Pom121 anchors core structures of the NPC to the membrane through its binding to the β-propeller domains of Nup155 and Nup160.
Abstract
Nuclear pore complexes (NPCs) control the movement of molecules across the nuclear envelope (NE). We investigated the molecular interactions that exist at the interface between the NPC scaffold and the pore membrane. We show that key players mediating these interactions in mammalian cells are the nucleoporins Nup155 and Nup160. Nup155 depletion massively alters NE structure, causing a dramatic decrease in NPC numbers and the improper targeting of membrane proteins to the inner nuclear membrane. The role of Nup155 in assembly is likely closely linked to events at the membrane as we show that Nup155 interacts with pore membrane proteins Pom121 and NDC1. Furthermore, we demonstrate that the N terminus of Pom121 directly binds the β-propeller regions of Nup155 and Nup160. We propose a model in which the interactions of Pom121 with Nup155 and Nup160 are predicted to assist in the formation of the nuclear pore and the anchoring of the NPC to the pore membrane.
Introduction
Nuclear pore complexes (NPCs) act as gateways that regulate the transport of macromolecules across the nuclear envelope (NE). In addition to their roles in controlling transport, NPCs also influence gene expression, chromatin organization, and chromosome inheritance. Although approaching ∼100 MDa in mass in vertebrate cells, NPCs are composed of only ∼30 distinct proteins termed nucleoporins (nups). Nups that form the core structural domain of the NPC are organized into distinct subcomplexes that are repetitively arranged throughout the pore, imparting on the structure eightfold rotational and twofold lateral symmetry in the plane of the NE (Unwin and Milligan, 1982; Akey, 1989; Akey and Radermacher, 1993). Attached to the core are fibrillar nups that form the nucleoplasmic basket and cytoplasmic filaments. Nups playing a central role in nuclear transport line the NPC channel and are composed of repetitive phenylalanine-glycine (FG) motifs (for review see Tran and Wente, 2006). Cargoes entering or leaving the nucleus bind transport receptors, many termed karyopherins, which escort the cargoes through the NPC. This process requires the interactions of transport factors with the FG-nups. How these interactions facilitate transport is actively debated (Lim et al., 2008). In addition to soluble proteins, recent evidence has also implicated karyopherins in the transport of membrane proteins to the inner nuclear membrane (INM; King et al., 2006). Whether membrane proteins use the same route through the NPC as soluble proteins has not been determined.
The cylindrical core of the NPC forms the superstructure on which the FG-nups are organized. About half of all nups appear to be part of the core, with most of these being assigned to three or four different subcomplexes (Tran and Wente, 2006). By analogy to their yeast counterparts (for review see Hetzer and Wente, 2009), components of two conserved vertebrate subcomplexes, the Nup107–160 complex (containing Nup37, Nup43, Nup85, Nup96, Nup107, Nup133, Nup160, Sec13, and Seh1; Belgareh et al., 2001; Loïodice et al., 2004) and the Nup53–Nup93 complex (including Nup53, Nup93, Nup155, Nup188, and Nup205; Grandi et al., 1997; Hawryluk-Gara et al., 2005), are presumed to form the primary scaffold of the NPC. Among these nups, Nup155 and several members of the Nup107–160 complex are predicted to contain two distinct fold types, an N-terminal β-propeller and a C-terminal α-solenoid domain (Berke et al., 2004; Devos et al., 2004, 2006; Schwartz, 2005; Brohawn et al., 2008). This organization is analogous to the molecular architecture of coat protein complexes (CPCs) that stabilize the sharp convex curvature of COPI, COPII, and clathrin-coated vesicles (for review see Stagg et al., 2007) and has led to the hypothesis that the β-propeller α-solenoid nups function similarly to induce curvature of the pore membrane (Devos et al., 2004, 2006; DeGrasse et al., 2009).
Several integral membrane proteins are also associated with the NPC and are predicted to both contribute to the core as well as anchor it to the pore membrane. In vertebrates, three pore membrane proteins have been identified: gp210 (Gerace et al., 1982), NDC1 (Mansfeld et al., 2006; Stavru et al., 2006), and Pom121 (Hallberg et al., 1993). Gp210 contains a single transmembrane domain with a short N-terminal region extending into the pore and available to bind the core (Wozniak et al., 1989; Greber et al., 1990). Pom121 also contains a single transmembrane segment but has a much larger, ∼120-kD domain extending into the NPC (Hallberg et al., 1993; Söderqvist and Hallberg, 1994). Finally, NDC1 is a multi-membrane spanning protein with an ∼45 kD C-terminal domain positioned in the pore (Lau et al., 2006; Mansfeld et al., 2006; Stavru et al., 2006). Although these proteins are likely to play an important role in NPC structure, how they interact with other nups is largely unknown. Moreover, no clues as to the structural and functional roles of Pom121 and gp210 have come from studies in yeast, as they contain no clear homologues (for review see Suntharalingam and Wente, 2003).
A number of important studies have revealed key roles for pore membrane proteins and components of the NPC core in NPC and NE biogenesis in metazoan cells (for review see Hetzer and Wente, 2009). Using Xenopus laevis in NE assembly assays in vitro, both Pom121 and NDC1 have been shown to be essential for this process (Antonin et al., 2005; Mansfeld et al., 2006). Similarly, Nup53, Nup155, and components of the Nup107–160 complex are also required for NPC and NE assembly. Importantly, they appear to function at distinct assembly steps. The Nup107–160 complex binds to chromatin in an early step of NPC assembly (Belgareh et al., 2001; Harel et al., 2003; Walther et al., 2003; Rasala et al., 2006, 2008; Franz et al., 2007; Gillespie et al., 2007; Dultz et al., 2008). Nup53 and Nup155 function later at a step required for the fusion of NE vesicles and the formation of the double membrane NE (Franz et al., 2005; Hawryluk-Gara et al., 2008). Their roles also seem to be linked to those of Pom121 and NDC1, both of which have been proposed to function at a similar point in NPC and NE assembly (Antonin et al., 2005; Franz et al., 2005; Mansfeld et al., 2006). Furthermore, the loss of Nup53, Nup155, Pom121, or NDC1 appears to activate a NE assembly checkpoint first proposed by Antonin et al. (2005), which arrests NE assembly after binding of membrane vesicles to chromatin and before formation of the double membrane NE. Importantly, a functional NE assembly checkpoint requires the Nup107–160 complex. Therefore, it seems reasonable to assume that termination of the checkpoint is linked to interactions of the Nup107–160 complex with pore membrane proteins and other members of the NPC scaffold.
In this study, we have examined the interactions between the NPC scaffold proteins Nup155 and Nup160 and the pore membrane. We show that Nup155 depletion causes alterations in NE structure, including a dramatic decrease in the number of NPCs and the improper targeting of membrane proteins to the INM. The role of Nup155 in maintaining normal NE function is potentially linked to its binding to the pore membrane proteins Pom121 and NDC1. In addition to binding Nup155, we also show that Pom121 interacts with the Nup107–160 complex through direct interactions with Nup160. We demonstrate that an N-terminal region of Pom121 binds directly to the N-terminal β-propeller regions of Nup155 and Nup160. We propose a model in which the NPC is anchored to the NE by multiple interactions between Pom121 and the surrounding nucleoporin scaffold, including Nup155 and Nup160, and we suggest these interactions play a critical role in NPC assembly.
Results
Loss of mammalian Nup155 alters NE structure
Studies in Caenorhabditis elegans and X. laevis suggest that Nup155 plays an essential role in NE and NPC assembly (Franz et al., 2005). Here we have used a variety of tools to investigate the function of mammalian Nup155 in establishing and maintaining the structure of the NE and NPC, with the goal of defining the mechanistic basis for the role of Nup155 in these processes. First, we examined the consequences of depleting Nup155 on the structure of HeLa cell nuclei. Several siRNAs directed against distinct regions of Nup155 mRNA were tested for their ability to deplete Nup155. Each of the siRNAs produced similar phenotypes (Fig. 1 and Fig. S1; unpublished data). By 72 h after siRNA transfection, the cellular levels of Nup155 were greatly reduced as determined by Western blotting (Fig. S1). Immunofluorescence analysis using anti-Nup155 antibodies showed that ∼50–70% of Nup155-specific siRNA-treated cells exhibited a decrease in Nup155 staining at the NE (Fig. 1 A and Fig. S1).
Figure 1.
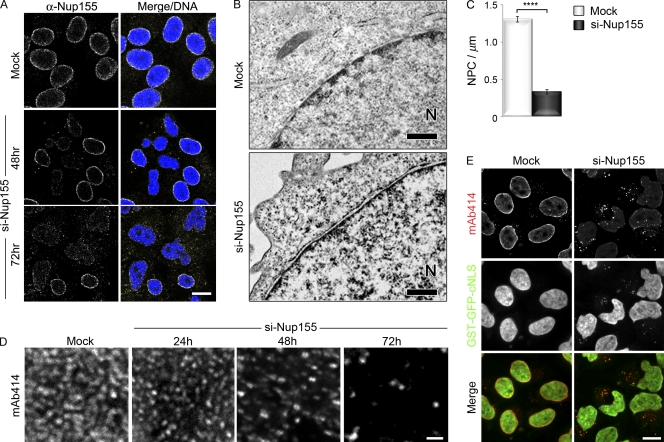
Depletion of Nup155 alters nuclear morphology and decreases NPC number. HeLa cells were treated with transfection reagent alone (Mock) or the Nup155-specific siRNA-targeting exon 27 (si-Nup155) as indicated. (A) Nup155 levels were assayed by indirect immunofluorescence using anti-Nup155 antibodies at the indicated times and DNA visualized with DRAQ5 (Merge/DNA). Bar, 10 µm. (B and C) Mock- or Nup155-depleted HeLa cells were processed for transmission electron microscopy 72 h after transfection. The position of the nucleoplasm is indicated (N). Bar, 0.5 µm. (C) The number of NPCs per micron of NE was determined by examining individual nuclear sections (n > 48 sections per condition). The average number of NPCs per micron of NE is shown. The P value (P < 0.0001, represented by asterisks) was calculated using a Welch corrected unpaired t test. Error bars indicate standard error. (D) NPCs at the nuclear surface of Nup155-depleted cells were visualized by indirect immunofluorescence using mAb414 at the indicated times after siRNA treatment. Bar, 1 µm. (E) HeLa cells expressing GST-GFP-cNLS were incubated in the presence of transfection reagent alone (Mock) or with Nup155-specific siRNAs. 72 h after transfection, GST-GFP-cNLS was visualized together with mAb414-reactive nups detected by immunofluorescence. Bar, 10 µm.
