The high-resolution structure of doublecortin-stabilized microtubules provides unprecedented insight into their in vivo architecture.
Abstract
Microtubule-associated proteins (MAPs) are essential for regulating and organizing cellular microtubules (MTs). However, our mechanistic understanding of MAP function is limited by a lack of detailed structural information. Using cryo-electron microscopy and single particle algorithms, we solved the 8 Å structure of doublecortin (DCX)-stabilized MTs. Because of DCX’s unusual ability to specifically nucleate and stabilize 13-protofilament MTs, our reconstruction provides unprecedented insight into the structure of MTs with an in vivo architecture, and in the absence of a stabilizing drug. DCX specifically recognizes the corner of four tubulin dimers, a binding mode ideally suited to stabilizing both lateral and longitudinal lattice contacts. A striking consequence of this is that DCX does not bind the MT seam. DCX binding on the MT surface indirectly stabilizes conserved tubulin–tubulin lateral contacts in the MT lumen, operating independently of the nucleotide bound to tubulin. DCX’s exquisite binding selectivity uncovers important insights into regulation of cellular MTs.
Introduction
Microtubules (MTs) are dynamic, polar polymers of αβ-tubulin heterodimers. Their polymerization is driven by GTP and they are selectively stabilized or destabilized by MT-associated proteins (MAPs) in different cell contexts (Desai and Mitchison, 1997). However, the mechanisms by which MAPs stabilize MTs remain poorly understood because of the lack of detailed structural information. αβ-tubulin heterodimers associate head to tail to form protofilaments (pfs) and laterally to form the cylindrical MT wall. In vivo, most MTs are built from 13 parallel pfs (Tilney et al., 1973). The dimers form a left-handed pseudo-helix, in which α lies next to α and β lies next to β-tubulin. The geometry of this so-called B lattice necessitates a single discontinuity called the seam, observed in vivo, where α- and β-tubulin lie next to each other and form A lattice contacts (Kikkawa et al., 1994; McIntosh et al., 2009). It has been proposed that some MAPs modify this canonical structure and thereby modulate MT dynamics in vivo (des Georges et al., 2008). Thus, detailed insight about MAP MT stabilization is essential for a general understanding of MT dynamics and regulation.
Doublecortin (DCX) is the best-described member of a growing family of stabilizing MAPs, with multiple homologues identified in eukaryotes (Reiner et al., 2006). Family members are involved in cell division, migration, and differentiation. Their MT binding region is built from a tandem repeat of so-called DC (DoubleCortin-like) domains with no homology to other MAPs. Mutations in DCX, often in its N- and C-DC domains, cause severe neuronal migration disorders in humans (des Portes et al., 1998; Gleeson et al., 1998). DCX binds between pfs on the MT lattice, which likely contributes to its unusual ability to preferentially nucleate and stabilize homogenous populations of 13-pf MTs (Moores et al., 2004). This contrasts with the heterogeneous MTs (typically 11–17 pfs) that are in in vitro preparations of tubulin alone (Chrétien et al., 1992).
To study the native structure of MAP-bound MTs, we exploited DCX’s selective polymerization and stabilization of 13-pf MTs and used cryo-EM and single particle algorithms to generate reconstructions of DCX–MTs. Because DCX–MTs support kinesin motility (Moores et al., 2006), we also added a kinesin-1 motor domain to the DCX–MTs (DCX-K-MTs) to facilitate image analysis. Single particle approaches have proven very successful in studying kinesin bound to 13-pf MTs, and kinesin provides a clear landmark to define the DCX binding site (Sindelar and Downing, 2007, 2010). Our cryo-EM map at sub-nanometer resolution brings new insights into the precise structural mechanism by which DCX binds to the B lattice and specifically stabilizes 13-pf MTs. Importantly, our work reveals the structure of MTs bound and stabilized exclusively by cellular ligands.
Results and discussion
DCX stabilizes lateral and longitudinal B lattice contacts between four tubulin dimers but does not bind the A lattice seam
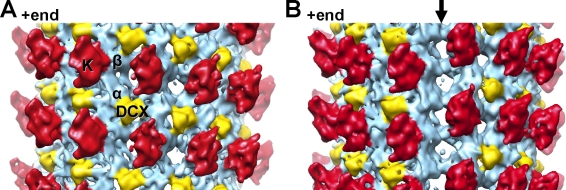
Our previous low-resolution reconstruction of DCX-bound paclitaxel-stabilized MTs was calculated using B lattice helical parameters (Moores et al., 2004). We verified that our 13-pf DCX-nucleated MTs were also built from a B lattice using cryo-electron tomography (Fig. S1); we collected low-dose images of DCX-K-MTs, and calculated an asymmetric reconstruction of the complex (Figs. 1 A and S1 C). This structure shows that density corresponding to DCX is wedged in the inter-pf valleys, with a spacing of 8 nm along the MT axis, as seen previously (Moores et al., 2004). Thus, despite the experimental differences, the DCX binding sites in both structures are the same. In addition, the presence of kinesin in our current reconstruction, with its known binding site on the αβ-tubulin dimer (Sindelar and Downing, 2007, 2010), enabled us to unambiguously locate the DCX binding site at the corner of four tubulin dimers, where it can stabilize both longitudinal and lateral interactions in the MT lattice (Fig. 1 A).
