Localization and activation of Elm1 at the bud neck coordinates SPC activity with mother–daughter polarity during cell division.
Abstract
How dividing cells monitor the effective transmission of genomes during mitosis is poorly understood. Budding yeast use a signaling pathway known as the spindle position checkpoint (SPC) to ensure the arrival of one end of the mitotic spindle in the nascent daughter cell. An important question is how SPC activity is coordinated with mother–daughter polarity. We sought to identify factors at the bud neck, the junction between mother and bud, which contribute to checkpoint signaling. In this paper, we show that the protein kinase Elm1 is an obligate regulator of the SPC, and this function requires localization of Elm1 to the bud neck. Furthermore, we show that Elm1 promotes the activity of the checkpoint kinase Kin4. These findings reveal a novel function for Elm1 in the SPC and suggest how checkpoint activity may be linked to cellular organization.
Introduction
The budding yeast Saccharomyces cerevisiae is a compelling model for the study of cell division because of its intrinsic geometric constraints: the daughter cell is formed by polarized growth at a specified site on the cortex of the mother, and the junction between the bud and mother, termed the bud neck, is the eventual site of cytokinesis. The success of mitosis, therefore, depends on positioning the mitotic spindle through the bud neck so that genomes lie on either side. Spindle positioning is accomplished by cytoplasmic microtubules that project outward from the spindle pole bodies (SPBs; the yeast centrosome equivalent) and interact with molecular motors at the cell cortex (Moore and Cooper, 2010). These interactions orient the spindle along the bud–mother axis and pull one end of the spindle through the neck with force provided by the microtubule motor dynein and its activator dynactin.
When the mechanisms that position the spindle are impaired, a cell may enter mitosis without moving the spindle into the bud neck. Under these conditions, however, the cell will remain in anaphase until the alignment of the spindle is corrected and one SPB moves through the bud neck. This delay is caused by a cell cycle checkpoint known as the spindle position checkpoint (SPC), which inhibits the mitotic exit network (MEN), thereby preventing the deactivation of cyclin-dependent kinase (Bardin et al., 2000; Bloecher et al., 2000; Daum et al., 2000; Pereira et al., 2000).
The activity of the SPC must be coordinated with mother–daughter polarity to prevent mitotic exit when the entire spindle is within the mother compartment and to permit mitotic exit once one SPB moves through the bud neck. This coordination depends on a Ras-like GTPase, Tem1, which localizes to the SPBs and activates the MEN (Bardin et al., 2000; Molk et al., 2004). Tem1 is negatively regulated by a bipartite putative GTPase-activating protein complex, Bub2–Bfa1, which also localizes to the SPBs (Pereira et al., 2000; Geymonat et al., 2002; Ro et al., 2002). The association of Bub2–Bfa1 with the SPBs is critical for SPC function, and it is regulated by another SPC component, the protein kinase Kin4 (Maekawa et al., 2007; Caydasi and Pereira, 2009). Tem1 is positively regulated by the putative guanine nucleotide exchange factor, Lte1, which localizes to the bud (Shirayama et al., 1994). Lte1 polarity is critical for the integrity of the SPC; mutations that allow Lte1 to access the mother compartment also disrupt the SPC (Bardin et al., 2000; Pereira et al., 2000; Castillon et al., 2003). Although genetic data indicate that Lte1 activates Tem1, the precise function of Lte1 has not been characterized. Importantly, Lte1 has not been shown to provide exchange activity toward Tem1 (Geymonat et al., 2009). Together, these results support a model in which Tem1 activity depends on the location of the SPBs; only when an SPB enters the bud does Tem1 encounter its activator and mitotic exit commence.
Although this model is consistent with the observed correlation between spindle position and the timing of mitotic exit, it does not explain similar correlations observed in mutants that lack Lte1. LTE1 is necessary for mitotic exit at low temperatures (<18°C), but lte1Δ-null mutant cells are viable at higher temperatures and also undergo mitotic exit after one SPB moves through the bud neck with kinetics similar to wild-type cells (Shirayama et al., 1994; Adames et al., 2001). SPC activity must therefore be coordinated with mother–daughter polarity via mechanisms that do not involve Lte1.
Several lines of evidence indicate that interactions between cytoplasmic microtubules and the bud neck are essential for this coordination. First, disrupting cytoplasmic microtubule interactions with the bud neck by mutations or laser cutting destabilizes the SPC (Adames et al., 2001; Moore et al., 2009). Second, the SPC depends on the network of septin filaments at the bud neck (Castillon et al., 2003). Third, interactions between cytoplasmic microtubules and unidentified factors associated with the bud neck and/or bud cortex regulate the dynamics of Bub2–Bfa1 association with the SPBs (Pereira et al., 2001; Fraschini et al., 2006; Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009). These results indicate that signaling factors at the bud neck promote SPC activity while the spindle is within the mother compartment.
To elucidate this signaling pathway, we screened mutants of bud neck–localized proteins for a loss-of-SPC phenotype. We find that the protein kinase Elm1 is necessary for the SPC and that this function requires its kinase activity and localization to the bud neck. Furthermore, our results indicate that the function of Elm1 in the SPC is independent of previously identified roles. Elm1 is closely related to the kinases Sak1 and Tos3, and we show that sak1 and tos3 mutants also exhibit disruption-of-SPC phenotypes, albeit with decreased penetrance. Finally, we provide evidence that the role of Elm1 in the SPC is to activate the Kin4 kinase. These findings uncover a novel step in the SPC and suggest how a polarized regulator may influence asymmetric SPC activity.
Results
ELM1 prevents spindle disassembly in the mother compartment
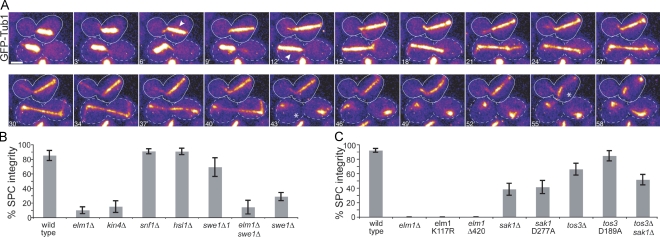
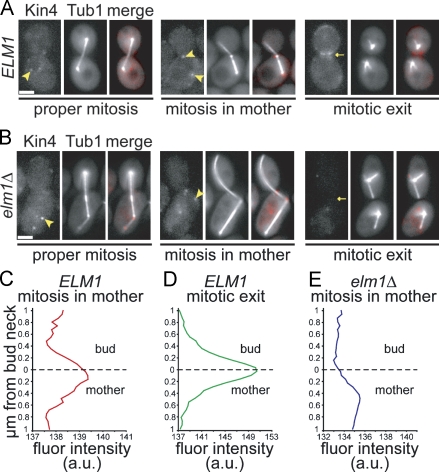
To identify factors at the bud neck that contribute to the SPC, we screened deletion mutants of neck-localized proteins for a loss-of-SPC phenotype. We combined these mutations with null mutations in dynein–dynactin to disrupt spindle positioning, and we recorded time-lapse videos of GFP-labeled microtubules to monitor spindle morphology as an indicator of mitotic exit. We found that elm1Δ mutant cells did not prolong mitosis when the spindle failed to move through the bud neck; instead, these spindles disassembled within the mother, resulting in binucleate mothers and anucleate daughters (Fig. 1 A). This phenotype is the defining characteristic of SPC loss-of-function mutants, such as kin4Δ (Fig. 1 B; Bloecher et al., 2000; Pereira et al., 2000; Castillon et al., 2003; D'Aquino et al., 2005; Pereira and Schiebel, 2005; Nelson and Cooper, 2007).
Figure 1.
A novel role for ELM1 in preventing spindle disassembly in the mother. (A) Time-lapse images of microtubules labeled with GFP-Tub1 in elm1Δ dyn1Δ double mutant cells. The cell outlined by the dashed lines undergoes a properly oriented anaphase (arrowhead), moving one end of the elongating spindle into the bud before spindle disassembly (asterisk). The cell outlined by the solid line undergoes a misoriented anaphase (arrowhead) and subsequently disassembles in the mother (asterisk). Each image is a projection of seven planes separated by 0.7 µm and was acquired on a confocal microscope. Bar, 2 µm. Strain: yJC4168. (B) The SPC phenotype is not caused by dysregulation of known Elm1-dependent pathways. Each indicated mutant was constructed in an arp1Δ mutant background to increase the frequency of misoriented mitoses and scored for SPC integrity in time-lapse video assays (see Materials and methods). “% SPC integrity” denotes the percentage of the cells exhibiting a misoriented mitosis that remained in mitosis while the spindle was within the mother. Strains: wild type, yJC3464, n = 27; elm1Δ, yJC2480, n = 40; kin4Δ, yJC7254, n = 20; snf1Δ, yJC3943 and 3944, n = 66; hsl1Δ, yJC2477, n = 43; swe1Δ1, yJC3464 with plasmid pBJ1492, n = 13; elm1Δ swe1Δ, yJC2738, n = 14; and swe1Δ, yJC3807, 3809, and 3810, n = 66. (C) EFKs promote the integrity of the SPC. Mutations that disrupt the function of EFKs were combined with a dyn1Δ mutation to increase the frequency of misoriented mitoses, and SPC integrity was assessed in time-lapse video assays. At least 13 cells were scored for each strain. Strains: wild type, yJC3871, n = 30; elm1Δ, yJC4168 and 4170, n = 20; elm1-K117R, yJC5363 with plasmid pBJ1606, n = 13; elm1Δ420, yJC6854, n = 15; sak1Δ, yJC4171, n = 34; sak1-D277A, yJC4171 with plasmid pBJ1691, n = 29; tos3Δ, yJC4174 and 4176, n = 43; tos3-D189A, yJC4174 with plasmid pBJ1689, n = 26; and tos3Δsak1Δ, yJC4484, n = 35. Error bars are the standard error of proportion.
