Abstract
Selenoprotein W (SelW) is expressed in various tissues, but it is especially high in the skeletal muscle of mammals. Such tissue-specific protein expression implies regulation by a tissue-specific factor. In this study, we investigated SelW expression during myogenic C2C12 cell differentiation using RT-PCR, quantitative PCR, and Western blot analysis. Both the protein and mRNA levels of SelW were increased during C2C12 cell differentiation, particularly during the early stage. Sequence analysis of the SelW promoter revealed four putative E-boxes, E1, E2, E3, and E4, which are known binding sites for MyoD, a myogenic transcriptional factor. Luciferase reporter assay showed that E1 and E4 were crucial for MyoD-dependent promoter activity. Using EMSA analysis, we observed that MyoD bound directly to E1 but not to E4, even though E4 mutation reduced SelW promoter activity in the luciferase reporter assay. Binding of MyoD to E1 was further investigated by ChIP assay. These results suggest that the SelW gene was activated by the binding of MyoD to a specific E-box during early skeletal muscle differentiation.
Keywords: Differentiation, Gene Regulation, Selenium, Transcription Factors, Transcription Regulation
Introduction
Selenoprotein W (SelW) is the smallest selenoprotein identified to date that contains the canonical amino acid selenocysteine (Sec).3 SelW is known to have an antioxidant effect on cells (1–3), a common feature of many selenoproteins due to the presence of Sec encoded by the stop codon UGA. Only a few selenoproteins, such as deiodinases and selenoprotein N (SelN), are known to lack this common feature (4). It was also found that 14-3-3 protein is a SelW-interacting protein with an unusual manner of redox regulation (5). NMR spectroscopy has been used to analyze the interaction of SelW with 14-3-3 protein as well as the structure of SelW containing Cys instead of Sec (6). SelW expression is increased by the knockdown of SelT in mouse fibroblast NIH 3T3 cells without any effect on the expression of other major selenoproteins such as thioredoxin reductase 1 (TrxR1 and Txnrd1), glutathione peroxidase 1 (Gpx1), glutathione peroxidase 4 (Gpx4), or selenoprotein 15 (Sep15) (7). This result implies that SelW might play a compensatory function for SelT.
SelW was first identified as a deficient protein in lambs suffering from white muscle disease, a selenium-responsive myopathy in livestock (8). SelW is expressed ubiquitously in various tissues, but it is specifically high in the skeletal muscle and brain of mammals (2, 9, 10). Tissue-specific protein expression implies regulation by a tissue-specific factor that plays a significant role in the tissue. Using Northern blot analysis, SelW expression was found to be regulated during C2C12 cell differentiation (2).
The expression of most proteins is regulated at the transcriptional level by mechanisms controlling complex transcriptional initiation networks. Therefore, elucidating the regulation of selenoprotein expression at the transcriptional level is vital to understanding how selenoproteins respond to environmental stimuli. Compared with the total number of selenoproteins actually identified, the number of studies focusing on the transcriptional regulation of selenoproteins is very low. Nonetheless, TrxR1, which is abundant in most tissues, is transcriptionally activated via binding of NF-E2-related factor-2 (Nrf2) to the antioxidant-responsive element in its promoter upon cadmium induction (11). Examination of point mutations and deletion fragments of FoxO-responsive elements in the promoter of selenoprotein P (SeP), a key selenium supplier produced in the liver, found that promoter activity and expression are enhanced by FoxO1a (forkhead box, class O) transcription factor in hepatoma cells (12). Selenoprotein S (SelS), a newly identified endoplasmic reticulum membrane protein responsive to endoplasmic reticulum stress (13, 14), contains putative binding sites for nuclear factor-κB as well as an endoplasmic reticulum stress-response element that increases expression and promoter activity upon activation by the pro-inflammatory cytokines TNF-α and IL-1β and the endoplasmic reticulum stress agents tunicamycin and thapsigargin (15).
There are few reports on the transcriptional regulation of SelW. It has been shown that metal-response element (MRE) is present in the SelW promoter, which is activated by exposure to copper and zinc ions but not cadmium (16). An in vitro binding assay found that a transcription factor, specificity protein 1 (Sp1), bound to a consensus Sp1 sequence in the SelW promoter as well as to the MRE sequence (17). In addition, another group observed the induction of the mouse SelW promoter by cadmium in a liver-specific metal-responsive transcription factor 1 (MTF-1) knock-out mouse model (18). In that particular study, MTF-1 was identified as a factor playing an important role in the regulation of SelW expression in liver-specific MTF-1 knock-out mice. MTF-1 is a zinc finger protein involved in the response to various stress stimuli, such as heavy metals and oxidative stress, which acts by binding to MRE consensus sequences in the promoters of its target genes. According to a recent publication, MTF-1 targets and represses the mRNA expression of SelH and Txnrd2 by binding to cis-acting MREs present in the coding regions (19). Taken together, MTF-1 might act as an activator or repressor of selenoprotein expression via binding to MREs located in the promoters or coding regions of its target genes.
To date, little is known about the transcriptional regulatory factors associated with tissue-specific SelW expression. Specifically, there is only limited information on the factors associated with transcriptional regulation of SelW in skeletal muscle, the major expression tissue. Skeletal muscle cells, which are derived from somites during embryogenesis, form precisely orchestrated physiological networks (20, 21). Myogenic precursor cells divide into proliferating myoblasts that then differentiate into multinucleated myotubes during cell elongation, alignment, and fusion. Myogenic regulatory factors (MRFs) composed of Myf5, MyoD, myogenin, and MRF4 play essential roles in skeletal muscle development. Myf5, MyoD, and MRF4 serve as myogenic determination factors that control the fate of proliferating myoblasts derived from myogenic precursor cells. The activation of myogenin and MRF4 is responsible for the differentiation of myotubes from proliferating myoblasts. The basic helix-loop-helix transcription factor MyoD is a member of the MRF family (22). Since its discovery, MyoD has been characterized as a key regulatory factor that controls the expression of many muscle-specific genes, such as muscle creatine kinase (MCK), myogenin, myosin alkaline light chains (MLC1), and MyoD itself during myogenesis (23–26). MyoD also regulates gene expression through the recognition of a consensus cis-acting element, called an E-box (CANNTG), which is located in the promoters or enhancers of its target genes.
As described above, some reports have achieved transcriptional regulation of SelW in C2C12 myoblast cells by using cis-acting elements with corresponding trans-acting factors to control SelW promoter activity. However, these reports are mainly limited to MRE sequences within the SelW promoter region between −40 and −20 bp. In this study, we investigated the tissue-specific regulation of SelW expression at the transcriptional level during skeletal muscle differentiation in mouse C2C12 cells, which are well established in muscle development (27, 28). The results show that SelW expression was up-regulated during the differentiation of C2C12 myoblasts into myotubes. Four putative E-boxes were identified in the SelW promoter, and the SelW promoter was activated by the binding of MyoD to at least one of these putative E-boxes during differentiation. Therefore, MyoD was required for transcriptional activation of SelW via putative E-boxes during skeletal muscle differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse skeletal muscle C2C12 (CRL-1772) myoblast and nonmyogenic mouse fibroblast C3H10T1/2 (CCL-226) cells were purchased from the American Type Culture Collection. C2C12 cells were cultured to ∼50% confluence in growth medium (GM) consisting of Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen). To induce differentiation, the cells were cultured to ∼100% confluence in GM, which is denoted as D0 in this study, followed by exchange with differentiation medium (DM) consisting of DMEM supplemented with heat-inactivated 2% horse serum (HS). DM was exchanged every day for the 5 days of culturing. To examine the effect of selenium status on SelW mRNA expression, confluent C2C12 cells at D0 were maintained in DMEM supplemented with different serum concentrations or in medium containing various levels of sodium selenite up to the indicated time point. AIM-V (AIM; Invitrogen) was also used as serum-free medium. C3H10T1/2 cells were grown in GM containing Basal Medium Eagle (JBI) supplemented with 10% FBS. For C3H10T1/2 differentiation, cells were transfected at ∼50% confluence with plasmid expressing MyoD, replaced with DM containing Basal Medium Eagle supplemented with 2% HS at ∼100% confluence, and then maintained. All culture media were supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (Invitrogen), and the cells were maintained at 37 °C under 5% CO2 atmosphere.