Figure 1.
DCX stabilizes lateral and longitudinal B lattice contacts between four tubulin dimers but does not bind the MT seam. (A) 13.5 Å reconstruction of DCX–MTs decorated with a kinesin-1 motor domain, low-pass filtered with a 13 Å cut-off, and thresholded at 3σ (Fig. S1). The reconstruction is displayed, with the MT +end oriented up. Each kinesin motor domain (red, K) binds one αβ-tubulin heterodimer (sky blue). DCX (yellow) binds at the corner between four tubulin dimers making B lattice contacts. (B) The same structure rotated 180°, revealing the seam of DCX–MTs (arrow). Density for DCX is found in the B lattice inter-pf valleys all around the 13-pf MT map (up to a density threshold of 5σ), but is absent at the seam where pfs make A lattice contacts.
The specificity with which DCX binds to MTs was further revealed in our asymmetric reconstruction by the observation that DCX does not bind at the MT seam (Fig. 1 B). Thus, the geometry of a 13-pf B-lattice MT nucleated by DCX preserves the A lattice seam. It was suggested that the fission yeast EB1 homologue, Mal3, binds specifically at MT seams (Sandblad et al., 2006). Our reconstruction will help to define more precisely how EB family members recognize an A lattice configuration, and emphasizes that EB and DCX family members could bind MTs simultaneously.
Molecular mechanism of DCX MT stabilization
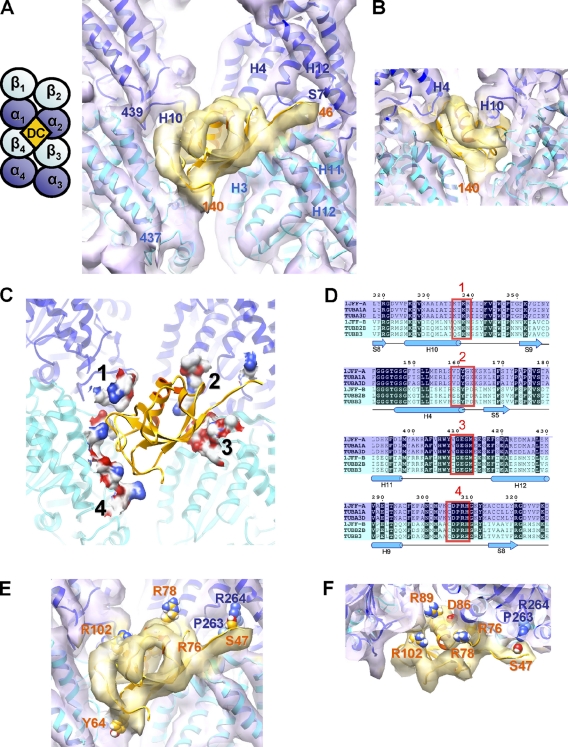
To gain further structural insight into the molecular mechanism of MT stabilization by DCX, we averaged together the B lattice contacts around the MT. This yielded an 8.2 Å resolution reconstruction of the DCX–tubulin interface in which secondary structures and several stable loops are clearly defined (Fig. 2, A and B; Fig. S2 A; and Video 1).
Figure 2.
The molecular basis of DCX selective stabilization of B-lattice 13-pf MTs. (A) Front view and schematic of the 8.2 Å resolution cryo-EM map of DCX–MTs fitted with structures of α-tubulin and β-tubulin (map, violet surface; 1JFF.pdb, α in blue, β in cyan ribbons; Löwe et al., 2001) and the solution structure of the N-DC domain of DCX (map, yellow surface; 1MJD.pdb, model 11, residues 46–140, orange; Kim et al., 2003). See also Fig. S2 and Video 1. The four dimers forming the DCX binding site are labeled αβ1–4. Tubulin helices α1-H10, α2-H4,H12, β3-H3,H11,H12, and tubulin strand α2-S7 are labeled, as are the aa numbers of the boundaries of N-DC and α1 and β4 C termini. (B) View from inside toward the outside of the MT, illustrating the quality of the fit of N-DC in the density. (C) Front view of tubulin aa <5 Å away from DCX displayed as a molecular surface colored by heteroatom in which four major binding patches can be identified. (D) Sections of a sequence alignment of the main isoforms of bovine tubulin used in our structure (1JFF-A and 1JFF-B), and of other human tubulin isoforms (TUBA1A, GenBank/EMBL/DDBJ accession no. 7846; TUBA3D, accession no. 113457; TUBB2B, accession no. 347733; TUBB3, accession no. 10381). DCX contacts are boxed in red. α-tubulins are highlighted in blue and β-tubulins in cyan; sequence identities are highlighted in black; secondary structures of 1JFF-A are depicted below. (E and F) Location of side chains of surface residues whose mutations cause neuronal migration disorders: S47R, Y64N, R76S, R78H/L, D86H, R89G, R102S in N-DC of DCX (orange text); P263T and R264C in TUBA1A, a human α-tubulin isoform (blue text).