Elm1 is a multifunctional kinase with roles in several signaling pathways. To determine whether the SPC phenotype of elm1Δ mutants could be attributed to defects in these pathways, we examined strains bearing mutations in known Elm1 substrates combined with the arp1Δ mutation, which abolishes dynein–dynactin function. One substrate of Elm1 is Snf1, the yeast AMP-activated protein kinase (AMPK; Hong et al., 2003; Sutherland et al., 2003). We analyzed snf1Δ mutants in our SPC integrity assay and found no detectable loss of SPC integrity (Fig. 1 B); thus, the function of Elm1 in the SPC does not involve AMPK/Snf1.
Elm1 also functions in a signaling cascade that controls morphogenesis by coupling bud growth with the G2/M transition. In this pathway, Elm1 phosphorylates Hsl1, promoting its interaction with Hsl7 (Szkotnicki et al., 2008). The Hsl1–Hsl7 complex recruits the Wee1-family kinase Swe1 to the bud neck (Longtine et al., 2000), where it is deactivated via phosphorylation by Cdc5/polo kinase (Sakchaisri et al., 2004). We hypothesized that the disruption of the SPC in elm1Δ mutants could be attributed to increased Swe1 activity. We tested this by first examining whether loss of Hsl1 disrupted the SPC similar to elm1 mutants. To the contrary, hsl1Δ mutants exhibited no detectable defect in SPC integrity (Fig. 1 B). Next, we examined SPC integrity in the presence of a stabilized mutant of Swe1, swe1Δ1 (McMillan et al., 2002). This mutant showed a slight disruption of the SPC but was not significantly different from wild-type cells (P = 0.4; Fig. 1 B). To test whether Swe1 was necessary for SPC failure in elm1Δ mutants, we combined the swe1Δ- and elm1Δ-null mutations. These cells exhibited checkpoint failure at a rate similar to elm1Δ mutants (P = 0.64; Fig. 1 B). Furthermore, the swe1Δ mutant alone exhibited impaired SPC integrity in the presence of wild-type ELM1, but this phenotype was less penetrant than elm1Δ (P = 0.03; Fig. 1 B). We conclude that the loss-of-SPC phenotype in elm1Δ mutants cannot be attributed to enhanced Swe1 activity.
Elm1-family kinases (EFKs) exhibit varying effects on SPC integrity
Elm1, Sak1, and Tos3 comprise the EFKs, which exhibit overlapping roles in the phosphoregulation of AMPK/Snf1 along with sequence similarity within the kinase domains (Hunter and Plowman, 1997; Hong et al., 2003; Sutherland et al., 2003; Rubenstein et al., 2006). We found that null mutants of SAK1 and TOS3 exhibit intermediate levels of SPC impairment (Fig. 1 C), indicating that each of the EFKs is important for the integrity of the SPC.
To determine whether the kinase activities of the EFKs are important for their function in the SPC, we introduced point mutations in ELM1, SAK1, and TOS3 designed to abrogate kinase activity (Blacketer et al., 1993; Nath et al., 2003). In each case, kinase-defective alleles exhibited defects in SPC integrity that were identical to the deletion mutants (Fig. 1 C). Thus, kinase activity appears to be necessary for the function of the EFKs in the SPC.
Next, we performed epistasis experiments to determine whether the EFKs exert redundant functions for the SPC. First, we generated sak1Δ tos3Δ double mutants in haploid cells; these exhibited a level of SPC integrity similar to either mutant alone (P = 0.34 compared with sak1Δ; P = 0.25 compared with tos3Δ; Fig. 1 C), suggesting that Sak1 and Tos3 are not functionally redundant and may act in the same pathway. The complete loss of SPC integrity in elm1Δ haploid mutants prevents the analysis of additive effects when combined with sak1Δ and tos3Δ. Furthermore, diploid strains heterozygous for elm1Δ did not exhibit defects in SPC integrity and did not confer an additive defect when combined with the homozygous null alleles of sak1 and tos3 (unpublished data). Thus, we cannot determine whether the function of Sak1 or Tos3 overlaps with Elm1. Nevertheless, our results support a prominent and necessary role for Elm1 in the SPC; in contrast, Sak1 and Tos3 serve minor roles and may act in a common pathway.
Structure function and localization analysis
Given the similarity among the EFKs, we asked what features of the Elm1 protein might confer its pronounced role in the SPC. The kinase domains of Elm1, Sak1, and Tos3 show sequence similarity (Elm1: 51% similarity to Sak1 and 53% similarity to Tos3), but the regions carboxy terminal to the kinase domains are highly divergent. A previous study demonstrated that the carboxy-terminal region of Elm1 was not necessary to regulate AMPK/Snf1; however, this region was essential for the unique function of Elm1 in regulating morphogenesis (Rubenstein et al., 2006). To test whether this carboxy-terminal region was required for the SPC, we generated a truncated allele similar to the one described by Rubenstein et al. (2006). The allele was expressed from the endogenous locus. This mutant, elm1Δ420, completely lost SPC integrity (Fig. 1 C). Therefore, the carboxy-terminal region of Elm1 confers a function that is necessary for the SPC.
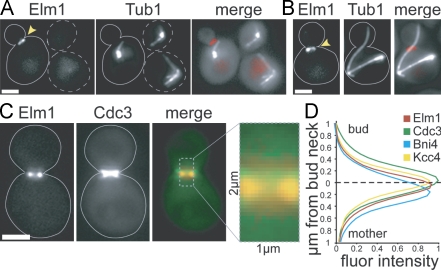
We considered that the localization of EFKs within the cell may be important for the SPC. Consistent with previous findings, we observed fluorescently tagged Elm1 at the bud neck in dividing cells (Fig. 2 A; Bouquin et al., 2000). Elm1 was also found at the neck in mitotic cells with aberrantly positioned spindles, indicating that Elm1 is enriched at the neck when the SPC is active (Fig. 2 B). In contrast, neither Sak1 nor Tos3 exhibited enrichment at the neck (Fig. S1). These results suggest that the presence of Elm1 at the bud neck may be important for its function in the SPC.
Figure 2.
Localization of Elm1. Images were collected using a wide-field microscope. Arrowheads point to Elm1-tdimer2 at the bud neck. Dashed lines indicate unbudded cells, and solid lines indicate budded cells. (A) Elm1 localizes to the bud neck. Tandem RFP/tdimer2 was fused to the carboxy terminus of Elm1 by integration at the endogenous ELM1 locus in arp1Δ mutant cells expressing GFP-Tub1. Strain: yJC6852. (B) Elm1 localizes to the neck in cells containing misoriented anaphase spindles. Strain: yJC6852. (C) Localization of Elm1 with respect to the septin network in cells expressing Elm1-tdimer2 and Cdc3-GFP. Merge image depicts Elm1 (red) and Cdc3 (green). Strain: yJC6848 with plasmid pBJ1488. (D) Quantification of normalized fluorescence intensities for Elm1, Cdc3, Bni4, and Kcc4 across the bud neck. Images were collected from cells simultaneously expressing Elm1-tdimer2 and either Cdc3-GFP, Bni4-GFP (yJC7251), or Kcc4-GFP (yJC7252). Fluorescence intensities were measured across a 2-µm region, depicted in C, extending from the mother into the bud and centered on the smallest diameter of the neck (dashed line). Intensities were measured in at least 10 large-budded cells. Values for Elm1 are compiled from the three strains, totaling 50 cells. Fluor intensity values are given in arbitrary units. Bars, 2 µm.
