Abstract
The current model for base excision repair (BER) involves two general sub-pathways termed single-nucleotide BER and long patch BER that are distinguished by their repair patch sizes and the enzymes/co-factors involved. Both sub-pathways involve a series of sequential steps from initiation to completion of repair. The BER sub-pathways are designed to sequester the various intermediates, passing them along from one step to the next without allowing these toxic molecules to trigger cell cycle arrest, necrotic cell death, or apoptosis. Although a variety of DNA-protein and protein-protein interactions are known for the BER intermediates and enzymes/co-factors, the molecular mechanisms accounting for step-to-step coordination are not well understood. In the present study we designed an in vitro assay to explore the question of whether there is a channeling or “hand-off” of the repair intermediates during BER in vitro. The results show that when BER enzymes are pre-bound to the initial single-nucleotide BER intermediate, the DNA is channeled from apurinic/apyrimidinic endonuclease 1 to DNA polymerase β and then to DNA ligase. In the long patch BER subpathway, where the 5′-end of the incised strand is blocked, the intermediate after DNA polymerase β gap filling is not channeled to the subsequent enzyme, flap endonuclease 1. Instead, flap endonuclease 1 must recognize and bind to the intermediate in competition with other molecules.
Keywords: ATP, DNA Damage, DNA Polymerase, DNA Repair, Nucleotide, DNA Polymerase β, LP BER, SN BER, Base Excision Repair, Substrate Channeling
Introduction
Mammalian genomic DNA is constantly exposed to a variety of physical and chemical agents, including ultraviolet light and ionization radiation, alkylating molecules, and endogenous reactive oxygen species that accumulate in cells due to environmental stress and natural metabolic processes (1–3). However, cells have evolved several specific pathways to remove such damage and maintain integrity of the genome. In mammalian cells the primary means for correcting discrete small DNA base lesions is base excision repair (BER)2 (4–7). The current and widely accepted working model for mammalian BER is that the process involves two sub-pathways that are differentiated by repair patch size and enzymes involved (8–11). These sub-pathways are termed “single-nucleotide BER” (SN BER) and “long-patch BER” (LP BER). SN BER involves removal of a single damaged nucleotide that is replaced with an undamaged nucleotide through template-directed DNA synthesis to fill the single-nucleotide gap, whereas in LP BER two or more nucleotides are replaced (8–13). Both of the BER sub-pathways are processed sequentially, one step to the next, and are initiated either by enzymatic removal of a damaged base or by spontaneous chemical hydrolysis of the glycosidic bond connecting the damaged base to the sugar phosphate backbone (1, 2, 14, 15). The resulting apurinic/apyrimidinic (AP) site is processed by AP endonuclease 1 (APE1) that incises the phosphodiester backbone 5′ to the abasic site, leaving a single-nucleotide gap with 3′-hydroxyl and 5′-deoxyribose phosphate at the gap margins (16, 17). DNA polymerase β (pol β), a multifunctional enzyme consisting of the 8-kDa amino-terminal domain with deoxyribose phosphate (dRP) lyase activity (18, 19) and the 31-kDa carboxyl-terminal domain with nucleotidyltransferase activity (20–22), then catalyzes template-guided gap filling and removal of the 5-dRP group to generate the substrate for the final BER step, DNA ligation (12, 13, 18, 19, 23–26). The ligation step is completed either by DNA ligase I or XRCC1-DNA ligase III complex (27–29).
The BER repair system and many of the BER enzymes are conserved in bacteria to humans, and these repair pathways have been reconstituted using purified natural and recombinant enzymes from various organisms (13, 23). From biochemical and structural studies, a common theme emerged that the individual steps in BER are sequentially ordered. For example, it is only after damaged base removal that the AP site-containing DNA strand is recognized by APE1. DNA polymerase β then processes the gapped intermediate, generating the substrate for DNA ligase. Thus, to prevent exposure of the intermediates to harmful nuclease activities, recombination events, or cell death signaling, it appears that the BER enzymes must coordinate with one another to efficiently receive the DNA substrates and pass the resulting DNA products along to the next enzyme in the sequence, just as a baton is passed from one individual to the next in a relay (30–32).
In the present study we tested the hypothesis that DNA can be channeled from one step to the next during BER. Steps in a reconstituted SN BER system were examined including APE1 strand incision, DNA synthesis, and dRP removal mediated by pol β, and ligation was mediated either by DNA ligase I or XRCC1-DNA ligase III complex. This was accomplished by preloading the enzymes onto BER substrates and then conducting the repair incubation in the presence of a trap such that any free enzyme or enzymes released from the DNA substrates during repair could no longer participate in the reactions. In the LP BER sub-pathway, we examined channeling of the DNA substrate from the DNA synthesis step to the FEN1 cleavage step. Our results show that the BER intermediates of the SN BER sub-pathway could be channeled from APE1 to pol β and to DNA ligase. In contrast, in the LP BER sub-pathway, the DNA product after gap filling by pol β was not channeled to FEN1.
EXPERIMENTAL PROCEDURES
Materials
High pressure liquid chromatography-purified oligodeoxynucleotides were from Oligos Etc., Inc. (Wilsonville, OR) and The Midland Certified Reagent Co. (Midland, TX). [α-32P]dCTP, [α-32P]cordycepin (3000 Ci/mmol), and [γ-32P]ATP (6000 Ci/mmol) were from PerkinElmer Life Sciences. Optikinase and terminal deoxynucleotidyltransferase were from United States Biochemical Corp. (Cleveland, OH) and Fermentas Inc. (Hanover, MD), respectively. Recombinant human pol β was overexpressed and purified as described previously (33). Human APE1, uracil-DNA glycosylase (UDG) with 84 amino acids deleted from the amino terminus, FEN1, DNA ligase I, and His-tagged DNA ligase III were purified as described previously (15, 34–37). The protein concentrations of enzyme samples specified below were measured by routine protein assays; the active fraction in each case corresponded to ∼50%.
Substrates for APE1 Incision, dRP Lyase, and Gap-filling Assays
Preparation of the 3′- or 5′-end-labeled dRP lyase substrate was as described previously (26). For the APE1 incision activity, the 32P-labeled duplex DNA was pretreated with UDG to generate an AP site. For the preparation of the 5′-end-labeled substrate, dephosphorylated 15-mer oligonucleotide (5′-CTG CAG CTG ATG CGC-3′) was phosphorylated with Optikinase and [γ-32P]ATP. A 34-mer (5′-GTA CCC GGG GAT CCG TAC GGC GCA TCA GCT GCA G-3′) template was then annealed with 32P-labeled 15-mer upstream and 19-mer (5′-pUGTACGGATCCCCGGGTAC-3′) downstream oligonucleotides by heating the solution at 90 °C for 3 min and allowing the solution to slowly cool to 25 °C. In some cases the 19-mer oligonucleotide was also 3′-labeled with [α-32P]ddATP before annealing. The 32P-labeled duplex DNA (labeled either at the 5′-end, 3′-end, or both ends) was treated with UDG to generate the 5′-dRP-containing single-nucleotide gapped substrate. In some experiments a 35-mer duplex DNA was used with uracil opposite guanine at position 15 in one strand and with the 3′-end blocked with cordycepin (3′-deoxyadenosine). In those experiments DNA was pretreated with UDG and APE1 to generate a single-nucleotide-gapped substrate with 3′-OH and 5′-dRP flap at the gap margins, and [α-32P]dCTP was used as radiolabel.