The density attributable to DCX at each binding site in our reconstruction represents only ∼1/4 of the protein, corresponding to a DC domain (also seen previously, Moores et al., 2004). The available atomic structure of a DC domain (N-DC, Leu46-Thr139) matches the visible DCX density in our reconstruction very well (Fig. 2, A and B; Kim et al., 2003). Our reconstruction also contains density for an extended chain of at least 5 aa upstream of the DC domain, running along the MT interdimer interface. We are unsure of the DC domain arrangement in our reconstruction, and it is possible that N-DC and C-DC of DCX bind at adjacent sites (ie 1:2 DCX/tubulin dimer). The 38-aa linker between the DC domains, the length of which is well-conserved, is long enough for this to be feasible, and although we find no corresponding density in our reconstruction, it might be averaged away during processing. Equivalent residues in N-DC and a homology-modeled C-DC that map to <5 Å from tubulin are compared in Fig. S2 C. Conversely, biochemical evidence suggests that MT binding saturates at 1:1 DCX/tubulin dimer (Moores et al., 2006), which supports the idea that ∼3/4 of the DCX molecule (a DC domain, Ser-Pro rich C terminus, and connecting linkers) in our reconstruction is flexible and thus not visible. This would also be consistent with DCX’s ability to bundle MTs (Kim et al., 2003). However, these binding modes for DCX are not mutually exclusive and more data are needed to probe this question.
The atomic coordinates of the tubulin dimer calculated from zinc-induced 2D sheets (1JFF) fit very well into our structure (Fig. 2 A; Löwe et al., 2001). All the α-helices have matching tubes of density both on the MT surface (e.g., H11 and H12; Fig. 2 A) and in the MT lumen (e.g., H6 and H7; Fig. 3 A and Video 1). In addition, density corresponding to the β-sheets and many of the loops are very clear. Mapping of residues <5 Å from DCX enabled us to identify the major binding contact on each of the four tubulin monomers (Fig. 2 and S2 C). Contacts 1 (H10-S9 loop) and 2 (H4-S5 loop) on adjacent α-tubulins (α1β1 and α2β2) are conserved in and specific for α-tubulins (Fig. 2 D), and the overall surface charge is different between α and β at these sites (Fig. S2 C). In contrast, contacts 3 (H11-H12 loop) and 4 (H9-S8 loop) on β-tubulins on the dimers below (α3β3 and α4β4) are conserved between α and β, as is the α2-H8-S7 loop, where the N-terminal extension of the DC domain binds. The precision with which DCX makes these contacts with tubulin further reinforces the selectivity of DCX for a B lattice architecture and explains why DCX cannot bind to the seam (Fig. S2 B).
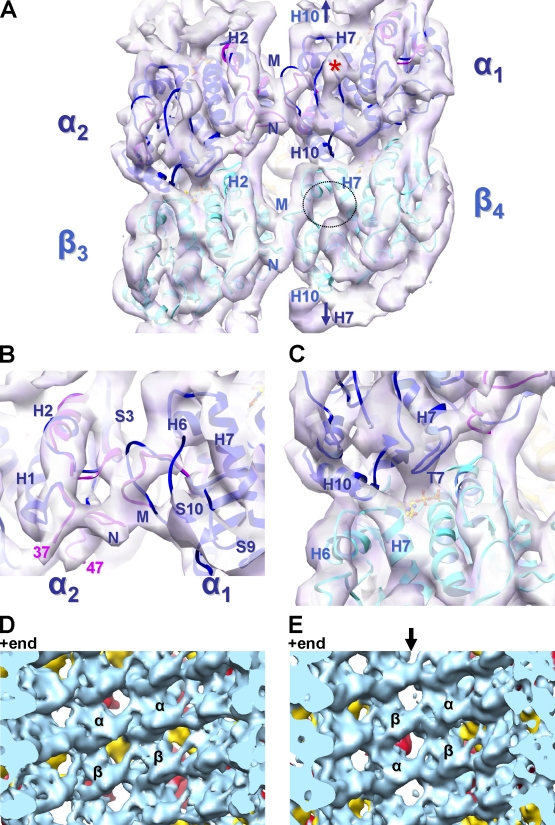
Figure 3.
Structural basis of MAP stabilization of the MT lattice. (A and B) View of the inside surface of the 8.2 Å cryo-EM map (violet surface) of B-lattice contacts between four dimers in DCX–MTs. The paclitaxel binding pocket in β-tubulin is empty (dotted circle; Fig. S3 A), whereas the equivalent area in α-tubulin is occupied by loop S9-S10 (density labeled with an asterisk; Fig. S3 A). The structures of α- and β-tubulin from zinc-induced sheets are fitted in the map (1JFF.pdb, α in blue and β in cyan). However, for α-tubulin, the N and M loops from a tubulin–stathmin complex match the reconstruction better (3HKE.pdb, pink; Dorléans et al., 2009). Arrows in A indicate equivalent H10-H7 contacts at the intradimer interfaces. (C) The reconstruction is well defined around the nucleotide (ball and stick) bound to β-tubulin, particularly loop T7. There is continuous density between α1-H10 and β4-H7 at the interdimer interface (also see Video 1), which is consistent with the straight conformation of β4-tubulin but is incompatible with a curved conformation (Fig. S3 B). (D and E) In the asymmetric reconstruction, these lateral contacts are present at both the B lattice (D) and A lattice (E) contacts, despite the absence of DCX on the outer surface of the seam (arrow).