The bud neck is surrounded by a network of septin filaments, which provide a scaffold for signaling proteins and may act as a diffusion barrier for components of the mother and bud cytosol (Barral et al., 2000; Takizawa et al., 2000). We examined the distribution of Elm1 at the neck closely to determine its location relative to the septin network. Consistent with previous results, Elm1 colocalized with the septin network, labeled by Cdc3, during bud growth, but Elm1 was absent once the septins split into two rings, which occurs during cytokinesis (Kim et al., 1991; Bouquin et al., 2000; Lippincott et al., 2001). In large-budded cells with an hourglass-shaped septin network, Elm1 occupies a narrow band within the septin ring and proximal to the mother side of the neck (Fig. 2 C). Quantitative comparison of the distributions of Elm1 and Cdc3 signal intensity revealed that Elm1 was enriched toward the mother relative to the distribution of Cdc3 (Fig. 2 D). We similarly compared Elm1 with Bni4 and Kcc4, which interact with the septin network and have been reported to localize to the mother and bud sides of the neck, respectively (Kozubowski et al., 2005). However, in our experiments, the mean distribution of Kcc4 signal peaked near the smallest diameter of the neck and was not clearly enriched in the bud of large-budded cells (Fig. 2 D). Here, Elm1 was enriched between the peaks of Bni4 and Kcc4 (Fig. 2 D). These data demonstrate that Elm1 is present on the mother side of the bud neck, where it may have access to components of the mother cytoplasm.
Placing Elm1 in the SPC pathway
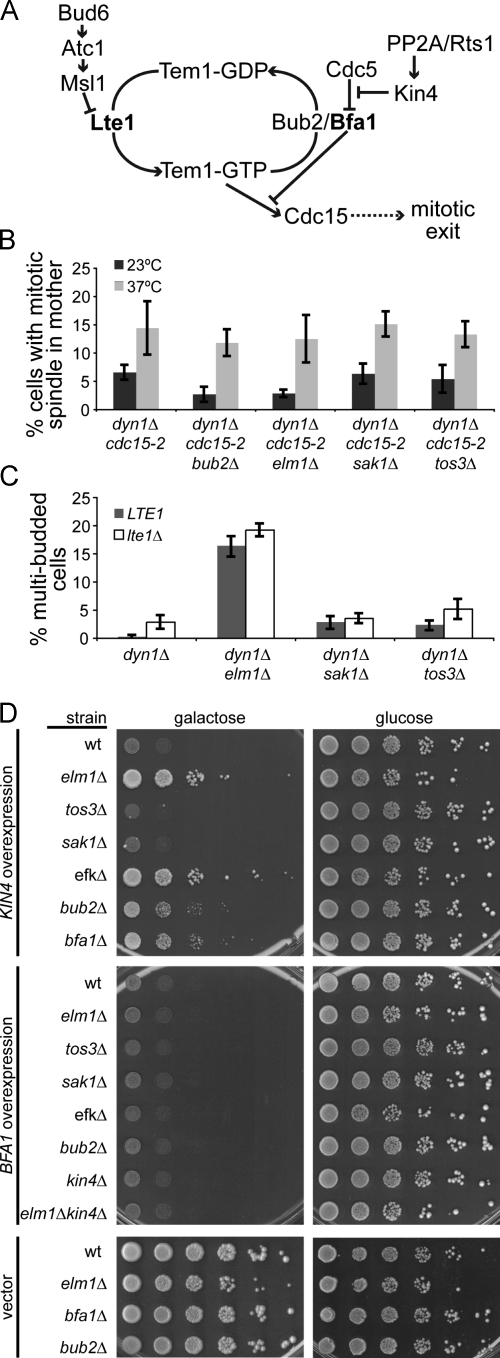
The SPC prolongs mitosis by inhibiting the activity of the MEN; therefore, we reasoned that Elm1 could either be important for inhibition of the MEN or for a later event within the MEN (Fig. 3 A). To determine whether Elm1 functions upstream of the MEN, we examined elm1Δ mutants under conditions in which the initiation of MEN signaling is abrogated. The temperature-sensitive cdc15-2 mutant disables the Cdc15 kinase, thereby blocking an early step in the MEN pathway (Hartwell et al., 1973; Mah et al., 2001), and we tested whether this block increases the frequency of anaphase spindles in the mother-compartment of elm1Δ mutant cells. When Cdc15 function was disrupted by shifting to the restrictive temperature (37°C), elm1Δ cdc15-2 dyn1Δ cells accumulated in anaphase, and the percentage of cells with intact mitotic spindles within the mother compartment increased to a level similar to that of cdc15-2 dyn1Δ cells expressing wild-type ELM1 (P = 0.66; Fig. 3 B). Loss-of-function mutations in bub2Δ, which are known to abolish the SPC, yielded results similar to the elm1Δ mutant (Fig. 3 B). We also tested the sak1Δ and tos3Δ mutants in this assay; however, our results were inconclusive because the decreased penetrance of these mutants failed to produce a phenotype distinguishable from the cdc15-2 dyn1Δ control cells (Fig. 3 B). We conclude that mitotic exit within the mother compartment of elm1Δ mutants requires the function of Cdc15, consistent with Elm1 acting upstream of the MEN as part of the SPC.
Figure 3.
Elm1 functions in the SPC. (A) Diagram of SPC regulation of mitotic exit. (B) Inappropriate mitotic exit in EFK mutants requires Cdc15 activity. Asynchronous cultures were grown to mid–log phase at 23°C, diluted 1:10 into new media, and incubated at either 23°C or 37°C for 3.5 h. SPC activity was measured by scoring the percentage of cells with intact mitotic spindles (>2 µm) within the mother compartment, based on GFP-labeled microtubules. Strains: dyn1Δ cdc15-2, yJC6380; bub2Δ dyn1Δ cdc15-2, yJC6497; elm1Δ dyn1Δ cdc15-2, yJC6926; sak1Δ dyn1Δ cdc15-2, yJC6493; and tos3Δ dyn1Δ cdc15-2, yJC7083. (C) Failure of the SPC (i.e., inappropriate mitotic exit) in EFK mutants does not require Lte1. Indicated strains expressing either wild-type LTE1 or the lte1Δ-null mutant were arrested at START by treatment with α factor and released into fresh media at 23°C. After 3 h, the percentage of cells exhibiting multiple buds, which is indicative of checkpoint failure, was determined. Strains: dyn1Δ, yJC4078; dyn1Δ lte1Δ, yJC7066; dyn1Δ elm1Δ, yJC7067; dyn1Δ elm1Δ lte1Δ, yJC7068; dyn1Δ sak1Δ, yJC7071; dyn1Δ sak1Δ lte1Δ, yJC7072; dyn1Δ tos3Δ, yJC7069; and dyn1Δ tos3Δ lte1Δ, yJC7070. (D) Elm1 is required for the growth inhibition caused by Kin4 overexpression. High-copy plasmids containing KIN4 or BFA1 under the control of a galactose-inducible promoter were transformed into the indicated strain background, and a 10-fold dilution series was spotted onto media selective for plasmid retention. Plates contained either galactose to induce expression or glucose to inhibit expression. Strains containing empty vector are shown as controls. Strains: wild type (wt), yJC2295; elm1Δ, yJC5254; tos3Δ, yJC6419; sak1Δ, yJC6492; elm1Δ sak1Δ tos3Δ, yJC6474; bub2Δ, yJC5251; bfa1Δ, yJC6447; kin4Δ, yJC6448; and kin4Δ elm1Δ, yJC6573. Plasmids: pGAL-KIN4, pBJ1651; pGAL-BFA1, pBJ1652; and vector, pBJ216. Values are the means of five counts of at least 50 cells. Error bars are the standard error of the means.
Two distinct biochemical pathways have been proposed to regulate the SPC: one attenuates the MEN activator Lte1 (Nelson and Cooper, 2007), and the other promotes the activity of the MEN inhibitor Bub2–Bfa1 (D'Aquino et al., 2005; Pereira and Schiebel, 2005; Maekawa et al., 2007; Chan and Amon, 2009). We performed a series of experiments to determine whether Elm1 functions upstream of Lte1 or Bub2–Bfa1. First, we asked whether Elm1 inhibits the activity of Lte1, in which case the SPC phenotype of elm1Δ mutants would be caused by hyperactive Lte1. We assayed elm1Δ lte1Δ double mutants for SPC integrity. We measured the frequency of multibudded cells, which are generated when cells exit mitosis with the spindle in the mother compartment and proceed to enter the next cell division without undergoing septation. Compared with the time-lapse video assay, this assay allows for rapid analysis of a greater number of cells. We found that elm1Δ lte1Δ double mutants generated multiple buds to the same extent observed in the elm1Δ single mutant (Fig. 3 C). Likewise, the defects of sak1Δ and tos3Δ mutants were not affected by the loss of Lte1. We also compared the localization of Lte1 in wild-type cells and elm1Δ mutants. In both cases, Lte1 was enriched in the bud, with no foci or accumulation of Lte1 seen in the mother compartment (Fig. S2). These data indicate that the role of the EFKs in the SPC is independent of Lte1.
Next, we asked whether Elm1 acts via regulation of Bub2–Bfa1. The SPC promotes the activity of Bub2–Bfa1 through the Kin4 kinase (Pereira and Schiebel, 2005; Maekawa et al., 2007). Whereas loss of Kin4 disables the SPC, overexpression of Kin4 constitutively inhibits mitotic exit, resulting in growth arrest (D'Aquino et al., 2005; Pereira and Schiebel, 2005). We examined whether Elm1 is necessary for the function of Kin4 using the growth arrest induced by Kin4 overexpression as a measure of activity. We found that elm1Δ mutants rescued the growth inhibition of Kin4 overexpression, similar to bub2Δ and bfa1Δ controls (Fig. 3 D). In contrast, sak1Δ and tos3Δ mutants were inhibited, similar to wild-type cells, and triple mutants lacking all three EFKs grew similar to the elm1Δ single mutant (Fig. 3 D).