Incision Activity of APE1 in the Presence of a Trap
To measure the incision activity of APE1 in the presence of a DNA trap, a reaction mixture containing 50 mm HEPES, pH 7.5, 20 mm KCl, 2 mm dithiothreitol, 0.5 mm EDTA, and 10 nm UDG-pretreated 32P-labeled or 40 μm unlabeled DNA trap (a synthetic AP site-containing oligonucleotide DNA with tetrahydrofuran (THF), mimicking the AP site) and 30 nm APE1 was assembled on ice. The reaction was initiated by adding a mixture of 40 μm DNA trap, 5 mm MgCl2, or 10 nm UDG-pretreated 5′-end 32P-labeled DNA substrate, 5 mm MgCl2 and transferring the reaction mixture to 30 °C. In some experiments when both incision and gap-filling DNA synthesis reactions were performed simultaneously, the reactions were initiated by adding a mixture of 40 μm DNA trap, 25 μm dCTP, and 5 mm MgCl2 or 10 nm UDG-pretreated 5′-end 32P-labeled DNA substrate, 25 μm dCTP, and 5 mm MgCl2. In Fig. 5, APE1 and pol β concentrations were 30 and 40 nm, respectively. Samples were withdrawn at 10 s and mixed with gel-loading dye solution. The mixtures were then heated at 75 °C for 2 min, and DNA products were separated by electrophoresis in a 15% polyacrylamide gel containing 8 m urea in 89 mm Tris-HCl, 89 mm boric acid, and 2 mm EDTA, pH 8.8. Imaging and data analysis were performed with phosphorimaging and ImageQuantTM software.
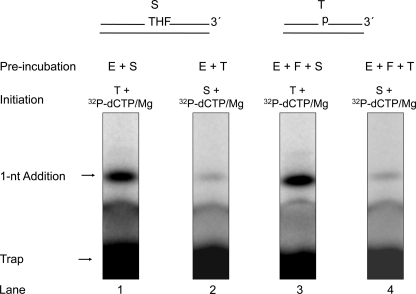
FIGURE 5.
Analysis of APE1-incision and gap-filling DNA synthesis steps in combination. Schematic representations of 32P-labeled AP site-containing DNA substrate (S) and DNA trap (T) are illustrated above the phosphorimage of the gels. The incision activity by APE1 and gap-filling DNA synthesis by pol β were examined in the presence of a DNA trap as described under “Experimental Procedures.” The reaction mixture was assembled on ice either with APE1, pol β, and 32P-labeled substrate DNA (a) or with APE1, pol β, and DNA trap (b). Reactions were then initiated by temperature jump and the addition of a mixture of DNA trap and dCTP/MgCl2 (a) or 32P-labeled substrate DNA and dCTP/MgCl2 (b), respectively. Samples were withdrawn at intervals and analyzed. The positions of the 32P-labeled substrate, APE1-incised product, and 1-nt gap-filling product are indicated. The APE1 product and gap-filling product corresponded to conversion of 24 and 9% of the substrate, respectively.
dRP Lyase in the Presence of a Trap
To measure the dRP lyase activity of pol β in the presence of a DNA trap, a reaction mixture containing 60 nm pol β, 50 mm HEPES, pH 7.5, 20 mm KCl, 2 mm dithiothreitol, 1 mm EDTA, and 20 nm 3′-end 32P-labeled DNA substrate or 10 μm unlabeled DNA trap was assembled on ice. The reaction was initiated by adding a mixture of 10 μm DNA trap or 20 nm 3′-end 32P-labeled DNA substrate and transferring the reaction mixture to 30 °C. Samples were withdrawn at 10 and 20 s, and the reaction products were stabilized by the addition of freshly prepared 20 mm NaBH4. Reaction mixtures then were put on ice and incubated 20 min. After this incubation an equal volume of gel-loading dye solution was added, and the mixtures were heated at 75 °C for 2 min. Reaction products were then separated by electrophoresis in a 15% polyacrylamide gel containing 8 m urea in 89 mm Tris-HCl, 89 mm boric acid, and 2 mm EDTA, pH 8.8. Imaging and data analysis were performed with phosphorimaging and ImageQuantTM software.
Gap-filling DNA Synthesis in the Presence of a Trap
To measure the gap-filling DNA synthesis activity of pol β in the presence of a DNA trap, reaction mixtures containing the same components as described for the dRP lyase assay were assembled with some exceptions; the substrate was 5′-end-labeled, instead of 3′-end-labeled, and 2 μm dCTP and 5 mm MgCl2 were added to initiate the reaction along with the DNA trap. After incubation, an equal volume of gel-loading dye was added, and the reaction mixtures were heated at 75 °C for 2 min. Reaction products were then separated by electrophoresis in a 15% polyacrylamide gel containing 8 m urea in 89 mm Tris-HCl, 89 mm boric acid, and 2 mm EDTA, pH 8.8. Imaging and data analysis were performed with phosphorimaging and ImageQuantTM software.
Gap-filling DNA Synthesis and Ligation in the Presence of a Trap
To perform combined gap-filling DNA synthesis and ligation reactions in the presence of a DNA trap, a reaction mixture containing 60 nm pol β, 50 mm HEPES, pH 7.5, 20 mm KCl, 2 mm dithiothreitol, 0.5 mm EDTA, 20 nm UDG/APE1-pretreated gapped DNA, or 10 μm unlabeled DNA trap was assembled on ice. The reaction was initiated by transferring the reaction mixture to 30 °C and adding a mixture of 10 μm unlabeled DNA trap or 20 nm UDG/APE1-pretreated gapped DNA substrate, 2.6 μm [α-32P]dCTP, and 5 mm MgCl2. Samples were withdrawn at 10 s, and the reaction was terminated by the addition of an equal volume of gel-loading dye solution. After incubation at 75 °C for 2 min, the reaction products were separated by electrophoresis and analyzed as above. However, in those cases when gap-filling, dRP lyase, and ligation steps were performed in combination, the reaction mixtures were supplemented with 4 mm ATP, 0.5 mm NAD, and 200 nm DNA ligase I.
LP BER Gap-filling DNA Synthesis, FEN1 Cleavage, and Ligation in the Presence of a Trap
To examine gap-filling DNA synthesis, FEN1 cleavage, and ligation reactions in the presence of a DNA trap, a reaction mixture containing 50 mm HEPES, pH 7.5, 20 mm KCl, 2 mm dithiothreitol, 1 mm EDTA, 20 nm APE1-pretreated THF-containing DNA or 10 μm unlabeled DNA trap, and 60 nm pol β and/or 25 nm FEN1 was assembled on ice. The reaction was initiated by adding a mixture of 10 μm unlabeled DNA trap or 20 nm APE1-pretreated THF-containing DNA substrate, 2.6 μm [α-32P]dCTP, 20 μm each dATP, dGTP, and TTP, and 5 mm MgCl2. When all three steps (gap-filling DNA synthesis, FEN1 cleavage, and ligation) were conducted in combination or in the same sample, a reaction mixture containing 60 nm pol β, 25 nm FEN1, and 200 nm DNA ligase I was incubated with DNA substrate or the trap as above. All incubations were at 30 °C. Samples were withdrawn at 10 s, and the reaction was terminated by the addition of an equal volume of gel-loading dye solution. After incubation at 75 °C for 2 min, the reaction products were separated by electrophoresis and analyzed as above. However, when gap-filling, FEN1 cleavage, and ligation steps were performed in combination, the reaction mixtures were also supplemented with 4 mm ATP and 200 nm DNA ligase I.