Mutations in DCX and α1a-, α8-, and β2B-tubulin cause different forms of neuronal migration disorders in humans (des Portes et al., 1998; Gleeson et al., 1998; Keays et al., 2007; Jaglin et al., 2009), and mutations in β3-tubulin have been associated with axonal growth defects (Tischfield et al., 2010). Several disease-causing mutations fall precisely at the four DCX–tubulin contacts, which suggests that these diseases arise because of disruption of their interaction (Fig. 2, E and F). In tubulin, Pro263 and Arg264 are in the H8-S7 loop of α2-tubulin (contact 3 extension; Fig. 2 A), and mutations of both these residues in the human isoform TUBA1A cause lissencephaly (Fig. 2, E and F; Keays et al., 2007; Poirier et al., 2007). Strikingly, DCX Ser47 (<5 Å from α-tubulin Arg264) is also mutated in patients with lissencephaly and is likely to be a site of phospho-regulation (Gleeson et al., 1998; Schaar et al., 2004). In DCX contact 1, the N-DC lissencephaly-linked mutation Arg78His is close to tubulin (Fig. 2, E and F), whereas the equivalent residue in C-DC is itself a histidine (His205; Fig. S2 C). In addition, the overall charge distribution at the four DCX–tubulin contacts suggests that N-DC matches the tubulin surface better than C-DC. However, the involvement of C-DC in MT binding (Kim et al., 2003) suggests that additional work is needed to precisely address the roles of each DC domain.
Tubulin–tubulin contacts in the MT lumen
Although no DCX density penetrates to the MT lumen, extensive lateral contacts are present at low radius between pairs of α- and β-tubulins, each centered ∼30 Å from the DCX density (Fig. 3 A). DCX contact 1 is located at the C terminus of α1-H10, which runs through the MT wall, with its N terminus emerging in the MT lumen (Fig. 2, C and D). Here, in our reconstruction, density for α1-H10 at the interdimer interface is continuous with β4-H7 (Fig. 3 A). The piston-like movement of H7 in tubulin, coupled to displacement of the preceding H6-H7 loop, has been shown to mediate curved-to-straight transitions and is thought to be sensitive to the nucleotide bound to β-tubulin (Ravelli et al., 2004). β4-H7 is clearly in a straight conformation in our reconstruction (Video 1), and docking of the curved tubulin conformation results in clashes between α1-H10 and the β4-H6-H7 loop (Fig. S3 B). Crucially, and as previously observed, the straight conformation of tubulin facilitates stabilization of the inter-pf contacts that define the MT polymer (Li et al., 2002; Ravelli et al., 2004). Accordingly, in our reconstruction, the β4 microtubule loop (M loop; aa 272–287) reaches across to the adjacent pf, to the β3-H2-S3 loop (aa 81–91) and β3-N loop (aa 28–64). β3-H3 binds against these lateral contact loops and also contacts β3-H11-H12. Together, these form the external contact 3 for DCX, emphasizing the cooperative network of interactions in the straight tubulin conformation that is stabilized by DCX (Fig. 2 A).
Lateral contacts between α1- and α2-tubulin are also visible (Fig. 3, A and B). In the Zn sheet structure, N loop residues 35–60 are absent and the M loop (aa 272–287) is artificially stabilized by a Zn2+ ion (Löwe et al., 2001); thus, they do not match the lateral α-α loops in our reconstruction as well as is seen for β-β (Fig. 3, A and B). Instead, we docked these loops from a tubulin-stathmin structure, which does fit our structure remarkably well (pink loops, Fig. 3, A and B; Dorléans et al., 2009). In the tubulin-stathmin structure, the N loop is more resolved, apart from aa 38–46, which was also not visible in our MT structure, and the M loop forms an arch that clearly fits our density (Fig. 3, A and B).
The α1-M loop reaches across to the adjacent pf to the α2-H2-S3 and α2-N-loop, forming very similar lateral contacts to those seen between β4 and β3. Although α- and β-tubulin share 40% sequence identity and a nearly identical fold, the sequences of the M and N loop are particularly divergent between α and β subunits. The observation that α-α and β-β lateral contacts are structurally similar is thus noteworthy, and consistent with what has been observed in paclitaxel-stabilized MTs, both in the presence and absence of kinesin (Bodey et al., 2009; Sindelar and Downing, 2010; Sui and Downing, 2010). However, density corresponding to the α-tubulin–specific insertion in loop α1-S9-S10 (residues 361–368; Fig. 3 A, asterisk) appears to collaborate with the α1-M loop region in providing additional stabilization for these lateral contacts, and in so doing, blocks the pocket on the α-tubulin equivalent to the β-tubulin paclitaxel-binding site (Figs. 3 A and S3 A). α1-H7 also adopts a straight conformation, and in structural parallel to the contacts observed at the interdimer interface, forms continuous density with β1-H10 in the same dimer (Fig. 3 A, arrows), further reinforcing its straight conformation.