Because the function of Kin4 is to activate Bub2–Bfa1, we next tested whether Elm1 was necessary for the function of Bfa1. Bfa1 overexpression, like Kin4 overexpression, inhibits mitotic exit and causes growth arrest (Li, 1999). Overexpression of Bfa1 inhibited the growth of elm1Δ mutants, similar to wild-type cells, indicating that Elm1 is not downstream of Bfa1 (Fig. 3 D). Furthermore, the Bfa1 overexpression phenotype was not suppressed by the loss of the two other EFKs, the loss of Bub2 or Kin4, or the simultaneous loss of Elm1 and Kin4 (Fig. 3 D). Together, these results suggest that Elm1 is necessary for the function of Kin4 and that Elm1 functions upstream of Bfa1 in the SPC.
If Elm1 acts in a common pathway with Kin4 and Bfa1, the overexpression of Elm1 might be expected to hyperactivate the SPC and delay mitotic exit. We tested this hypothesis using growth assays and cell cycle analysis under conditions in which ELM1 was ectopically expressed at high levels. Neither assay showed evidence of impaired mitotic exit; overexpression of ELM1 did not suppress growth, nor did it cause an accumulation of mitotic cells over time (Fig. S3). These results indicate that, unlike Kin4 and Bfa1, increased levels of Elm1 activity are not sufficient to inhibit mitotic exit.
Elm1 is not necessary for the spindle assembly checkpoint (SAC)
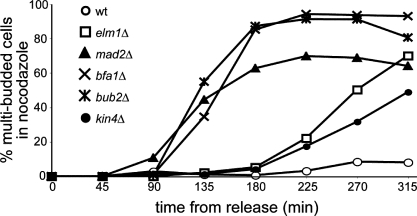
In addition to the SPC, Bub2 and Bfa1 function in cell cycle checkpoints that respond to spindle assembly errors and DNA damage (Hoyt et al., 1991; Li, 1999; Wang et al., 2000; Hu et al., 2001). We tested whether Elm1 might also have checkpoint functions beyond the SPC. Treatment with nocodazole destabilizes the yeast microtubule cytoskeleton and triggers cell cycle arrest by the SAC. Whereas wild-type cells delay cell cycle progression in the presence of nocodazole, SAC mutants (mad2Δ, bfa1Δ, and bub2Δ) complete the cell cycle and enter another round of division, forming an additional bud (Fig. 4). We found that elm1Δ mutants delay cell cycle progression in nocodazole (Fig. 4). Consistent with previous results, kin4Δ mutants also exhibit cell cycle delay in nocodazole (Fig. 4; D'Aquino et al., 2005; Pereira and Schiebel, 2005). This suggests that Elm1, like Kin4, is not necessary for the SAC. We did, however, observe an increase in the frequency of multibudded elm1Δ and kin4Δ cells after several hours in nocodazole. Although the basis for this increase is not clear, we have observed similar effects in other mutants that promote the activity of the MEN (unpublished data). We conclude that Elm1 functions primarily in the SPC.
Figure 4.
ELM1 is not necessary for the SAC. Cells were arrested in G1 by treatment with mating pheromone (α factor) and then released into media containing 15 µg/ml nocodazole. Samples were collected at 45-min intervals, and the percentages of cells exhibiting multiple buds were scored. Each data point represents the mean percentage of multibudded cells from three separate experiments. At least 200 cells were scored per time point in each experiment. Strains: wild type (wt), yJC2295; elm1Δ, yJC5254; mad2Δ, yJC6497; bfa1Δ, yJC6447; bub2Δ, yJC5251; and kin4Δ, yJC6448.
Does Elm1 influence the localization of Kin4?
Kin4 localizes to the mother cell cortex, SPBs, and bud neck, and its function in the SPC requires exchange between these sites (D'Aquino et al., 2005; Pereira and Schiebel, 2005; Maekawa et al., 2007; Chan and Amon, 2009). To determine whether Elm1 regulates the localization of Kin4, we visualized Kin4 in living cells by generating a functional fusion of dimeric RFP (tdimer2) to the carboxy terminus of the endogenous gene. Although we did not observe a distinct enrichment of the Kin4-tdimer2 signal on the mother cortex in our experiments, we did detect Kin4 at the SPB in the mother compartment during mitosis (Fig. 5 A). When the mitotic spindle was entirely within the mother, Kin4 prominently localized to both SPBs, consistent with previous studies (Fig. 5 A; Pereira and Schiebel, 2005; Maekawa et al., 2007). In elm1Δ mutants, Kin4 was present at the SPB in the mother compartment during mitosis (Fig. 5 B). We conclude that Elm1 is not necessary for the localization of Kin4 to the SPBs.
Figure 5.
Kin4 localization in elm1Δ mutants. Images were collected using a wide-field microscope. (A) Kin4 localization in wild-type cells. Tandem RFP/tdimer2 was fused to the carboxy terminus of Kin4 by integration at the endogenous KIN4 locus in arp1Δ mutant cells that also express GFP-Tub1. In mitotic cells, Kin4 localizes to SPBs within the mother compartment (arrowheads). After spindle disassembly, Kin4 accumulates at the bud neck (arrow). (B) In elm1Δ arp1Δ mutant cells, Kin4 localizes to the SPBs in the mother compartment during mitosis (arrowheads) but does not accumulate at the bud neck after spindle disassembly (arrow). (C) Distribution of mean Kin4-tdimer2 fluorescence intensity across the bud neck in SPC active cells. Fluorescence intensities were measured across a 2 × 1–µm region centered on the smallest diameter of the neck (dashed line) in 14 cells with mitotic spindles in the mother. (D) Kin4-tdimer2 fluorescence intensity across the bud neck in postmitotic cells. Mean values from 10 cells with disassembled spindles are shown. X axis is not equivalent to C. (E) Kin4-tdimer2 fluorescence intensity across the bud neck in elm1Δ cells when the spindle remains in the mother. Mean values from six cells are shown. X axis is not equivalent to C. Strains: ELM1 arp1Δ, yJC6498 and elm1Δ arp1Δ, yJC6499. a.u., arbitrary units. Bars, 2 µm.
We then measured the fluorescence intensity of the Kin4-tdimer2 signal at the neck in wild-type and elm1Δ mutant cells. Kin4-tdimer2 fluorescence was enriched at the bud neck in cells with mitotic spindles in the mother compartment, suggesting that Kin4 associates with the neck when the SPC is active (Fig. 5 C). Once cells exited mitosis, which is indicated by the disassembly of the spindle, the enrichment of Kin4 at the neck increased 10-fold (Fig. 5, A and D). In elm1Δ cells, Kin4 was not enriched at the bud neck when the spindle was in the mother, nor did we detect Kin4 at the neck in postmitotic elm1Δ cells (Fig. 5, B and E). These results suggest that Elm1 may contribute to the accumulation of Kin4 at the bud neck.
Is Elm1 necessary for activation of the Kin4 kinase?
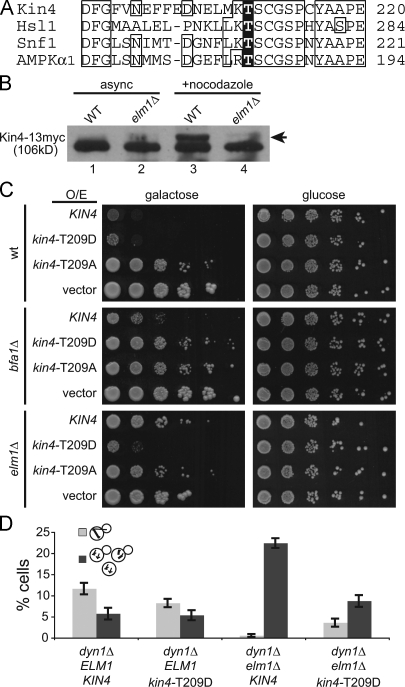
Next, we asked whether Elm1 is important for activation of the Kin4 kinase. Elm1 activates the kinases Snf1 and Hsl1 by phosphorylating a threonine residue within the activation loop of the kinase domain (Fig. 6 A; Hong et al., 2003; Sutherland et al., 2003; Szkotnicki et al., 2008). The analogous region of Kin4 contains a threonine at position 209, and preventing phosphorylation of this residue by substitution with alanine ablates Kin4 kinase activity and function in the SPC (D'Aquino et al., 2005; Maekawa et al., 2007). We therefore hypothesized that Elm1 may activate the Kin4 kinase by phosphorylating residue T209.
Figure 6.