Plasmids
SelW promoter fragments were generated by PCR using genomic DNA extracted from rat muscle, as described previously (16), and then cloned into pGL3-basic vector (Promega) to construct SelW promoter luciferase reporter plasmids. The resulting constructs, −973SelW/Luc and −240SelW/Luc, contained SelW promoter regions from −973 and −240 to +32 bp, respectively. Using a QuikChange site-directed mutagenesis kit (Stratagene), E-box mutants of −973SelW/Luc were constructed in which the conserved E-box sequence CANNTG was converted to CGNNAG, as described previously (29). Myogenesis-dependent luciferase plasmid MCK-Luc was a kind gift from Prof. Da-Zhi Wang (University of North Carolina) and 4RSV-Luc a kind gift from Prof. Andrew Lassar (Harvard Medical School) and Prof. Makoto Inui (Yamaguchi University School of Medicine), and all pcDNA3 plasmids expressing MRFs and MEF2 family proteins were kindly provided by Prof. Young-Gyu Ko (Korea University). All primers used for the construction of plasmids are represented in Table 1. Enzyme restriction sites for the cloning of SelW promoter plasmids were KpnI for the forward primer and HindIII for the reverse primer, as underlined in the table. The sequences of all DNA constructs were confirmed by sequencing.
TABLE 1.
Oligonucleotide sequences for PCR and EMSA used in this study
F indicates forward, and R indicates reverse.
| Target gene (product size) | Primer (5′ → 3′) |
|---|---|
| Cloning for rat SelW promoter | |
| −973SelW/Luc, F | GCGCGGTACCGCCTTGCGCTTCCTAGGC |
| −240SelW/Luc, F | GCGCGGTACCGAAGGGACAGCGAGGGGC |
| +32, R | CCGCAAGCTTGCACAAAGCGAGGACCCG |
| Site-directed mutagenesis for rat SelW promoter | |
| E1m, F | GATTTTTTGTTTTGAGACGACAGCTCTTGTAGCCCAG |
| E1m, R | CTGGGCTACAAGAGCTGTCGTCTCAAAACAAAAAATC |
| E2m, F | CTTCAGTCTCCTCTTCCCGAAAGCTGGGATTAGAGGC |
| E2m, R | GCCTCTAATCCCAGCTTTCGGGAAGAGGAGACTGAAG |
| E3m, F | GGCAAGTCCTCTGCGCCGACAGCGCTACAGCCCAAGC |
| E3m, R | GCTTGGGCTGTAGCGCTGTCGGCGCAGAGGACTTGCC |
| E4m, F | GGAGGAGAGATCCATCCCGATAGTCTCCCGATGCGTG |
| E4m, R | CACGCATCGGGAGACTATCGGGATGGATCTCTCCTCC |
| Gene amplification | |
| SelW, F (232 bp) | GTGTATTGTGGAGCTTGAGGC |
| SelW, R | CCAAGGCAGCTTTGATGGCGG |
| MyoD, F (482 bp) | CATCCGCTACATCGAAGGTC |
| MyoD, R | TCGCATTGGGGTTTGAGCC |
| p21, F (295 bp) | GTCCAATCCTGGTGATGTCC |
| p21, R | CAGGGCAGAGGAAGTACTGG |
| Myogenin, F (430 bp) | AGGAGAGAAAGATGGAGTCCAGAG |
| Myogenin, R | TAACAAAAGAAGTCACCCCAAGAG |
| MyHC, F (625 bp) | AGAAGGAGGAGGCAACTTCTG |
| MyHC, R | ACATACTCATTGCCGACCTTG |
| TrxR1, F (876 bp) | TGGATTTTGTCACACCGACTCC |
| TrxR1, R | CGATGGCGTAGATGTAAGGCAC |
| Hprt1, F (225 bp) | GCAAACTTTGCTTTCCCTGG |
| Hprt1, R | GCTTTGTATTTGGCTTTTCC |
| Gapdh, F (369 bp) | CATGACAACTTTGGCATTGTG |
| Gapdh, R | GTTGAAGTCGCAGGAGACAAC |
| EMSA | |
| E1 | GTTTTGAGACAACTGCTCTTG |
| E2 | CTCTTCCCAAATGCTGGGAT |
| E3 | CTCTGCGCCAACTGCGCTACAGCC |
| E4 | GATCCATCCCAATTGTCTCCCGAT |
| MCK | CCCCCCAACACCTGCTGCCTGA |
| ChIP | |
| E1, F | CCTCGAACCCACAGAGATTTC |
| E1, R | GAAGAGGAGACTGAAGGATTG |
| Gapdh, F | AGCTACTCGCGGCTTTACG |
| Gapdh, R | TCACCTGGCACTGCACAAG |
RT-PCR and Quantitative PCR
Total RNA was isolated from C2C12 cells at the indicated time points during differentiation using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. One microgram of total RNA was reverse-transcribed to cDNA using SuperScriptaseTM III reverse transcriptase and oligo(dT) (Invitrogen) in a 50-μl total reaction volume, followed by PCR. For each exponential amplification, PCR was performed for 20–28 cycles, after which each product was visualized by ethidium bromide staining following 1.2% agarose gel electrophoresis or 2% in the case of the SelW, p21, and hypoxanthine phosphoribosyltransferase 1 (Hprt1) genes. All specific primers are shown in Table 1. Real time PCR was performed to measure the expression of SelW. One hundred nanograms of SelW cDNA and 150 ng of Hprt1 cDNA were amplified with LightCycler 480 SYBR Green I Master (Roche Diagnostics). Amplification included a denaturation step at 95 °C for 5 min, followed by 45 cycles at 95 °C for 10 s, 55 °C for 15 s, and 72 °C for 20 s. To verify amplification specificity, melting curves of the PCR products of each primer set were analyzed. The specific primers used were identical to those used in RT-PCR. Each experiment was prepared in triplicate, and data are represented as means ± S.D. of four independent experiments.
Western Blotting and Antibodies
C2C12 cells were washed twice with PBS, harvested by scraping in lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride (PMSF), 5 μg/ml aprotinin, 5 μg/ml leupeptin, 3 mm dithiothreitol (DTT), 1 mm NaF, 1 mm Na3VO4), and incubated on ice about for 30 min with periodic vortexing. After centrifugation, the protein concentration was determined by Bradford assay (Sigma) with bovine serum albumin as a standard. To detect protein expression, 30 μg of total protein was separated by 8–15% SDS-PAGE, except for SelW, in which case 60 μg of total protein was analyzed by 12% NuPAGE (Invitrogen). Protein was transferred to polyvinylidene difluoride (PVDF; Millipore) membranes, blocked for 1–2 h with 5% skim milk, and then probed overnight with primary antibodies as follows: anti-MyoD, anti-p21, and anti-myogenin from Pharmingen; anti-MyHC from Sigma; anti-phosphohistone H3 from Upstate; anti-tubulin from AbFrontier; and rabbit anti-TrxR1 kindly provided by Prof. Seung-Rock Lee (Chonnam National University Medical School). To observe SelW expression, we used rabbit polyclonal anti-SelW raised against the peptide HSKKKGDGYVDTESK (synthesized and purified by Invitrogen). HRP-conjugated secondary antibodies (anti-mouse from Calbiochem and anti-rabbit from Invitrogen) were incubated with the membranes for 1 h, followed by visualization using SuperSignal West Pico chemiluminescent substrate (Pierce).
Transfections
To estimate the activity of the SelW promoter, C2C12 cells were seeded onto a 60-mm total area at ∼30% confluence, followed by transfection with 1 μg of SelW firefly luciferase plasmids and 0.15 μg of control Renilla luciferase plasmid pRL-TK (Promega) using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's recommendations. Following 6 h of transfection, C2C12 cells were cultured in GM for 1 day and then harvested for the measurement of SelW promoter activity in proliferating myoblasts. To measure SelW promoter activity in differentiating myotubes, cells were cultured in GM up to ∼100% confluence, followed by transfer into DM for 1 day to induce differentiation. C3H10T1/2 cells were transfected with 0.3 μg of SelW luciferase plasmids, the same amount of MRFs or MEF2 family of transcription factors, as well as 0.15 μg of pRL-TK. The cells were cultured in GM and then harvested for Dual-Luciferase reporter assay. To observe further the effect of MyoD on SelW promoter activity during muscle differentiation, MyoD-transfected C3H10T1/2 cells were grown in GM up to ∼100% confluence, switched to DM, and then harvested after 1 day of culture.