The symmetry-breaking A lattice seam is built from mismatches of these lateral contacts, α- with β-tubulin and β- with α-tubulin. The roles of these seams have been the subject of much speculation, and some evidence has suggested that they are the weakest point in in vitro polymerized MTs (Sandblad et al., 2006). One hypothesis is that the seam must be specifically stabilized for effective MT polymerization. However our DCX–MT structure, where DCX density is absent at the seam, suggests instead an indirect mechanism of stabilization. At the resolution of our 13.5 Å asymmetric reconstruction, the heterotypic lateral contacts at the seam appear as well-defined and stabilized as the rest of the MT lattice (Fig. 3, D and E), which is in agreement with recent modeling experiments (Sui and Downing, 2010).
Our structure reveals that the lateral contacts formed in DCX-stabilized MTs are very similar to those seen in paclitaxel-stabilized MTs, underlining the fundamental nature of these contacts for MT formation and stabilization (Sindelar and Downing, 2010; Sui and Downing, 2010). Remarkably, this is despite the fact that DCX acts at a distance on the outer MT surface, whereas paclitaxel directly contacts the β-tubulin lateral loops. Paclitaxel is a promiscuous stabilizer of tubulin polymers and can stabilize MTs built from a variety of pf numbers as well as the antiparallel pfs of Zn-induced sheets (Nogales et al., 1998; Sui and Downing, 2010). The triangular paclitaxel binding site formed by β-H7, H6, H1, and M loop is visible in our structure (Fig. 3, A, D, and E; and Fig. S3 A). The existence of this binding site in the absence of drugs suggests that paclitaxel-like compounds recognize and stabilize the native configuration of β-H7, H6, and M loop in polymerized tubulin, presumably with either A or B lattice architectures.
DCX binds at the interdimer longitudinal junction between β- and α-tubulin, which forms the exchangeable GTP site (Fig. 3 C; Nogales et al., 1998). We previously showed that DCX overrides the nucleotide dependence of MT polymerization and architecture, inducing GTP-, GDP-, and GMPCPP-tubulin, to form 13-pf MTs (Moores et al., 2006). GTP is hydrolyzed in DCX–MTs (Fig. S3 C) but our data also show that DCX–MTs can accommodate a range of tubulin dimer lengths, which is characteristic of the bound nucleotide (Fig. S3 D; Vale et al., 1994; Hyman et al., 1995). Thus, the DCX MT stabilization mechanism described above is sufficiently plastic to accommodate small changes in the lattice, independent of the tubulin-bound nucleotide. This is distinct from the behavior of paclitaxel, which can force a longer conformation of tubulin both in the MT lattice and in solution (Fig. S3 D; Vale et al., 1994; Elie-Caille et al., 2007).
Implications for DCX function in vivo
In cells, the 13-pf B-lattice architecture is thought to be imposed by γ-tubulin ring complexes, which act as templates for MT minus ends and propagate the MT architecture during elongation (Kollman et al., 2010). However, transitions in pf number have been observed in MTs polymerized from cell extracts (Chrétien et al., 1992). It therefore seems possible that cells might use MAPs like DCX to maintain a constant architecture along MTs, an activity that might be particularly important in long neuronal processes devoid of γ-tubulin (Baas and Joshi, 1992). DCX is essential for neuronal migration and differentiation during development, and although its exact role is unknown, it is enriched in neuronal processes (Moores et al., 2006; Bielas et al., 2007). It is tempting to speculate that binding by DCX to the corner of four tubulin dimers could represent a minimal oligomer for DCX-induced MT nucleation. With both longitudinal and lateral contacts being stabilized, DCX could act as an on switch for nucleation independently of the bound nucleotide. The importance of noncentrosomal sites of MT nucleation is increasingly being highlighted, particularly in the extended processes of neurons (Stiess et al., 2010). The molecular properties of DCX suggest that it could play an important role in regulation of MT dynamics, nucleation, and architecture in these regions of the cell.
The specific effect of DCX on MTs highlights the importance of MT architecture, the in vivo significance of which is unclear. A distinctive property of 13-pf MTs is their straight pfs, and, although difficult to test, it is expected that such a pf trajectory is optimized for cellular function. Non–13-pf architectures are also very precisely defined in a variety of systems (e.g., Chalfie and Thomson, 1982), and although the functional significance of the architecture of these MTs is poorly understood, it will be fascinating to discover if there are molecular equivalents of DCX that can specify alternative MT architectures.