DNA Synthesis and dRP Lyase Kinetic Assays
Single-nucleotide gap filling reactions utilized the same UDG-processed nicked substrate that was prepared for dRP lyase assay reaction mixtures. In these present reactions, however, the primer strand was 5′-end 32P-labeled. Single turnover assays (E > S) were performed to measure insertion of dCTP (i.e. templating dG in the gap) using a KinTek Model RQF-3 rapid quench-flow system (Austin, TX). Unless noted otherwise, all concentrations refer to the final reaction concentrations after mixing. The final concentrations were 100 nm DNA, 500 nm pol β, 100 μm dCTP, 5 mm MgCl2, 50 mm HEPES-KOH, pH 7.5, 20 mm KCl, 0.5 mm EDTA, 1 mm dithiothreitol, 50 μg/ml bovine serum albumin, and 10% glycerol. After various time periods at 15 °C, reactions were quenched with 0.25 m EDTA. Quenched reactions were mixed with formamide dye, heated (5 min at 95 °C), and separated on a 20% polyacrylamide gel containing 8 m urea. Substrate and product bands were quantified after exposure to a phosphor screen using ImageQuantTM software. Time courses were fitted to a rising exponential, yielding the rate of single nucleotide insertion. The first-order rate constant is equivalent to the insertion rate (kpol).
Steady-state dRP lyase reactions used the 3′-end 32P-labeled gapped DNA with a 5′-dRP group in the gap. The preparation of this substrate is described above. Reactions were initiated with pol β (50, 100, or 150 nm) and 500 nm labeled DNA substrate in 50 mm HEPES-KOH, pH 7.5, 20 mm KCl, 0.5 mm EDTA, 1 mm dithiothreitol, 50 μg/ml bovine serum albumin, and 10% glycerol. After various time periods at 15 °C, 10 μl of the reaction mixture was removed and quenched with an equal volume of freshly prepared 0.4 m NaBH4. Quenched reactions were mixed with formamide dye, heated, and resolved on 20% polyacrylamide gels containing 8 m urea. Bands were visualized by exposure to a phosphorimaging screen and analyzed using ImageQuantTM software.
Single turnover dRP lyase reactions utilized the 3′-end 32P-labeled gapped DNA with a 5′-dRP group in the gap. The preparation of this substrate is described above. Time courses were performed using the chemical quench-flow system. In these assays, pol β and pretreated 3′-end 32P-labeled DNA were rapidly mixed for various times at 15 °C, and the reactions quenched by collection into tubes containing freshly prepared 0.4 m NaBH4. The final concentrations were 500 nm pol β, 100 nm DNA substrate, 50 mm HEPES-KOH, pH 7.5, 20 mm KCl, 0.5 mm EDTA, 1 mm dithiothreitol, 50 μg/ml bovine serum albumin, and 10% glycerol. Quench flow parameters were input manually to obtain the desired time points when quenching occurred in the collection tubes, thereby correcting for the time that each reaction mixture spent in the exit loop. This was necessary due to the volatile nature of the high concentration of NaBH4. The DNA substrate and product were ethanol-precipitated in the presence of linear polyacrylamide carrier (10–15 μg) and re-suspended in formamide gel loading buffer. After brief heating (5 min at 95 °C), the DNA was resolved on a 20% polyacrylamide gel containing 8 m urea. The dried gel was exposed to a phosphorimaging screen and quantified using ImageQuantTM software. Time courses were fitted to a rising exponential with a base-line term.
RESULTS AND DISCUSSION
Biochemical and structural studies of BER enzymes suggested that BER intermediates could be passed sequentially from one enzyme to the next in a coordinated fashion, in contrast to a model where all substrates and products are in equilibrium with free enzymes. To examine the hypothesis that the product of one step can be channeled as substrate to the next enzyme in the SN BER pathway, we studied AP site incision, gap filling, removal of the 5′-dRP group, and ligation. These steps are mediated by APE1, pol β, and DNA ligase I, respectively, and were studied here either individually or in combination after preloading the enzyme(s) onto DNA and initiating the reactions in the presence of a trap.
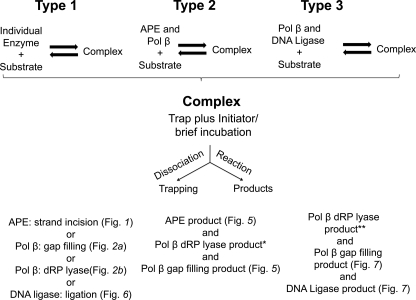
The design for these “single turnover” repair experiments is summarized in Scheme 1. Either an individual BER enzyme or a mixture of enzymes was first preincubated with the respective substrate DNA, and then the enzyme-substrate complex was mixed with a DNA trap plus an initiator for the reaction(s). After a brief incubation, products were recovered by gel electrophoresis and quantified. Scheme 1 illustrates the different types of incubations used; type 1 involved individual BER enzymes and steps as shown, and types 2 and 3 involved mixtures of BER enzymes and more than one reaction product. In the preincubations, the substrate DNA concentrations were in the range of the dissociation constants for the respective enzymes (10–20 nm). The reaction products observed in the presence of trap resulted from turnover of enzyme that was bound during the preincubation. The incubations were for 10 s, the shortest period we could manage using manual methods but much longer than the catalytic rate constants for each enzyme.
SCHEME 1.
Illustration of different types of incubation protocols used in the single turnover experiments to examine substrate channeling during SN BER. Purified human BER enzymes were preincubated with substrates either individually (Type 1) or as mixtures of enzymes (Types 2 and 3). The enzyme-substrate complexes formed during the preincubation were then mixed with a DNA trap plus initiator of the individual reaction. The trap was designed to remove any unbound free enzyme left in solution after the preincubation along with any enzyme that might dissociate from the complex after initiation of the reaction. After addition of the trap and initiator, reaction mixtures were incubated for 10 s. In the type 3 incubation, 32P-labeled dNTP was the labeled substrate, whereas in types 1 and 2 incubations 32P-labeled substrate DNA was used; the asterisks indicate that the respective products were not measured. Figures illustrating the corresponding results are designated.
In LP BER, the DNA synthesis reaction may occur as a series of single-nucleotide gap filling steps in conjunction with FEN1 5′-tailoring (38). Accordingly, we also examined whether FEN1 can preload onto its substrate, conduct flap removal, and then pass the product to next step, gap-filling DNA synthesis.
