Abstract
The mechanism underlying the interaction of the Escherichia coli signal recognition particle receptor FtsY with the cytoplasmic membrane has been studied in detail. Recently, we proposed that FtsY requires functional interaction with inner membrane lipids at a late stage of the signal recognition particle pathway. In addition, an essential lipid-binding α-helix was identified in FtsY of various origins. Theoretical considerations and in vitro studies have suggested that it interacts with acidic lipids, but this notion is not yet fully supported by in vivo experimental evidence. Here, we present an unbiased genetic clue, obtained by serendipity, supporting the involvement of acidic lipids. Utilizing a dominant negative mutant of FtsY (termed NG), which is defective in its functional interaction with lipids, we screened for E. coli genes that suppress the negative dominant phenotype. In addition to several unrelated phenotype-suppressor genes, we identified pgsA, which encodes the enzyme phosphatidylglycerophosphate synthase (PgsA). PgsA is an integral membrane protein that catalyzes the committed step to acidic phospholipid synthesis, and we show that its overexpression increases the contents of cardiolipin and phosphatidylglycerol. Remarkably, expression of PgsA also stabilizes NG and restores its biological function. Collectively, our results strongly support the notion that FtsY functionally interacts with acidic lipids.
Keywords: Membrane Lipids, Membrane Proteins, Phosphatidylglycerol, Protein Targeting, Protein Translocation, Acidic Lipids, ffh, ftsy, Signal Recognition Particle, SRP
Introduction
Membrane-bound ribosomes are responsible for the biosynthesis of many integral membrane proteins that insert into the membrane in a co-translational manner (1, 2). Targeting of ribosomes to the cytoplasmic membrane in Escherichia coli requires the signal recognition particle (SRP)3 receptor, FtsY (3).
Many reports have suggested that FtsY functions as a membrane-bound receptor (4, 5), despite the fact that it has no known membrane anchor partner homologous to the mammalian β-subunit of the SRP receptor (SR-β). In agreement with this, previous studies showed that FtsY interacts with membrane-bound ribosomes (6) and the translocon (7) and that its membrane localization is required for its function (4, 5, 8). In addition to convincing evidence for FtsY-membrane protein interaction(s), other studies demonstrated unequivocally that the receptor also interacts with lipids (9, 10) and more specifically acidic lipids (11, 12). However, the identified interactions probably do not dictate docking of the receptor (13). In direct support of a functional interaction between FtsY and lipids, our recent studies revealed that the receptor contains a short and conserved lipid-binding amphipathic α-helix at the N-terminal edge of the N domain, which affects the enzymatic behavior of FtsY upon interaction with lipids (13, 14), and recent studies further confirmed our conclusions (15). Modeling of the lipid-binding helix of FtsY and its similarity to the MinD membrane targeting sequence (14, 16) suggest that the short helix interacts with acidic lipids, as proposed previously (11). Through the following in vivo studies, we obtained genetic evidence that lends support to this notion.
EXPERIMENTAL PROCEDURES
Materials
Antibodies to FtsY were described previously (6). India HisProbe-horseradish peroxidase was purchased from Pierce. Arabinose and isopropyl β-thiogalactoside (IPTG) were purchased from Sigma. n-Dodecyl β-d-maltopyranoside was from Anatrace. Nickel-nitrilotriacetic acid beads were from Qiagen.
Strains and Plasmids
E. coli Top10 (Invitrogen) was used for the propagation and preparation of various plasmid constructs as well as screening for genes suppressing the NG effect. E. coli K12 BW25113 (Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514) and Top10 were used for in vivo suppression assays. For complementation assays, we used E. coli IY28 (BW25113-Kan-araCFUP-FtsY), in which the endogenous promoter of FtsY was replaced by the arabinose promoter.4 E. coli MG1655 was used for the construction of genomic library. The PgsA mutant T60P and insertion of a 6-histidine (His6) tag into PgsA were constructed by PCR.
