACCAGACTTGCCCTC-3′; Trim16 forward, 5′-AACAAAGAGGTGGACCTTCG-3′; Trim16 reverse, 5′-TTCACCTCTGACACCGACAC-3′; Usp20 forward, 5′-TACCTGTGACCGGGTGTCTA-3′; Usp20 reverse, 5′-TGGTAGATGGCAGAGTGGAG-3′; Fbxo25 forward, 5′-GTCGGGAACATCAACATCTG-3′; and Fbxo25 reverse, 5′-TACCTGCTTTGTCACCTGGA-3′.
Predicted transcription factor–binding site enrichment of Ub-associated genes
Transcription factor–binding sites were predicted by using the algorithm as described in Benita et al. (2009). In brief, we ran STORM (Smith et al., 2006; Schones et al., 2007) to match position weight matrices (PWMs) in the JASPAR and TRANSFAC collections to genomic DNA. Conservation information was also incorporated into the prediction using multiSTORM (Smith et al., 2006) to achieve PWM matches across at least four organisms. In addition, site-cons algorithm (Smith et al., 2006) was applied to compute the level of conservation at each binding site compared with the flanking 100 bases. Conservation information was obtained from genome alignments available from the University of California, Santa Cruz genome browser project (Karolchik et al., 2008). The predicted Nrf2 sites have a central core motif GGAAG, flanked by sequences that are more highly variable as specified by the PWM. Thus, there is no exact ARE sequence but rather a weighted consensus motif (TRANSFAC accession M00108 and ID V$NRF2_01). Enrichment for the occurrence of transcription factor–binding sites in the promoter regions of Ub-associated genes against its representation genome wide was assessed using the hypergeometric distribution and implemented in the R language. P-values were corrected for multiple testing using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). The set of Ub-associated genes was identified from annotation information from the UniProt database (UniProt Consortium, 2008), Gene Ontology (Ashburner et al., 2000), and Panther classification (Thomas et al., 2003) systems.
Data analysis and linear modeling of gene expression microarray analysis
The microarray data analyzed have been deposited in NCBI Gene Expression Omnibus (GEO; Edgar et al., 2002) and are accessible through GEO series accession number GSE20415. Data analysis and linear modeling were performed in the R statistical programming language, using functions from linear models for microarray data (limma; Smyth, 2005). The moderated t-statistic for each probe was computed using a Bayesian model. The associated p-value was adjusted to control the false discovery rate in multiple testing using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). Differentially expressed genes were identified with an adjusted p-value <0.05. Hierarchical clustering (centroid linkage method) was performed with Cluster 3.0 (Eisen et al., 1998) using Pearson’s correlation as the similarity metric. Heat maps were generated using JavaTreeview (Saldanha, 2004).
Network analysis
A second-order network anchoring on core autophagy components was constructed from protein–protein interaction (PPI) data assembled from a large number of PPI data sources, including IntAct, BIND/BOND, GRID, MINT, HPRD, and DIP, and filtered for high confidence interactions using network topology and a set of reliability scoring metric as detailed in Lage et al. (2007). Enrichment of Ub-associated Nrf2 targets in the autophagy network was assessed using the hypergeometric distribution.
Statistical analysis
Single, double, and triple asterisks denote P ≤ 0.05, P ≤ 0.01, and P ≤ 0.005, respectively. The significance levels were determined using the unpaired t test.