APE1 Strand Incision
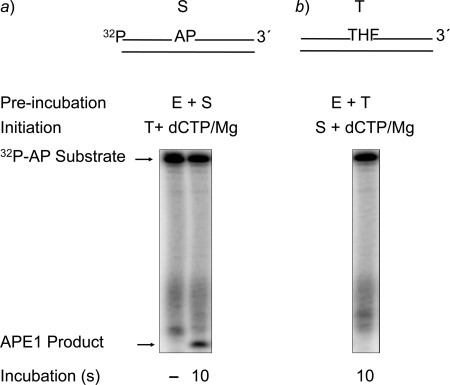
To study the APE1-mediated BER step, the reaction mixtures were assembled and incubated as shown in Scheme 1. The 32P-labeled DNA containing a lone uracil residue at position 15 was pretreated with UDG to generate an AP site-containing substrate. A 34-bp unlabeled DNA containing a synthetic AP site (THF) was used to trap any unbound APE1 in the reaction mixture. The reaction mixture was assembled on ice and initiated by transferring the reaction mixture to 30 °C and adding a mixture of MgCl2 and ∼4000-fold excess DNA trap. In another set of control reaction mixtures, APE1 was preincubated with trap first, and the reaction was initiated as above. The results showed that APE1 prebound to the DNA substrate was able to incise the AP site (Fig. 1a); unbound APE1 in solution was quenched by the trap, as no increase in product formation was observed with a 20-s incubation (not shown). In contrast, when APE1 was first preincubated with DNA trap and then the reaction was initiated by adding a mixture of DNA substrate and MgCl2, very little product was observed. This indicated that the trap was able to capture almost all of APE1 in the reaction mixture before it could bind and incise DNA substrate (Fig. 1b). Under these conditions 22% of the substrate was converted to product.
FIGURE 1.
Incision of the AP site-containing DNA by APE1 in the presence of a DNA trap. Schematic representations of 5′-end 32P-labeled AP site-containing DNA substrate (S) and DNA trap (T) are illustrated above the phosphorimage of the gels. The incision activity APE1 in the presence of a DNA trap was examined as described in detail under “Experimental Procedures.” The reaction mixture was assembled on ice either with 30 nm APE1 and 10 nm 32P-labeled UDG-treated DNA (a) or with APE1 and DNA trap (b). Reactions were then initiated by temperature jump and the addition of a mixture of DNA trap and MgCl2 (a) or with the addition of 32P-labeled UDG-treated DNA and MgCl2 (b), respectively. Samples were withdrawn after 10 s and analyzed. The positions of the 32P-labeled substrate and APE1-incised product are indicated. In a, 22% of the substrate was converted into product.
Gap-filling and dRP Lyase Steps
To study these two pol β-mediated BER steps, reaction mixtures were assembled and incubated for 10 and 20 s. To examine DNA synthesis, a 34-mer nicked DNA was first prepared by annealing 5′-end-labeled 15- and 19-mer oligonucleotides to a complementary 34-mer DNA strand. The 19-mer oligonucleotide contained a 5′-phosphorylated uracil nucleotide. This duplex DNA was pretreated with UDG, resulting in a single-nucleotide gapped DNA with 3′-OH and 5′-dRP groups at the gap margins, mimicking the APE1-incised BER intermediate. A 21-bp unlabeled gapped DNA was used to trap any unbound pol β in the reaction mixture.
To examine use of this BER intermediate by pol β in the DNA synthesis step, the reaction mixture was assembled on ice, and the reaction was initiated by transferring the tubes to 30 °C and adding a mixture of dCTP, MgCl2, and a ∼500-fold excess of trap. In another set of reaction mixtures, pol β was preincubated with the trap first, and the reaction was initiated by adding a mixture of DNA substrate, dCTP, and MgCl2. The reaction mixtures were incubated at 30 °C, and samples were withdrawn for products analysis. The results showed that pol β pre-bound to the DNA substrate was able to incorporate one nucleotide (Fig. 2a, lane 2), i.e. incorporation of dCMP into DNA; unbound pol β in solution was trapped completely as no further increase in dCMP incorporation was observed with a 20-s incubation (Fig. 2a, lane 3). In contrast, when pol β was first preincubated with the trap and then the reaction was initiated by adding a mixture of DNA substrate, dCTP, and MgCl2, no incorporation of dCMP was observed. This indicated that the enzyme was productively bound during the preincubation and that the trap was able to capture all of the pol β in the reaction mixture (Fig. 2a, lanes 4 and 5). Under these conditions 25% of the substrate was converted to product.
FIGURE 2.
Gap-filling DNA synthesis and removal of the dRP group by pol β in the presence of a DNA trap. Schematic representations of 32P-labeled DNA substrate (S) and DNA trap (T) are illustrated above the phosphorimage of the gels. E denotes pol β. a, gap-filling DNA synthesis by pol β in the presence of a DNA trap was examined as described under “Experimental Procedures.” The reaction mixture was assembled on ice either with 60 nm pol β and 20 nm 5′-end 32P-labeled UDG/APE1-treated DNA (lanes 1–3) or with pol β and DNA trap (lanes 4 and 5). Reactions were then initiated by temperature jump and the addition of a mixture of dCTP, DNA trap, and MgCl2 (lanes 1–3) or dCTP, 32P-labeled UDG/APE1-treated DNA, and MgCl2 (lanes 4 and 5), respectively. Samples were withdrawn at 10 and 20 s and analyzed. The positions of the 32P-labeled primer and 1-nt gap-filling product are indicated. b, for analyzing the dRP lyase activity of pol β in the presence of a DNA trap, the reaction mixture was assembled on ice with either 60 nm pol β and 20 nm 3′-end 32P-labeled UDG/APE1-treated DNA (lanes 1–3),or pol β and the trap (lanes 4 and 5). Reactions then were initiated by temperature jump and the addition of a DNA trap (lanes 1–3) or 32P-labeled substrate (lanes 4 and 5), respectively. Samples were withdrawn at 10 and 20 s. The reaction products were stabilized by the addition of NaBH4, and the reaction products were analyzed. The positions of the 32P-labeled dRP substrate and the product are indicated. For gap-filling, 25% of the substrate was converted to product, and for dRP lyase, 37% of the substrate was converted to product.
Using the same reaction mixture assembly protocol as above, we examined the 5′-dRP lyase step that also is catalyzed by pol β. In this case the DNA substrate was 3′-end-labeled. The removal of the dRP group from the substrate strand was monitored in a denaturing gel as a radiolabeled band migrating faster than the substrate. The reaction mixture was assembled on ice, and the reaction was initiated by transferring the reaction mixture to 30 °C and adding the trap. The results of this analysis showed that pol β prebound to the substrate excised the 5′-dRP group (Fig. 2b); 37% of the substrate was converted to product in the 10-s incubation, and there was no increase in product with the longer incubation. This indicated that the trap quenched free pol β in solution. To confirm the trapping of free enzyme, a reciprocal experiment was conducted where pol β was first preincubated with the trap, and the reaction was then initiated along with the addition of the radiolabeled substrate. No radiolabeled product above the background level was observed (Fig. 2b, lanes 4 and 5). These results indicated that after enzyme-substrate complex formation during the preincubation, dRP lyase removal is more rapid that substrate dissociation.