Elm1 is necessary for Kin4 activation. (A) Alignment of the activation-loop regions of Kin4 and three known substrates of the Elm1 kinase. Phosphorylation of each highlighted threonine residue is necessary for kinase activation. Boxes indicate the majority sequence identity at a given residue position. (B) The presence of slow-migrating species of Kin4 depends on Elm1. Immunoblots of lysates from wild-type and elm1Δ cells expressing Kin4 tagged with a 13myc epitope, either grown asynchronously or arrested at metaphase by treatment with nocodazole. Arrow denotes slower-migrating species of Kin4-13myc. Strains: wild type (WT), yJC6382 and elm1Δ, yJC6396. (C) Phosphomimetic kin4-T209D is functional in the absence of Elm1. High-copy plasmids containing KIN4, kin4-T209D, or kin4-T209A under the control of a galactose-inducible promoter were transformed into either wild-type, bfa1Δ, or elm1Δ strain backgrounds. 10-fold dilution series were spotted onto media selective for plasmid retention and containing either galactose to induce expression or glucose to inhibit expression. Strains containing empty vector are shown as controls. Strains: wild type (wt), yJC2295; bfa1Δ, yJC6447; and elm1Δ, yJC5254. Plasmids: pGAL-KIN4, pBJ1651; pGAL-kin4-T209D, pBJ1896; pGAL-kin4-T209A, pBJ1840; and vector, pBJ216. (D) Phosphomimetic Kin4-T209D rescues the SPC defect of elm1Δ mutants. Indicated strains were arrested by treatment with α factor and released into fresh media at 14°C for 20 h. Cells were then briefly fixed in formaldehyde, and the percentage of cells exhibiting intact anaphase spindles within the mother compartment (light gray bars) or multiple MTOCs resulting from checkpoint failure (dark gray bars) was determined based on observation of GFP-tubulin. Values are the means of 10 counts of at least 50 cells from two separate experiments. Error bars are the standard error of the means. Strains: dyn1Δ, yJC5603; dyn1Δ kin4-T209D, yJC7080; dyn1Δ elm1Δ, yJC7079; and dyn1Δ elm1Δ kin4-T209D, yJC7081.
To determine whether Elm1 is necessary for the phosphorylation of Kin4, we first compared the phosphorylation of Kin4 in wild-type and elm1Δ mutants. Consistent with previous results, we observed Kin4 from extracts of asynchronously growing wild-type cells migrating as a primary band with a faint slower-migrating species on 1D SDS-PAGE (Fig. 6 B, lane 1; D'Aquino et al., 2005). The slower-migrating species was enriched when cells were arrested in metaphase by nocodazole treatment (Fig. 6 B, lane 3). In separate experiments, the slower-migrating species was depleted by treatment with phosphatase, confirming that this band represents phosphorylated Kin4 (unpublished data). The slower-migrating species was not detected in extracts from elm1Δ mutant cells, even during metaphase arrest (Fig. 6 B, lanes 2 and 4). Thus, phosphorylated species of Kin4 are diminished in the absence of Elm1, consistent with the notion that Elm1 phosphorylates Kin4.
If the critical function of Elm1 in the SPC is to phosphorylate T209 of Kin4, constitutive phosphorylation of this residue might obviate the need for Elm1. To test this hypothesis, we generated alleles of KIN4 that either mimic the negative charge of phosphorylation (kin4-T209D) or prevent phosphorylation (kin4-T209A). Overexpression of phosphomimetic Kin4-T209D strongly inhibited cell growth in a wild-type strain background, indicating that this mutant acts as a functional Kin4 kinase (Fig. 6 C). Furthermore, growth inhibition by Kin4-T209D overexpression required the downstream target of the Kin4 kinase, Bfa1, confirming that Kin4-T209D functions by activating the SPC (Fig. 6 C). In contrast, overexpression of the phosphoablated Kin4-T209A mutant did not inhibit growth, confirming that phosphorylation at T209 is necessary for function (Fig. 6 C; D'Aquino et al., 2005; Maekawa et al., 2007). To test whether Elm1 is necessary for the function of Kin4-T209D, we repeated the overexpression experiment in elm1Δ mutant cells. Overexpression of Kin4-T209D inhibited the growth of elm1Δ mutants, similar to wild-type cells (Fig. 6 C). Thus, Elm1 is not necessary for the function of Kin4 when T209 is constitutively phosphorylated, consistent with the hypothesis that Elm1 phosphorylates T209 of Kin4.
Next, we tested whether constitutive phosphorylation of Kin4 at T209 restored SPC function in elm1Δ mutants. For this, we constructed a phosphomimetic kin4-T209D allele at the chromosomal KIN4 locus, replacing the wild-type allele. Cells expressing kin4-T209D grew normally and did not exhibit prolonged mitoses (Fig. S4). We assayed SPC integrity by measuring the frequency of multinucleate cells and found that these were rare in the kin4-T209D mutant, similar to the level seen for wild-type KIN4 (Fig. 6 D; P = 0.84 when comparing ELM1 KIN4 cells with ELM1 kin4-T209D cells). This suggests that Kin4-T209D promotes the proper function of the SPC. When combined with the elm1Δ mutation, kin4-T209D suppressed the generation of multinucleate cells, indicating a restoration of SPC activity (Fig. 6 D). The frequency of multinucleate elm1Δ kin4-T209D cells was slightly greater than that of wild-type cells, but not significantly different (P = 0.14). Thus, constitutive phosphorylation of Kin4 at residue T209 eliminates the requirement for Elm1 in the SPC.
Is the accumulation of Elm1 at the bud neck necessary for the SPC?
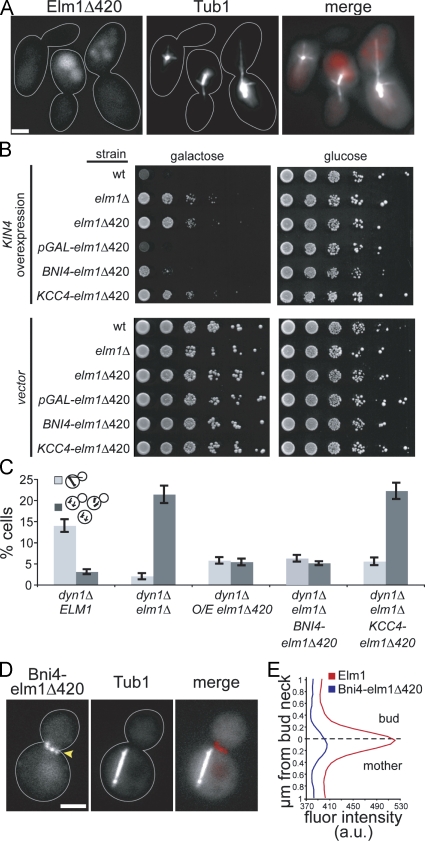
Having identified a role for Elm1 in the SPC, we returned to the question of how the association of Elm1 with the bud neck might influence this function. Elm1 is enriched at the neck when the SPC is active (Fig. 2) and during nocodazole arrest (not depicted). We showed that Elm1Δ420, which retains the kinase domain but lacks the carboxy-terminal domain, is deficient in the SPC (Fig. 1 C). We found that Elm1Δ420-tdimer2 showed no fluorescence accumulation at the bud neck, suggesting that the carboxy-terminal domain is necessary to target Elm1 to the neck (Fig. 7 A). We also considered that truncation of the carboxy-terminal domain may disrupt other functions of Elm1 critical for the SPC. To assess the functionality of Elm1Δ420-tdimer2, we tested whether overexpression might restore function. Indeed, overexpressed Elm1Δ420-tdimer2 did support Kin4's growth inhibition activity (Fig. 7 B), and it suppressed the accumulation of multiple microtubule-organizing centers (MTOCs) seen in elm1Δ dyn1Δ–null mutants (Fig. 7 C).
Figure 7.
The accumulation of Elm1 at the bud neck promotes its function in the SPC. (A) The localization of Elm1 to the bud neck requires its carboxy-terminal region. Tandem RFP/tdimer2 was integrated behind codon 420 of ELM1. Cells also express GFP-Tub1. Images were collected on a wide-field microscope. Strain: yJC6854. (B) Kin4 activity is supported by overexpressed elm1Δ420 or chimeric Bni4-elm1Δ420. Strains with high-copy plasmids containing KIN4 under the control of a galactose-inducible promoter and 10-fold dilution series were spotted onto media selective for plasmid retention and containing either galactose to induce expression or glucose to inhibit expression. Strains containing empty vector are shown as controls. Strains: wild type (wt), yJC2295; elm1Δ, yJC5254; elm1Δ420-tdimer2, yJC6849; PGAL1-elm1Δ420-tdimer2, yJC7286; BNI4-elm1Δ420-tdimer2, yJC7292; and KCC4-elm1Δ420-tdimer2, yJC7293. Plasmids: pGAL-KIN4, pBJ1651 and vector, pBJ216. (C) Multi-MTOC assay for SPC function. Mother compartment is indicated by light gray bars, and multiple MTOCs resulting from checkpoint failure are indicated by dark gray bars. Values are the means of 10 counts of at least 50 cells from two separate experiments. Error bars are the standard error of the means. Strains: dyn1Δ, yJC5603; dyn1Δ elm1Δ, yJC7079; dyn1Δ PGAL1-elm1Δ420-tdimer2, yJC7299,7300; dyn1Δ elm1Δ BNI4-elm1Δ420-tdimer2, yJC7296,7297; and dyn1Δ elm1Δ KCC4-elm1Δ420-tdimer2, yJC7298. (D) Bni4-elm1Δ420-tdimer2 localizes to the bud neck when the mitotic spindle is in the mother compartment. Cells are dyn1Δ mutants that also express GFP-Tub1. Images were collected on a wide-field microscope. Arrowhead points to Bni4-elm1Δ420-tdimer2 at the bud neck. Strain: yJC7296. (E) Quantification of fluorescence intensities for Elm1-tdimer2 and Bni4-elm1Δ420-tdimer2 across the bud neck. Fluorescence intensities were measured (see Materials and methods) in cells with mitotic spindles in the mother based on GFP-tubulin. Plotted values are the means from 12 cells for each strain. Solid lines indicate outlines of cells. Strains: Elm1-tdimer2, yJC6852 and Bni4-elm1Δ420-tdimer2, yJC7296. a.u., arbitrary units. Bars, 2 µm.