Dual-Luciferase Reporter Assay
SelW firefly luciferase activity was determined using the Dual-Luciferase reporter assay system (Promega) with a Junior LB 9509 luminometer (Berthold Technologies) according to the manufacturer's instructions with minor modification. Transfected cells were washed twice with PBS and lysed with 1× passive lysis buffer (Promega). Ten microliters of cell lysates was added to 50 μl of luciferase assay reagent II, followed by measurement of firefly activity for 10 s. Fifty microliters of Stop and Glo reagent was also added to quench firefly activity, after which Renilla activity was measured for 10 s. To control transfection efficiency, firefly activity was normalized to Renilla activity. Each experiment was prepared in triplicate, and data are expressed as means ± S.D. of three to four separate experiments.
MyoD Knockdown with siRNA
MyoD siRNA duplexes against mouse MyoD, designated as siMyoD in this study, were designed and synthesized by Invitrogen (Stealth RNAi). Two target sequences for siMyoD were as follows: siMyoD 1, 5′-GACGACTTCTATGATGATCCGTGTT-3′, siMyoD 2, 5′-CCAATGCGATTTATCAGGTGCTTTG-3′. Stealth RNAi negative control duplexes (Invitrogen) served as a negative control. To determine the most effective target sequence for the two siMyoDs, C2C12 myoblasts cultured in 60-mm dishes were transiently transfected with 50 nm of each siRNA using Lipofectamine 2000 transfection reagent. After 6 h of transfection, the cells were maintained in GM up to ∼90% confluence and then transferred into DM for 1 day to induce differentiation. The expression levels of MyoD protein and mRNA were analyzed in C2C12 cells by Western blotting and RT-PCR, respectively.
EMSA
Nuclear extracts were prepared from C2C12 myotubes cultured in DM for 1 day. Briefly, C2C12 cells were washed twice with ice-cold PBS, lysed in buffer A (10 mm Hepes, pH 7.8, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 0.5 mm PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin), incubated on ice for 15 min, and then centrifuged at 5,000 rpm for 5 min. The pellet was then resuspended in buffer B (20 mm Hepes, pH 7.8, 1.5 mm MgCl2, 0.5 mm DTT, 25% glycerol, 420 mm NaCl, 0.2 mm EDTA, 0.5 mm PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin), incubated on ice for 40 min with periodic vortexing, and then centrifuged at 13,000 rpm for 15 min. The supernatants were used as nuclear extracts for DNA binding assays. Sequences of the oligonucleotide sense strands used in the EMSA are listed in Table 1 (E-box is underlined). To construct radiolabeled probes, oligonucleotides were annealed, end-labeled with [γ-32P]dATP using T4 polynucleotide kinase (Takara), and then purified with MicroSpin G-25 columns (GE Healthcare). A total of 30 μg of nuclear proteins along with 1 μg of poly(dI-dC)·poly(dI-dC) were incubated in 1× binding buffer (15 mm Hepes, pH 7.8, 1 mm EDTA, 40 mm KCl, 0.5 mm DTT, 5% glycerol) for 20 min at room temperature, followed by the addition of radiolabeled, double-stranded oligonucleotide probes. After incubation for 20 min, the samples were resolved by 5% nondenaturing PAGE with 0.5× TBE (1× TBE is 45 mm Tris borate and 1 mm EDTA) run at 200 V for 2 h at room temperature. The resulting gel was dried, and the radioactive bands were developed using a Fuji BAS 2500 phosphorimager (Fujifilm Corp.). For supershift assays, reaction mixtures were incubated with 5 μg of anti-MyoD prior to the addition of labeled probes. For competition experiments, various amounts of unlabeled oligonucleotide competitors were added (10-, 50-, and 100-fold excess) to the reaction mixtures before incubation with the labeled probes.
ChIP
For ChIP assay, C3H10T1/2 cells grown in 100-mm culture dishes were co-transfected with the expression plasmids HA-MyoD and SelW promoter −973SelW/Luc. Following transfection, the cells were maintained in GM up to ∼100% confluence and then transferred into DM for 1 day, after which ChIP assay was performed. To isolate cross-linked protein-DNA complexes, a ChIP assay kit (Upstate) was used according to the manufacturer's instructions with minor modification. Briefly, transfected C3H10T1/2 cells were cross-linked by the direct addition of 1% formaldehyde to the culture medium, followed by incubation for 10 min at 37 °C. The cross-linking reaction was stopped by the addition of 125 mm glycine. The cells were washed three times with ice-cold PBS, collected, and then resuspended in SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.0) containing protease inhibitors (1 mm PMSF, 2 μg/ml aprotinin, 2 μg/ml leupeptin). Sonication was then performed to shear 200–1000-bp DNA fragments. Precleared chromatin was immunoprecipitated using 2 μg of mouse monoclonal anti-HA antibody (Santa Cruz Biotechnology) per sample. For input DNA, 1% of the sample was removed prior to immunoprecipitation (IP). Following IP and vigorous washing, reverse cross-linking and DNA purification were performed according to the kit protocol. Immunoprecipitated fragments were analyzed by PCR, and the primers used are presented in Table 1.
RESULTS
SelW Expression Is Up-regulated during Differentiation of Myogenic C2C12 Cells into Myotubes
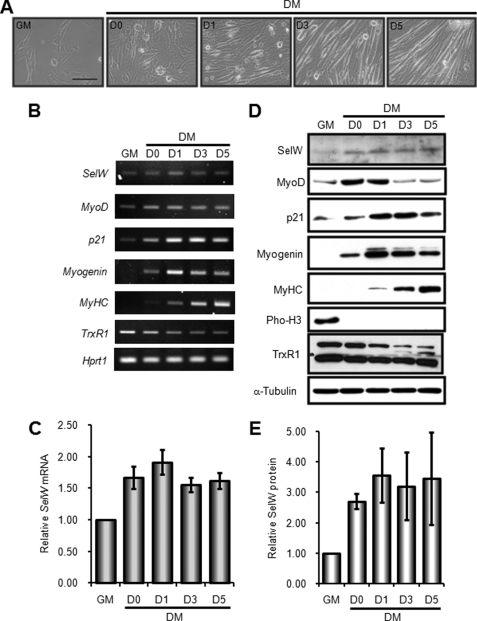
Using the mouse skeletal muscle cell line C2C12, we analyzed the expression of SelW during myogenic differentiation. The C2C12 cell line is a well established in vitro model of muscle development. Fig. 1A represents the morphological changes C2C12 cells undergo during differentiation under culture conditions. To induce differentiation, proliferating C2C12 myoblasts were grown up to ∼50% confluence in GM, after which they were maintained in the same medium up to ∼100% confluence (D0). Myogenic differentiation was then induced in confluent cells upon exchange of GM with DM (D1). Multinucleated C2C12 myotubes were observed 3 days after initial differentiation (D3), after which the cells were further cultured for 5 days to obtain full differentiation (D5). At each indicated time point, we measured the expression of SelW mRNA by RT-PCR analysis. As shown in Fig. 1B, the low level of SelW mRNA in proliferating C2C12 myoblasts increased from D0 and reached a maximum at D1. Thereafter, the level of mRNA expression was maintained with only a slight reduction. The level of SelW protein was also examined by Western blot analysis using polyclonal SelW peptide antibody. The pattern of SelW protein expression was consistent with that of mRNA expression (Fig. 1, D and E). Specificity of the polyclonal antibody was confirmed by Western blot analysis using C2C12 cells transfected with wild-type SelW plasmid and Sec mutant SelW-Cys plasmid (data not shown). Myogenic differentiation was monitored by measuring the mRNA and protein expression levels of well known muscle-specific markers such as MyoD, p21, myogenin, and MyHC. Phosphohistone H3 (Pho-H3), a cell proliferation marker, was detected in C2C12 myoblasts cultured in GM, but its expression disappeared at D0. Therefore, confluent C2C12 cells at D0 had already begun the process of cell cycle arrest and differentiation. It is known that the expression of TrxR1 mRNA decreases upon the differentiation of C2C12 myoblasts to myotubes (30). Accordingly, we observed that the mRNA and protein expression of TrxR1 slightly and gradually reduced during the differentiation process (Fig. 1, B and D). The expression of SelW mRNA was further analyzed quantitatively by real time PCR (Fig. 1C). Similar to the results obtained by RT-PCR, the expression of SelW mRNA was rapidly elevated at the early stage of differentiation, after which it was maintained at a constant level. Taken together, SelW was expressed both in proliferating C2C12 myoblasts and differentiating C2C12 myotubes, although it was specifically up-regulated at the early stage of differentiation and thereafter constantly maintained. These results allow us to speculate that the up-regulation of SelW expression in the early stage of C2C12 cell differentiation may have been due to transcriptional activation of SelW early in myogenesis.