Construction of a Chromosomal Library
Genomic DNA of E. coli MG1655 was partially digested with Sau3AI. Following size selection (2–4 kb), the digested DNA was ligated into BamHI-digested plasmids pCV3 or pT7-5.
Screening Strategy
E. coli Top10 harboring either pT7-5-tacP-NG or pCV3-araP-NG was transformed with the genomic library and plated on LB plates containing ampicillin (200 μg/ml) and kanamycin (30 μg/ml), in the absence (as a control) or presence of a lethal concentration of the NG inducer (0.1 mm IPTG or 0.5% arabinose). Viable colonies from plates with the NG inducer were further analyzed.
Growth Experiments, NG Toxicity, and FtsY Complementation Studies
For growth experiments with NG and FtsY, cells were co-transformed with the compatible plasmids pCV3-araP-NG and pT7-5-tacP-FtsY. For growth experiments with NG and PgsA and for purification of NG, cells were co-transformed with pT75-tacP-NG and either pCV3, pCV3-pgsA (+tRNAs, see under “Results”), or pCV3-pgsA(T60P)(+tRNAs). For growth experiments with NG and PgsA cloned under the regulation of the lac promoter (lacP His6-pgsA) (Aska clone JW1897) (17), cells were co-transformed with pCV3-araP-NG and either pCA24N or pCA24N-His6-pgsA. Cultures were grown overnight at 37 °C in LB medium and supplemented with either ampicillin (200 μg/ml) or chloramphenicol (30 μg/ml) and kanamycin (30 μg/ml). Cells were diluted to A600 0.015 induced at A600 0.03 with either 0.2–0.5 mm IPTG or 1% arabinose. Cultures were then grown for 5 or 2 h for protein expression or purification studies, respectively. For FtsY complementation experiments in broth, cells harboring pT7-5-tacP-NG and either pCV3 or pCV3-pgsA(+tRNAs) were grown overnight at 37 °C in LB, supplemented with ampicillin (200 μg/ml), kanamycin (30 μg/ml), and arabinose (0.2%), the inducer of the chromosomal ftsY. Cells were washed four times and diluted in LB broth to A600 0.0005 without arabinose, with or without IPTG (10 μm) and grown for 10 h.
Cell Fractionation
Harvested cultures were washed in 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA and resuspended in the same buffer. The cell suspensions were sonicated, and cell debris was removed by centrifugation (10 min at 10,000 × g). Membranes were collected by ultracentrifugation (45 min at 150,000 × g) and resuspended in the same buffer. The supernatant was also collected and used for analysis of soluble proteins.
Protein Purification from Membrane and Cytosolic Fractions
Cells were resuspended in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl (buffer S), supplemented with 8% sucrose and 1 mm phenylmethylsulfonyl fluoride, and sonicated. Cell debris was removed by centrifugation (10 min at 10,000 × g). Membranes were isolated by ultracentrifugation (45 min at 150,000 × g). Membranes were solubilized by 1% n-dodecyl β-d-maltopyranoside in buffer S, and insoluble materials were removed by ultracentrifugation. For the purification of membrane-bound NG, 0.05% n-dodecyl β-d-maltopyranoside was added to all the buffers. Cytosolic proteins and solubilized membrane proteins were incubated with pre-equilibrated nickel-nitrilotriacetic acid beads in buffer S supplemented with 5 mm imidazole for 1 h at 4 °C with agitation. The beads were washed once with 10 column volumes of 20 mm Tris-HCl, pH 8.0, 500 mm NaCl (buffer B) supplemented with 5 mm imidazole and two more times with 10 column volumes of buffer B supplemented with 10 mm imidazole. Bound proteins were eluted using 1 column volume of buffer S containing 150 mm imidazole.
Membrane Interaction Studies Using Liposome Flotation
Flotation experiments were performed as described previously (14). Briefly, purified NG and NG+1 proteins (20 μg) were incubated with large unilamellar vesicles of different composition in assay buffer for 20 min at 37 °C. Details of the individual lipid composition are given in the legend to Fig. 6D.