Materials and methods
Sample preparation
Human DCX (1–366) was expressed in Spodoptera frugiperda Sf9 cells and purified as described previously (Moores et al., 2004). DCX–MTs were polymerized by incubating 10 µM of bovine brain tubulin (Cytoskeleton, Inc.) and 10 µM DCX in 80 mM Pipes, pH 6.8, 1 mM EGTA, 3 mM MgCl2, 1 mM TCEP, and 0.5 mM GTP for 1 h at 37°C. DCX–MTs were diluted 1:1 in BRB20 (20 mM Pipes, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) and 2 mM TCEP, and adsorbed to glow-discharged copper grids with lacey carbon film (Agar). Grids were washed with 30 µM DCX and 5 µM monomeric rat kinesin (aa 1–340, P-loop mutant T93N-K340T93N, a generous gift from R. Cross and M. Alonso, University of Warwick, Coventry, England, UK) in BRB20 and 2 mM TCEP, and immediately transferred into a Vitrobot (FEI Company) at 37°C and 100% humidity, blotted for 2 s, and vitrified.
Cryo-EM data collection and image processing
Low-dose images were collected on a microscope (Tecnai F20 FEG; FEI Company) operating at 200 kV, 50,000×, and 0.8–2.9 µm defocus. Micrographs were recorded on films (SO-163; Kodak) and digitized (SCAI scanner; Carl Zeiss, Inc.) to a final sampling of 1.4 Å/pixel. 63 films containing 172 MTs (∼186,000 tubulin dimers) were selected. 3D reconstructions were generated using a previously described custom single particle procedure (Sindelar and Downing, 2007, 2010). For each MT, the orientation of the seam was determined by projection matching using a B lattice 13-pf MT decorated with kinesin-1 motor domain, lacking DCX, as the reference. Two rounds of reference alignment and reconstruction using custom SPIDER scripts and five refinement rounds using FREALIGN (Grigorieff, 2007) were performed. ∼168,000 tubulin dimers (90% of the data) went into the final reconstruction. 3D reconstruction yielded an asymmetric map with a resolution of 13.5 Å according to the Fourier shell correlation (FSC) 0.5 criterion (Fig. S1 C) calculated from half-dataset models (van Heel, 1987). Subsequently, the 12 B lattice contacts around the MT were averaged during 3D reconstruction. The resolution was determined by two methods: 8.2 Å from the FSC between the half dataset (Fig. S2 A), and 8.3 Å from Rmeasure (Sousa and Grigorieff, 2007). This map was sharpened using a band-pass filter (8–20 Å). The asymmetric map and the 8.2 Å map were deposited in the EMDB with accession nos. 1787 and 1788, respectively.
Atomic model building
UCSF Chimera (Pettersen et al., 2004) was used for visualization of 3D models and rigid-body fitting of atomic structures in the cryo-EM volumes. Crystal structures of α- and β-tubulin (1JFF.pdb, Löwe et al., 2001; 3HKE.pdb, Dorléans et al., 2009) were fitted separately. The quality of the fit was assessed by cross-correlation between our structure and simulated 8 Å maps for each PDB: 1JFF β-tubulin gave the best score of 0.651 in our β-tubulin density (3HKE β-tubulin chain B gave 0.599); α-tubulin from 1JFF and 3HKE chain A scored similarly, 0.605 and 0.607, against 0.614 for a chimeric structure (3HKE chain A residues 31–61, 69–92, and 275–298 substituted to corresponding ones in 1JFF chain A). The DCX–MT atomic model was refined by multiple-subunit fitting in Flex-EM (Topf et al., 2008) with four tubulin subunits and the N-DC domain of DCX (1MJD.pdb, model 11, residues 46–140). The best multiple-subunit fit had a cross-correlation value of 0.819. The atomic model was deposited in the Protein Data Bank (accession no. 2XRP).
Tubulin GTPase assay
Mixtures of proteins and nucleotides were prepared in polymerization buffer, on ice, in a final volume of 10 µl. Where present, tubulin and DCX were at 20 µM and 30 µM final concentrations, respectively. MgGTP was present at 0.5 mM along with 500,000 dpm [8-3H]GTP (GE Healthcare) per reaction. After incubating at 0°C or 37°C for 30 min, the reactions were stopped by returning the mixtures to ice. To ensure linearity, no more than 5% of the GTP was hydrolyzed. Excess carrier GDP and GTP was added in 1 µl to give 0.5 mM final concentration each, and protein was then precipitated with 11 µl of ice-cold formamide. Precipitated protein was removed by centrifugation, the supernatant was recovered, and the volume was reduced under vacuum. Final samples were spotted onto PEI-cellulose TLC plates, which were then dried under vacuum and developed in 1.5 M LiCl. After drying and marking the positions of the authentic nucleotides under UV illumination, the radioactivity in GTP and GDP was measured by phosphorimaging.
Dimer axial repeat measurements
DCX–MTs were polymerized in the presence of GTP, GDP, or GMPCPP (Jena Biosciences), then supplemented by an excess of DCX (30 µM), vitrified, and imaged at 68,000× on a 4,000 × 4,000 charge-coupled device (Gatan, Inc.) calibrated using tobacco mosaic virus. The dimer axial repeat was derived from the height of the 8-nm layer line in power spectra computed using PHOELIX (Whittaker et al., 1995).