FIGURE 1.
SelW expression is up-regulated in the early stage of C2C12 cell differentiation. A, phase-contrast microscopic images of proliferating and differentiating C2C12 cells cultured under the experimental conditions of this study. Proliferating C2C12 myoblasts were cultured in GM up to ∼50% confluence. To induce differentiation, C2C12 myoblasts were maintained in the same medium up to ∼100% confluence (D0). After exchanging the culture medium with DM, differentiating C2C12 cells were observed at 1, 3, and 5 days of differentiation (D1, D3, and D5). Bar, 100 μm. B, RT-PCR analysis of SelW and muscle-specific marker genes during C2C12 cell differentiation. Total RNA was isolated from C2C12 cells at the indicated time points and subjected to reverse transcription, followed by PCR with specific primers. Hprt1 was used as a loading control. A representative result of three independent experiments is shown. C, real time PCR analysis for the quantification of SelW mRNA expression during C2C12 cell differentiation. Using cDNA prepared in B, real time PCR was carried out with SelW- and Hprt1-specific primers. Expression level of SelW mRNA in proliferating C2C12 cells in GM was arbitrarily set at 1. Data are represented as means ± S.D. of four independent experiments performed in triplicate. D, expression of SelW and muscle-specific marker proteins during C2C12 cell differentiation. Western blotting was performed with cell lysates cultured at the indicated time points using specific antibodies and the generated polyclonal peptide antibody anti-SelW. α-Tubulin served as a loading control. The result shown is representative of three separate experiments. E, relative expression of SelW protein. The expression of SelW protein was quantified by scanning the SelW blots shown in D and was normalized based on the expression level of α-tubulin. Expression level of normalized SelW in proliferating C2C12 cells in GM was arbitrarily set at 1. Data are represented as means ± S.D. of three independent experiments.
SelW Promoter Activity Is Up-regulated upon Differentiation of C2C12 Myoblasts into Myotubes
SelW is expressed well in various tissues, particularly in the skeletal muscle of mammals. In Fig. 1, we demonstrate that the expression of both SelW mRNA and protein was up-regulated in the early stage of C2C12 cell differentiation. To determine whether or not this up-regulation was due to the transcriptional activation of SelW during myogenesis, C2C12 cells were transfected with a rat SelW firefly luciferase plasmid, −973SelW/Luc, after which SelW promoter activity was determined in proliferating cells in GM and in differentiating cells in DM after 1 day. As shown in Fig. 2, differentiating C2C12 myotubes had ∼5-fold higher activity than proliferating C2C12 myoblasts, suggesting that SelW transcription was up-regulated by the activation of muscle-specific transcription factors during myogenesis. The MCK reporter gene (MCK-Luc), which is dramatically activated during muscle cell differentiation into myotubes, was used as a positive control. Along with the expression of SelW mRNA and protein, the promoter activity of SelW was up-regulated in the early stage of C2C12 cell differentiation into myotubes.
FIGURE 2.
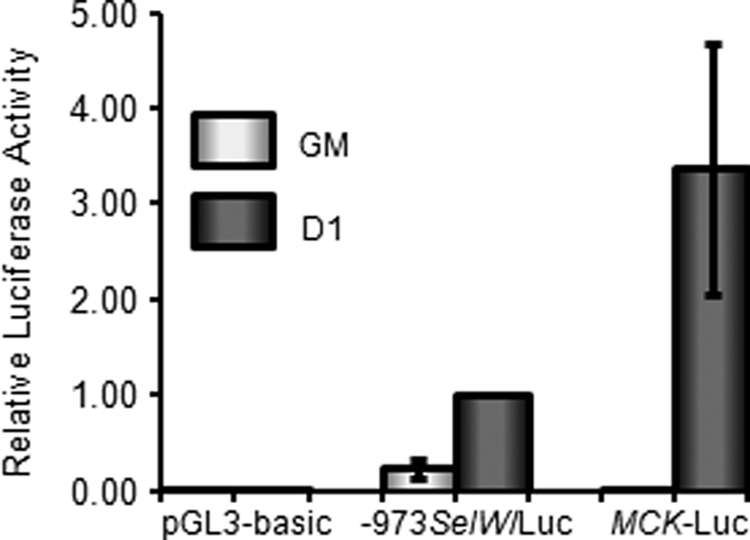
SelW promoter activity is up-regulated early in C2C12 myoblast differentiation. C2C12 myoblasts were transfected with −973SelW/Luc, MCK-Luc, or pGL3-basic and pRL-TK. Transfected cells were cultured in GM up to ∼50% confluence, followed by harvesting for SelW promoter analysis. For SelW promoter activity in DM, transfected C2C12 cells were grown up to ∼100% confluence and then transferred into DM to induce differentiation for 1 day prior to SelW luciferase analysis. Firefly luciferase activity was normalized to Renilla activity, and data are represented as means ± S.D. of three independent experiments performed in triplicate.
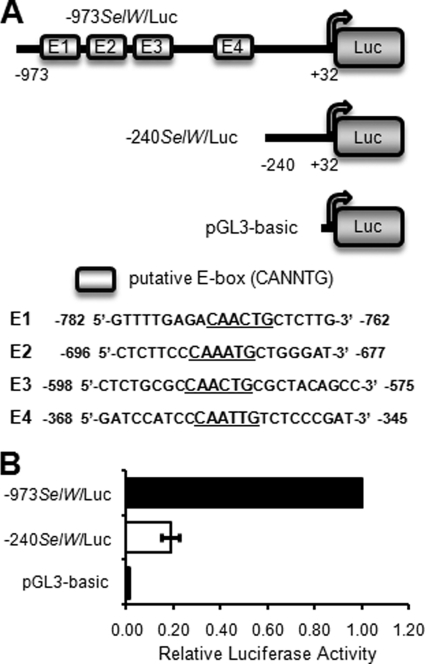
Putative E-boxes Are Associated with SelW Gene Promoter Activation in Differentiating C2C12 Cells
The transcription of many muscle-specific genes is regulated by MRFs or MEF2 factors during myogenesis. In particular, the MRF family of transcription factors activates its target genes using specific partner proteins such as E proteins (E12 and E47), which bind to conserved E-boxes located in target gene promoters or enhancers. The E-box is a representative binding element for muscle-specific transcription factors as well as non-muscle transcription factors. Sequence analysis found that there were four putative E-boxes in the SelW promoter from −973 to +32 bp, hereafter designated as E1, E2, E3, and E4 (Fig. 3A). To confirm that the presence of E-boxes in the SelW promoter is associated with promoter activation, C2C12 cells were transfected with −973SelW/Luc or −240SelW/Luc, after which SelW promoter activity was measured after 1 day of culture in DM. −240SelW/Luc is a luciferase reporter plasmid containing a SelW promoter deletion fragment without any E-boxes, as shown in Fig. 3A. As shown in Fig. 3B, the activity of −240SelW/Luc decreased by ∼80% compared with that of −973SelW/Luc, which contains all of the E-boxes. This result suggests that the putative E-boxes in the SelW promoter caused promoter activation in the early stage of differentiation.
FIGURE 3.