The loss of Nup155 was accompanied by a decrease in the number of NPCs. Transmission electron microscopy (TEM) revealed a decrease in the number of NPCs per micrometer of sectioned NE, from ∼1.31 NPCs/µm in mock-treated samples to ∼0.31 NPCs/µm in Nup155-depleted cells (Fig. 1, B and C). This decrease in NPCs was also revealed by immunofluorescence microscopy using the monoclonal antibody mAb414, which binds several FG-nups, including Nup62, Nup153, Nup214, and Nup358. In cells treated with Nup155 siRNA, the number of foci recognized by mAb414 at the NE progressively decreased during the time course of the experiment (Fig. 1 D). Those NPCs that remained, even after 72 h of Nup155-specific siRNA treatment, appeared to be functional, as a GST-GFP-cNLS import reporter continued to accumulate in the nucleus (Fig. 1 E).
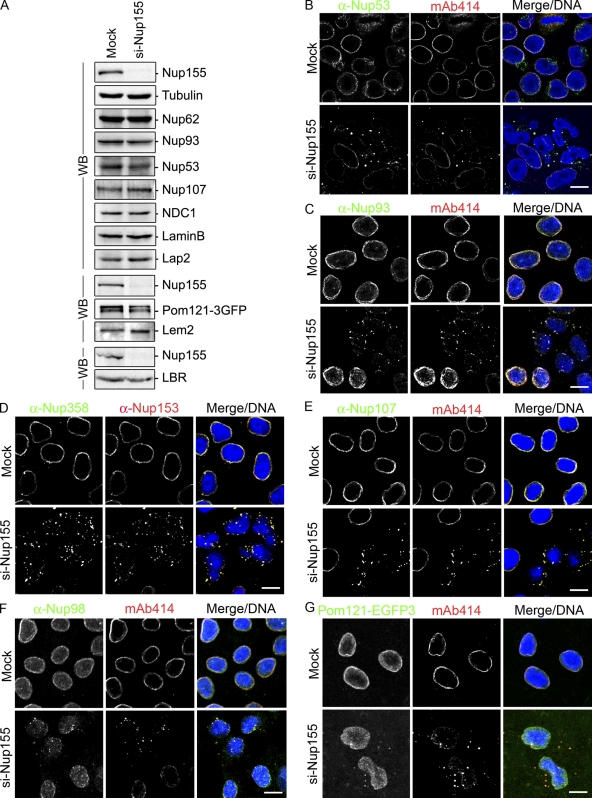
Although Nup155 depletion causes a decrease in the number of NPCs, we detected little or no change in the cellular levels of various nups including Nup62, Nup93, Nup107, and NDC1 by Western blot analysis (Fig. 2 A). Moreover, Nup53, a binding partner of Nup155 (see below), showed only a slight decrease in cellular levels. The apparent discrepancy between the decrease in NPC numbers and the unchanged levels of nups in Nup155-depleted cells was explained by immunofluorescence analysis. All of the nups examined, including Nup53, Nup93, Nup98, Nup107, Nup153, Nup358, and the pore membrane protein Pom121, accumulated within cytoplasm foci concomitant with a decrease in their levels at the NE (Fig. 2, B–G). Moreover, dual-labeling analysis revealed overlapping signals between the mAb414 antibody and that produced by the anti-Nup53, -Nup93, -Nup98, -Nup358, or -Nup107 antibodies or Pom121-EGFP3, suggesting that each individual focus contains multiple nups.
Figure 2.
Depletion of Nup155 in HeLa cells leads to the mislocalization of nucleoporins. HeLa cells were incubated in the presence of transfection reagent alone (Mock) or Nup155-specific siRNA (si-Nup155) for 72 h. (A) Total cell extracts from Mock- and Nup155-depleted cells were assayed by Western blot (WB) analysis using antibodies directed against the indicated proteins. Note: the bottom blot panels are from separate experiments. (B–F) The cellular distribution of various nups was visualized by indirect immunofluorescence using the indicated polyclonal anti-nup antibodies and the mouse monoclonal mAb414 (mAb414 detects Nup62, Nup153, Nup214, and Nup358) or mouse monoclonal anti-Nup153. (G) In Mock- or Nup155-specific siRNA-transfected cells, Pom121-EGFP3 was visualized directly and compared with mAb414-reactive nups detected by immunofluorescence. In all panels merged fluorescence images and DNA visualized with DRAQ5 are shown (Merge/DNA). Bar, 10 µm.
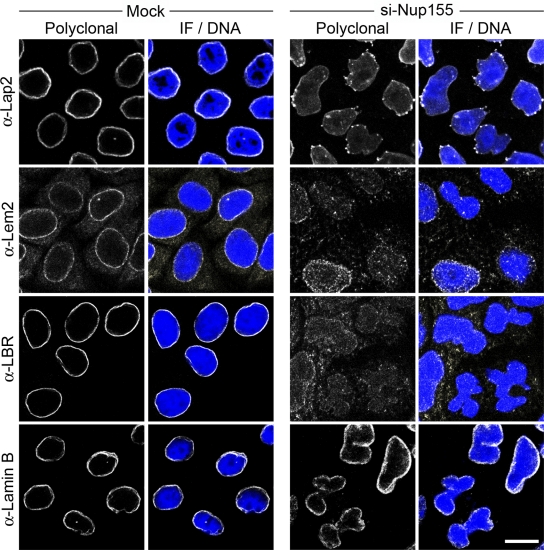
Accompanying the loss of NPCs, essentially all cells depleted of Nup155 exhibited alterations in the morphology of the nucleus after 72 h of siRNA treatment. Nuclei were observed that were kidney shaped or contained multiple lobes connected by bridges of NE membrane (Figs. 1–3, S1, and S2). These changes in the contour of depleted nuclei did not appear to be due to an overt change in the structure of the lamina, as both lamin B and lamins A/C retained their normal peripheral nuclear localization in these cells (Fig. 3; unpublished data). This abnormal nuclear morphology also prompted us to examine the distribution of several INM proteins including Lap2 (lamina-associated polypeptide 2), Lem2 (lamina-associated polypeptide–emerin–MAN1), and LBR (lamin B receptor; for review see Schirmer and Gerace, 2005). Depleting Nup155 did not affect total cellular levels of these proteins (Fig. 2 A); however, NE levels of all three proteins were decreased (Fig. 3), with Lap2 also accumulating in multiple foci adjacent to the surface of the NE (Fig. 3 and Fig. S2). This latter observation prompted us to examine the localization of Lap2 in cells permeabilized with digitonin. This detergent perforates the plasma membrane but leaves the NE intact. In mock-treated cells, anti-Lap2 antibodies failed to detect Lap2, as the NE blocked their accessibility to Lap2 in the INM (Fig. S2 B). However, in Nup155-depleted cells, Lap2 was detected in the ER-like structures and the outer nuclear membrane, suggesting that it failed to efficiently target to the INM. On the basis of these results, we conclude that, in addition to its role in NPC assembly, Nup155 is likely required for the targeting of at least some integral membrane proteins to the INM. This function appears to be specific for Nup155, as depletion of its interacting partner, Nup53, did not alter the INM localization of Lap2 (Fig. S2 C) or emerin (Hawryluk-Gara et al., 2005).
Figure 3.
Nup155 depletion alters targeting of INM proteins. HeLa cells were incubated in the presence of transfection reagent alone (Mock) or Nup155-specific siRNA (si-Nup155) for 72 h and then processed for immunofluorescence microscopy. The indicated INM proteins and lamin B were visualized using specific polyclonal antibodies and DNA was visualized using DRAQ5 (Merge/DNA). Bar, 10 µm.
Nup155 is anchored to the pore membrane by Pom121 and NDC1
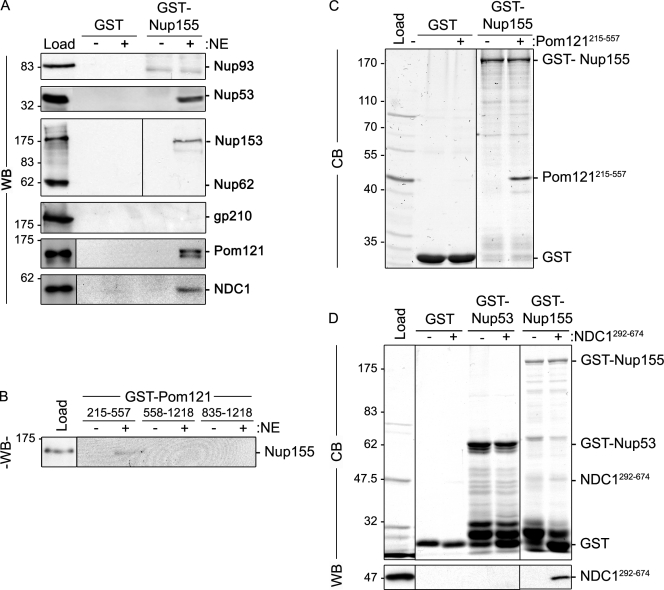
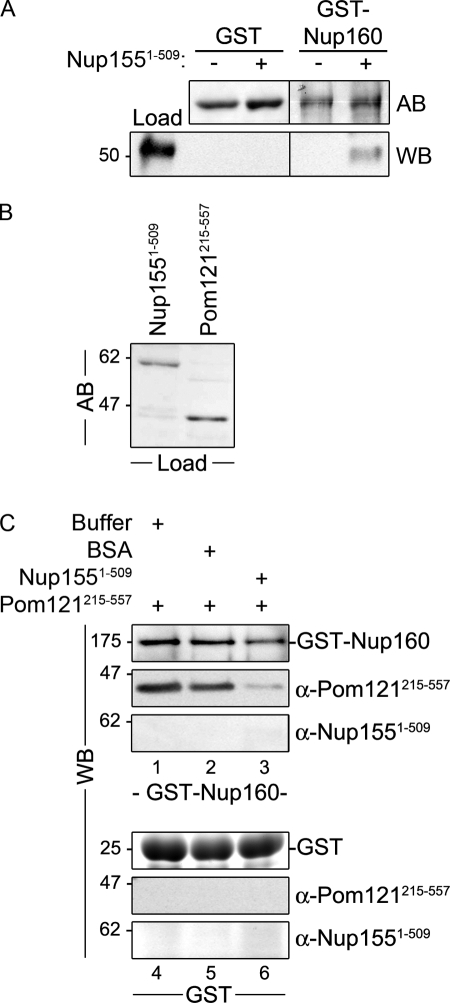
To further understand the mechanistic basis for the requirement of Nup155 in NPC and NE structure, we used GST pull-down assays to define its interacting partners. To facilitate this analysis, purified rat liver NEs were used as an abundant source of mammalian proteins. During extract preparation, conditions were used that disrupt the NE and solubilize most nups, lamins, and integral membrane proteins. These extracts were then incubated with GST-Nup155 immobilized on glutathione–Sepharose beads. Bound protein species were analyzed by SDS-PAGE and Western blotting (Fig. 4). Our results show that many nups failed to bind Nup155 and were absent from the bound fraction, including Nup93 and Nup62. However, as anticipated based on previous studies (Hawryluk-Gara et al., 2005), Nup53 was detected bound to GST-Nup155 but not GST alone. In addition, Nup153 was also detected bound to Nup155. Bound and unbound fractions were also probed with antibodies directed against each of the three known mammalian pore membrane proteins, gp210, NDC1, and Pom121. As shown in Fig. 4, both NDC1 and Pom121 specifically bound Nup155, whereas gp210 was absent from the bound fraction. To extend these results, pull-down experiments were performed using GST fusion proteins containing the pore-facing domain of Pom121. GST fusions to the entire pore-exposed region of Pom121 were largely insoluble and heavily degraded after expression in Escherichia coli (unpublished data), necessitating the construction of smaller, more stable GST truncations. As shown in Fig. 4 B, a GST-Pom121 fusion containing amino acid residues 215–557 (Pom121215–557) was capable of binding Nup155 in rat liver NE extracts, whereas more C-terminal pore-exposed regions, including residues 558–1218 and 835–1218 (Pom121558–1218 and Pom121835–1218), did not. Of note, the Pom121215–557 construct lacks FG-repeats.