Because pol β conducts both DNA synthesis and dRP lyase steps in the SN BER scheme, we next explored whether pol β bound to DNA could catalyze both steps before dissociating from the DNA. Because pol β is a distributive enzyme and inserts one nucleotide at a time, our assumption was that if pol β incorporated dNMP first and then dissociated from the DNA, the trap in the reaction mixture would capture it. Hence, pol β would not be able to perform the next step in SN BER, i.e. the 5′-dRP lyase. We tested the possibility of processive DNA synthesis and dRP lyase activities. A double-labeled 34-mer nicked DNA was prepared by annealing a 5′-end-labeled 15-mer oligonucleotide and a 3′-end-labeled 19-mer oligonucleotide to a complementary 34-mer DNA strand. The duplex DNA was pretreated with UDG, resulting in a single-nucleotide gapped DNA with 3′-OH and 5′-dRP groups at the gap margins and radiolabels on both ends.
The reaction mixture was assembled on ice as above, and the reactions were initiated by temperature jump along with the addition of a mixture of dCTP, MgCl2, and a ∼500-fold excess of unlabeled trap DNA. The gap-filling and dRP lyase activities were measured in the same reaction mixture. Samples were withdrawn at 10 s, and the results shown in Fig. 3 revealed that pol β prebound to the DNA substrate incorporated one nucleotide and also excised the 5′-dRP group. In the control experiment, where pol β was preincubated first with the DNA trap, neither gap-filling nor 5′-dRP lyase activities were observed (Fig. 3b). For gap filling and dRP lyase, respectively, 26 and 46% of the substrate was converted to product.
FIGURE 3.
Analysis of gap-filling DNA synthesis and removal of the dRP group steps by pol β simultaneously. Schematic representations of a [32P]DNA substrate labeled at both ends (S) and the DNA trap (T) are illustrated above the phosphorimage of the gels. E denotes pol β. Double-labeled 34-bp DNA was prepared by annealing a 5′-end-labeled 15-mer oligonucleotide and a 3′-end-labeled 19-mer oligonucleotide to their complementary 34-mer DNA strand. The 19-mer oligonucleotide also contained a 5′-end phosphate and uracil. The duplex DNA was pretreated with UDG, resulting in a single-nucleotide gapped DNA with 3′-OH and 5′-dRP groups at the margins and radiolabels on both ends. Gap-filling DNA synthesis and dRP lyase reactions were performed by pol β in the presence of excess DNA trap as described under “Experimental Procedures.” The repair reaction mixture was assembled on ice either with pol β and 32P-labeled substrate (a) or pol β and DNA trap (b). Reactions then were initiated by temperature jump and the addition of a mixture of dCTP, DNA trap, and MgCl2 (a) or dCTP, 32P-labeled substrate DNA, and MgCl2 (b), respectively. Samples were withdrawn at 10 s and then analyzed. The positions of the 32P-labeled primer, 1-nt gap-filling DNA synthesis product, 32P-labeled dRP substrate, and the dRP lyase product are indicated. Product formation corresponded to 25% of the substrate and 46% of the substrate, respectively, for gap-filling DNA synthesis and dRP lyase.
Rapid dRP Lyase Reaction
From the results in Fig. 3, the prebound pol β was able to perform both gap-filling synthesis and lyase without releasing the DNA substrate. However, from these experiments it was not obvious which step occurred first. Previously we had characterized the 5′-dRP lyase activity of pol β using steady-state kinetic methods (26), and it appeared that the DNA synthesis step was faster than the lyase step and presumably occurred first.
To further dissect kinetic features of the 5′-dRP lyase reaction, a quantitative pre-steady-state approach was undertaken to see whether intermediates accumulated during the course of the reaction. In the presence of high enzyme concentrations, time courses were biphasic (Fig. 4a). A rapid burst of product formation was followed by a linear slow phase. Extrapolation of the linear portion of the time course to the y axis (t = 0) indicated that a burst of product formation occurred that was too fast to measure by manual sampling. The amplitude of this rapid unresolved phase was proportional to the concentration of enzyme in the reaction mixture (Fig. 4b). Normalizing the time courses for the differing enzyme concentrations indicates that the burst amplitude represented 70% of the added enzyme and that the linear phase corresponded to a turnover number of 0.12/min (Fig. 4c). Note that these time course experiments were performed at 15 °C to limit the slope of the linear phase, thereby providing a better estimate of the extrapolated burst amplitude. The biphasic nature of the time course indicated that a step after chemistry (i.e. product release) limits the steady-state linear phase of the time course. The observation that 70% of the enzyme rapidly excised the dRP group in the gap also indicates that only one molecule of pol β is bound/gap. If two or more molecules of pol β bound to a single-nucleotide gap, the burst amplitude would be ≤50%.
FIGURE 4.
Transient-state kinetic analysis of pol β dRP lyase reaction. Time courses were determined as described under “Experimental Procedures.” a, reactions were initiated by adding enzyme (50 (open), 100 (half-filled), or 150 nm (filled squares)) to 500 nm DNA substrate, and product formation was determined at the indicated time points. b, a shown is a secondary plot of the burst amplitudes (y intercept) determined from an extrapolated linear fit for each enzyme concentration. The solid line is a linear fit of the data with a y intercept of 0 and slope of 0.69 nm product per 1 nm added enzyme. c, shown is a re-plot of the data in panel a normalized for enzyme concentration. Accordingly, the ordinate scale provides the number of enzyme turnovers and indicates that 0.7 nm product is formed during the burst, indicating that ∼70% of the added enzyme is productively bound. The turnover number for the linear phase is 0.12/min. d, single turnover analysis of the dRP lyase reaction is shown. Pol β (1 μm) was rapidly mixed with 200 nm DNA substrate, and time points were collected. The time course exhibits two rapid phases; that is, a fast phase that was too rapid to measure (amplitude ∼ 20 nm) and a slower exponential phase (kobs ∼ 120/min). Both of these phases were considerably more rapid that the linear phase determined in panel c.
To measure the rate of the burst phase, single-turnover reactions were measured. In this situation the enzyme concentration (500 nm after mixing) exceeds the substrate concentration (100 nm after mixing), and enzyme cycling (i.e. product release) does not confound the time course. The rapid rate of the burst required that time points be gathered with a rapid-mixing and quenching instrument as outlined under “Experimental Procedures.” The volatile nature of the NaBH4-quenching agent required that the reaction be stopped in the reaction collection tube rather than the quench syringe of the instrument. Accordingly, the earliest time point that could be collected safely and reliably was 15 ms. A typical time course is illustrated in Fig. 4d. Again, the time course was biphasic; a rapid phase (∼20 nm) was lost in the dead time of the instrument followed by a single-exponential time course of product formation (kobs = 120/min). When gap-filling DNA synthesis was measured under these identical conditions and with this same substrate, the rate of dNTP insertion was 7/min (not shown). Thus, the dRP lyase activity of pol β was at least 20-fold faster than the DNA synthesis activity. The biphasic nature of the time course (Fig. 4d) suggested that enzyme bound DNA substrate was partitioned between two forms; that is, an active form that is poised for reaction (∼25%) and a population where the substrate is bound in an inappropriate conformation. Crystallographic structures of pol β with substrate analogues indicated that the 5′-sugar-phosphate moiety binds in a non-productive mode that must undergo a conformational change before chemistry can occur (39).