Putative E-boxes enhance SelW promoter activity in differentiating C2C12 cells. A, schematic illustration of SelW promoter fragments. Four putative E-boxes are present in the promoter region from −973 to −240 bp, and the reporter plasmid constructs used in the luciferase analysis are shown (upper panel). DNA sequences in each E-box are shown in the lower panel. B, SelW promoter activity in C2C12 cells using a deletion mutant after 1 day of culture in DM. C2C12 cells were transfected with SelW promoter reporter plasmids (−973SelW/Luc or −240SelW/Luc) and pRL-TK. After cultivation up to ∼100% confluence in GM, the cells were cultured in DM for 1 day. SelW gene promoter activity was measured and normalized to the Renilla activity of pRL-TK. Results are expressed as means ± S.D. of three independent experiments performed in triplicate.
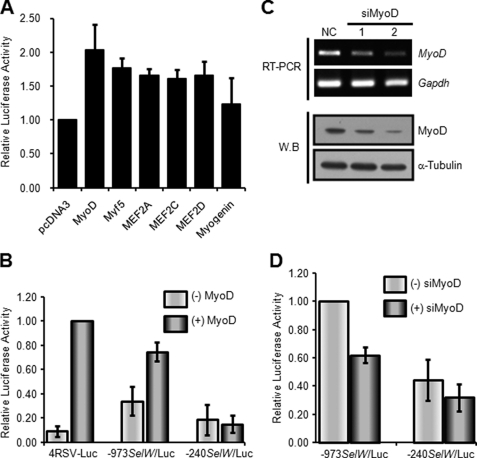
MyoD Regulates Promoter Activation of SelW in the Early Stage of C2C12 Cell Differentiation
To investigate whether or not the E-boxes present in the SelW gene promoter associate with muscle-specific transcription factors, we used the non-muscle cell line C3H10T1/2, which is an embryonic mouse fibroblast cell line that is converted to skeletal myoblasts by MRFs such as MyoD (31). C3H10T1/2 cells were transiently co-transfected with −973SelW/Luc and a plasmid expressing one of three MRF family members (MyoD, Myf5, and myogenin) or one of three MEF2 family members (MEF2A, MEF2C, and MEF2D) (Fig. 4A). −973SelW/Luc promoter activity was increased by the co-expression of MRFs or MEF2s, except myogenin. Among them, the highest luciferase activity was an ∼2.5-fold increase observed when the cells were co-expressed with MyoD. However, the activity of C3H10T1/2 cells transfected with −240SelW/Luc was not affected as shown in Fig. 4B. In the experiment, 4RSV-Luc was used as a positive control. These results suggest that MyoD was a transcriptional activator for the SelW promoter. To further confirm the results, C2C12 cells were transiently transfected with siRNA against MyoD (siMyoD). We designed two siMyoDs (siMyoD 1 and siMyoD 2) and tested their knockdown efficiencies. As shown in Fig. 4C, the mRNA and protein levels of endogenous MyoD were reduced remarkably in C2C12 cells by siMyoD 2 transfection. Of the two siMyoDs examined, we therefore selected siMyoD 2 for our next experiments. To examine the effect of endogenous MyoD on SelW promoter activity in myogenic C2C12 cells, we measured −973SelW/Luc activity or −240SelW/Luc activity in differentiating C2C12 cells after knockdown of MyoD expression with siMyoD. As shown in Fig. 4D, siMyoD decreased −973SelW/Luc activity but had no significant effect on −240SelW/Luc activity. These results suggest that MyoD was involved in the regulation of SelW promoter activity.
FIGURE 4.
MyoD enhances SelW gene promoter activity. A, activity of the SelW promoter by MRFs and MEF2 factors. Nonmyogenic C3H10T1/2 cells were co-transfected with −973SelW/Luc and pRL-TK, together with an expression plasmid containing a MRF (MyoD, Myf5, and myogenin) or MEF2 (MEF2A, MEF2C, and MEF2D) factor. The cells were cultured in GM and harvested for Dual-Luciferase reporter analysis. Results are expressed as means ± S.D. of four independent experiments in triplicate. B, effect of exogenous MyoD on SelW gene promoter activity. SelW promoter plasmids and 4RSV-Luc plasmid were co-transfected with or without an expression plasmid containing MyoD into C3H10T1/2 cells. The cells were cultured in GM up to ∼100% confluence, maintained, and harvested after 1 day of culture in DM. Results are expressed as means ± S.D. of four independent experiments performed in triplicate. C, knockdown of MyoD expression by siRNA in C2C12 cells. C2C12 myoblasts were separately transfected with two distinct siRNAs specific for MyoD (siMyoD 1 and 2) along with a negative control (NC). The cells were maintained in GM up to ∼90% confluence, followed by cultivation in DM for 1 day. MyoD expression was monitored by RT-PCR and Western blot analysis. The Gapdh gene and α-tubulin protein were used as loading controls for each analysis. D, effect of MyoD depletion on SelW promoter activity in differentiating C2C12 cells. C2C12 cells transfected with siMyoD 2 were analyzed for SelW gene promoter activity after 1 day of culture in DM. Data are represented as means ± S.D. of four independent experiments performed in triplicate.
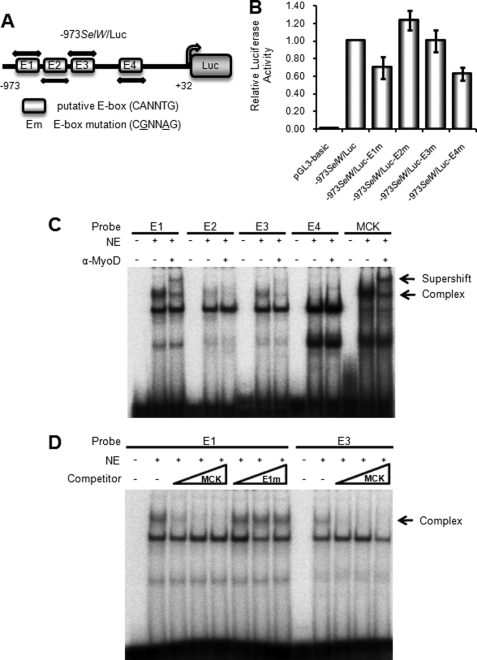
MyoD Binds to a Putative SelW Promoter E-box
As described above, we found that the SelW gene promoter was activated by the myogenic basic helix-loop-helix protein MyoD in the early stage of C2C12 cell differentiation. This activation may be due to the binding of MyoD to putative E-boxes located in the SelW promoter region from −973 to −240 bp. To determine the biological role of each E-box in the SelW gene promoter, the E-box consensus sequence CANNTG was mutated to CGNNAG using −973SelW/Luc (Fig. 5A). The resulting reporter plasmids were designated as −973SelW/Luc-E1m, −973SelW/Luc-E2m, −973SelW/Luc-E3m, and −973SelW/Luc-E4m and were introduced into C2C12 cells. Luciferase activity was measured after 1 day of culture in DM (Fig. 5B). The activities of −973SelW/Luc-E1m and −973SelW/Luc-E4m were reduced by ∼60% compared with wild-type −973SelW/Luc, whereas those of −973SelW/Luc-E2m and −973SelW/Luc-E3m were slightly increased or did not change. Therefore, these data show that each E-box differentially contributed to SelW promoter activity. Nevertheless, the E1 and E4 E-boxes might have played important roles in MyoD-dependent SelW promoter activation during C2C12 cell differentiation. Next, EMSA was performed with [γ- 32P]dATP-labeled oligonucleotide probes for each E-box to confirm MyoD binding during C2C12 cell differentiation. Of the four labeled E-box probes, E1 probe formed the strongest complex with nuclear extracts of differentiating C2C12 cells (Fig. 5C). Furthermore, this complex supershifted in the presence of anti-MyoD antibody, indicating that MyoD bound directly to the E1 E-box of the SelW promoter. The E3 probe also showed complex formation with the nuclear extracts, but to a lesser extent, and this complex also supershifted in response to anti-MyoD antibody. Labeled MCK probe was used as a positive control. The E1 and E3 E-boxes were further examined using specific unlabeled competitors (Fig. 5D). The amount of E1-MyoD complex was reduced in a dose-dependent manner by unlabeled MCK probe, but complex formation was not inhibited by unlabeled E1 mutant probe. Weak formation of the E3-MyoD complex was also reduced dose-dependently upon the addition of unlabeled MCK probe. The in vivo interaction of MyoD with the E1 E-box was confirmed by ChIP analysis (Fig. 6) after the expression of HA-tagged MyoD plasmid was analyzed by Western blotting (Fig. 6A). For ChIP, C3H10T1/2 cells were co-transfected with HA-tagged MyoD and SelW promoter −973SelW/Luc. After 1 day of culture in DM, the protein-DNA complexes were cross-linked, immunoprecipitated, and amplified. Meanwhile, anti-HA immunoprecipitated complexes were confirmed by immunoblotting with anti-MyoD antibody (Fig. 6B). As shown in Fig. 6C, anti-HA immunoprecipitated DNA corresponded to the E1 E-box of the SelW promoter but not the Gapdh promoter. Taken together, our study shows that MyoD directly bound to the E1 E-box of the SelW promoter. MyoD bound very weakly to E3 and thus had a lesser effect on SelW promoter activity. Unexpectedly, E4 did not bind to MyoD even though its mutation reduced SelW promoter activity as much as the E1 mutation (Fig. 5B).