Figure 4.
Identification of Nup155-interacting proteins. (A) GST-Nup155 or GST alone were bound to glutathione–Sepharose beads and then incubated in the presence (+) or absence (−) of solubilized rat liver NE extracts. Bound proteins were eluted from beads using SDS-sample buffer. Interacting nups were detected by Western blot (WB) using antibodies directed against the indicated proteins. mAb414 was used to detect Nup62 and Nup153. Approximately 5% of the NE extract loaded on each column was resolved in the lane marked Load. (B) GST-tagged truncations of the pore-facing domain of Pom121 (amino acid residues 215–557, 558–1218, or 835–1218) were incubated with (+) or without (−) rat liver NE extracts. Interacting proteins were eluted with SDS-sample buffer. Polypeptides were resolved by SDS-PAGE and analyzed by Western blotting (WB) using a Nup155-specific polyclonal antibody. Approximately 5% of the NE extract loaded on each column was resolved in the lane marked Load. (C and D) Glutathione–Sepharose beads with attached GST-Nup155, GST-Nup53, or GST alone were incubated with (+) or without (−) purified recombinant Pom121215–557 (C) or Ndc1292–674 (D). Bound proteins were processed as described in A and polypeptides were detected using Coomassie blue staining (CB) or Western blot (WB) using anti-NDC1 antibodies. The lanes marked Load contain ∼50% of the total purified Pom121215–557 (C) or NDC1292–674 (D) loaded on the beads. To the right of each panel, the point at which the named recombinant protein migrates in the appropriate lane is indicated. Mass markers are in kilodaltons.
It was unclear from our pull-down experiments whether Nup155 directly bound Pom121 and NDC1, or instead their interactions were mediated by other nups. To test for direct interactions, GST-Nup155 was immobilized on glutathione–Sepharose beads and then incubated with purified recombinant Pom121215–557 or a fragment of NDC1 consisting of its C-terminal residues 292–674 positioned on the cytoplasmic face of the pore membrane. As shown in Fig. 4 C, Pom121215–557 binds to GST-Nup155 but not GST alone. Similarly, the NDC1292–674 fragment was also capable of binding GST-Nup155 while failing to bind GST or GST-Nup53 (Fig. 4 D).
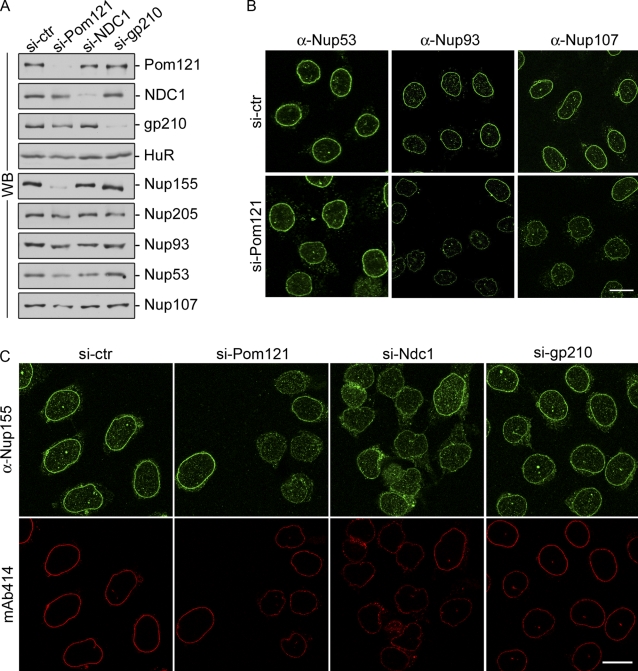
On the basis of our results, we conclude that Nup155 directly interacts with both Pom121 and NDC1 near the pore membrane. We infer from these data that Pom121 and NDC1 may function in the assembly and anchoring of Nup155 at the NPC. To test this idea, we used siRNAs to deplete NDC1, Pom121, or gp210 in HeLa cells and then examined the levels and localization of Nup155 and other nups. Western blot analysis revealed that each of these membrane proteins could be efficiently depleted (Fig. 5 A). The depletion of gp210 or NDC1 had little effect on cellular levels of Nup155 or other nups examined (Fig. 5 A). Moreover, the NE association of Nup155 was unaffected by depletion of gp210 (Fig. 5 C). Nup155 was also detected at the NE of cells depleted of NDC1, albeit at reduced levels relative to untreated cells. The most striking effects on Nup155 were seen in cells depleted of Pom121, where both the NE localization and total cellular levels of Nup155 were greatly reduced as detected by Western blotting and immunofluorescence microscopy. We also observed that depletion of Pom121 caused slight reductions in total cellular levels and reduced NE association of Nup53, Nup93, and Nup107 (Fig. 5 B).
Figure 5.
Depletion of Pom121 reduces cellular levels of Nup155. HeLa cells were incubated in the presence of control siRNAs (si-ctr) or siRNAs targeting Pom121, NDC1, or gp210 as indicated. (A) Cells were harvested and analyzed by Western blotting (WB) using antibodies directed against the indicated proteins. α-HuR was used as a loading control. (B) HeLa cells were processed for immunofluorescence microscopy using anti-Nup53, anti-Nup93, or anti-Nup107 polyclonal antibodies. Bar, 10 µm. (C) Cells were processed for immunofluorescence microscopy and interrogated using anti-Nup155 or mAb414 antibodies. Bar, 10 µm.
Pom121 directly interacts with Nup160 and Nup98
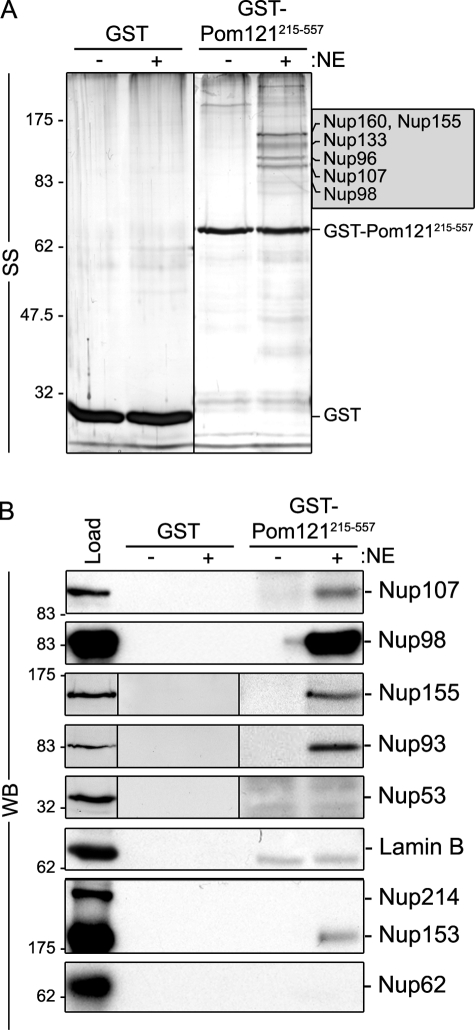
In addition to Nup155, further analysis of GST-Pom121215–557–interacting partners revealed a more complex set of interacting nups. Several abundant polypeptides with masses ranging from ∼90 to 160 kD bound GST-Pom121215–557, but not GST alone (Fig. 6). Less abundant, but specific, interactors were also detected at higher and lower molecular masses. Mass spectrometry (MS) analysis of five of the most abundant polypeptides identified peptides derived from Nup155, Nup98, Nup96, Nup107, Nup133, and Nup160, the latter four being members of the Nup107–160 complex (Fig. 6 A and Fig. S3). The presence of Nup107 and Nup98 in the GST-Pom121215–557–bound fraction was further confirmed by Western blotting (Fig. 6 B). The identities of several less-intensely staining species were also determined by MS analysis or Western blotting, including the interacting nups Nup93 and Nup205, additional members of the Nup107–160 complex (Nup85/75, Nup43), Nup153, Nup358, and ELYS (Fig. 6 B and Fig. S3). Western blotting failed to detect various other nups including Nup53, Nup62, and Nup214.
Figure 6.
Pom121 interacts with Nup155 and the Nup107–160 complex in vitro. Bead-bound GST-Pom121215–557 or GST alone was incubated with (+) or without (−) rat liver NE extracts. Interacting proteins were eluted with SDS-sample buffer. Polypeptides were resolved by SDS-PAGE and visualized by silver staining (SS; A) or analyzed by Western blotting (WB; B) using antibodies directed against the indicated proteins. mAb414 was used to detect Nup62, Nup153, and Nup214. Approximately 5% of the NE extract loaded on each column was resolved in the lane marked Load. Prominent bands were analyzed by mass spectrometry (see Fig. S3) and the predominant species identified in the 90–160-kD range are indicated in the shaded box. Molecular mass markers are indicated in kilodaltons.