Coordinated APE1 Strand Incision and Gap-filling Steps
To examine whether the APE1-incised BER intermediate can be channeled to the next enzyme, pol β, we repeated assays for the incision and gap-filling synthesis steps with a 5′-end-labeled AP site-containing DNA substrate. The incision activity of APE1 and the gap-filling activity of pol β were measured in the same reaction mixture using the type 2 protocol in Scheme 1. The results shown in Fig. 5 revealed that APE1 prebound to the substrate was able to incise the DNA, and then the product was channeled to pol β for gap-filling DNA synthesis (Fig. 5a). In a control experiment, where APE1 and pol β were preincubated first with the trap and then the reaction was initiated by adding a mixture of 32P-labeled DNA substrate, dCTP, and MgCl2, gap filling was not observed. Under these conditions, 33% of the labeled substrate was incised by APE1, and 27% of this product was used in gap filling. Thus, these results showed that a portion of the BER intermediate after the APE1 incision step was channeled to pol β. This coordination between APE1 and pol β appears to be consistent with the complex formation and functional interaction between these enzymes and substrate DNA that was observed earlier (40).
Ligation Step
To explore whether the nicked DNA after the gap-filling and dRP lyase steps in SN BER can be channeled to DNA ligase, experiments were performed with a 5′-end-labeled nicked DNA substrate. The strategy for study of the ligation step was type 1 protocol shown in Scheme 1. An unlabeled nicked DNA was used as the trap. The reaction mixture was assembled on ice with DNA ligase I and 32P-labeled nicked substrate, and then the reaction was initiated by adding a mixture of ATP, MgCl2, and ∼500-fold excess of unlabeled trap (nicked 30-mer DNA duplex). The reaction mixtures were transferred to 30 °C for ligation to occur. After 10 and 20 s, samples were withdrawn, and the reaction products were analyzed. The results shown in Fig. 6 indicate that the substrate was converted into a fully ligated product. With the control experiment, where DNA ligase I was first preincubated with the trap and the reaction then initiated, no ligated product was observed (Fig. 6, lanes 3 and 4). These results indicated that ligation occurred more rapidly than nicked DNA dissociation. Next, we asked whether other DNA ligases, such as DNA ligase III and T4 DNA ligase, have a preference over DNA ligase I for use of the nicked substrate, as it had been reported that DNA ligase III is the preferred ligase in the SN BER pathway (41). Using the same reaction conditions, we found that DNA ligase III and T4 ligase functioned with a similar outcome as that seen with DNA ligase I (Fig. 6, lanes 5 and 6 and lanes 9 and 10, respectively).
FIGURE 6.
Analysis of the ligation step in the BER scheme conducted by purified DNA ligases. Schematic representations of 32P-labeled nicked DNA substrate (S) and the DNA trap (T) are illustrated above the phosphorimage of the gel. The ligation reaction was performed with DNA ligase I (lanes 1–4), DNA ligase III (lanes 5–8), or T4 DNA ligase (lanes 9–12) in the presence of a DNA trap as described under “Experimental Procedures.” The ligation reactions were assembled either with 32P-labeled DNA substrate and a DNA ligase or with DNA trap and a DNA ligase. Reactions were then initiated by temperature jump and the addition of a mixture of ATP, MgCl2, and DNA trap or ATP, MgCl2, and 32P-labeled DNA substrate, as indicated at the top of each lane. Samples were withdrawn at 10- and 20-s intervals and analyzed. The positions of the 32P-labeled primer and ligated product are indicated. L1, L3, and T4 denote DNA ligase I, DNA ligase III, and T4 DNA ligase, respectively.
Gap-filling, dRP Lyase, and Ligation Steps
So far, we have demonstrated that the BER intermediates, after the base removal step, can be channeled from initial DNA binding to catalysis for the APE1, pol β 5′-dRP lyase, and gap-filling DNA synthesis, when these steps were analyzed either individually or with APE1 and pol β together. To gain insight on whether the BER intermediates can be channeled throughout a complete BER reaction, we examined all three steps in a single reaction mixture. The complete BER reaction was assembled as above, with UDG- and APE1-pretreated DNA substrate, pol β, and DNA ligase I (Scheme 1, Type 3). The reaction mixture was assembled on ice, and the reaction was initiated by transferring the tubes to 30 °C and adding a mixture of [α-32P]dCTP, MgCl2, ATP, and an ∼500-fold excess of trap. The reaction mixtures were incubated at 30 °C, and a sample was withdrawn after 10 s (Fig. 7). The results revealed that when the enzymes were preincubated with the substrate, a complete or ligated repaired product was observed (Fig. 7, lane 1). When the enzymes were first preincubated with the trap, the complete repair product was not observed, and similarly, when ligase was not included, the gap-filling product was observed but not the complete product (Fig. 7, lanes 2 and 3). This indicated that the initial BER intermediate was subjected to dRP lyase and gap-filling and then channeled to DNA ligase. The efficiency of transition from gap filling to ligation can be estimated by comparing the products with and without ligase (Fig. 7, lanes 1 and 3); 65% of the gap-filling product was converted to the final product in the presence of trap.
FIGURE 7.
Analysis of gap-filling, dRP lyase, and ligation steps in combination. Schematic representations of UDG/APE1-treated DNA substrate (S) and the DNA trap (T) are illustrated above the phosphorimage of the gels. A complete BER reaction containing the pretreated DNA substrate or trap DNA, pol β, and DNA ligase I was assembled on ice. The reaction was initiated by a temperature jump and the addition of a mixture of [α-32P]dCTP, MgCl2, ATP, and DNA trap or [α-32P]dCTP, MgCl2, ATP, and DNA substrate as indicated at the top of each lane. Samples were withdrawn at 10 s and then analyzed. The positions of the 1-nt gap-filling product, ligated complete BER product, free trap, and the ligated trap are indicated. E and L denote pol β and DNA ligase I, respectively. In lane 1, the ligated product corresponded to 56% of the 1-nt gap-filling product found in lane 3. A minor amount of gap-filling product was observed in the presence of the trap.
“Hit and Run” Mechanism in the LP-BER Sub-pathway
Earlier, a working model for LP-BER was proposed where pol β conducts strand displacement DNA synthesis (2–16 nucleotides) leaving a long DNA flap; FEN1 then removes the DNA flap, and a DNA ligase seals the resulting nick. However, Liu et al. (38) recently suggested an alternate Hit and Run mechanism for LP BER. In this model pol β fills the initial one-nucleotide gap in LP BER, leaving a lesion-containing flap. FEN1 then removes the flap plus one nucleotide in the damaged strand to create a 1-nt gap, and finally, the gap is filled and sealed by pol β and DNA ligase, respectively. Furthermore, pol β and FEN1 can cooperate with each other in their 1-nt gap-filling and gap-forming activities, respectively, to form a longer repair patch (38). Here, we explored the possibility in LP BER that the DNA product after gap filling by pol β was passed along to FEN1 for flap removal.