FIGURE 5.
MyoD binds to E-boxes of the SelW promoter in differentiating C2C12 cells. A, schematic representation of SelW promoter −973SelW/Luc harboring four putative E-boxes. B, SelW promoter activity of −973SelW/Luc and E-box mutant plasmids in differentiating C2C12 cells. C2C12 cells were transfected with SelW promoter −973SelW/Luc or E-box mutants and pRL-TK. Luciferase activity was measured by harvesting of transfected C2C12 cells after 1 day of culture in DM. Data are expressed as means ± S.D. of three or four independent experiments in triplicate. C and D, EMSA of the putative E-boxes in the SelW promoter. [γ- 32P]dATP-labeled oligonucleotide probes for the consensus E-boxes were incubated with nuclear extracts (NE) of differentiating C2C12 cells in DM for 1 day. For supershift analysis (C), anti-MyoD antibody was incubated with nuclear extracts prior to addition of labeled probes. For competition assays of the E1 and E3 E-boxes (D), unlabeled competitors were added (10-, 50-, and 100-fold excess) in a dose-dependent manner to nuclear extracts prior to incubation with labeled probes. The unlabeled competitor MCK was the same oligonucleotide used in C, and E1m was the oligonucleotide used for the mutagenesis of E1.
FIGURE 6.
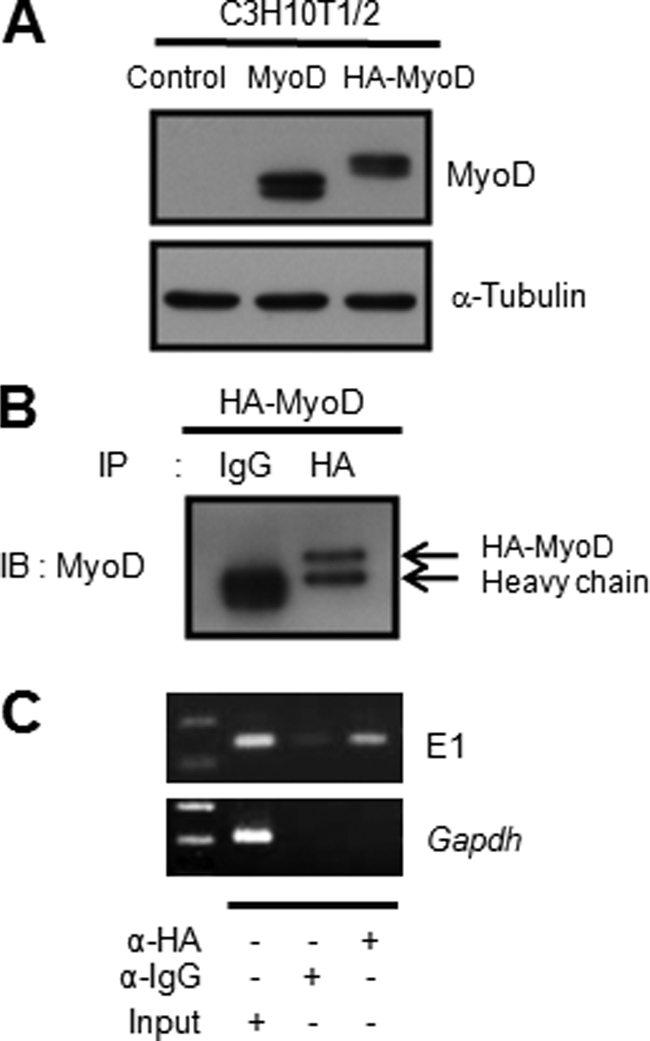
MyoD binds to an E-box of the SelW promoter in vivo. A, expression of HA-tagged MyoD plasmid. Prior to ChIP, HA-tagged MyoD expression plasmid was confirmed by Western blotting with anti-MyoD antibody after transfection into C3H10T1/2 cells. B, confirmation of immunoprecipitated (IP) HA-MyoD complexes by ChIP. For the ChIP assay, C3H10T1/2 cells were co-transfected with HA-MyoD and −973SelW/Luc, cultured in GM and then transferred into DM. After 1 day of culture in DM, the cells were subjected to ChIP analysis. After immunoprecipitation with anti-HA, the immunoprecipitated HA-MyoD complexes were confirmed by immunoblotting with anti-MyoD antibody. C, MyoD binding to the E1 E-box of the SelW promoter by ChIP. The immunoprecipitated complexes were amplified by PCR with specific primers. Gapdh promoter was used as a negative control for specificity.
Selenium Is Not Involved in SelW Promoter Activation during Myogenic C2C12 Differentiation
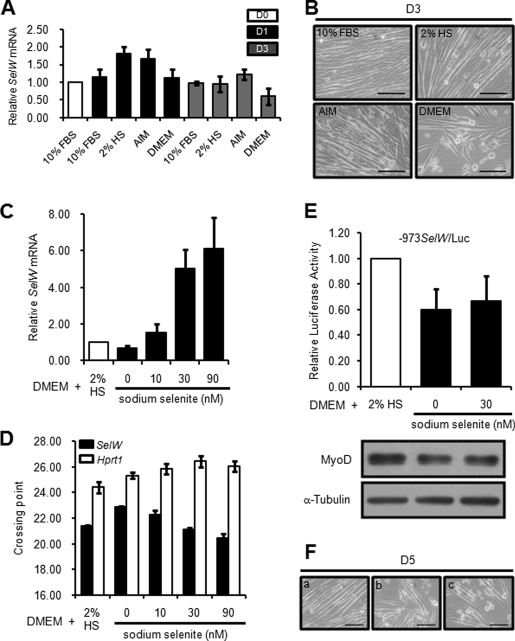
SelW expression did not prominently decrease after exchange of GM containing 10% FBS with DM supplemented with 2% HS, as shown in Fig. 1. It was also reported that 2% fetal calf serum is adequate for SelW mRNA expression in L8 rat myoblasts (32). Selenium is known as one of the most important factors regulating selenoprotein expression. In this study, we induced the differentiation of C2C12 cells by culturing in GM or DM containing different concentrations of serum as well as in serum-free DMEM to examine the effect of selenium on SelW expression during differentiation. AIM, which is a type of serum-free medium for C2C12 differentiation (33, 34) containing an unknown component, was also used. As shown in Fig. 7A, SelW mRNA expression in C2C12 cells cultured in DMEM was lower than that in cells cultured in medium containing 2% HS or AIM at D1. Interestingly, the expression level of SelW mRNA in C2C12 cells grown in DMEM was comparable with that grown in GM with 10% FBS. However, the expression level of SelW mRNA was relatively lower in C2C12 cells cultured in DMEM without serum at D3. Furthermore, because the number of adherent C2C12 cells cultured in DMEM was remarkably reduced compared with cells cultured in other media (Fig. 7B), further experiments were carried at the time point D1. Using real time PCR, it was found that SelW mRNA expression was up-regulated by increased selenium concentration in DMEM at D1 (Fig. 7C). Fig. 7D shows the mean values of PCR crossing points for mRNA expression of SelW and Hprt1. The value of SelW decreased by increasing the selenium concentration, whereas the value of Hprt1 increased. Thus, the relative mRNA level of SelW further increased at higher selenium content. The mRNA of Hprt1 was measured for normalization purposes. To determine how the SelW mRNA level was increased by selenium, SelW promoter activity of −973SelW/Luc was analyzed. According to a recent report (35), culture medium containing ∼10% FBS has ∼29 nm selenium. Thus, we maintained C2C12 cells transfected with −973SelW/Luc in DMEM without or with 30 nm sodium selenite. As shown in Fig. 7E, SelW promoter activity of C2C12 cells cultured in DMEM alone was lower than that of cells cultured with 2% HS. Furthermore, the addition of selenium to DMEM had no effect on the promoter activity of differentiating C2C12 cells. MyoD expression also decreased in cells cultured in DMEM without 2% HS (Fig. 7E, bottom). Thus, SelW promoter activity was dependent on MyoD expression as described above. Therefore, increased SelW mRNA expression at D1 by addition of selenium to DMEM, as shown in Fig. 7C, might have been due to increased SelW mRNA stabilization, as reported previously (32), and not transcriptional regulation. In this study, expression of SelW mRNA during differentiation was determined in the presence of 2% HS. When the cells were cultured in DMEM without 2% HS, cell attachment and the extent of myotube formation were abnormally low, even though selenium was added as shown in Fig. 7F. The differentiation conditions used are not physiological as they are artificially selenium-deficient, and thus the regulation observed may only be relevant under conditions of selenium starvation.