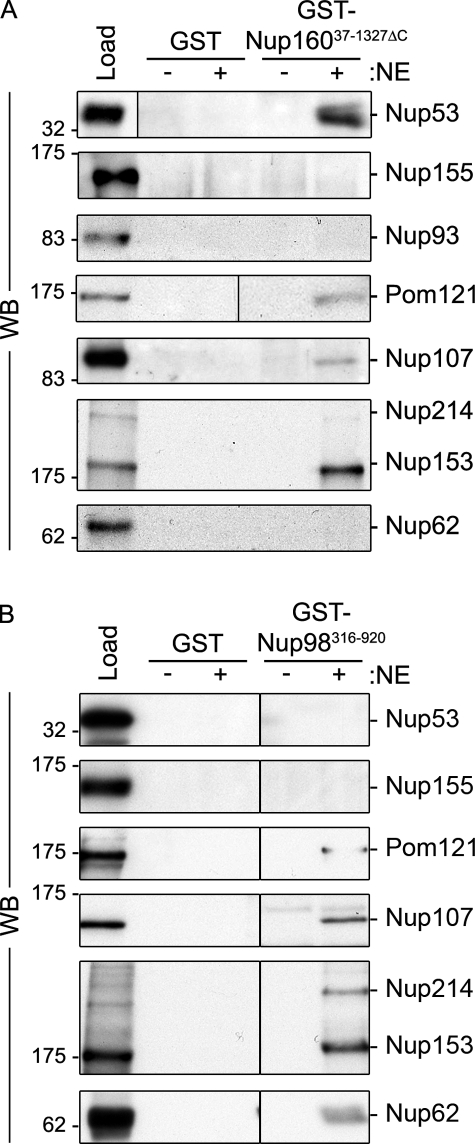
Members of the Nup107–160 complex and Nup98 represent the abundant species in Pom121 pull-downs, and as such were strong candidates for directly binding Pom121. Moreover, Nup160 reproducibly represented the most abundant component of the Nup107–160 complex present in the GST-Pom121215–557 pull-downs. Thus, we further investigated these interactions in pull-down experiments using recombinant GST-Nup160 and GST-Nup98 (Fig. 7). For Nup98, a truncation lacking most of the N-terminal FG repeats (amino acid residues 316–920; Nup98316–920) was tested, as full-length Nup98 was largely insoluble (unpublished data). Similar to the experiments described above for GST-Nup155 and GST-Pom121215–557, GST-Nup160 and GST-Nup98 were incubated with NE extract and proteins in the bound fraction were interrogated by Western blot analysis. As predicted, GST-Nup160 bound to Nup107. Furthermore, we detected Nup53, Nup153, and, importantly, Pom121 bound to GST-Nup160. These interactions appeared to be specific as various other nups examined, including Nup155, Nup93, and Nup62, failed to bind GST-Nup160. Similar experiments using GST-Nup98316–920 also detected a subset of bound NPC proteins, including Pom121, Nup107, and various other nups (Nup214, Nup153, and p62), but not Nup155 or Nup53.
Figure 7.
Identification of nups that bind Nup160 and Nup98. GST-Nup16037–1327ΔC (A), GST-Nup98316–920 (B), or GST alone were bound to glutathione–Sepharose beads and incubated in the presence (+) or absence (−) of solubilized rat liver NE extracts. Bound proteins were eluted from beads using SDS-sample buffer. Interacting nups were detected by Western blot (WB) using antibodies directed against the indicated proteins. mAb414 was used to detect Nup62, Nup153, and Nup214. Approximately 5% of the NE extract loaded on each column was resolved in the lane marked Load. Molecular mass markers are indicated in kilodaltons.
To understand the molecular basis for these interactions, we tested whether Nup160 and Nup98316–920 were capable of directly binding Pom121215–557. Using recombinant proteins and in vitro–binding assays, we observed that both GST-Nup160 and GST-Nup98316–920 bound specifically to Pom121215–557 (Fig. 8 B and Fig. S3). These data, and those discussed above, suggest that at least three nups, Nup98, Nup155, and Nup160, are capable of directly binding Pom121.
Figure 8.
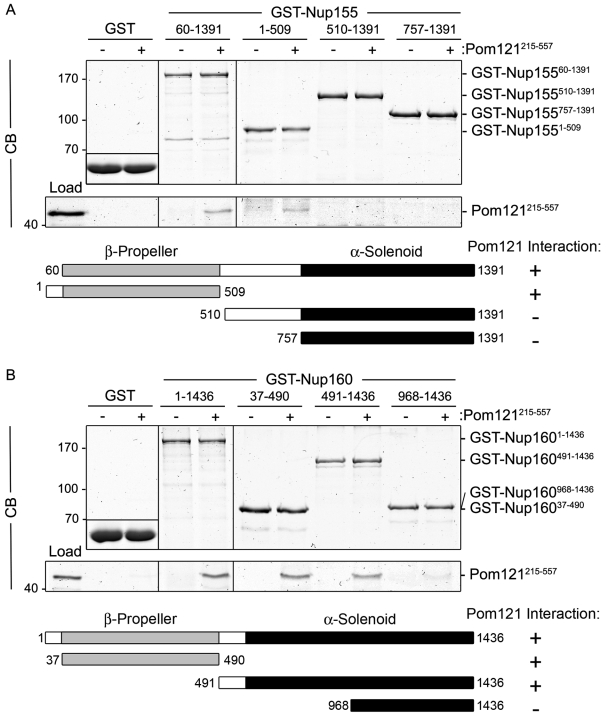
Pom121 interacts directly with Nup98 and N-terminal domains of Nup155 and Nup160. Purified Pom121215–557 was incubated with bead-bound (A) GST-Nup15560–1391, GST-Nup1551–509, GST-Nup155510–1391, and GST-Nup155757–1391, or (B) GST-Nup1601–1436, GST-Nup16037–490, GST-Nup160491–1436, and GST-Nup160968–1436. Shown in each panel are controls where Pom121215–557 was incubated with GST alone (shown as inset). To obtain the results shown in each panel, bead-bound proteins were eluted using SDS-sample buffer, resolved by SDS-PAGE, and visualized by Coomassie blue (CB) staining. The lane marked Load contains ∼10% of the total purified Pom121215–557 loaded on the beads. To the right of each panel, the point at which the named protein migrates in the appropriate lane is indicated. Schematic representations of Nup155 (A) and Nup160 (B) fragments and their observed binding to Pom121215–557 are summarized. Mass markers are in kilodaltons.
Predicted β-propeller domains of Nup155 and Nup160 bind Pom121
Nup155 and Nup160 are predicted to share a similar structural organization consisting of an N-terminal β-propeller and C-terminal α-solenoid domain (Berke et al., 2004; Devos et al., 2004, 2006; Schwartz, 2005). We investigated the role of these domains in binding to Pom121 using in vitro–binding assays with GST fusions containing the β-propeller or α-solenoid domains of Nup155 or Nup160 (Fig. 8). Similar to full-length Nup155, a GST fusion containing the Nup155 β-propeller region (GST-Nup1551–509) robustly bound to Pom121215–557 (Fig. 8 A); however, no interaction was observed with truncations of the α-solenoid domain (GST-Nup155510–1391 and GST-Nup155757–1391). The interaction of Nup160 with Pom121 was more complex. The predicted β-propeller of Nup160 (GST-Nup16037–490) also interacted with Pom121215–557 (Fig. 8 B). However, unlike Nup155, a GST fusion containing a portion of the α-solenoid region of Nup160 (residues 491–1436) also bound Pom121. Further truncation of this fragment suggested the binding activity is likely contained within residues 491–967, as GST-Nup160968–1436 showed only trace levels of bound Pom121215–557. Together, these results suggest that Pom121 binds the β-propeller domains of Nup155 and Nup160, with Nup160 containing additional Pom121-interacting determinants. These former interactions appear to be specific for the β-propeller regions of Nup155 and Nup160, as the β-propeller domains of two other Nup107–160 complex subunits, Sec13 and Seh1, did not significantly bind Pom121215–557 (Fig. S3 C; unpublished data).
The ability of similarly structured regions of Nup155 and Nup160 to bind to Pom121215–557 raised the question of whether their β-propeller domains can bind simultaneously to Pom121 or whether their binding is mutually exclusive. To test this, we examined the ability of a preformed complex of purified, recombinant Pom121215–557 and the β-propeller region of Nup155 (Nup1551–509) to bind full-length Nup160. Individually, Pom121215–557 or Nup1551–509 was capable of binding bead-bound GST-Nup160 (Fig. 8 B; Fig. 9, A and C). However, when Pom121215–557 was first incubated with Nup1551–509, and the two proteins allowed to interact, the binding of these proteins to Nup160 was greatly inhibited (Fig. 9 C). These results suggest that the binding of Pom121215–557 to Nup1551–509 inhibits their interactions with Nup160.
Figure 9.
Binding of Nup155 to Pom121 prevents their interactions with Nup160. (A) Bead-bound GST-Nup160 or GST alone was incubated with purified Nup1551–509. Bound proteins were eluted using SDS-sample buffer and resolved by SDS PAGE. Levels of GST and GST-Nup160 were visualized using amido black (AB). Nup1551–509 was detected using a Nup155-specific polyclonal antibody. The lane marked Load contains ∼6% of the total purified Nup1551–509 loaded on the beads. (B and C) Purified Pom121215–557 and Nup1551–509 (∼25% of the total Load protein is shown in B after amido black [AB] staining) were mixed and allowed to interact for 30 min before addition to bead-bound GST-Nup160 or GST alone (lanes 3 and 6). Note: to perform the binding reactions under similar conditions, Pom121215–557 was supplemented with BSA to a total protein concentration similar to that of combined Pom121215–557–Nup1551–509. Bound proteins were eluted using SDS-sample buffer and resolved by SDS-PAGE. Western blotting (WB) using anti-Pom121, anti-Nup155, or anti-GST-Nup160 antibodies was used to detect the presence of each protein. Mass markers are indicated in kilodaltons.
Discussion
We have demonstrated that Nup155 plays an essential role in the assembly of the mammalian NPC and is required for normal nuclear morphology. Depletion of Nup155 from HeLa cells results in a dramatic reduction in NPC number and the accumulation of nups in cytoplasmic foci. Loss of Nup155 is also accompanied by massive changes in NE structure and the improper targeting of several integral membrane proteins to the INM. The critical role of Nup155 in NPC assembly and NE structure is likely played at the nuclear pore membrane domain where it interacts directly with the membrane nups Pom121 and NDC1. We show that the N-terminal β-propeller region of Nup155 binds to the N-terminal third of Pom121. This same region of Pom121 also binds directly to the Nup107–160 complex through an interaction with the β-propeller region of Nup160. These results support a model in which the N terminus of Pom121 anchors the NPC core through its binding to the β-propeller regions of Nup155 and Nup160. These interactions are likely to play an important role in the formation of the pore membrane and the assembly of the NPC core.
Nup155 is essential for mammalian NPC assembly and nuclear morphology
Through the use of siRNA-mediated depletion assays in human HeLa cells, we have shown that reducing cellular levels of Nup155 causes a drastic decrease in the number of NPCs and massive changes in nuclear structure (Figs. 1–3, S1, and S2). Changes in nuclear morphology in response to the loss of an individual nup have been documented in various organisms, most notably in yeast (for review see Doye and Hurt, 1995 and Hetzer and Wente, 2009). Although the underlying cause of this general phenotype is unclear, its occurrence in response to different mutations suggests that it likely reflects a systemic response to the loss of NPC numbers or function that includes an increase in nuclear surface area and, perhaps, an attempt to increase total NPC numbers.
The depletion of Nup155 also causes a general decrease in the NE association of at least three INM proteins, LBR, Lem2, and Lap2, and a corresponding increase in the cytoplasmic pool of these proteins, presumably within the ER or other endomembranes (Fig. 3 and Fig. S2). Moreover, Lap2 appears to concentrate in numerous foci along the NE, which are most clear in cells extracted with detergent before fixation (Fig. 3). These Lap2 foci appear to accumulate in the outer nuclear membrane (ONM), as they are accessible to “cytoplasmically positioned” antibodies in cells where the NE membrane is intact (Fig. S2). The ability of Lap2 to accumulate in the ONM would suggest binding sites for Lap2 are present there. One possibility is that Lap2, like another INM protein, emerin, can interact with LINC (linkers of the nucleoskeleton to the cytoskeleton) complexes, macromolecular assemblies that span the double membrane of the NE (Haque et al., 2010).