Next, we asked whether targeting the Elm1 kinase domain to the bud neck might rescue its function in the SPC. To test this, we created chimeras by fusing Elm1Δ420 to the neck-localized proteins Bni4 and Kcc4. Expression of either chimera rescued the elongated cell morphology phenotype of Elm1 mutants, indicating that both are at least partially functional. Both chimeras exhibited localization to the neck, but only Bni4-elm1Δ420-tdimer2 was enriched at the neck during mitosis (Fig. 7, D and E; and not depicted). Furthermore, only Bni4-elm1Δ420-tdimer2 supported Kin4 growth inhibition activity (Fig. 7 B) and suppressed the generation of multi-MTOC cells in an elm1Δ dyn1Δ–null mutant background (Fig. 7 C). These results indicate that accumulation of Elm1 at the bud neck promotes its function in the SPC.
Discussion
In a previous study, we demonstrated that the activity of the SPC depends on the septin network at the bud neck, and, based on these results, we proposed that the neck serves as a platform for signaling mechanisms that inhibit mitotic exit when the spindle is delayed within the mother compartment (Castillon et al., 2003). This model could explain why movement of one spindle pole beyond the neck and into the bud is a critical event: this allows MEN components at that pole to escape the inhibitory activity of the SPC. In the current study, we seek to identify molecules involved in the sensing or signaling mechanism at the neck, using localization to the neck and requirement for SPC activity as criteria for our screen. Elm1 meets these criteria, and our results indicate that Elm1 contributes a specific function to the SPC that is independent of its known roles in the activation of AMPK/Snf1 and the regulation of Swe1.
Elm1 is involved in organizing the septin network at the bud neck (Bouquin et al., 2000; Gladfelter et al., 2004); therefore, the disruption of the SPC in elm1Δ mutants could be an indirect effect of altering septin organization. Our results argue against this possibility. Elm1 contributes to septin organization via a pathway that includes the protein kinase Gin4 and the cyclin-binding protein Nap1 (Gladfelter et al., 2004). Loss of this pathway, however, does not account for the elm1Δ phenotype. We previously showed that gin4Δ mutants exhibit a mild SPC phenotype (Castillon et al., 2003). We have also found that loss-of-function mutations in nap1 exhibit a similarly mild SPC phenotype (unpublished data). These intermediate phenotypes stand in contrast to the total loss of SPC function in elm1-null mutants. Furthermore, the SPC relies on the septin network to restrict Lte1 to the bud (Castillon et al., 2003), but the disruption of the SPC in elm1Δ mutants is not attributable to aberrant Lte1 activity (Figs. 3 C and S2). Together, these data support our conclusion that Elm1 has a primary role in the SPC that is separate from its role in septin organization.
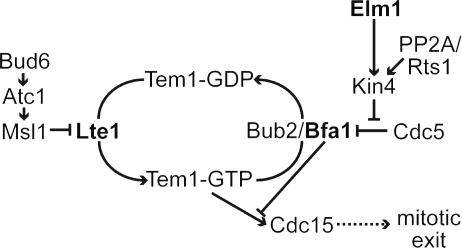
We propose that the role of Elm1 in the SPC is to activate the Kin4 kinase (Fig. 8). Elm1 is necessary for the function of Kin4 in our epistasis experiments (Fig. 3 D), suggesting that Elm1 either activates Kin4 or functions downstream between Kin4 and Bfa1. Previous studies have shown that Kin4 kinase is activated by phosphorylation of residue T209 (D'Aquino et al., 2005; Cayadasi et al., 2010). We found that phosphorylated isoforms of Kin4 were absent or greatly diminished in elm1Δ mutants (Fig. 6 B). Furthermore, Elm1 was not necessary for the function of phosphomimetic Kin4-T209D, indicating that Elm1 acts upstream of Kin4 phosphorylation (Fig. 6 C). Consistent with this notion, phosphomimetic Kin4-T209D restored SPC integrity in elm1Δ mutants (Fig. 6 D). Therefore, Elm1 appears to activate Kin4 by phosphorylating residue T209. Our results are consistent with those of Cayadasi et al. (2010), who further demonstrate that Elm1 directly phosphorylates Kin4 at T209. This mechanism is akin to the documented roles of Elm1 in activating AMPK/Snf1 and Hsl1 by phosphorylating residues analogous to T209 of Kin4 (Fig. 6 A; Hong et al., 2003; Sutherland et al., 2003; Szkotnicki et al., 2008).
Figure 8.
Elm1 activates Kin4 kinase in the SPC pathway.
In addition to Elm1, we found that loss-of-function mutations in the related kinases Sak1 and Tos3 also disrupt the integrity of the SPC; however, the effects of these mutations were less penetrant than those observed for elm1Δ, indicating a lesser role in the SPC (Fig. 1 C). The differences in penetrance between the elm1Δ, sak1Δ, and tos3Δ mutants may be explained by each kinase targeting discrete substrates in redundant pathways or exhibiting differential activity toward a common substrate. At this point, we cannot discriminate between these possibilities. Elm1, Sak1, and Tos3 are known to serve overlapping roles in regulating the yeast AMPK/Snf1, and each is sufficient to phosphorylate the activation loop threonine of AMPK/Snf1 (Hong et al., 2003; Sutherland et al., 2003). Thus, it remains possible that Sak1 and Tos3 can activate Kin4, but at a lower rate than Elm1.
Given the redundant biochemical activity of the EFKs, why does Elm1 exhibit a more pronounced role in the SPC? The answer is likely to involve the carboxy-terminal region of Elm1, which is not conserved in Sak1 or Tos3. This region of Elm1 is not necessary for the redundant function of Elm1 in regulating Snf1 (Rubenstein et al., 2006) nor the phosphorylation of Kin4 in vitro (Caydasi et al., 2010); however, it is necessary for the function of Elm1 in the SPC (Fig. 1 C). Furthermore, removal of this region abolishes the localization of Elm1 to the bud neck (Fig. 7 A). The SPC function of Elm1 can be rescued by fusing the Elm1 kinase domain to the neck-localized protein Bni4 (Fig. 7, B–E). Therefore, we speculate that the carboxy-terminal region of Elm1 may confer a more pronounced role in the SPC by targeting Elm1 to the mother side of the neck.
Our findings raise the question of whether the Elm1 mechanism acts as a sensor of spindle position. SPC activity is coordinated with spindle position through the interactions of cytoplasmic microtubules with the bud neck, which occur when the mitotic spindle fails to move into the neck (Adames et al., 2001; Moore et al., 2009). If the Elm1 mechanism acts as a sensor of cytoplasmic microtubules at the bud neck, we would expect Elm1 to activate Kin4 only when the spindle is inappropriately positioned. To the contrary, several lines of evidence suggest that the Elm1 activates Kin4 during every mitosis, even when the spindle is properly positioned. First, Kin4 appears to be phosphorylated—and the kinase activated—in a cell cycle–dependent manner, regardless of spindle position (D'Aquino et al., 2005; Caydasi et al., 2010). Second, Kin4 is phosphorylated during nocodazole treatment (Fig. 6 B; D’Aquino et al., 2005; Caydasi et al., 2010) and, therefore, is not dependent on a microtubule-sensing mechanism. Third, phosphomimetic Kin4-T209D, which is constitutively active and functions in the absence of Elm1, does not prolong mitosis when spindle positioning is unperturbed (Fig. S4). Fourth, overexpression of Elm1 does not itself inhibit mitotic exit, suggesting that SPC is not hyperactivated by high levels of Elm1 activity (Fig. S3). Based on these data, we conclude that Elm1 does not act in a sensor mechanism that detects aberrant spindle position.