Protein expression, purification, and p62-Ub binding assay
Pet28a-6xHis-p62(298–440) was a gift from J. Shin and was verified by sequencing. p62(298–440) was expressed in Escherichia coli BL21(DE3) (EMD) cells by the addition of 1 mM IPTG after the temperature was shifted from 37°C to 30°C at OD600 1.0. After 6 h of induction, cells were harvested, flash frozen in liquid nitrogen, and stored at −80°C for later use. For protein purification, frozen bacterial cells were resuspended in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 0.5% Triton X-100, 10 mM imidazole, and 1 mM β-mercaptoethanol) and lysed using a French press at 12,000 PSI (Thermo Fisher Scientific). Lysate was then centrifuged in a SS-34 rotor (Sorval) at 18,500 rpm for 60 min, and the supernatant was incubated in batch with Ni-Sepharose resin (GE Healthcare) at 4°C for 4 h with gentle stirring. 6xHis-p62(298–440) was retained on the beads and washed three times with 4 bed vol of lysis buffer before elution with 50 mM Tris, pH 7.5, 500 mM NaCl, 0.5% Triton X-100, 250 mM imidazole, and 1 mM β-mercaptoethanol. p62-containing elutions were dialyzed into 50 mM of Tris buffer, pH 8, 50 mM NaCl, and 10 mM DTT and were loaded onto a hiTrap Q sepharose Fast Flow column (GE Healthcare) and eluted with a linear gradient of NaCl from 50 to 500 mM. p62 was further purified by gel filtration chromatography on a Sephacryl S-200 column (GE Healthcare) and dialyzed into 20 mM Hepes, pH 7.0, and 100 mM NaCl before adding 1 mM Tris(2-carboxyethyl)phosphine and flash freezing aliquots in liquid nitrogen for storage at −80°C.
For p62-UBA binding assays, brain hemispheres were homogenized in 600 µl of lysis buffer (1% Triton X-100, 150 mM NaCl, 20 mM Tris, pH 8.0, 10% glycerol, 16 mM NEM, and complete protease inhibitor tablets, no EDTA [Roche]), followed by sonication (3×) for 10–15 s. Lysates were then centrifuged (2×) for 10 min at 14,000 rpm in a microcentrifuge at 4°C then quantified using BCA. 1.2 mg of protein was diluted to a final volume of 450 µl of the aforementioned lysis buffer (no NEM or protease inhibitors), to which 50 µl of resin (1:1 slurry) was added and incubated overnight at 4°C with constant rotation. Resins used were blocked Affigel-10 (Bio-Rad Laboratories), blocked Sulfolink (Thermo Fisher Scientific), Affigel-P2UBA(575–624) (1 mg protein/ml resin), Affigel-p62UBA(298–440) (1 mg protein/ml resin), and Sulfolink-p62UBA(298–440). Samples were washed, eluted, and prepared for mass spectrometry as described in Poly-Ub affinity capture.
[35S]Met incorporation
For radioactive labeling of newly synthesized proteins, cells were washed twice before starvation in Cys-/Met-free media (15 min), followed by labeling with [35S]Met (250 µCi/10-cm dish) for 1, 2, and 3 h. At the different time points, the plates were washed with 3× ice-cold PBS, and cells were scraped off plates and harvested at 1,000 g for 5 min. Lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, and 0.1% SDS containing 1× protease inhibitors [Roche]) was added, and the cells were vortexed and incubated on ice for 15 min followed by syringe lysis (10x, 21cc, 4x, and 25cc) followed by centrifugation at 4°C (10 min at 14,000 rpm in a microcentrifuge). The resulting supernatants were spotted on Whatman paper and boiled in 10% TCA to remove tRNAs. The [35S]Met incorporation was then measured by scintillation counting, and equal amounts of [35S]Met were used for the 1-, 2-, and 3-h immunoprecipitations.
Northern blotting
Total RNA was prepared using RNeasy Plus (QIAGEN) followed by mRNA purification (2×) from 125 µg of total RNA using Oligo25 beads (Invitrogen). Northern blot analysis was performed as described previously (Streit et al., 2009) with the exception that probes were digoxigenin (DIG) labeled (Roche). To determine the size of the transcript, we used a single-stranded RNA ladder (New England Biolabs, Inc.). The actin DIG-labeled RNA probe (588 bases) was purchased from Roche and used to detect actin (1.8 kb). A chFP probe (584 bases) was constructed by PCR using pcDNA3.1-chFP as a template and 5′-GTGAGCAAGGGCGAGGAG-3′ as the forward primer and 5′-TAATACGACTCACTATAGGGCTTGTACAGCTCGTCCATG-3′ as the reverse primer (t7 tag is underlined). The resulting PCR product was used in the in vitro transcription reaction with DIG-11-UTP (Roche) to make the DIG-labeled chFP probe used to detect htt(Q47)-GFP-IRES-chFP (∼2.4 kb).