The inhibition of membrane protein targeting to the INM appears to be a phenotype specific to the depletion of Nup155. Depletion of several other nups, including Nup53 (Hawryluk-Gara et al., 2005; Fig. 3), Mel28 (Franz et al., 2007), or the core nup Nup188 (Theerthagiri et al., 2010) does not inhibit access of membrane proteins to the INM. In contrast, X. laevis nuclei assembled in vitro in the absence of Nup188 showed increased accessibility of membrane proteins to the INM (Theerthagiri et al., 2010). Furthermore, as depletion of Nup53 or Mel28 inhibits NPC formation, merely reducing the number of NPCs is unlikely to limit the movement of membrane proteins to the INM. Instead, Nup155 may play a more direct role as suggested for its counterpart, Nup170p, in Saccharomyces cerevisiae (King et al., 2006). In yeast, karyopherins have been shown to play an active role in the targeting of at least a subset of membrane proteins to the INM. These studies also showed that INM protein targeting is compromised by the loss of Nup170p. Although the mechanistic basis for these observations is unclear, our data are consistent with a similar pathway functioning in higher eukaryotes.
The β-propeller regions of Nup155 and Nup160 bind directly to Pom121
On the basis of several observations, we conclude that Nup155 is positioned in close proximity to the pore membrane. Our pull-down analysis using GST-Nup155 revealed binding to Pom121 and NDC1, and, importantly, direct interactions between domains of these membrane proteins and Nup155 could be reconstituted in vitro using recombinant proteins (Fig. 4). Our proposed localization of Nup155 adjacent to the pore membrane is similar to that suggested for its yeast counterparts, Nup157p and Nup170p (Tcheperegine et al., 1999; Alber et al., 2007b; Flemming et al., 2009; Makio et al., 2009; Onischenko et al., 2009). Nup170p directly interacts with the yeast pore membrane protein Pom152p (Makio et al., 2009) and is positioned in the vicinity of Ndc1p in architectural models of the yeast NPC (Alber et al., 2007a,b). However, no direct interaction has been reported between Nup170p and Ndc1p.
The ability of Pom121 and Pom152p to bind Nup155 and Nup170p, respectively, would suggest they share similar functions in the NPC. In support of this idea, we also observed that the Nup155-binding region of Pom121 (residues 215–557) can directly bind yeast Nup170p (Fig. S4 E). Similarly, a pore-exposed region of Pom152p (residues 1–111) can bind Nup155, as well as members of the Nup107–160 complex, in pull-down assays (Fig. S4). Thus, although Pom121 and Pom152p fail to exhibit any extensive sequence similarity, their ability to bind similar nups across species lead us to conclude that they contain conserved nup-binding regions. However, these domains remain difficult to pinpoint. They may be small, or contained within uncharacterized secondary or tertiary structural features.
In addition to Nup155, our GST-Pom121 pull-down experiments also detected components of the Nup53–Nup93 complex, the Nup107–160 complex, and EYLS (a binding partner of the Nup107–160 complex; Fig. 6 and Fig. S3). These results are in agreement with previously reported data obtained from pull-down experiments using X. laevis Pom121 and egg cytosol (Rasala et al., 2008). Importantly, we have now shed light on the molecular basis of these associations by establishing the nature of these interactions, most notably the identification of Nup160 as a direct binding partner of Pom121 within the Nup107–160 complex (Figs. 8 and 9). Through further analysis of the interactions of Pom121 with Nup160 and Nup155, we have determined that the primary Pom121-binding region in each nup is contained within an N-terminal β-strand–rich region predicted to form a β-propeller (Berke et al., 2004). This structural feature is known to mediate many protein–protein interactions (Hudson and Cooley, 2008). Within the NPC, β-propeller motifs are most prevalent in the core scaffold nups. In particular, six of the nine members of the Nup107–160 complex contain predicted β-propeller structures. However, little is known about their interacting partners or their functions. In the yeast counterpart of the Nup107–160 complex (the Nup84p complex), binding partners for two β-propeller proteins, Seh1p and Sec13p, have been identified as Nup85p (Brohawn et al., 2008; Debler et al., 2008) and Nup145Cp (Hsia et al., 2007), both of which are components of the Nup84p complex. Our studies now establish the β-propeller regions of Nup160 and Nup155 as key structural elements in linking their respective protein complexes to Pom121.
A role for Nup160 in positioning the Nup107–160 complex near the pore membrane is also inferred by studies of its yeast counterpart, Nup120p. Like Nup160, Nup120p has been proposed to lie adjacent to the pore membrane (Alber et al., 2007a,b). Moreover, other observations suggest Nup120p is strategically positioned to influence the structure and oligomeric state of the Nup84p complex. Elegant studies have shown that Nup120p contributes one arm to the Y-shaped Nup84p complex (Siniossoglou et al., 2000; Lutzmann et al., 2002, 2005; Kampmann and Blobel, 2009). Recently, it has also been proposed that Nup120p may link consecutive Nup84p complexes by binding Nup133p, positioned at the foot of the Y complex, to form rings consisting of eight Nup84p complexes arranged head to tail (Seo et al., 2009). The C-terminal, α-helical region of Nup120p mediates binding to Nup133p. This would position the N-terminal β-propeller of Nup120p near the junction between Nup84p complexes but free to interact with other binding partners, including membrane proteins such as Pom152p. It is intriguing to consider that this conformation of Nup120p molecules, established by the formation of a ring of Nup84p complexes, may facilitate their membrane association. By analogy, Nup160 could contribute to the interactions of similar higher-order structures to Pom121.
We also observe that Pom121 binds to the predicted β-propeller region of Nup155. These results lead us to conclude that the N-terminal region of Pom121 (within residues 215–557) contains one or more binding sites for the β-propeller regions of at least two nups (Nup155 and Nup160). The extent to which Pom121 binds other β-propeller–containing nups has not been fully explored. However, in examining a subset of these nups, we have observed specificity for Nup155 and Nup160, as the β-propeller proteins Sec13 and Seh1, as well as the COPII protein Sec31, show only negligible binding to Pom121 (Fig. S3; unpublished data). Importantly, our analysis has also led us to conclude that, although Pom121 can bind the β-propeller regions of both Nup155 and Nup160, it cannot interact with them both simultaneously (Fig. 9). This could be explained by direct competition between these nups for a single site on Pom121 or by one β-propeller inducing a conformational change in Pom121 that blocks a second β-propeller binding site.
Our results may reflect the presence of separate Pom121–Nup155 and Pom121–Nup160 complexes within the NPC. A single nup in two distinct complexes is not unprecedented. For example, yeast Nsp1p appears to be part of two distinct complexes (Grandi et al., 1993, 1995; Bailer et al., 2001). Another possibility is that the interactions between individual β-propeller proteins and Pom121 are dynamic and change under different conditions. For instance, interactions between Pom121 and a specific nup could be established and later broken during the course of NPC assembly or be altered in response to changes in the transport properties of the NPC (Feldherr et al., 1998, 2002; Makhnevych et al., 2003). Plasticity in molecular interactions between nups and Pom121 is supported by our experiments showing that full-length Nup160 can bind to Pom121 and the β-propeller of Nup155 if each is presented individually (Fig. 9). However, Nup160 does not bind a preformed Nup155–Pom121 complex. These results would suggest that conditions that disrupt the interactions of Nup155 and Pom121 could facilitate their interactions with Nup160.
Implications for the role of Nup155 and Nup160 in NPC assembly
The molecular architecture of Nup155 and Nup160, consisting of an N-terminal β-propeller and a C-terminal α-solenoid, is similar to the outer cage components of CPCs functioning in vesicular transport, including clathrin and Sec31 (Berke et al., 2004; Schwartz, 2005; Stagg et al., 2007). Based on this similarity, it has been hypothesized that the β-propeller α-solenoid nups play a role at the pore membrane similar to their counterparts in the CPC (Devos et al., 2004, 2006; Schwartz, 2005), including inducing and maintaining membrane curvature during and after NPC formation. How nups such as Nup155 and Nup160 might perform these functions, and the degree to which they are similar to those performed by CPCs, is unclear. Based on our data, we believe that these nups play a more direct role in interfacing with membranes and membrane proteins than clathrin or Sec31. For instance, clathrin and Sec31 do not bind directly to membranes and both require adaptor proteins to bridge their interactions with integral membrane cargo proteins (Gurkan et al., 2007; Pucadyil and Schmid, 2009). By contrast, our data indicate that Nup155 and Nup160 directly interact with membrane proteins, which could be viewed analogously to “cargo” proteins of CPCs. Moreover, certain β-propeller α-solenoid nups also contain predicted amphipathic ALPS motifs, including Nup133, Nup120p, and Nup170p (Drin et al., 2007), that may allow them to directly bind the pore membrane. In fact, in vitro binding studies have demonstrated that the ALPS motif of Nup133 is capable of directly binding vesicles with a defined membrane curvature (Drin et al., 2007). ALPS motifs and other amphipathic features are present in some components of the adaptor layer of CPCs but have not been detected in clathrin or Sec31. In addition to their ability to interface with the membrane, members of β-propeller α-solenoid nups also interact with other nups that appear capable of inducing and/or stabilizing membrane curvature. Nup155 interacts with Nup53 (Fig. 4; Hawryluk-Gara et al., 2005) and the Nup107–160 complex binds Nup153 (Fig. 7; Vasu et al., 2001). The overproduction of both Nup153 and the yeast counterpart of Nup53 induces membrane proliferation and the formation of tubular membranes (Bastos et al., 1996; Marelli et al., 2001). Thus, the Pom121-mediated juxtaposition of Nup155 and Nup160, as well as Nup98, to the membrane may aid in the association of additional nups with the pore membrane.
We envisage a model in which the pore-exposed region of Pom121 extends toward the central axis of the NPC with its N-terminal region positioned nearest the membrane and functioning as a binding site for Nup155 and Nup160 and their respective complexes. As is the case in yeast, how these complexes are configured relative to one another and the membrane is open to speculation (Alber et al., 2007b; Hsia et al., 2007; Brohawn and Schwartz, 2009). More C-terminal regions of Pom121 containing its FG-repeat domain are predicted to extend beyond this coat into the central channel of the NPC, where it would join other FG-nups to facilitate the movement of transport factors through the NPC.