The repair reaction mixture was assembled as above with the APE1-pretreated THF-containing DNA substrate, pol β, and FEN1. The reaction was initiated by adding [α-32P]dCTP, dATP, dGTP, TTP, MgCl2, and ∼500-fold excess of unlabeled DNA trap. In another set of reaction mixtures, pol β and FEN1 were mixed with the trap first, and then the reaction was initiated by adding the DNA substrate, [α-32P]dCTP, dATP, dGTP, TTP, and MgCl2. After 10 s, samples were withdrawn for product analysis. The assumption tested was that pol β will incorporate one nucleotide, and the product will be passed to FEN1 for flap cleavage. FEN1 cleavage will create a one-nucleotide gap for pol β to insert the second nucleotide in LP BER. Interestingly, the results of this analysis showed that pol β prebound to the DNA substrate incorporated only one nucleotide whether or not FEN1 was in the preincubation (Fig. 8). We explained these results as follows; once pol β filled the one-nucleotide gap, it dissociated from the DNA and was captured by the trap. It was possible that the nicked-flap DNA product, thus, formed was passed to FEN1 for cleavage of the THF-flap structure and to create one-nucleotide-gapped DNA for pol β to insert the second nucleotide in the LP BER gap. Because pol β was trapped before formation of the second nucleotide gap, we did not expect to observe a second nucleotide insertion into the DNA (Fig. 8). To our surprise, however, in an experiment where a 3′-end-labeled THF-containing strand was used, we failed to observe cleavage of the THF flap (data not shown). These results indicated that the product after gap-filling DNA synthesis was not passed to FEN1. Another explanation could be that the trap captured FEN1 even before its DNA substrate was formed. In other words, the enzymes were not preassembled on the BER substrate before the pol β gap-filling step. To further investigate this point, experiments were repeated in the presence of DNA ligase. The results of this analysis revealed gap-filling incorporation of only one nucleotide without formation of ligated product (Fig. 9). Thus, after gap filling by pol β, the BER intermediate still contained the THF flap and was not a substrate for DNA ligase. These results confirmed that the THF-flap product formed after gap-filling DNA synthesis by pol β was not channeled to FEN1. Instead, FEN1 action on this intermediate involved binding by free FEN1 in solution to the product of the pol β gap-filling reaction.
FIGURE 8.
Analysis of the DNA synthesis step in LP BER. Schematic representations of APE1-treated THF-DNA substrate (S), a LP BER intermediate, and the DNA trap (T) are illustrated above the phosphorimage of the gels. E and F denote pol β and FEN1, respectively. DNA synthesis reaction was performed by pol β alone or pol β and FEN1 in the presence of excess DNA trap as described under “Experimental Procedures.” The repair reaction mixture was assembled on ice either with pol β alone (lane 1) and substrate DNA or with pol β and FEN1 (lane 3) and substrate DNA, respectively. The reactions were initiated by a temperature jump and the addition of a mixture of [α-32P]dCTP, dATP, dGTP, TTP, DNA trap, and MgCl2 (lanes 1 and 3). In another set of reaction mixtures, pol β or pol β and FEN1 were mixed with the DNA trap first, and then the reactions were initiated by adding a mixture of [α-32P]dCTP, dATP, dGTP, TTP, substrate DNA, and MgCl2 (lanes 2 and 4), respectively. Reaction mixtures were incubated for 10 s and analyzed. The positions of the 1-nt addition and free trap are indicated.
FIGURE 9.
Analysis of gap-filling, FEN1 cleavage, and ligation steps in LP BER. Schematic representations of APE1-treated THF-DNA substrate (S), LP BER intermediate, and the DNA trap (T) are illustrated above the phosphorimage of the gels. E, F, and L denote pol β, FEN1, and DNA ligase I, respectively. The repair reaction mixture was assembled on ice either with pol β, FEN1 substrate DNA (lane 1), or with pol β, FEN1, DNA ligase I, and substrate DNA (lane 3). The reactions were initiated by a temperature jump and the addition of a mixture of [α-32P]dCTP, dATP, dGTP, TTP, ATP, DNA trap, and MgCl2 (lanes 1 and 3). In another set of reaction mixtures, pol β and FEN1 or pol β, FEN1, and DNA ligase I were preincubated with the DNA trap first, and then the reactions were initiated by adding a mixture of [α-32P]dCTP, dATP, dGTP, TTP, ATP, substrate DNA, and MgCl2 (lanes 2 and 4). Reaction mixtures were incubated for 10 s and analyzed. The positions of 1-nt gap-filling product, ligated BER product, free trap, and the ligated trap are indicated.
Concluding Remarks
Based on structural studies of APE1-DNA cocrystals by Tainer and co-workers (30, 31), it has been suggested that enzymatic steps in BER may involve recognition of a product-enzyme complex by the next enzyme in the pathway rather than binding to an intermediate that is free in solution. Wilson and Kunkel (32) referred to these coordinated events of processing DNA repair intermediates as “passing the baton.” For the first time, we tested the hypothesis that the BER intermediates can be channeled from one step to the next.
Purified human BER enzymes APE1, pol β, and DNA ligase I, when prebound to their respective DNA substrates, were able to conduct their respective enzymatic reactions in the presence of a DNA trap. This was not surprising, as these DNA enzymes are known to conduct activity by a sequential mechanism with addition to the DNA substrate to form the first kinetic intermediate. A new finding here was that mixtures of these enzymes could conduct a sequence of BER reactions in the presence of a DNA trap when the enzymes were prebound to the initial BER intermediate (Table 1). This indicated that substrates and products of the BER steps could be handed off from one enzyme to the next during the SN BER process, reducing the possibility that a sequestered intermediate could trigger the DNA damage surveillance system. The possibility that an ensemble of BER enzymes is recruited to the site of a BER lesion is consistent with the rapid recruitment of the enzymes studied here to sites of DNA damage in living cells (42, 43). On the other hand, rapid recruitment in cells does not always correlate with hand-off for purified enzymes, as FEN1 and pol β did not exhibit hand-off in the studies reported here but are rapidly recruited to sites of LP BER intermediates in living cells (44). Perhaps accessory proteins might influence the behavior of the purified enzymes, but this possibility has not been studied as yet.
TABLE 1.
Summary of substrate channeling
| BER step | Transfer in the presence of trapa |
|---|---|
| % | |
| APE1 to pol β gap filling | 50% (Fig. 5) |
| Pol β gap filling to DNA ligase I | 53% (Fig. 7) |
a Percent transfer of product in the preceding step to product in the subsequent step, as specified. The corresponding figure is indicated in parentheses.
Finally, another new finding was the very rapid enzymatic processing of the 5′-dRP lyase step of SN BER. In addition, the order of the gap-filling and lyase steps was found to be different from our previous understanding (25, 26). The 5′-dRP lyase rate constant was much faster (at least 20-fold) than the DNA polymerase gap-filling rate constant. As found earlier for the gap-filling reaction by pol β, the product release step for the 5′-dRP lyase reaction is rate-limiting under steady-state reaction conditions.
- BER
- base excision repair
- SN BER
- single-nucleotide BER
- LP BER
- long-patch BER
- dRP
- deoxyribose phosphate
- UDG
- uracil-DNA glycosylase
- AP
- apurinic/apyrimidinic
- APE1
- AP endonuclease 1
- pol β
- DNA polymerase β
- THF
- tetrahydrofuran
- nt
- nucleotide.