Microscopy
For direct fluorescence, cells were grown on glass coverslips, fixed in 4% paraformaldehyde, permeabilized by 0.2% Triton X-100, and stained for 20 min in 50 µg/ml bisbenzimide (Sigma-Aldrich) for nuclear staining and mounted using glycerol/gelatin N-propyl gallate. For immunofluorescence, cells were fixed, permeabilized, and blocked with 5% BSA in PBS followed by primary antibody incubation at 37°C for 1 h, washing in PBS, incubation for 1 h at 37°C with secondary antibodies (Alexa Fluor 488 or 594; Invitrogen) and 50 µg/ml bisbenzimide and finally mounted using glycerol/gelatin N-propyl gallate. Conventional epifluorescence micrographs were obtained on a microscope (Axiovert 200M; Carl Zeiss, Inc.) with a 100× NA 1.4 oil lens (Carl Zeiss, Inc.) or 40× NA 0.75 air objection (Carl Zeiss, Inc.). Digital (12 bit) images were acquired with a cooled charge-coupled device (Roper Industries) and processed by using MetaMorph software (Universal Imaging). For bisbenzimide (Ex. 365/50, Em. 450/65), GFP (Ex.490/20, Em. 535/20) and chFP (Texas red; Ex.560/55, Em. 645/75) the dichroics were: 400/LP (bisbenzimide), 500/LP (GFP or Alexa Fluor 488), and 595/LP (chFP or Alexa Fluor 594). Scale bars equal 1 µm.
Genotype of animals
The genotypes of the animals used in this study are shown in Table I.
Table I.
Genotype of animals
| Name | Genotype | n | Tissue | Age |
| wk | ||||
| control (Atg5) | F/+:Nes | 4 | brain | 16 |
| control (Atg5) | F/+:Nes | 3 | brain | 26 |
| Atg5 KO | F/F:Nes | 4 | brain | 16 |
| Atg5 KO | F/F:Nes | 3 | brain | 26 |
| control (Atg7) | F/+:Nes:p62+/− | 2 | brain | 6 |
| control (Atg7) | F/F:p62+/− | 1 | brain | 6 |
| control (Atg7) | F/F:p62+/−:GFP-LC3+/− | 1 | brain | 6 |
| Atg7 KO | F/F:Nes | 1 | brain | 6 |
| Atg7 KO | F/F:Nes:p62+/−:GFP-LC3+/− | 2 | brain | 6 |
| p62 KO | F/+:p62−/− | 1 | brain | 6 |
| p62 KO | F/F:p62−/− | 1 | brain | 6 |
| p62 KO | F/F:p62−/−:GFP-LC3+/− | 1 | brain | 6 |
| Atg7-/p62-DKO | F/F:Nes:p62−/− | 2 | brain | 6 |
| Atg7-/p62-DKO | F/F:Nes:p62−/−:GFP-LC3+/− | 1 | brain | 6 |
| control (Atg7) | F/F | 2 | liver | 6 |
| control (Atg7) | F/F:Nrf2+/− | 2 | liver | 6 |
| Atg7 KO | F/F:Mx1 | 2 | liver | 6 |
| Atg7 KO | F/F:Mx1:Nrf2+/− | 3 | liver | 6 |
| Atg7 KO | F/F:Mx1:Nrf2+/−:GFP-LC3+/− | 1 | liver | 6 |
| Nrf2 KO | F/F:Nrf2−/− | 3 | liver | 6 |
| Nrf2 KO | F/F:Nrf2−/−:GFP-LC3 | 1 | liver | 6 |
| Atg7-/Nrf2-DKO | F/F:Mx1:Nrf2−/− | 5 | liver | 6 |
| Atg7-/Nrf2-DKO | F/F:Mx1:Nrf2−/−:GFP-LC3+/− | 1 | liver | 6 |
Online supplemental material
Fig. S1 shows the comparison of p62-UBA and P2UBA Ub binding. Fig. S2 shows quantitative real-time PCR validation of the Nrf2 Ub-associated genes identified by microarray. Fig. S3 shows that selective accumulation of htt(Q47)-GFP over chFP after autophagy shutoff is not caused by differences in cap-dependent versus cap-independent translation or proteasome inhibition. Fig. S4 shows Western blot and fluorescence microscopy analysis of the bicistronic constructs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005012/DC1.