Online supplemental material
Fig. S1 shows data that DCX–MTs have a B lattice 13-pf architecture and the FSC of the asymmetric map. Fig. S2 shows the FSC of the symmetrized structure and defines the specificity of the DCX–MT interface. Fig. S3 provides further analysis of MT stabilization by DCX. Video 1 shows the DCX–MT interface. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007081/DC1.
Acknowledgments
We thank Maya Topf and Daven Vasishtan for advice.
C.A. Moores and F.J. Fourniol thank the Wellcome Trust, Biotechnology and Biological Sciences Research Council, New Life and the French Ministère de la Recherche for funding. F. Francis was supported by the Institut National de la Santé et de la Recherche Médicale Avenir program and the Fondation Bettencourt Schueller; F. Francis and A. Houdusse are grateful to the Federation pour la Recherche sur le Cerveau for support.
Footnotes
Abbreviations used in this paper:
- DCX
- doublecortin
- FSC
- Fourier shell correlation
- MAP
- microtubule-associated protein
- M loop
- microtubule loop
- MT
- microtubule
- pf
- protofilament
References
- Baas P.W., Joshi H.C. 1992. Gamma-tubulin distribution in the neuron: implications for the origins of neuritic microtubules. J. Cell Biol. 119:171–178 10.1083/jcb.119.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas S.L., Serneo F.F., Chechlacz M., Deerinck T.J., Perkins G.A., Allen P.B., Ellisman M.H., Gleeson J.G. 2007. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell. 129:579–591 10.1016/j.cell.2007.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodey A.J., Kikkawa M., Moores C.A. 2009. 9-Angström structure of a microtubule-bound mitotic motor. J. Mol. Biol. 388:218–224 10.1016/j.jmb.2009.03.008 [DOI] [PubMed] [Google Scholar]
- Chalfie M., Thomson J.N. 1982. Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. J. Cell Biol. 93:15–23 10.1083/jcb.93.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D., Metoz F., Verde F., Karsenti E., Wade R.H. 1992. Lattice defects in microtubules: protofilament numbers vary within individual microtubules. J. Cell Biol. 117:1031–1040 10.1083/jcb.117.5.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A., Katsuki M., Drummond D.R., Osei M., Cross R.A., Amos L.A. 2008. Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat. Struct. Mol. Biol. 15:1102–1108 10.1038/nsmb.1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Portes V., Pinard J.M., Billuart P., Vinet M.C., Koulakoff A., Carrié A., Gelot A., Dupuis E., Motte J., Berwald-Netter Y., et al. 1998. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 92:51–61 10.1016/S0092-8674(00)80898-3 [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- Dorléans A., Gigant B., Ravelli R.B., Mailliet P., Mikol V., Knossow M. 2009. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc. Natl. Acad. Sci. USA. 106:13775–13779 10.1073/pnas.0904223106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elie-Caille C., Severin F., Helenius J., Howard J., Muller D.J., Hyman A.A. 2007. Straight GDP-tubulin protofilaments form in the presence of taxol. Curr. Biol. 17:1765–1770 10.1016/j.cub.2007.08.063 [DOI] [PubMed] [Google Scholar]
- Gleeson J.G., Allen K.M., Fox J.W., Lamperti E.D., Berkovic S., Scheffer I., Cooper E.C., Dobyns W.B., Minnerath S.R., Ross M.E., Walsh C.A. 1998. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 92:63–72 10.1016/S0092-8674(00)80899-5 [DOI] [PubMed] [Google Scholar]
- Grigorieff N. 2007. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157:117–125 10.1016/j.jsb.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Chrétien D., Arnal I., Wade R.H. 1995. Structural changes accompanying GTP hydrolysis in microtubules: information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J. Cell Biol. 128:117–125 10.1083/jcb.128.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin X.H., Poirier K., Saillour Y., Buhler E., Tian G., Bahi-Buisson N., Fallet-Bianco C., Phan-Dinh-Tuy F., Kong X.P., Bomont P., et al. 2009. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet. 41:746–752 10.1038/ng.380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays D.A., Tian G., Poirier K., Huang G.-J., Siebold C., Cleak J., Oliver P.L., Fray M., Harvey R.J., Molnár Z., et al. 2007. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 128:45–57 10.1016/j.cell.2006.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M., Ishikawa T., Nakata T., Wakabayashi T., Hirokawa N. 1994. Direct visualization of the microtubule lattice seam both in vitro and in vivo. J. Cell Biol. 127:1965–1971 10.1083/jcb.127.6.1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Cierpicki T., Derewenda U., Krowarsch D., Feng Y., Devedjiev Y., Dauter Z., Walsh C.A., Otlewski J., Bushweller J.H., Derewenda Z.S. 2003. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat. Struct. Biol. 10:324–333 10.1038/nsb918 [DOI] [PubMed] [Google Scholar]