The interactions of Pom121 with Nup155 and Nup160 and their juxtaposition to the pore membrane are consistent with several observations that place these proteins at critical, and temporally related, steps in NPC assembly. As we have shown here, siRNA depletion of Nup155 in HeLa cells leads to a reduction of NPCs and the mislocalization of all nups examined, including Pom121, into cytoplasmic foci (Fig. 2). These phenotypes are consistent with those observed upon depletion of the counterparts of human Nup155 in C. elegans and S. cerevisiae (Franz et al., 2005; Flemming et al., 2009; Makio et al., 2009; Onischenko et al., 2009). Efficient depletion of Pom121 also leads to a decrease in NPC numbers and an accompanying decline in the NE association of several nups including Nup155 (Fig. 5; Mansfeld et al., 2006; Funakoshi et al., 2007). Consistent with these data, several observations indicate that Pom121 and Nup155 may function at a similar step in NPC assembly that is closely linked to formation and stabilization of the pore membrane. Beyond the aforementioned physical relationship to the membrane, studies examining the effects of depleting these proteins on NE and NPC assembly in X. laevis egg extracts point to a similar role for these proteins in the membrane dynamics required for NE and NPC assembly (Antonin et al., 2005; Franz et al., 2005). In fact, depletion of either Nup155 or Pom121 appears to activate a NE assembly checkpoint, which inhibits the formation of the double membrane NE (Antonin et al., 2005; Franz et al., 2005). Interestingly, NE assembly arrest is dependent on the Nup107–160 complex, suggesting that this complex plays a key role in sensing the completion of critical NE and NPC assembly steps necessary to inactivate the checkpoint. Among these steps may be the interactions of Pom121 and/or Nup155 with Nup160.
Materials and methods
Subcloning E. coli expression constructs
The open reading frame (ORF) of human Nup155 (GenBank/EMBL/DDBJ accession no. NM_153485.1) was amplified using the Expand High Fidelity PCR system (Roche) using the cDNA clone KIAA0791 (Kazusa DNA Research Institute, Chiba, Japan) as a template. This cDNA clone encodes a protein product lacking the N-terminal 59 amino acid residues of Nup155 isoform 1 (NCBI Protein database accession no. NP_705618.1); therefore, its protein product is referred to as Nup15560–1391. cDNA encoding Nup15560–1391 was inserted into the EcoRI site of pGEX-6P-1 (GE Healthcare). Truncations of Nup155 (encoding amino acid residues 510–1391 and 757–1391) were amplified from KIAA0791 and directionally inserted into the EcoRI (5′) and NotI (3′) sites of pGEX-6P-1. cDNA encoding the Nup155 β-propeller domain (residues 1–515) was obtained from HeLa total RNA using the SuperScript One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen) and Nup155-specific primers. This PCR product was directionally inserted into the EcoRI (5′) and NotI (3′) sites of pGEX-6P-1.
The ORF of human Nup160 (GenBank accession no. NM_015231.1) was amplified using the Phusion High-Fidelity PCR kit (New England BioLabs Inc.) from IMAGE cDNA clone MGC:150678 (Clone ID 40124592; Thermo Fisher Scientific). cDNAs encoding Nup160 amino acid residues 1–1436, 491–1436, and 968–1436 were directionally cloned into the BamHI (5′) and NotI (3′) sites of pGEX-6P-1. cDNAs encoding additional regions of Nup160 (amino acid residues 37–490 and 37–1327ΔC, see below) were amplified using the Phusion High-Fidelity PCR kit, using cDNA clone KIAA0197 (Kazusa DNA Research Institute) as a template. PCR products were directionally inserted into the BamHI (5′) and NotI (3′) sites of pGEX-6P-1. This cDNA clone contains small deletions of the both the N and C termini, corresponding to amino acid residues 1–36, 1328–1402, and 1410–1420 of full-length Nup160 (1436 amino acid residues in length; NCBI Protein database accession no. NP_056046.1). Nup160 constructs created using this cDNA as a template are referred to as Nup16037–490 and Nup16037–1327ΔC.
Truncations of human Pom121 (Pom121A; GenBank accession no. NM_172020.1) were produced using cDNA clone KIAA0618 (Kazusa DNA Research Institute) as a template. cDNAs corresponding to amino acid residues 215–557, 364–458, and 558–1218 of Pom121A were amplified using the Phusion High-Fidelity PCR kit and inserted into the NotI site of pGEX-6P-1.
cDNAs corresponding to regions of human NDC1 (amino acid residues 292–674; GenBank accession no. NM_018087.4) and human Nup98 (amino acid residues 316–920 and 496–855; GenBank accession no. NM_016320.3) were amplified using the Expand High Fidelity PCR system and Human Fetal Kidney cDNA library (BD) as a template. All PCR products were directionally inserted into pGEX-6P-1; the PCR product encoding NDC1292–674 was inserted into EcoR1 (5′) and Not (3′) sites, the Nup98316–920 PCR product was inserted into BamHI (5′) and NotI (3′) sites, and the Nup98496–855 PCR product was inserted into BamHI (5′) and EcoRI (3′) sites.
Recombinant protein purification and antibody production
pGEX-6P-1–based plasmids were transformed into E. coli BL21-CodonPlus(DE3)-RIL cells (Agilent Technologies). Cells were grown to an OD260 of 0.6–0.8, and production of GST fusion proteins was induced using 1 mM IPTG for 4 h at 20°C. Cells were collected by centrifugation and resuspended in lysis buffer (LB) consisting of 50 mM Tris, pH 7.5, 300 mM NaCl, 150 mM KOAc, 2 mM MgOAc, 10% glycerol, 0.1% Igepal (Sigma-Aldrich), 1 mM DTT, and complete protease inhibitor cocktail tablets (Roche; 60 ml of LB/g pellet). Cell suspensions were sonicated and then clarified by centrifugation at 27,000 g for 20 min. GST fusion proteins were isolated using glutathione–Sepharose 4B Media (GE Healthcare) according to the manufacturer’s instructions. After repeated washing, bead-bound recombinant proteins were incubated with LB containing 2 mM ATP and 10 mM MgSO4 for 10 min at 37°C. Protein yield was estimated using SDS-PAGE followed by staining with Bio-Safe Coomassie (Bio-Rad Laboratories). Bead-bound recombinant proteins were used directly in pull-down assays with NE extracts or for binding purified recombinant proteins (see below).
Recombinant proteins used for the purpose of eliciting antibodies in rabbits (including Nup98496–855, Nup1551–509, Nup155757–1391, Nup16037–490, Ndc1292–449, and Pom121215–557) were released from glutathione–Sepharose media by proteolysis using PreScission Protease (GE Healthcare) for 16 h at 4°C. After repeated washing, the pooled eluates were concentrated to 0.1 mg of protein/ml. Approximately 100 µg of purified protein was injected into rabbits concomitantly with Freund’s Adjuvant (Sigma-Aldrich), and sera were subsequently collected at 4-wk intervals. Purified Pom121215–557 and NDC1292–674 were additionally used in direct in vitro binding experiments (see below).
Subfractionation and extraction of NEs
Subfractionation of rat liver nuclei was performed as described previously (Blobel and Potter, 1966; with modifications outlined by Wozniak et al., 1989). Livers harvested from Sprague-Dawley rats were washed in 0.25 M STEAKM (0.25 M sucrose, 50 mM triethanolamine, 25 mM KCl, 5 mM MgCl2, 0.5 mM PMSF, and 1 mM DTT) and minced using ethanol-washed razor blades. The triturate was further homogenized in 2 × 0.25 M STEAKM (vol/wt) using a chilled Potter homogenizer, and filtered through cheesecloth before centrifugation at 4°C for 10 min at 800 g. Pellet fractions were collected, briefly homogenized, and diluted with 2 × 2.3 M STEAKM (vol/vol; 2.3 M sucrose, 50 mM triethanolamine, 25 mM KCl, 5 mM MgCl2, 0.5 mM PMSF, and 1 mM DTT). The diluted homogenate was layered over a cushion of 2.3 M STEAKM and centrifuged at 4°C for 75 min at 140,000 g. Nuclei in the pellet fraction were harvested, resuspended in 0.25 M STEAKM, and stored at −80°C.
Nuclei were ruptured by successive rounds of nuclease digestion, as described by Dwyer and Blobel (1976). NEs obtained from this procedure were pelleted in 100 A260 unit fractions (1 A260 unit represents the amount of material derived from ∼3 × 106 nuclei; Aaronson and Blobel, 1974), and proteins were extracted from membranes as described previously (Radu et al., 1993). In brief, NEs were extracted with 1% Triton X-100, 400 mM NaCl, 20 mM Tris, pH 7.5, 1 mM DTT, and complete protease inhibitor cocktail tablets. Insoluble material was pelleted by centrifugation at 20,000 g for 20 min at 4°C. The supernatant fraction was further clarified by syringe filtration using a 0.22-µm filter (Millipore), diluted 3.75-fold in 20 mM Tris, pH 7.5, and supplemented with Tween 20 (final concentration: 20 mM Tris, pH 7.5, 106 mM NaCl, 0.3% Triton X-100, 0.1% Tween 20, and 66 A260 of NE/ml). These NE extracts were immediately used in GST pull-down assays.
GST pull-down assays and in vitro–binding experiments
GST pull-down assays were performed as described previously (Hawryluk-Gara et al., 2005). NE extracts derived from ∼30 A260 units of nuclei were incubated with bead-bound recombinant proteins (∼5–10 µg) for 4 h at 4°C. After binding, beads were extensively washed in a buffer containing 20 mM Tris, pH 7.5, 106 mM NaCl, 0.3% Triton X-100, 0.1% Tween 20, 1 mM DTT, and complete protease inhibitor cocktail tablets. Bound proteins were eluted from beads using SDS-sample buffer and resolved by SDS-PAGE. Proteins were visualized using Bio-Safe Coomassie blue stain or silver staining, or were transferred to nitrocellulose for Western blot analysis (see below).
To assess direct protein–protein interactions using recombinant proteins, ∼1–5 µg of purified recombinant protein was incubated with ∼1 µg of the bead-bound GST fusion protein for 30 min at 4°C. After repeated washing, proteins were eluted from beads using SDS-sample buffer and resolved by SDS-PAGE. Proteins were visualized using Bio-Safe Coomassie blue stain. For experiments using purified NDC1292–674 and those presented in Fig. 9, bound proteins were transferred to nitrocellulose after SDS-PAGE and detected using specific polyclonal antibodies.
RNA interference
Silencer pre-designed siRNAs targeting exon 2 (#138652, 5′-r[CCGUUUCUGGCAUGUCAGA]dTT-3′), the exon 8/9 junction (#110275, 5′-r[GGAGUAAUACAGGUGUAUG]dTT-3′), and exon 27 (#110276, 5′-r[GGAUGAGCUCUUUAGUAUU]dTT-3′) of Nup155 mRNA were obtained from Applied Biosystems. siRNAs were also designed to target the Nup155 exon 4/5 junction (5′-r[GCAGGCAUCUUUCAACCUC]dTT-3′), Pom121 (5′-r[CAGUGGCAGUGGACAUUCA]dTT-3′), NDC1 (5′-r[CUGCACCACAGUAUUUAUU]dTT-3′), and gp210 (5′-r[GAACCUCCAUUCACUACAA]dTT-3′) (QIAGEN). Non-specific siRNAs used as controls in siRNA experiments were designated nonsense (5′-r[AAAGCGCAUUGCGCAUACG]dTT-3′) and control (5′-r[CUGUGCAAGCCGUUGUGUA]dTT-3′) (QIAGEN).