REFERENCES
- 1.Lindahl T. (1982) Annu. Rev. Biochem. 51, 61–87 [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 3.Loeb L. A., Preston B. D. (1986) Annu. Rev. Genet. 20, 201–230 [DOI] [PubMed] [Google Scholar]
- 4.Kubota Y., Nash R. A., Klungland A., Schär P., Barnes D. E., Lindahl T. (1996) EMBO J. 15, 6662–6670 [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson S. H. (1998) Mutat. Res. 407, 203–215 [DOI] [PubMed] [Google Scholar]
- 6.Wilson D. M., 3rd, Thompson L. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12754–12757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindahl T., Wood R. D. (1999) Science 286, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 8.Frosina G., Fortini P., Rossi O., Carrozzino F., Raspaglio G., Cox L. S., Lane D. P., Abbondandolo A., Dogliotti E. (1996) J. Biol. Chem. 271, 9573–9578 [DOI] [PubMed] [Google Scholar]
- 9.Klungland A., Lindahl T. (1997) EMBO J. 16, 3341–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortini P., Pascucci B., Parlanti E., Sobol R. W., Wilson S. H., Dogliotti E. (1998) Biochemistry 37, 3575–3580 [DOI] [PubMed] [Google Scholar]
- 11.Biade S., Sobol R. W., Wilson S. H., Matsumoto Y. (1998) J. Biol. Chem. 273, 898–902 [DOI] [PubMed] [Google Scholar]
- 12.Singhal R. K., Prasad R., Wilson S. H. (1995) J. Biol. Chem. 270, 949–957 [DOI] [PubMed] [Google Scholar]
- 13.Dianov G., Price A., Lindahl T. (1992) Mol. Cell. Biol. 12, 1605–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosbaugh D. W., Bennett S. E. (1994) Prog. Nucleic Acid Res. Mol. Biol. 48, 315–370 [DOI] [PubMed] [Google Scholar]
- 15.Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N. M., Haug T., Levine D. W., Krokan H. E. (1995) Biochemistry 34, 128–138 [DOI] [PubMed] [Google Scholar]
- 16.Doetsch P. W., Helland D. E., Haseltine W. A. (1986) Biochemistry 25, 2212–2220 [DOI] [PubMed] [Google Scholar]
- 17.Doetsch P. W., Cunningham R. P. (1990) Mutat. Res. 236, 173–201 [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto Y., Kim K. (1995) Science 269, 699–702 [DOI] [PubMed] [Google Scholar]
- 19.Piersen C. E., Prasad R., Wilson S. H., Lloyd R. S. (1996) J. Biol. Chem. 271, 17811–17815 [DOI] [PubMed] [Google Scholar]
- 20.Casas-Finet J. R., Kumar A., Morris G., Wilson S. H., Karpel R. L. (1991) J. Biol. Chem. 266, 19618–19625 [PubMed] [Google Scholar]
- 21.Kumar A., Abbotts J., Karawya E. M., Wilson S. H. (1990) Biochemistry 29, 7156–7159 [DOI] [PubMed] [Google Scholar]
- 22.Kumar A., Widen S. G., Williams K. R., Kedar P., Karpel R. L., Wilson S. H. (1990) J. Biol. Chem. 265, 2124–2131 [PubMed] [Google Scholar]
- 23.Dianov G., Lindahl T. (1994) Curr. Biol. 4, 1069–1076 [DOI] [PubMed] [Google Scholar]
- 24.Sobol R. W., Horton J. K., Kühn R., Gu H., Singhal R. K., Prasad R., Rajewsky K., Wilson S. H. (1996) Nature 379, 183–186 [DOI] [PubMed] [Google Scholar]
- 25.Srivastava D. K., Berg B. J., Prasad R., Molina J. T., Beard W. A., Tomkinson A. E., Wilson S. H. (1998) J. Biol. Chem. 273, 21203–21209 [DOI] [PubMed] [Google Scholar]
- 26.Prasad R., Beard W. A., Strauss P. R., Wilson S. H. (1998) J. Biol. Chem. 273, 15263–15270 [DOI] [PubMed] [Google Scholar]
- 27.Prasad R., Singhal R. K., Srivastava D. K., Molina J. T., Tomkinson A. E., Wilson S. H. (1996) J. Biol. Chem. 271, 16000–16007 [DOI] [PubMed] [Google Scholar]
- 28.Prigent C., Satoh M. S., Daly G., Barnes D. E., Lindahl T. (1994) Mol. Cell. Biol. 14, 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldecott K. W., McKeown C. K., Tucker J. D., Ljungquist S., Thompson L. H. (1994) Mol. Cell. Biol. 14, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mol C. D., Izumi T., Mitra S., Tainer J. A. (2000) Nature 403, 451–456 [DOI] [PubMed] [Google Scholar]
- 31.Parikh S. S., Mol C. D., Hosfield D. J., Tainer J. A. (1999) Curr. Opin. Struct. Biol. 9, 37–47 [DOI] [PubMed] [Google Scholar]
- 32.Wilson S. H., Kunkel T. A. (2000) Nat. Struct. Biol. 7, 176–178 [DOI] [PubMed] [Google Scholar]
- 33.Beard W. A., Wilson S. H. (1995) Methods Enzymol. 262, 98–107 [DOI] [PubMed] [Google Scholar]
- 34.Prasad R., Dianov G. L., Bohr V. A., Wilson S. H. (2000) J. Biol. Chem. 275, 4460–4466 [DOI] [PubMed] [Google Scholar]
- 35.Strauss P. R., Beard W. A., Patterson T. A., Wilson S. H. (1997) J. Biol. Chem. 272, 1302–1307 [DOI] [PubMed] [Google Scholar]
- 36.Wang Y. C., Burkhart W. A., Mackey Z. B., Moyer M. B., Ramos W., Husain I., Chen J., Besterman J. M., Tomkinson A. E. (1994) J. Biol. Chem. 269, 31923–31928 [PubMed] [Google Scholar]
- 37.Chen X., Pascal J., Vijayakumar S., Wilson G. M., Ellenberger T., Tomkinson A. E. (2006) Methods Enzymol. 409, 39–52 [DOI] [PubMed] [Google Scholar]
- 38.Liu Y., Beard W. A., Shock D. D., Prasad R., Hou E. W., Wilson S. H. (2005) J. Biol. Chem. 280, 3665–3674 [DOI] [PubMed] [Google Scholar]
- 39.Prasad R., Batra V. K., Yang X. P., Krahn J. M., Pedersen L. C., Beard W. A., Wilson S. H. (2005) DNA Repair 4, 1347–1357 [DOI] [PubMed] [Google Scholar]
- 40.Wong D., Demple B. (2004) J. Biol. Chem. 279, 25268–25275 [DOI] [PubMed] [Google Scholar]
- 41.Tomkinson A. E., Chen L., Dong Z., Leppard J. B., Levin D. S., Mackey Z. B., Motycka T. A. (2001) Prog. Nucleic Acid Res. Mol. Biol. 68, 151–164 [DOI] [PubMed] [Google Scholar]
- 42.Zolghadr K., Mortusewicz O., Rothbauer U., Kleinhans R., Goehler H., Wanker E. E., Cardoso M. C., Leonhardt H. (2008) Mol. Cell Proteomics 7, 2279–2287 [DOI] [PubMed] [Google Scholar]
- 43.Lan L., Nakajima S., Oohata Y., Takao M., Okano S., Masutani M., Wilson S. H., Yasui A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asagoshi K., Liu Y., Masaoka A., Lan L., Prasad R., Horton J. K., Brown A. R., Wang X. H., Bdour H. M., Sobol R. W., Taylor J. S., Yasui A., Wilson S. H. (2010) DNA Repair 9, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]