Acknowledgments
We thank N. Hosokawa for the m5-7 cells, J. Shin for the p62 antibody, T. Ishii and T. Yanagawa for the p62−/− animals, R. Tsien for the chFP plasmid, and B. Chen and E. Sofos for generating constructs. We acknowledge the technical assistance of T. Knaak and C. Crumpton at the Stanford University FACS facility, and are thankful to members of the Kopito laboratory, Autophagy Biologists of the Bay Area, and David Madden for helpful discussion.
This work was supported by a fellowship award from the Hereditary Disease Foundation (to B.E. Riley), a fellowship award from the Crohn’s and Colitis Foundation of America (to A.C.Y. Ng), and grants from the Huntington’s Disease Society of America, Cure Huntington’s Disease, Forskningsrådet for Sundhed og Sygdom (to K. Lage), Eunice Kennedy Shriver National Institute of Child Health and Human Development (to K. Lage), and National Institute of Neurological Disease and Stroke (to R.R. Kopito and R.J. Xavier).
B.E. Riley and R.R. Kopito devised the bicistronic cell-based strategy. B.E. Riley designed, performed, and analyzed all cell-based experiments. B.E. Riley and S.E. Kaiser performed all P2UBA pull-down experiments and analyzed all of the mass spectrometry data. T.A. Shaler obtained all of the mass spectrometry data and contributed mass spectrometry technical discussion. T. Hara performed all of the Atg5−/− brain dissections. M.S. Hipp contributed to the discussion of the results. A.C.Y. Ng and R.J. Xavier performed the computational analyses of transcription factor–binding site enrichment, gene expression microarrays, and the autophagy PPI network. K. Lage contributed the integrated PPI data. K.-Y. Ryu contributed the real-time RT-PCR data, and B.E. Riley prepared the RNA for the Ub genes. K. Taguchi performed the gene expression microarrays and the real-time RT-PCR data for Nqo1 and the Nrf2 Ub-associated genes, and M. Yamamoto supervised this work. M. Yamamoto provided the Nrf2−/− animals. N. Mizushima supervised the Atg5−/− experiments and discussed the results. M. Komatsu performed all of the Atg7, p62, and Nrf2 brain/liver dissections and discussed the results, and K. Tanaka supervised this work. B.E. Riley and R.R. Kopito wrote the manuscript, and S.E. Kaiser contributed to editing.
Footnotes
Abbreviations used in this paper:
- AQUA
- absolute quantification
- ARE
- antioxidant response element
- ASI
- autophagic selectivity index
- BCA
- bicinchoninic acid
- chFP
- cherry fluorescent protein
- DIG
- digoxigenin
- DKO
- double KO
- dox
- doxycycline
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HD
- Huntington’s disease
- htt
- huntingtin
- IRES
- internal ribosome entry site
- KO
- knockout
- LDH
- lactate dehydrogenase
- NEM
- N-ethylmaleimide
- pIpC
- polyinosinic acid–polycytidylic acid
- PMW
- position weight matrix
- PPI
- protein–protein interaction
- Ub
- ubiquitin
- UBA
- Ub association
References
- Alves-Rodrigues A., Gregori L., Figueiredo-Pereira M.E. 1998. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 21:516–520 10.1016/S0166-2236(98)01276-4 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. 2008. Adapting proteostasis for disease intervention. Science. 319:916–919 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- Benita Y., Kikuchi H., Smith A.D., Zhang M.Q., Chung D.C., Xavier R.J. 2009. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 37:4587–4602 10.1093/nar/gkp425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- Bennett E.J., Shaler T.A., Woodman B., Ryu K.Y., Zaitseva T.S., Becker C.H., Bates G.P., Schulman H., Kopito R.R. 2007. Global changes to the ubiquitin system in Huntington’s disease. Nature. 448:704–708 10.1038/nature06022 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Engemann S., Lurz R., Okazawa H., Lehrach H., Wanker E.E. 2003. Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington’s disease. J. Biol. Chem. 278:41452–41461 10.1074/jbc.M303354200 [DOI] [PubMed] [Google Scholar]
- Carra S., Brunsting J.F., Lambert H., Landry J., Kampinga H.H. 2009. HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2alpha phosphorylation. J. Biol. Chem. 284:5523–5532 10.1074/jbc.M807440200 [DOI] [PubMed] [Google Scholar]
- Chau V., Tobias J.W., Bachmair A., Marriott D., Ecker D.J., Gonda D.K., Varshavsky A. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 243:1576–1583 10.1126/science.2538923 [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Sun L.J. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 33:275–286 10.1016/j.molcel.2009.01.014 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., Terlecky S.R., Dice J.F., Knecht E. 1994. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J. Biol. Chem. 269:26374–26380 [PubMed] [Google Scholar]