To deplete Nup155, HeLa cells were transfected with 480 nM siRNA using Oligofectamine (Invitrogen), according to the manufacturer’s instructions. Cells were harvested at 24-, 48-, and 72-h time points and analyzed by immunofluorescence or Western blot analysis (see below). As a control, cells were incubated with nonsense siRNA or with transfection reagent alone. This protocol was additionally used to deplete Nup155 from HeLa cells expressing ectopic GST-GST-cNLS (Erkmann et al., 2005) and from HeLa cells stably expressing Pom121-EGFP3 (Pom121-EGFP3 expression vector was kindly provided by Jan Ellenberg, EMBL, Heidelberg, Germany; Rabut et al., 2004).
To deplete the pore membrane proteins Pom121, NDC1, and gp210, HeLa cells were transfected with 35-nM concentrations of the indicated siRNAs using Oligofectamine. As a control, cells were alternatively left untreated or incubated with control siRNA. 72 h after transfection, cells were analyzed by immunofluorescence or Western blot analysis (see below).
Western blotting
Sample preparation.
Before Western blotting, HeLa cells grown in 35-mm dishes were detached using 0.05% Trypsin-EDTA (Invitrogen) and then washed with PBS. Cell pellets were then resuspended in SDS-sample buffer at a concentration of ∼104 cells/µl, yielding a total protein concentration of ∼1 mg/ml. Samples were briefly sonicated and denatured at 95°C for 2 min; ∼10 µg of total protein was resolved by SDS-PAGE and transferred to nitrocellulose. Proteins extracted from NEs were precipitated in 10% TCA and resuspended in SDS-sample buffer (0.1 A260 units/µl). 1 A260 unit was resolved by SDS-PAGE and transferred to nitrocellulose (Load fractions). Membranes were blocked with 5% skim milk powder in PBS-T (PBS containing 0.1% Tween 20).
Antibodies.
Membranes were incubated overnight at 4°C with anti-serum diluted in 5% milk-PBS-T. Antibodies directed against Nup155, NDC1, Pom121, and Nup98 are described above. Antibodies directed against Nup53 (Hawryluk-Gara et al., 2005), Nup107 (kindly provided by V. Doye, Institue Curie, Paris, France; Belgareh et al., 2001), Nup205 (kindly provided by U. Kutay; Mansfeld et al., 2006), gp210 (IQ294; ImmuQuest Ltd.), and Lamin B (Chaudhary and Courvalin, 1993) were previously described. Polyclonal guinea pig antibodies directed against Nup93 were kindly provided by V. Cordes (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). Commercially available monoclonal antibodies were used to detect Nup62, Nup153, Nup214, and Nup358 (using mAb414 [MMS-120p]; Covance), tubulin (T9026; Sigma-Aldrich), Lap2 (611000; BD), and HuR (3A2; Santa Cruz Biotechnology, Inc.). HRP-conjugated secondary antibodies used to detect primary antibodies included goat anti–guinea pig IgG (Sigma-Aldrich), donkey anti–rabbit IgG, and sheep anti–mouse IgG (GE Healthcare). Chemiluminescence was initiated using ECL detection reagent (GE Healthcare) and the signal was detected using Fuji RX film (Fujifilm).
Immunofluorescence, image acquisition, and image processing
Immunofluorescence was performed as described previously (Hawryluk-Gara et al., 2005). HeLa cells grown on coverslips were rinsed with PBS and then treated with 0.2% Triton X-100 in PBS for 2 min. Coverslips were gently washed to remove detergent and fixed with 3.75% formaldehyde in PBS for 10 min. After a PBS wash, coverslips were incubated with 2% skim milk powder in PBS-T for 30 min, followed by incubation with primary antibodies diluted in 2% milk/PBS-T for 2 h at room temperature. Primary antibodies used for immunofluorescence detection are listed above. In addition, antibodies used to detect the INM proteins Lem2 (Ulbert et al., 2006) and LBR (Buendia and Courvalin, 1997) were kindly provided by I. Mattaj (EMBL, Heidelberg, Germany) and H. Herrmann (German Cancer Center, Heidelberg, Germany), respectively. Primary antibody binding was detected with fluorophore-conjugated secondary antibodies, including Cy3-conjugated donkey anti–mouse IgG, Cy3-conjugated donkey anti–guinea pig IgG (Jackson ImmunoResearch Laboratories, Inc.), and Alexa Fluor 488 donkey anti–rabbit IgG (Invitrogen). Secondary antibodies were diluted in PBS-T containing the nuclear DNA stain DRAQ5 (1 µM; Biostatus Ltd.). Coverslips were mounted on glass slides using Fluoromount-G (Electron Microscopy Sciences).
Some deviations from the protocol outlined above were used. (1) For the results presented in Fig. 1 E, HeLa cells expressing ectopic GST-GFP-cNLS were first fixed with 3.75% formaldehyde in PBS and then permeabilized with 0.2% Triton X-100 in PBS. (2) For the results presented in Fig. 5, 1% paraformaldehyde in PBS was used in place of formaldehyde, and 2% BSA was used in place of skim milk powder. (3) To further investigate the localization of Lap2 after Nup155 depletion (Fig. S2), 0.005% Digitonin (EMD) was used in place of Triton X-100, and PBS was used in place of PBS-T (Joseph et al., 2002).
Micrographs were obtained using a laser scanning system (LSM510; Carl Zeiss, Inc.) coupled with a microscope (Axiovert 100M; Carl Zeiss, Inc.) and 63x Plan-Apochromat objective (Carl Zeiss, Inc.). Images were acquired using LSM 5 software (Carl Zeiss, Inc.) and processed using ImageJ 1.42q software (National Institutes of Health, Bethesda, MD; Abramoff et al., 2004).
For visualization of the GST-GFP-cNLS reporter protein after Nup155 depletion, images were obtained using a cell observer (Observer.Z1; Carl Zeiss, Inc.) coupled with a camera (AxioCam MRm; Carl Zeiss, Inc.). Images were acquired as a series of Z-stacks (∼15 sections), and were deconvolved using Axiovision software (Carl Zeiss, Inc.). Subsequent image processing was performed using ImageJ software. Z-series are displayed as average intensity Z-projections.
Transmission electron microscopy
HeLa cells analyzed by transmission electron microscopy were released from culture dishes using 0.05% trypsin, pelleted, and then washed with PBS. Cell pellets were fixed with 2.5% gluteraldehyde, 0.1 M cacodylate, pH 7.4, for 24 h. Pellets were washed repeatedly with 0.1 M cacodylate, followed by staining with 1% OsO4 in 0.1 M cacodylate for 1 h. Pellets were rinsed with distilled water and dehydrated though progressive ethanol washes of increasing concentration (10 min each; 60%, 80%, 95%, and 100% ethanol). Pellets were further treated with 100% propylene oxide (3 × 10-min washes), incubated in propylene oxide containing epon (1:1), and finally embedded in 100% epon at 65°C for 24 h. Once cured, blocks were sectioned. These resulting sections were examined using a transmission electron microscope (model 410; Philips/FEI Corporation) and images were acquired using a CCD camera (Megaview III; Soft Imaging System/Olympus) and AnalySIS software (Soft Imaging System). Images were subsequently processed using ImageJ. The number of NPCs per micrometer of distance along the NE was manually counted and data was presented using Excel. Data from three independent experiments were statistically analyzed using InStat (GraphPad Software, Inc.).
Mass spectrometry
Proteins detected by Coomassie blue staining in pull-down experiments using GST-Pom121364–458 or GST-Pom152p1–111 were excised from SDS-polyacrylamide gels and subjected to in-gel trypsin digestion followed by LC-MS/MS using a mass spectrometer (Q-TOF Premier; Waters Corp.). Protein identification was performed by peptide mass fingerprinting using PEAKS mass spectrometry software (Bioinformatics Solutions, Inc.).
Online supplemental material
Fig. S1 shows that multiple siRNAs targeting distinct regions of Nup155 mRNA efficiently deplete Nup155 and similarly alter nuclear morphology. Fig. S2 shows that depletion of Nup155 affects the targeting of Lap2 to the INM leading to its relocalization to cytoplasmic membranes. Fig. S3 shows the results of mass spectrometry analysis of Pom121215–557–interacting proteins. Fig. S4 shows the binding of yeast Pom152p1–111 to Nup155 and the Nup107–160 complex, and the binding of Pom121215–557 to yeast Nup170p. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007098/DC1.
Supplementary Material
Acknowledgments
We would like to thank Paul LaPointe and members of the Wozniak Laboratory for helpful discussions, Honey Chan for assistance in EM experiments, and Günter Blobel for his generous assistance during revisions.
Funds for this work were provided to R.W. Wozniak by the Canadian Institutes of Health Research, the Alberta Innovation Health Solutions, and the Howard Hughes Medical Institute; and to U. Kutay by the Swiss National Science Foundation.