We favor an alternative model in which Elm1 is one component in a network that creates a region of high SPC activity within the mother compartment. Two critical features of this network are the localization and activity of Kin4. Kin4 localizes to the mother cortex and bud neck before mitosis and accumulates at the SPB in the mother compartment during mitosis with concomitantly decreased localization at the cortex and neck (D’Aquino et al., 2005; Pereira and Schiebel, 2005). Preventing exchange between these sites by constitutively targeting Kin4 to the cortex or SPBs disrupts SPC activity, suggesting that Kin4 must visit both sites to function (Maekawa et al., 2007; Chan and Amon, 2009). We propose that exchange is necessary for Kin4 to be activated by Elm1 at the bud neck before being targeted to the SPBs by an independent regulatory module involving PP2A-Rts1 (Chan and Amon, 2009). Our findings also reveal that the localization of Elm1 to the bud neck is critical for Kin4 activation, even when high levels of Kin4 are present in the cell (Fig. 7 B). Why is the localization of Elm1 critical? Because SPC activity can be restored by elevating expression levels of the Elm1 kinase domain or by exogenously targeting this domain to the neck (Fig. 7 C), we hypothesize that the SPC requires a threshold of Elm1 kinase activity at the neck to function. It is tempting to speculate that Elm1 could be linked at the neck to additional SPC-regulating mechanisms that control Kin4 localization and sense spindle positioning errors; further study will be required.
Materials and methods
Yeast strains and manipulation
Chemicals and reagents were obtained from Sigma-Aldrich or Thermo Fisher Scientific unless stated otherwise. General yeast manipulation, media, and transformation were performed by standard methods (Amberg et al., 2005). Strains and plasmids are listed in Table S1. Gene deletions were constructed by PCR product–mediated transformation (Petracek and Longtine, 2002). Fluorescent chimeras of Elm1, Kin4, Sak1, and Tos3 were generated by fusion of PCR-amplified fluor-marker cassettes to the 3′ end of the chromosomal locus (Sheff and Thorn, 2004). Fusion proteins were assayed for functionality by scoring the fidelity of the SPC in arp1Δ haploids that expressed tagged versions of each fusion. To construct elm1Δ420, the tdimer2 fluor-marker cassette was integrated behind codon 420 of chromosomal ELM1. For overexpression of ELM1 or elm1Δ420-tdimer2, the TRP1-PGAL1 cassette (Petracek and Longtine, 2002) was integrated at the 5′ end, replacing the endogenous promoter. To construct plasmid-borne kin4 mutants, substitution mutations were introduced into pBJ1651 by site-directed mutagenesis and verified by DNA sequencing. To generate kin4-T209D at the endogenous chromosomal locus, we used a site-specific genomic mutagenesis strategy (Gray et al., 2005). In brief, the URA3 marker was amplified from pRS306 with oligonucleotides containing flanking sequences homologous to the KIN4 locus and integrated into a wild-type strain. The URA3 marker was then excised by transformation with a PCR product containing point mutations and flanking the KIN4 sequence. Excision of the URA3 marker was verified by PCR of genomic DNA. The presence of the T209D substitution and absence of other mutations were confirmed by sequencing the genomic locus. Western blot analysis confirmed that kin4-T209D is expressed at levels similar to wild-type Kin4. To construct Bni4-elm1Δ420-tdimer2 and Kcc4-elm1Δ420-tdimer2 chimeras, elm1Δ420-tdimer2::KanMX6 was amplified from the chromosomal locus of yJC6849 using oligos with flanking homology to the 3′ end of the BNI4 or KCC4 loci. Integration was confirmed by diagnostic PCR and sequencing; expression was confirmed by detection of a tdimer2/RFP signal at the bud neck.
Plasmids used in this study were provided by the following people: Dr. D. Lew (Duke University, Durham, NC) provided the swe1Δ1 expression plasmid (pBJ1492/pJM1139). Dr. J. Pringle (Stanford University, Stanford, CA) provided the CDC3-GFP expression plasmid (pBJ1488). Dr. K. Lee (National Institutes of Health, Bethesda, MD) provided the TUB1-GFP::LEU2 integration plasmid (pBJ1351/pSK1050). Drs. A. Khmelinskii and E. Schiebel (Universitat Heidelberg, Heidelberg, Germany) provided the mCherry-TUB1::URA3 plasmid (pAK011).
Assay for SPC integrity
The integrity of the SPC was analyzed by time-lapse microscopy of GFP-labeled microtubules in living cells as previously described (Castillon et al., 2003). In brief, living cells from asynchronous cultures grown to early log phase were suspended in nonfluorescent medium, mounted on a slab of 2% agarose, and sealed beneath a coverslip with paraffin wax. Images were captured at 30°C on a microscope (BX52; Olympus) equipped with a 1.35 NA 100× UPlan Apo objective, a spinning disc confocal scanner unit (CSU10; Yokogawa), a 488-nm cyan laser (Picarro), and an intensified charged-coupled device camera (XR/MEGA-10; Stanford Photonics) using QED software (Media Cybernetics). During acquisition, the temperature of the stage was maintained at 30°C. Microtubules were visualized with GFP-Tub1 expressed from the TUB1 promoter integrated at either the LEU2 locus (plasmid pSK1050) or URA3 locus (plasmid pAFS125). SPC integrity was determined by monitoring the morphology of anaphase spindles (those longer than 2 µm) that failed to move through the bud neck and remained in the mother compartment. The percentage of SPC integrity is defined as the number of checkpoint-positive cells (anaphase spindles that remain intact within the mother) divided by the sum of checkpoint-positive and checkpoint-failure cells. Cells that corrected spindle orientation defects during video acquisition by moving one spindle pole through the bud neck were excluded from the analysis. Statistical significance between rates of SPC integrity was determined by Fisher’s exact test.
Fluorescence microscopy
For still wide-field images, cells were suspended in nonfluorescent medium at 25°C, and images were collected on an inverted fluorescence microscope (IX70; Olympus) with a 1.40 NA 100× Plan Apo objective lens and a camera (CoolSNAP HQ; Roper Industries) using QED software. Images were processed using Photoshop (Adobe) to adjust levels and ImageJ (National Institutes of Health) to apply color look-up tables.
Fluorescence intensities were measured across a 2 × 1–µm region of interest, perpendicular to the bud neck and centered on the smallest diameter of the cell. The mean signal intensities for each pixel column along the axis perpendicular to the neck were measured using the Plot Profile function of ImageJ. Normalized values were determined by first subtracting the mean of the first and last points on the axis and then dividing by the maximum intensity within the region. Elm1-tdimer2 fluorescence was measured in large-budded mitotic cells, evident by single rings of Cdc3-GFP, Bni4-GFP, or Kcc-GFP. Kin4-tdimer2 fluorescence was measured in mitotic and postmitotic cells, indicated by the morphology of spindles labeled with GFP-tubulin.
Overexpression analysis
For the cell growth assay, liquid cultures of cells carrying high-copy 2µ plasmids with indicated genes under galactose-inducible promoters were grown to saturation in media selective for plasmid retention and containing 2% glucose to inhibit expression. Cell density was normalized by OD600. 10-fold dilution series were spotted to media selective for plasmid retention and containing either 2% galactose and 2% raffinose to induce expression or 2% glucose to inhibit expression. Images of plates were captured after 4 d of growth at 30°C.
For cell cycle analysis, liquid cultures of cells carrying high-copy 2µ plasmids with ELM1 or BFA1 under the control of a galactose-inducible promoter were grown to early log phase in media selective for plasmid retention and neutral sugar (2% sucrose) and then were arrested at START by treatment with 0.6 µM α factor. Cells were pelleted, washed, and resuspended in new media containing 2% sucrose and 2% galactose to induce expression. Samples were collected at 30-min intervals, fixed in 3.7% formaldehyde and 100 mM potassium phosphate for 5 min, and washed once with quencher solution (100 mM potassium phosphate, 0.1% Triton X-100, and 10 mM ethanolamine) and once with 100 mM potassium phosphate. The proportion of mitotic cells was determined by the presence of an anaphase-length spindle (>2 µm) based on fluorescence imaging of GFP-labeled microtubules.
Western blotting
Kin4 tagged with a 13myc epitope at the endogenous chromosomal locus was detected in lysates of cells grown asynchronously or arrested in metaphase. Asynchronous cells were grown to mid–log phase. For metaphase arrest, cells were grown to early log phase and then treated with 15 µg/ml nocodazole for 2 h. Cell number was normalized by OD600. 5% trichloroacetic acid was added to each culture and mixed for 10 min. Cells were then pelleted and washed with cold acetone. Cell pellets were resuspended in equal parts lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, and 2.75 mM DTT and supplemented with yeast protease inhibitor cocktail) and acid-washed glass beads and were lysed by bead beating for six cycles of 2 min each. Crude lysates were supplemented with Laemmli SDS-PAGE buffer, boiled, spun briefly, and then run on 8% SDS-PAGE and transferred to a nitrocellulose membrane. Blots were probed with mouse anti-myc 9E10 (Covance) at 1:2,000.
Online supplemental material
Fig. S1 shows the localization of Sak1 and Tos3. Fig. S2 shows Lte1 localization in elm1Δ mutants. Fig. S3 shows that ELM1 overexpression does not inhibit mitotic exit. Fig. S4 shows that phosphomimetic Kin4-T209D is not sufficient to prolong mitosis. Table S1 shows strains and plasmids used in this study and in four figures. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006092/DC1.