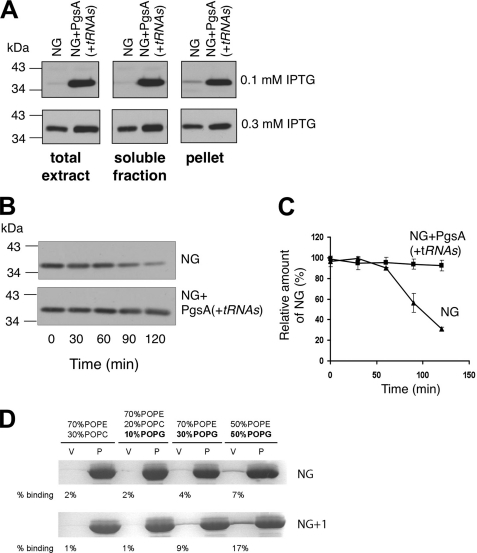
FIGURE 6.
PgsA expression stabilizes NG and restores membrane localization. A, E. coli BW25113 cells expressing NG alone (NG) or with PgsA (NG+ pgsA(+tRNAs)) were induced using 0.1 or 0.3 mm IPTG for 2 h. B, cells were induced using 0.5 mm IPTG for 30 min, rinsed four times to remove the inducer, and grown again. Samples were taken at different time points, and cells were fractionated. NG expression was analyzed by Western blotting using antibodies against FtsY. C, amount of NG was quantified by densitometry, and the average of three independent experiments is shown, with error bars representing standard deviations. D, flotation experiments with anionic phospholipids. In presence of zwitterionic phospholipids (phosphatidylethanolamine/phosphatidylcholine ratio of 70:30), no interaction was seen. Lipid interaction could be restored with increasing amounts of anionic phospholipids (10–50% phosphatidylglycerol). The vesicle fraction is marked by “v” and the pellet fraction by “p.” POPE, palmitoyloleoylphosphatidylethanolamine; POPG, palmitoyloleoylphosphatidylglycerol; POPC, palmitoyloleoylphosphatidylcholine.
Determination of Acidic Lipids
To analyze changes in the content of various phospholipids, cells were grown in LB medium containing 1 μCi/ml [32P]phosphoric acid for 3 h at 37 °C. For pCA24N (empty vector) and pCA24N-His6-PgsA, 50 μm IPTG was included in the incubation media. The cultures (25 ml) were harvested by a 15-min centrifugation at 5000 × g, and the pellet was resuspended in 0.8 ml of PBS, after which 2 ml of methanol and 1 ml of chloroform were added with thorough mixing. After a 1-h incubation at room temperature, PBS and chloroform (1 ml each) were added and mixed thoroughly. Following a brief, low speed centrifugation, the resulting chloroform phase was analyzed by thin layer chromatography with Silica Gel 60 (20 × 20 cm) (Merck), utilizing chloroform, methanol, water, 25% NH4OH (120:75:5.6:2.4, v/v) as the developing solvent. Phospholipids were identified on the chromatogram by phosphorimaging (Fuji FLA7000), and the resulting bands were quantified by densitometry using ImageJ software (rsb.info.nih.gov). Phospholipid identities were confirmed by comparison with known reference lipids: dioleoylphosphatidylethanolamine (DOPE), dioleoylphosphatidylglycerol (DOPG), and tetraoleoyl cardiolipin (TOCL) (Avanti Polar Lipids), which were visualized by 20% phosphomolybdic acid solution.
N-terminal Amino Acid Sequence Analysis
Purified NG proteins were subjected to SDS-PAGE, electroblotted to a polyvinylidene fluoride (PVDF) membrane, and sequenced by Protein Sequencer Procise 491 (Applied Biosystems).
SDS-PAGE and Immunoblotting
Membrane or cytosolic fractions (10–15 μg of protein) were subjected to 12% SDS-PAGE. Proteins were electroblotted to nitrocellulose membranes and probed with rabbit anti-FtsY antibodies or HisProbe-HRP-conjugated goat anti-rabbit immunoglobulin antibodies.