FIGURE 7.
SelW promoter is not activated by selenium in differentiating C2C12 cells. A, real time PCR analysis of SelW gene expression in different culture media after induction of C2C12 differentiation. Expression level of SelW mRNA in D0 was arbitrarily set at 1. Data are represented as means ± S.D. of three independent experiments performed in triplicate. B, phase-contrast microscopic images of differentiating C2C12 cells cultured under the experimental conditions at D3 shown in A. Bar, 100 μm. C, relative expression of SelW mRNA in culture media supplemented with different levels of sodium selenite. The data are represented as means ± S.D. of three independent experiments performed in triplicate. D, real time PCR crossing point values of SelW and Hprt1 mRNAs shown in C. The values are expressed as means ± S.D. E, SelW promoter activity and MyoD expression in differentiating C2C12 cells in different culture media. C2C12 cells were transfected with −973SelW/Luc and pRL-TK, maintained in GM up to ∼100% confluence, followed by cultivation in culture media without or with selenium for 1 day. SelW gene promoter activity was measured and normalized to the Renilla activity of pRL-TK. MyoD expression was analyzed by Western blotting. F, phase-contrast microscopic images of differentiating C2C12 cells for 5 days under the same medium conditions shown in E. Panel a, DMEM with 2% HS; panel b, DMEM; panel c, DMEM with 30 nm sodium selenite. Bar, 100 μm.
DISCUSSION
For decades, selenoproteins, which contain selenium in their protein sequences, have been studied for their beneficial biological effects. SelW is a selenoprotein containing the thioredoxin reductase-like motif CXXU and is mainly understood in terms of its antioxidant effects. In this study, we investigated its high tissue-specific expression in skeletal muscle. Tissue-specific expression of a protein implies regulation by a tissue-specific factor as well as a significant role for the protein. Thus, we hypothesized that high SelW expression in skeletal muscle may be regulated by a muscle-specific factor.
To analyze SelW expression in mouse skeletal muscle C2C12 cells during differentiation, we performed RT-PCR and real time PCR with a specific primer set for SelW, along with Western blot analysis with SelW peptide antibody. We found that SelW expression was up-regulated during C2C12 myogenesis, particularly in the early stage of differentiation (Fig. 1). According to a previous report by Loflin et al. (2), SelW is involved in muscle growth and proliferation based on its strong expression in proliferating C2C12 myoblasts but not differentiating C2C12 myotubes. However, they only examined SelW expression by Northern blot analysis and therefore did not investigate any muscle-specific marker proteins expressed during C2C12 myogenesis for confirmation of the differentiation conditions. Therefore, we considered the optimal cell density for the induction of C2C12 cell differentiation. It was important that C2C12 cells be maintained at a low density so that their myoblast properties are not lost under proliferation conditions. This is because confluent C2C12 cells slowly differentiate even if grown in GM supplemented with serum, which affects protein expression. For example, the transcription factor c-Myb shows differential expression depending on the density of C2C12 cells during differentiation (36). Therefore, at the time point mentioned in the previous report, C2C12 cells might have already begun to differentiate even in GM-containing serum.
There are two main pieces of evidence supporting the up-regulation of SelW expression in differentiating C2C12 cells. One is our previous report that found SelW mRNA is not expressed in back muscle during rat embryogenesis (10). However, SelW expression could not be ruled out in the skeletal muscle of neonatal or adult animals (9–10). The other was observed by in situ hybridization during zebrafish embryogenesis (37). There are three SelW genes in zebrafish called SePW1, SePW2a, and SePW2b. Interestingly, none of these SelW genes are expressed in the muscle of zebrafish embryos.
We also examined SelW promoter activity during myogenesis using luciferase reporter analysis. Consistent with the expression levels of SelW mRNA and protein, the SelW promoter activity of −973SelW/Luc was markedly increased in the early stage of differentiation (Fig. 2). This activity was associated with the presence of four putative E-boxes located in the SelW promoter (Fig. 3). Many muscle-specific proteins are transcriptionally regulated by the binding of MRFs to E-boxes within their promoters or enhancers during muscle differentiation. The E-box is one of the most understood motifs in the promoters and enhancers of many genes, not just in those related to muscle. According to a review by Berkes and Tapscott (22), an E-box occurs about once every 256 bases throughout the whole genome. Thus, the E-box is physiologically important to the recognition of distinct cis-acting elements by transcription factors such as MRFs. In this study, the SelW promoter was significantly activated by MyoD during C2C12 differentiation. Using point mutagenesis, we found that SelW promoter activation was associated with two of its four E-boxes, namely E1 and E4 (Fig. 5B). However, this does not rule out the participation of other factors. In addition, EMSA revealed that E1 directly interacted with MyoD (Fig. 5, C and D), as did E3, but to a much lesser extent. Noticeably, E4 did not interact directly with MyoD, although its mutation resulted in reduced activity similar to that of E1 mutant in differentiating C2C12 cells, as shown in Fig. 5. It is known that in vivo transcriptional activation and in vitro binding affinities for E-boxes by MyoD do not always correlate (38). Therefore, to test the biological significance of the E4 E-box, we constructed deletion mutants of the SelW promoter containing only E4 or an E4 mutant, −585SelW/Luc and −585SelW/Luc-E4m, respectively (data not shown). We found that −585SelW/Luc activity was increased about ∼2.5-fold by MyoD compared with control pcDNA3. However, the mutant −585SelW/Luc-E4m was not affected by MyoD, indicating that E4 might be activated indirectly via a MyoD-dependent downstream factor.
Selenium is an important factor regulating the expression of the selenoproteome. It has been reported that the expression level of SelW mRNA is increased in selenium-supplemented medium due to SelW mRNA stabilization but not its transcriptional rate (32). In this study, we show that transcriptional regulation of the SelW gene was not affected by addition of selenium into serum-free DMEM but was affected by the expression of the myogenic factor MyoD in differentiating C2C12 cells (Fig. 7). As described above, selenium is an important factor for the stability of SelW mRNA. Myogenic differentiation of C2C12 cells was induced by culturing in DMEM containing 2% HS, namely DM in this study. As C2C12 cells were cultured without 2% HS, impaired differentiation was observed, even though the cells were cultured with additional selenium (Fig. 7F). Thus, the effect of selenium on stability of SelW mRNA during myogenic differentiation could not be determined in this study. C2C12 cells are capable of differentiating in serum-free media. Choosing an ideal serum-free medium should be considered to preserve the important characteristics of muscle during differentiation. In addition, to precisely study the effect of selenium in differentiating cells, moderate concentrations of selenium are recommended.
In addition to SelW, TrxR1 and SelN are also expressed in skeletal muscle. In contrast to SelW, their expression is down-regulated in C2C12 myotubes, as was shown in this study (SelN data not shown) (30, 39). Indeed, there was a study on SelN expression in mouse and human as well as in cells (39, 40). SelN shows a dynamic expression pattern in skeletal muscle during development, decreasing in amount from the fetus to adult. Based on this developmental expression pattern, it is suggested that SelN may have a role in maturation. The expression pattern of SelW in this study, which displays increased expression upon differentiation and constant expression thereafter, suggests that SelW probably participates in the maintenance of muscle differentiation.