Footnotes
Abbreviations used in this paper:
- CPC
- coat protein complex
- FG
- phenylalanine-glycine
- INM
- inner nuclear membrane
- LBR
- lamin B receptor
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- nup
- nucleoporin
- ONM
- outer nuclear membrane
References
- Aaronson R.P., Blobel G.. 1974. On the attachment of the nuclear pore complex. J. Cell Biol. 62:746–754. 10.1083/jcb.62.3.746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff M.D., Magelhaes P.J., Ram S.J.. 2004. Image processing with ImageJ. Biophotonics International. 11:36–42. [Google Scholar]
- Akey C.W. 1989. Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy. J. Cell Biol. 109:955–970. 10.1083/jcb.109.3.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey C.W., Radermacher M.. 1993. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122:1–19. 10.1083/jcb.122.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007a. Determining the architectures of macromolecular assemblies. Nature. 450:683–694. 10.1038/nature06404 [DOI] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007b. The molecular architecture of the nuclear pore complex. Nature. 450:695–701. 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Antonin W., Franz C., Haselmann U., Antony C., Mattaj I.W.. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell. 17:83–92. 10.1016/j.molcel.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Bailer S.M., Balduf C., Hurt E.. 2001. The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport. Mol. Cell. Biol. 21:7944–7955. 10.1128/MCB.21.23.7944-7955.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R., Lin A., Enarson M., Burke B.. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141–1156. 10.1083/jcb.134.5.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N., Rabut G., Baï S.W., van Overbeek M., Beaudouin J., Daigle N., Zatsepina O.V., Pasteau F., Labas V., Fromont-Racine M., et al. 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147–1160. 10.1083/jcb.200101081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke I.C., Boehmer T., Blobel G., Schwartz T.U.. 2004. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J. Cell Biol. 167:591–597. 10.1083/jcb.200408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Potter V.R.. 1966. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 154:1662–1665. 10.1126/science.154.3757.1662 [DOI] [PubMed] [Google Scholar]
- Brohawn S.G., Schwartz T.U.. 2009. A lattice model of the nuclear pore complex. Commun Integr Biol. 2:205–207. 10.4161/cib.2.3.7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., Leksa N.C., Spear E.D., Rajashankar K.R., Schwartz T.U.. 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 322:1369–1373. 10.1126/science.1165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia B., Courvalin J.C.. 1997. Domain-specific disassembly and reassembly of nuclear membranes during mitosis. Exp. Cell Res. 230:133–144. 10.1006/excr.1996.3395 [DOI] [PubMed] [Google Scholar]
- Chaudhary N., Courvalin J.C.. 1993. Stepwise reassembly of the nuclear envelope at the end of mitosis. J. Cell Biol. 122:295–306. 10.1083/jcb.122.2.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler E.W., Ma Y., Seo H.S., Hsia K.C., Noriega T.R., Blobel G., Hoelz A.. 2008. A fence-like coat for the nuclear pore membrane. Mol. Cell. 32:815–826. 10.1016/j.molcel.2008.12.001 [DOI] [PubMed] [Google Scholar]
- DeGrasse J.A., DuBois K.N., Devos D., Siegel T.N., Sali A., Field M.C., Rout M.P., Chait B.T.. 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics. 8:2119–2130. 10.1074/mcp.M900038-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B.T., Sali A., Rout M.P.. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2:e380. 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B.T., Rout M.P., Sali A.. 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 103:2172–2177. 10.1073/pnas.0506345103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V., Hurt E.C.. 1995. Genetic approaches to nuclear pore structure and function. Trends Genet. 11:235–241. 10.1016/S0168-9525(00)89057-5 [DOI] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., Antonny B.. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146. 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J.. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857–865. 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N., Blobel G.. 1976. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J. Cell Biol. 70:581–591. 10.1083/jcb.70.3.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann J.A., Wagner E.J., Dong J., Zhang Y., Kutay U., Marzluff W.F.. 2005. Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol. Biol. Cell. 16:2960–2971. 10.1091/mbc.E04-11-1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr C., Akin D., Moore M.S.. 1998. The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J. Cell Sci. 111:1889–1896. [DOI] [PubMed] [Google Scholar]
- Feldherr C., Akin D., Littlewood T., Stewart M.. 2002. The molecular mechanism of translocation through the nuclear pore complex is highly conserved. J. Cell Sci. 115:2997–3005. [DOI] [PubMed] [Google Scholar]
- Flemming D., Sarges P., Stelter P., Hellwig A., Böttcher B., Hurt E.. 2009. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J. Cell Biol. 185:387–395. 10.1083/jcb.200810016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C., Askjaer P., Antonin W., Iglesias C.L., Haselmann U., Schelder M., de Marco A., Wilm M., Antony C., Mattaj I.W.. 2005. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 24:3519–3531. 10.1038/sj.emboj.7600825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., Antonin W.. 2007. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 8:165–172. 10.1038/sj.embor.7400889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T., Maeshima K., Yahata K., Sugano S., Imamoto F., Imamoto N.. 2007. Two distinct human POM121 genes: requirement for the formation of nuclear pore complexes. FEBS Lett. 581:4910–4916. 10.1016/j.febslet.2007.09.021 [DOI] [PubMed] [Google Scholar]
- Gerace L., Ottaviano Y., Kondor-Koch C.. 1982. Identification of a major polypeptide of the nuclear pore complex. J. Cell Biol. 95:826–837. 10.1083/jcb.95.3.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie P.J., Khoudoli G.A., Stewart G., Swedlow J.R., Blow J.J.. 2007. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 17:1657–1662. 10.1016/j.cub.2007.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E.C.. 1993. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 12:3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Emig S., Weise C., Hucho F., Pohl T., Hurt E.C.. 1995. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J. Cell Biol. 130:1263–1273. 10.1083/jcb.130.6.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Dang T., Pané N., Shevchenko A., Mann M., Forbes D., Hurt E.. 1997. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell. 8:2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber U.F., Senior A., Gerace L.. 1990. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 9:1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan C., Koulov A.V., Balch W.E.. 2007. An evolutionary perspective on eukaryotic membrane trafficking. Adv. Exp. Med. Biol. 607:73–83. 10.1007/978-0-387-74021-8_6 [DOI] [PubMed] [Google Scholar]
- Hallberg E., Wozniak R.W., Blobel G.. 1993. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J. Cell Biol. 122:513–521. 10.1083/jcb.122.3.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F., Mazzeo D., Patel J.T., Smallwood D.T., Ellis J.A., Shanahan C.M., Shackleton S.. 2010. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 285:3487–3498. 10.1074/jbc.M109.071910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D.J.. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 11:853–864. 10.1016/S1097-2765(03)00116-3 [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara L.A., Shibuya E.K., Wozniak R.W.. 2005. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol. Biol. Cell. 16:2382–2394. 10.1091/mbc.E04-10-0857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara L.A., Platani M., Santarella R., Wozniak R.W., Mattaj I.W.. 2008. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol. Biol. Cell. 19:1753–1762. 10.1091/mbc.E07-08-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M.W., Wente S.R.. 2009. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell. 17:606–616. 10.1016/j.devcel.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia K.C., Stavropoulos P., Blobel G., Hoelz A.. 2007. Architecture of a coat for the nuclear pore membrane. Cell. 131:1313–1326. 10.1016/j.cell.2007.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A.M., Cooley L.. 2008. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell. Biochem. 48:6–19. 10.1007/978-0-387-09595-0_2 [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S.H., Karpova T.S., McNally J.G., Dasso M.. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595–602. 10.1083/jcb.200110109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M., Blobel G.. 2009. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol. 16:782–788. 10.1038/nsmb.1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Lusk C.P., Blobel G.. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007. 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- Lau C.K., Delmar V.A., Forbes D.J.. 2006. Topology of yeast Ndc1p: predictions for the human NDC1/NET3 homologue. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288:681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R.Y., Aebi U., Fahrenkrog B.. 2008. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 129:105–116. 10.1007/s00418-007-0371-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loïodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J.B., Doye V.. 2004. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 15:3333–3344. 10.1091/mbc.E03-12-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E.. 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21:387–397. 10.1093/emboj/21.3.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Stangl K., Stelter P., Tóth K.F., Böttcher B., Hurt E.. 2005. Reconstitution of Nup157 and Nup145N into the Nup84 complex. J. Biol. Chem. 280:18442–18451. 10.1074/jbc.M412787200 [DOI] [PubMed] [Google Scholar]
- Makhnevych T., Lusk C.P., Anderson A.M., Aitchison J.D., Wozniak R.W.. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 115:813–823. 10.1016/S0092-8674(03)00986-3 [DOI] [PubMed] [Google Scholar]
- Makio T., Stanton L.H., Lin C.C., Goldfarb D.S., Weis K., Wozniak R.W.. 2009. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 185:459–473. 10.1083/jcb.200810029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J., Güttinger S., Hawryluk-Gara L.A., Panté N., Mall M., Galy V., Haselmann U., Mühlhäusser P., Wozniak R.W., Mattaj I.W., et al. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 22:93–103. 10.1016/j.molcel.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Marelli M., Lusk C.P., Chan H., Aitchison J.D., Wozniak R.W.. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. 10.1083/jcb.153.4.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E., Stanton L.H., Madrid A.S., Kieselbach T., Weis K.. 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 185:475–491. 10.1083/jcb.200810030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil T.J., Schmid S.L.. 2009. Conserved functions of membrane active GTPases in coated vesicle formation. Science. 325:1217–1220. 10.1126/science.1171004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J.. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Radu A., Blobel G., Wozniak R.W.. 1993. Nup155 is a novel nuclear pore complex protein that contains neither repetitive sequence motifs nor reacts with WGA. J. Cell Biol. 121:1–9. 10.1083/jcb.121.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Orjalo A.V., Shen Z., Briggs S., Forbes D.J.. 2006. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 103:17801–17806. 10.1073/pnas.0608484103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Ramos C., Harel A., Forbes D.J.. 2008. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell. 19:3982–3996. 10.1091/mbc.E08-01-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E.C., Gerace L.. 2005. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 30:551–558. 10.1016/j.tibs.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Schwartz T.U. 2005. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 15:221–226. 10.1016/j.sbi.2005.03.003 [DOI] [PubMed] [Google Scholar]
- Seo H.S., Ma Y., Debler E.W., Wacker D., Kutik S., Blobel G., Hoelz A.. 2009. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc. Natl. Acad. Sci. USA. 106:14281–14286. 10.1073/pnas.0907453106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Lutzmann M., Santos-Rosa H., Leonard K., Mueller S., Aebi U., Hurt E.. 2000. Structure and assembly of the Nup84p complex. J. Cell Biol. 149:41–54. 10.1083/jcb.149.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderqvist H., Hallberg E.. 1994. The large C-terminal region of the integral pore membrane protein, POM121, is facing the nuclear pore complex. Eur. J. Cell Biol. 64:186–191. [PubMed] [Google Scholar]
- Stagg S.M., LaPointe P., Balch W.E.. 2007. Structural design of cage and coat scaffolds that direct membrane traffic. Curr. Opin. Struct. Biol. 17:221–228. 10.1016/j.sbi.2007.03.010 [DOI] [PubMed] [Google Scholar]
- Stavru F., Hülsmann B.B., Spang A., Hartmann E., Cordes V.C., Görlich D.. 2006. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173:509–519. 10.1083/jcb.200601001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M., Wente S.R.. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 4:775–789. 10.1016/S1534-5807(03)00162-X [DOI] [PubMed] [Google Scholar]
- Tcheperegine S.E., Marelli M., Wozniak R.W.. 1999. Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem. 274:5252–5258. 10.1074/jbc.274.8.5252 [DOI] [PubMed] [Google Scholar]
- Theerthagiri G., Eisenhardt N., Schwarz H., Antonin W.. 2010. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J. Cell Biol. 189:1129–1142. 10.1083/jcb.200912045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.J., Wente S.R.. 2006. Dynamic nuclear pore complexes: life on the edge. Cell. 125:1041–1053. 10.1016/j.cell.2006.05.027 [DOI] [PubMed] [Google Scholar]
- Ulbert S., Antonin W., Platani M., Mattaj I.W.. 2006. The inner nuclear membrane protein Lem2 is critical for normal nuclear envelope morphology. FEBS Lett. 580:6435–6441. 10.1016/j.febslet.2006.10.060 [DOI] [PubMed] [Google Scholar]
- Unwin P.N., Milligan R.A.. 1982. A large particle associated with the perimeter of the nuclear pore complex. J. Cell Biol. 93:63–75. 10.1083/jcb.93.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W.H., Forbes D.J.. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339–354. 10.1083/jcb.200108007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T., et al. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 113:195–206. 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- Wozniak R.W., Bartnik E., Blobel G.. 1989. Primary structure analysis of an integral membrane glycoprotein of the nuclear pore. J. Cell Biol. 108:2083–2092. 10.1083/jcb.108.6.2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.