Acknowledgments
We are grateful to Drs. Scott Nelson and Mark Longtine and other members of the Cooper laboratory for advice and suggestions.
This research was supported by a grant from the National Institutes of Health to J.A. Cooper (GM47337). J.K. Moore was supported by a postdoctoral fellowship from the Molecular Oncology program of the Siteman Cancer Center at Washington University, funded by the National Institutes of Health (T-32-CA113275).
Footnotes
Abbreviations used in this paper:
- AMPK
- AMP-activated protein kinase
- EFK
- Elm1-family kinase
- MEN
- mitotic exit network
- MTOC
- microtubule-organizing center
- SAC
- spindle assembly checkpoint
- SPB
- spindle pole body
- SPC
- spindle position checkpoint
References
- Adames N.R., Oberle J.R., Cooper J.A. 2001. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 153:159–168 10.1083/jcb.153.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D.C., Burke D., Strathern J. 2005. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 203 pp. [Google Scholar]
- Bardin A.J., Visintin R., Amon A. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 102:21–31 10.1016/S0092-8674(00)00007-6 [DOI] [PubMed] [Google Scholar]
- Barral Y., Mermall V., Mooseker M.S., Snyder M. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 5:841–851 10.1016/S1097-2765(00)80324-X [DOI] [PubMed] [Google Scholar]
- Blacketer M.J., Koehler C.M., Coats S.G., Myers A.M., Madaule P. 1993. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol. Cell. Biol. 13:5567–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A., Venturi G.M., Tatchell K. 2000. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat. Cell Biol. 2:556–558 10.1038/35019601 [DOI] [PubMed] [Google Scholar]
- Bouquin N., Barral Y., Courbeyrette R., Blondel M., Snyder M., Mann C. 2000. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 113:1435–1445 [DOI] [PubMed] [Google Scholar]
- Castillon G.A., Adames N.R., Rosello C.H., Seidel H.S., Longtine M.S., Cooper J.A., Heil-Chapdelaine R.A. 2003. Septins have a dual role in controlling mitotic exit in budding yeast. Curr. Biol. 13:654–658 10.1016/S0960-9822(03)00247-1 [DOI] [PubMed] [Google Scholar]
- Caydasi A.K., Pereira G. 2009. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev. Cell. 16:146–156 10.1016/j.devcel.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Caydasi A.K., Kurtulmus B., Orrico M.I., Hofmann A., Ibrahim B., Pereira G. 2010. Elm1 kinase activates the spindle position checkpoint kinase Kin4. J. Cell Biol. 190:975–989 10.1083/jcb.201006151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L.Y., Amon A. 2009. The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes Dev. 23:1639–1649 10.1101/gad.1804609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aquino K.E., Monje-Casas F., Paulson J., Reiser V., Charles G.M., Lai L., Shokat K.M., Amon A. 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell. 19:223–234 10.1016/j.molcel.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Daum J.R., Gomez-Ospina N., Winey M., Burke D.J. 2000. The spindle checkpoint of Saccharomyces cerevisiae responds to separable microtubule-dependent events. Curr. Biol. 10:1375–1378 10.1016/S0960-9822(00)00780-6 [DOI] [PubMed] [Google Scholar]
- Fraschini R., D’Ambrosio C., Venturetti M., Lucchini G., Piatti S. 2006. Disappearance of the budding yeast Bub2–Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 172:335–346 10.1083/jcb.200507162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S.J., Wheatley E., Rittinger K., Johnston L.H., Sedgwick S.G. 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277:28439–28445 10.1074/jbc.M202540200 [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., de Bettignies G., Sedgwick S.G. 2009. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J. Cell Biol. 187:497–511 10.1083/jcb.200905114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A.S., Zyla T.R., Lew D.J. 2004. Genetic interactions among regulators of septin organization. Eukaryot. Cell. 3:847–854 10.1128/EC.3.4.847-854.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., Piccirillo S., Honigberg S.M. 2005. Two-step method for constructing unmarked insertions, deletions and allele substitutions in the yeast genome. FEMS Microbiol. Lett. 248:31–36 10.1016/j.femsle.2005.05.018 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Mortimer R.K., Culotti J., Culotti M. 1973. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics. 74:267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.P., Leiper F.C., Woods A., Carling D., Carlson M. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA. 100:8839–8843 10.1073/pnas.1533136100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S.J. 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 107:655–665 10.1016/S0092-8674(01)00580-3 [DOI] [PubMed] [Google Scholar]
- Hunter T., Plowman G.D. 1997. The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22:18–22 10.1016/S0968-0004(96)10068-2 [DOI] [PubMed] [Google Scholar]
- Kim H.B., Haarer B.K., Pringle J.R. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112:535–544 10.1083/jcb.112.4.535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L., Larson J.R., Tatchell K. 2005. Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell. 16:3455–3466 10.1091/mbc.E04-09-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. 1999. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl. Acad. Sci. USA. 96:4989–4994 10.1073/pnas.96.9.4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Shannon K.B., Shou W., Deshaies R.J., Li R. 2001. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 114:1379–1386 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., Theesfeld C.L., McMillan J.N., Weaver E., Pringle J.R., Lew D.J. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049–4061 10.1128/MCB.20.11.4049-4061.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Priest C., Lechner J., Pereira G., Schiebel E. 2007. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 179:423–436 10.1083/jcb.200705197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A.S., Jang J., Deshaies R.J. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 98:7325–7330 10.1073/pnas.141098998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J.N., Theesfeld C.L., Harrison J.C., Bardes E.S., Lew D.J. 2002. Determinants of Swe1p degradation in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:3560–3575 10.1091/mbc.E02-05-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk J.N., Schuyler S.C., Liu J.Y., Evans J.G., Salmon E.D., Pellman D., Bloom K. 2004. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol. Biol. Cell. 15:1519–1532 10.1091/mbc.E03-09-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F., Amon A. 2009. Cell polarity determinants establish asymmetry in MEN signaling. Dev. Cell. 16:132–145 10.1016/j.devcel.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Cooper J.A. 2010. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin. Cell Dev. Biol. 21:283–289 10.1016/j.semcdb.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Magidson V., Khodjakov A., Cooper J.A. 2009. The spindle position checkpoint requires positional feedback from cytoplasmic microtubules. Curr. Biol. 19:2026–2030 10.1016/j.cub.2009.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N., McCartney R.R., Schmidt M.C. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909–3917 10.1128/MCB.23.11.3909-3917.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S.A., Cooper J.A. 2007. A novel pathway that coordinates mitotic exit with spindle position. Mol. Biol. Cell. 18:3440–3450 10.1091/mbc.E07-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2005. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell. 19:209–221 10.1016/j.molcel.2005.05.030 [DOI] [PubMed] [Google Scholar]
- Pereira G., Höfken T., Grindlay J., Manson C., Schiebel E. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 6:1–10 10.1016/S1097-2765(00)00002-2 [DOI] [PubMed] [Google Scholar]
- Pereira G., Tanaka T.U., Nasmyth K., Schiebel E. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20:6359–6370 10.1093/emboj/20.22.6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek M.E., Longtine M.S. 2002. PCR-based engineering of yeast genome. Methods Enzymol. 350:445–469 10.1016/S0076-6879(02)50978-2 [DOI] [PubMed] [Google Scholar]
- Ro H.S., Song S., Lee K.S. 2002. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 99:5436–5441 10.1073/pnas.062059999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein E.M., McCartney R.R., Schmidt M.C. 2006. Regulatory domains of Snf1-activating kinases determine pathway specificity. Eukaryot. Cell. 5:620–627 10.1128/EC.5.4.620-627.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakchaisri K., Asano S., Yu L.R., Shulewitz M.J., Park C.J., Park J.E., Cho Y.W., Veenstra T.D., Thorner J., Lee K.S. 2004. Coupling morphogenesis to mitotic entry. Proc. Natl. Acad. Sci. USA. 101:4124–4129 10.1073/pnas.0400641101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M.A., Thorn K.S. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Toh-E A. 1994. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 14:7476–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C.M., Hawley S.A., McCartney R.R., Leech A., Stark M.J., Schmidt M.C., Hardie D.G. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299–1305 10.1016/S0960-9822(03)00459-7 [DOI] [PubMed] [Google Scholar]
- Szkotnicki L., Crutchley J.M., Zyla T.R., Bardes E.S., Lew D.J. 2008. The checkpoint kinase Hsl1p is activated by Elm1p-dependent phosphorylation. Mol. Biol. Cell. 19:4675–4686 10.1091/mbc.E08-06-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., DeRisi J.L., Wilhelm J.E., Vale R.D. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 290:341–344 10.1126/science.290.5490.341 [DOI] [PubMed] [Google Scholar]
- Wang Y., Hu F., Elledge S.J. 2000. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr. Biol. 10:1379–1382 10.1016/S0960-9822(00)00779-X [DOI] [PubMed] [Google Scholar]