In a recent review on transcriptional regulation of mammalian selenoprotein expression, putative binding sites for transcription factors that control the transcription of SelW were predicted in the promoters of human selenoprotein genes, with MyoD one of the notable candidates (41). In this same manner, we suggest that MyoD was a transcriptional activator of SelW expression in skeletal muscle cells. This is the first evidence for the transcriptional regulation of SelW expression by a tissue-specific factor.
Acknowledgments
We thank Soon-Young Jung and Yoonhee Bae (Korea University, Seoul, Korea) for technical support.
This work was authored, in whole or in part, by National Institutes of Health staff. This work was funded by Korea Research Foundation Grant from the Korean Government (MOEHRD) KRF-2005-070-C00086 (to I. Y. K.) and in part by a Korea University grant.
- Sec
- selenocysteine
- MRE
- metal response element
- MRFs
- myogenic regulatory factors
- MCK
- muscle creatine kinase
- DM
- differentiation medium
- GM
- growth medium
- HS
- horse serum.
REFERENCES
- 1.Jeong D., Kim T. S., Chung Y. W., Lee B. J., Kim I. Y. (2002) FEBS Lett. 517, 225–228 [DOI] [PubMed] [Google Scholar]
- 2.Loflin J., Lopez N., Whanger P. D., Kioussi C. (2006) J. Inorg. Biochem. 100, 1679–1684 [DOI] [PubMed] [Google Scholar]
- 3.Chung Y. W., Jeong D., Noh O. J., Park Y. H., Kang S. I., Lee M. G., Lee T. H., Yim M. B., Kim I. Y. (2009) Mol. Cells 27, 609–613 [DOI] [PubMed] [Google Scholar]
- 4.Flohé L. (2009) Biochim. Biophys. Acta 1790, 1389–1403 [DOI] [PubMed] [Google Scholar]
- 5.Dikiy A., Novoselov S. V., Fomenko D. E., Sengupta A., Carlson B. A., Cerny R. L., Ginalski K., Grishin N. V., Hatfield D. L., Gladyshev V. N. (2007) Biochemistry 46, 6871–6882 [DOI] [PubMed] [Google Scholar]
- 6.Aachmann F. L., Fomenko D. E., Soragni A., Gladyshev V. N., Dikiy A. (2007) J. Biol. Chem. 282, 37036–37044 [DOI] [PubMed] [Google Scholar]
- 7.Sengupta A., Carlson B. A., Labunskyy V. M., Gladyshev V. N., Hatfield D. L. (2009) Biochem. Cell Biol. 87, 953–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whanger P. D. (2000) Cell. Mol. Life Sci. 57, 1846–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh J. Y., Beilstein M. A., Andrews J. S., Whanger P. D. (1995) FASEB J. 9, 392–396 [DOI] [PubMed] [Google Scholar]
- 10.Jeong D. W., Kim E. H., Kim T. S., Chung Y. W., Kim H., Kim I. Y. (2004) Mol. Cells 17, 156–159 [PubMed] [Google Scholar]
- 11.Sakurai A., Nishimoto M., Himeno S., Imura N., Tsujimoto M., Kunimoto M., Hara S. (2005) J. Cell. Physiol. 203, 529–537 [DOI] [PubMed] [Google Scholar]
- 12.Walter P. L., Steinbrenner H., Barthel A., Klotz L. O. (2008) Biochem. Biophys. Res. Commun. 365, 316–321 [DOI] [PubMed] [Google Scholar]
- 13.Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigó R., Gladyshev V. N. (2003) Science 300, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 14.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 15.Gao Y., Hannan N. R., Wanyonyi S., Konstantopolous N., Pagnon J., Feng H. C., Jowett J. B., Kim K. H., Walder K., Collier G. R. (2006) Cytokine 33, 246–251 [DOI] [PubMed] [Google Scholar]
- 16.Amantana A., Vorachek W. R., Butler J. A., Costa N. D., Whanger P. D. (2002) J. Inorg. Biochem. 91, 356–362 [DOI] [PubMed] [Google Scholar]
- 17.Amantana A., Vorachek W. R., Butler J. A., Ream W., Whanger P. D. (2004) J. Inorg. Biochem. 98, 1513–1520 [DOI] [PubMed] [Google Scholar]
- 18.Wimmer U., Wang Y., Georgiev O., Schaffner W. (2005) Nucleic Acids Res. 33, 5715–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoytcheva Z. R., Vladimirov V., Douet V., Stoychev I., Berry M. J. (2010) Biochim. Biophys. Acta 1800, 416–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tajbakhsh S. (2003) Curr. Opin. Genet. Dev. 13, 413–422 [DOI] [PubMed] [Google Scholar]
- 21.Shi X., Garry D. J. (2006) Genes Dev. 20, 1692–1708 [DOI] [PubMed] [Google Scholar]
- 22.Berkes C. A., Tapscott S. J. (2005) Semin. Cell Dev. Biol. 16, 585–595 [DOI] [PubMed] [Google Scholar]
- 23.Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. (1989) Cell 58, 823–831 [DOI] [PubMed] [Google Scholar]
- 24.Tapscott S. J. (2005) Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 25.Asakura A., Fujisawa-Sehara A., Komiya T., Nabeshima Y., Nabeshima Y. (1993) Mol. Cell. Biol. 13, 7153–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thayer M. J., Tapscott S. J., Davis R. L., Wright W. E., Lassar A. B., Weintraub H. (1989) Cell 58, 241–248 [DOI] [PubMed] [Google Scholar]
- 27.Blau H. M., Chiu C. P., Webster C. (1983) Cell 32, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 28.Blau H. M., Pavlath G. K., Hardeman E. C., Chiu C. P., Silberstein L., Webster S. G., Miller S. C., Webster C. (1985) Science 230, 758–766 [DOI] [PubMed] [Google Scholar]
- 29.Parker M. H., Perry R. L., Fauteux M. C., Berkes C. A., Rudnicki M. A. (2006) Mol. Cell. Biol. 26, 5771–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catani M. V., Savini I., Duranti G., Caporossi D., Ceci R., Sabatini S., Avigliano L. (2004) Free Radic. Biol. Med. 37, 1024–1036 [DOI] [PubMed] [Google Scholar]
- 31.Davis R. L., Weintraub H., Lassar A. B. (1987) Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 32.Gu Q. P., Ream W., Whanger P. D. (2002) Biometals 15, 411–420 [DOI] [PubMed] [Google Scholar]
- 33.Lawson M. A., Purslow P. P. (2000) Cells Tissues Organs 167, 130–137 [DOI] [PubMed] [Google Scholar]
- 34.Fujita H., Endo A., Shimizu K., Nagamori E. (2010) Biotechnol. Bioeng. 107, 894–901 [DOI] [PubMed] [Google Scholar]
- 35.Budiman M. E., Bubenik J. L., Miniard A. C., Middleton L. M., Gerber C. A., Cash A., Driscoll D. M. (2009) Mol. Cell 35, 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaspar P., Pajer P., Sedlak D., Tamaoki T., Dvorak M. (2005) Exp. Cell Res. 309, 419–428 [DOI] [PubMed] [Google Scholar]
- 37.Thisse C., Degrave A., Kryukov G. V., Gladyshev V. N., Obrecht-Pflumio S., Krol A., Thisse B., Lescure A. (2003) Gene Expr. Patterns 3, 525–532 [DOI] [PubMed] [Google Scholar]
- 38.Huang J., Blackwell T. K., Kedes L., Weintraub H. (1996) Mol. Cell. Biol. 16, 3893–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castets P., Maugenre S., Gartioux C., Rederstorff M., Krol A., Lescure A., Tajbakhsh S., Allamand V., Guicheney P. (2009) BMC Dev. Biol. 9, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petit N., Lescure A., Rederstorff M., Krol A., Moghadaszadeh B., Wewer U. M., Guicheney P. (2003) Hum. Mol. Genet. 12, 1045–1053 [DOI] [PubMed] [Google Scholar]
- 41.Stoytcheva Z. R., Berry M. J. (2009) Biochim. Biophys. Acta 1790, 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]