- Cuervo A.M., Dice J.F., Knecht E. 1997. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 272:5606–5615 10.1074/jbc.272.9.5606 [DOI] [PubMed] [Google Scholar]
- de Vries H.E., Witte M., Hondius D., Rozemuller A.J., Drukarch B., Hoozemans J., van Horssen J. 2008. Nrf2-induced antioxidant protection: a promising target to counteract ROS-mediated damage in neurodegenerative disease? Free Radic. Biol. Med. 45:1375–1383 10.1016/j.freeradbiomed.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Doherty F.J., Osborn N.U., Wassell J.A., Heggie P.E., Laszlo L., Mayer R.J. 1989. Ubiquitin-protein conjugates accumulate in the lysosomal system of fibroblasts treated with cysteine proteinase inhibitors. Biochem. J. 263:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 95:14863–14868 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J., Yaman I., Sarnow P., Snider M.D., Hatzoglou M. 2002. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J. Biol. Chem. 277:19198–19205 10.1074/jbc.M201052200 [DOI] [PubMed] [Google Scholar]
- Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477–513 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S.A., Rush J., Stemman O., Kirschner M.W., Gygi S.P. 2003. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA. 100:6940–6945 10.1073/pnas.0832254100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hayes J.D., McMahon M. 2009. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 34:176–188 10.1016/j.tibs.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., Mizushima N. 2006. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580:2623–2629 10.1016/j.febslet.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. 2008. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283:22847–22857 10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- Ikeda F., Dikic I. 2008. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 9:536–542 10.1038/embor.2008.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., Yamamoto M. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023–16029 10.1074/jbc.275.21.16023 [DOI] [PubMed] [Google Scholar]
- Iwata A., Christianson J.C., Bucci M., Ellerby L.M., Nukina N., Forno L.S., Kopito R.R. 2005. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl. Acad. Sci. USA. 102:13135–13140 10.1073/pnas.0505801102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A., McMahon M., Hayes J.D., Johansen T. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285:22576–22591 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.A., Johnson D.A., Kraft A.D., Calkins M.J., Jakel R.J., Vargas M.R., Chen P.C. 2008. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann. N. Y. Acad. Sci. 1147:61–69 10.1196/annals.1427.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. 2008. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 36:D773–D779 10.1093/nar/gkm966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. 1999. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. USA. 96:11404–11409 10.1073/pnas.96.20.11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.T., Kim K.P., Lledias F., Kisselev A.F., Scaglione K.M., Skowyra D., Gygi S.P., Goldberg A.L. 2007. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282:17375–17386 10.1074/jbc.M609659200 [DOI] [PubMed] [Google Scholar]
- Kim J., Scott S.V., Oda M.N., Klionsky D.J. 1997. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137:609–618 10.1083/jcb.137.3.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Johansen T., Dikic I. 2009a. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 5:732–733 10.4161/auto.5.5.8566 [DOI] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009b. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kirkin V., McEwan D.G., Novak I., Dikic I. 2009c. A role for ubiquitin in selective autophagy. Mol. Cell. 34:259–269 10.1016/j.molcel.2009.04.026 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick D.S., Denison C., Gygi S.P. 2005. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol. 7:750–757 10.1038/ncb0805-750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12:213–223 [DOI] [PubMed] [Google Scholar]
- Kopito R.R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524–530 10.1016/S0962-8924(00)01852-3 [DOI] [PubMed] [Google Scholar]
- Kozak M. 2005. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 33:6593–6602 10.1093/nar/gki958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Matsui M., Mizushima N. 2007. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 3:323–328 [DOI] [PubMed] [Google Scholar]
- Lage K., Karlberg E.O., Størling Z.M., Olason P.I., Pedersen A.G., Rigina O., Hinsby A.M., Tümer Z., Pociot F., Tommerup N., et al. 2007. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25:309–316 10.1038/nbt1295 [DOI] [PubMed] [Google Scholar]