- Kollman J.M., Polka J.K., Zelter A., Davis T.N., Agard D.A. 2010. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 466:879–882 10.1038/nature09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., DeRosier D.J., Nicholson W.V., Nogales E., Downing K.H. 2002. Microtubule structure at 8 A resolution. Structure. 10:1317–1328 10.1016/S0969-2126(02)00827-4 [DOI] [PubMed] [Google Scholar]
- Löwe J., Li H., Downing K.H., Nogales E. 2001. Refined structure of alpha beta-tubulin at 3.5 A resolution. J. Mol. Biol. 313:1045–1057 10.1006/jmbi.2001.5077 [DOI] [PubMed] [Google Scholar]
- McIntosh J.R., Morphew M.K., Grissom P.M., Gilbert S.P., Hoenger A. 2009. Lattice structure of cytoplasmic microtubules in a cultured Mammalian cell. J. Mol. Biol. 394:177–182 10.1016/j.jmb.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores C.A., Perderiset M., Francis F., Chelly J., Houdusse A., Milligan R.A. 2004. Mechanism of microtubule stabilization by doublecortin. Mol. Cell. 14:833–839 10.1016/j.molcel.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Moores C.A., Perderiset M., Kappeler C., Kain S., Drummond D., Perkins S.J., Chelly J., Cross R., Houdusse A., Francis F. 2006. Distinct roles of doublecortin modulating the microtubule cytoskeleton. EMBO J. 25:4448–4457 10.1038/sj.emboj.7601335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Wolf S.G., Downing K.H. 1998. Structure of the α β tubulin dimer by electron crystallography. Nature. 391:199–203 10.1038/34465 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Poirier K., Keays D.A., Francis F., Saillour Y., Bahi N., Manouvrier S., Fallet-Bianco C., Pasquier L., Toutain A., Tuy F.P., et al. 2007. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A). Hum. Mutat. 28:1055–1064 10.1002/humu.20572 [DOI] [PubMed] [Google Scholar]
- Ravelli R.B., Gigant B., Curmi P.A., Jourdain I., Lachkar S., Sobel A., Knossow M. 2004. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 428:198–202 10.1038/nature02393 [DOI] [PubMed] [Google Scholar]
- Reiner O., Coquelle F.M., Peter B., Levy T., Kaplan A., Sapir T., Orr I., Barkai N., Eichele G., Bergmann S. 2006. The evolving doublecortin (DCX) superfamily. BMC Genomics. 7:188 10.1186/1471-2164-7-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Sandblad L., Busch K.E., Tittmann P., Gross H., Brunner D., Hoenger A. 2006. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 127:1415–1424 10.1016/j.cell.2006.11.025 [DOI] [PubMed] [Google Scholar]
- Schaar B.T., Kinoshita K., McConnell S.K. 2004. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 41:203–213 10.1016/S0896-6273(03)00843-2 [DOI] [PubMed] [Google Scholar]
- Sindelar C.V., Downing K.H. 2007. The beginning of kinesin’s force-generating cycle visualized at 9-A resolution. J. Cell Biol. 177:377–385 10.1083/jcb.200612090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar C.V., Downing K.H. 2010. An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. USA. 107:4111–4116 10.1073/pnas.0911208107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa D., Grigorieff N. 2007. Ab initio resolution measurement for single particle structures. J. Struct. Biol. 157:201–210 10.1016/j.jsb.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Stiess M., Maghelli N., Kapitein L.C., Gomis-Rüth S., Wilsch-Bräuninger M., Hoogenraad C.C., Tolić-Nørrelykke I.M., Bradke F. 2010. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 327:704–707 10.1126/science.1182179 [DOI] [PubMed] [Google Scholar]
- Sui H., Downing K.H. 2010. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure. 18:1022–1031 10.1016/j.str.2010.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L.G., Bryan J., Bush D.J., Fujiwara K., Mooseker M.S., Murphy D.B., Snyder D.H. 1973. Microtubules: evidence for 13 protofilaments. J. Cell Biol. 59:267–275 10.1083/jcb.59.2.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield M.A., Baris H.N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W.M., Andrews C., Demer J.L., Robertson R.L., et al. 2010. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 140:74–87 10.1016/j.cell.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M., Lasker K., Webb B., Wolfson H., Chiu W., Sali A. 2008. Protein structure fitting and refinement guided by cryo-EM density. Structure. 16:295–307 10.1016/j.str.2007.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D., Coppin C.M., Malik F., Kull F.J., Milligan R.A. 1994. Tubulin GTP hydrolysis influences the structure, mechanical properties, and kinesin-driven transport of microtubules. J. Biol. Chem. 269:23769–23775 [PubMed] [Google Scholar]
- van Heel M. 1987. Similarity measures between images. Ultramicroscopy. 21:95–100 10.1016/0304-3991(87)90010-6 [DOI] [Google Scholar]
- Whittaker M., Carragher B.O., Milligan R.A. 1995. PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy. 58:245–259 10.1016/0304-3991(95)00057-8 [DOI] [PubMed] [Google Scholar]