- Lamark T., Kirkin V., Dikic I., Johansen T. 2009. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 8:1986–1990 [DOI] [PubMed] [Google Scholar]
- Larsen K.E., Sulzer D. 2002. Autophagy in neurons: a review. Histol. Histopathol. 17:897–908 [DOI] [PubMed] [Google Scholar]
- Li W., Kong A.N. 2009. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 48:91–104 10.1002/mc.20465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kern J.T., Walker J.R., Johnson J.A., Schultz P.G., Luesch H. 2007. A genomic screen for activators of the antioxidant response element. Proc. Natl. Acad. Sci. USA. 104:5205–5210 10.1073/pnas.0700898104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Gallagher T.R., Cavey J.R., Sheppard P.W., Ralston S.H., Layfield R., Searle M.S. 2008. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J. Biol. Chem. 283:5427–5440 10.1074/jbc.M704973200 [DOI] [PubMed] [Google Scholar]
- Meierhofer D., Wang X., Huang L., Kaiser P. 2008. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J. Proteome Res. 7:4566–4576 10.1021/pr800468j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668 10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka U., Kim K., Jana N.R., Doi H., Maruyama M., Mitsui K., Oyama F., Nukina N. 2004. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J. Neurochem. 91:57–68 10.1111/j.1471-4159.2004.02692.x [DOI] [PubMed] [Google Scholar]
- Nair U., Klionsky D.J. 2005. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 280:41785–41788 10.1074/jbc.R500016200 [DOI] [PubMed] [Google Scholar]
- Nezis I.P., Simonsen A., Sagona A.P., Finley K., Gaumer S., Contamine D., Rusten T.E., Stenmark H., Brech A. 2008. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180:1065–1071 10.1083/jcb.200711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoi S.M., Chien A.C., Lee C.G. 2004. Exploiting internal ribosome entry sites in gene therapy vector design. Curr. Gene Ther. 4:15–31 10.2174/1566523044578095 [DOI] [PubMed] [Google Scholar]
- Nguyen T., Nioi P., Pickett C.B. 2009. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 284:13291–13295 10.1074/jbc.R900010200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J.D. 2003. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 374:337–348 10.1042/BJ20030754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R.A. 2006. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 29:528–535 10.1016/j.tins.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Chin L.S. 2008. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 4:85–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S., Aebersold R., Chen R., Rush J., Goodlett D.R., McIntosh M.W., Zhang J., Brentnall T.A. 2009. Mass spectrometry based targeted protein quantification: methods and applications. J. Proteome Res. 8:787–797 10.1021/pr800538n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921–926 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA. 98:7029–7036 10.1073/pnas.111145798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M., Fushman D. 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8:610–616 10.1016/j.cbpa.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Pohl C., Jentsch S. 2009. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat. Cell Biol. 11:65–70 10.1038/ncb1813 [DOI] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., Balch W.E. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78:959–991 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Raasi S., Varadan R., Fushman D., Pickart C.M. 2005. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 12:708–714 10.1038/nsmb962 [DOI] [PubMed] [Google Scholar]
- Rangasamy T., Cho C.Y., Thimmulappa R.K., Zhen L., Srisuma S.S., Kensler T.W., Yamamoto M., Petrache I., Tuder R.M., Biswal S. 2004. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J. Clin. Invest. 114:1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C.A., Poirier M.A. 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10:S10–S17 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- Rushmore T.H., Morton M.R., Pickett C.B. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 266:11632–11639 [PubMed] [Google Scholar]
- Ryu K.Y., Sinnar S.A., Reinholdt L.G., Vaccari S., Hall S., Garcia M.A., Zaitseva T.S., Bouley D.M., Boekelheide K., Handel M.A., et al. 2008. The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol. Cell. Biol. 28:1136–1146 10.1128/MCB.01566-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A.J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics. 20:3246–3248 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Santacruz K., Lewis J., Spires T., Paulson J., Kotilinek L., Ingelsson M., Guimaraes A., DeTure M., Ramsden M., McGowan E., et al. 2005. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 309:476–481 10.1126/science.1113694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones D.E., Smith A.D., Zhang M.Q. 2007. Statistical significance of cis-regulatory modules. BMC Bioinformatics. 8:19 10.1186/1471-2105-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A.L., Ciechanover A., Brandt R.A., Geuze H.J. 1988. Immunoelectron microscopic localization of ubiquitin in hepatoma cells. EMBO J. 7:2961–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Guan J., Hutchins M.U., Kim J., Klionsky D.J. 2001. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 7:1131–1141 10.1016/S1097-2765(01)00263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Babu J.R., Geetha T., Wong H.C., Krishna N.R., Wooten M.W. 2004. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 24:8055–8068 10.1128/MCB.24.18.8055-8068.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Geetha T., Wooten M.W. 2007. Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 581:175–179 10.1016/j.febslet.2006.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Steinbach P.A., Tsien R.Y. 2005. A guide to choosing fluorescent proteins. Nat. Methods. 2:905–909 10.1038/nmeth819 [DOI] [PubMed] [Google Scholar]
- Smith A.D., Sumazin P., Xuan Z., Zhang M.Q. 2006. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc. Natl. Acad. Sci. USA. 103:6275–6280 10.1073/pnas.0508169103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. 2005. limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Gentleman R., Carey V., Huber W., Irizarry R., Dudoit S., Springer, New York: 397–420 [Google Scholar]
- Streit S., Michalski C.W., Erkan M., Kleeff J., Friess H. 2009. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat. Protoc. 4:37–43 10.1038/nprot.2008.216 [DOI] [PubMed] [Google Scholar]
- Szeto J., Kaniuk N.A., Canadien V., Nisman R., Mizushima N., Yoshimori T., Bazett-Jones D.P., Brumell J.H. 2006. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2:189–199 [DOI] [PubMed] [Google Scholar]
- Tan J.M., Wong E.S., Kirkpatrick D.S., Pletnikova O., Ko H.S., Tay S.P., Ho M.W., Troncoso J., Gygi S.P., Lee M.K., et al. 2008. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 17:431–439 10.1093/hmg/ddm320 [DOI] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13:2129–2141 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium 2008. The universal protein resource (UniProt). Nucleic Acids Res. 36:D190–D195 10.1093/nar/gkm895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roon-Mom W.M., Pepers B.A., ’t Hoen P.A., Verwijmeren C.A., den Dunnen J.T., Dorsman J.C., van Ommen G.B. 2008. Mutant huntingtin activates Nrf2-responsive genes and impairs dopamine synthesis in a PC12 model of Huntington’s disease. BMC Mol. Biol. 9:84 10.1186/1471-2199-9-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R., Jaiswal A.K. 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA. 93:14960–14965 10.1073/pnas.93.25.14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A., Jahreiss L., Sarkar S., Saiki S., Menzies F.M., Ravikumar B., Rubinsztein D.C. 2006. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr. Top. Dev. Biol. 76:89–101 10.1016/S0070-2153(06)76003-3 [DOI] [PubMed] [Google Scholar]
- Xu P., Cheng D., Duong D.M., Rush J., Roelofs J., Finley D., Peng J. 2006. A proteomic strategy for quantifying polyubiquitin chain topologies. Isr. J. Chem. 46:171–182 10.1560/1JVL-J4EE-FVW4-MXRE [DOI] [Google Scholar]
- Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 137:133–145 10.1016/j.cell.2009.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Lucas J.J., Hen R. 2000. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 101:57–66 10.1016/S0092-8674(00)80623-6 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Cremona M.L., Rothman J.E. 2006. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J. Cell Biol. 172:719–731 10.1083/jcb.200510065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H.C., Xu Q., Chou D.M., Zhao Z., Elledge S.J. 2008. Global protein stability profiling in mammalian cells. Science. 322:918–923 10.1126/science.1160489 [DOI] [PubMed] [Google Scholar]
- Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., Denk H. 2002. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160:255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., Zhao Y., Li Z., Song B., Han J., et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 136:308–321 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
- Zu T., Duvick L.A., Kaytor M.D., Berlinger M.S., Zoghbi H.Y., Clark H.B., Orr H.T. 2004. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J. Neurosci. 24:8853–8861 10.1523/JNEUROSCI.2978-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]