RESULTS AND DISCUSSION
An Unbiased Genetic Search for Factors That Interact with FtsY
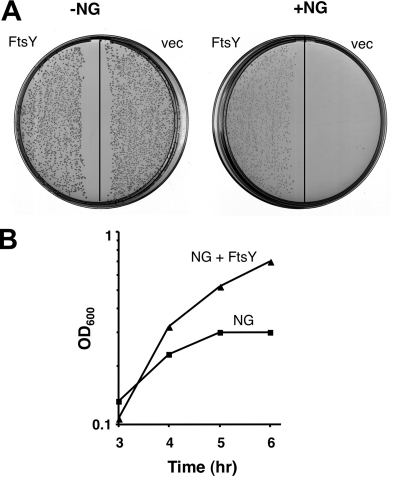
Previous studies have shown that the biological activity of E. coli FtsY is mediated through its C-terminal domain (termed NG+1) (18). When NG+1 is truncated by one amino acid in its N terminus (NG), it loses activity and becomes highly toxic upon overexpression (13).5 To test the possibility that NG confers a dominant negative effect, we co-expressed wild-type FtsY and NG. The results show that FtsY expression in trans relieves the toxicity of NG (Fig. 1), suggesting that the nonfunctional NG mutant might compete with the wild-type receptor for binding to an unknown site. To search for proteins that functionally interact with NG (and FtsY), we utilized a genetic screen that is based on NG toxicity. Briefly, we anticipated that overexpression of any protein that interacts with NG might dilute the amount of free NG and thus relieve its toxicity. E. coli harboring a plasmid encoding NG under a strong promoter (either tacP or araP) was transformed with a chromosomal library, and the transformants were plated on LB agar containing the NG inducer, IPTG, or arabinose. In the absence of phenotype suppressors, no colonies were formed under conditions of IPTG induction (Fig. 1). In contrast, in plates with the library-transformed cells, several colonies appeared under these conditions. Following re-evaluation of the results by plasmid preparation and retransformation, the positive clones were sequenced, and the results are shown in Table 1. Not surprisingly, clones harboring putative sugar efflux transporters were identified. Although this assumption was not fully validated by deletion analysis, it is plausible that these transporters would rescue the cells by extruding the NG inducer IPTG, which is a sugar analog (e.g. 19), or arabinose. In sharp distinction to the efflux transporters that one would have expected to identify, the fourth suppressor clone was both surprising and potentially relevant and informative. This clone harbored the pgsA gene, which encodes the enzyme phosphatidylglycerophosphate synthase (PgsA) (Fig. 2A, GS100-3). PgsA is an integral membrane protein that catalyzes the committed step in the biosynthesis of acidic phospholipids (20). The importance of PgsA and consequently of acidic lipids in recruiting and regulating the activity of peripheral membrane enzymes has been investigated extensively by Dowhan and co-workers (20–22).
FIGURE 1.
FtsY relieves the toxic effect of NG. A, E. coli cells harboring pCV3-araP-NG were co-transformed either with plain vector (vec) or pT75-tacP-FtsY (FtsY). The transformants were plated on LB agar without (as a control) or with 0.5% arabinose for NG induction. B, overnight cultures were diluted and grown in LB broth. At A600 (OD600) of 0.03, the cultures were induced with 1% arabinose and grown for additional 4 h. Growth was followed by measuring the optical density of the cultures.
TABLE 1.
Proteins identified as suppressors of the NG toxicity
| Protein | Inducer useda | Cellular location | Function |
|---|---|---|---|
| PgsAb | IPTG/arabinose | Inner membrane | Biosynthesis of acidic phospholipids |
| MdfAc | IPTG | Inner membrane | Multidrug transporter |
| SetAc | IPTG | Inner membrane | Sugar efflux transporter |
| YdeAc | IPTG/arabinose | Inner membrane | Sugar efflux transporter |
a The screening was performed in two different systems, in which the NG was expressed under different inducible promoter (tac and ara promoter) utilizing different inducers (IPTG or arabinose, respectively).
b Protein was identified.
c Protein was uncharacterized.
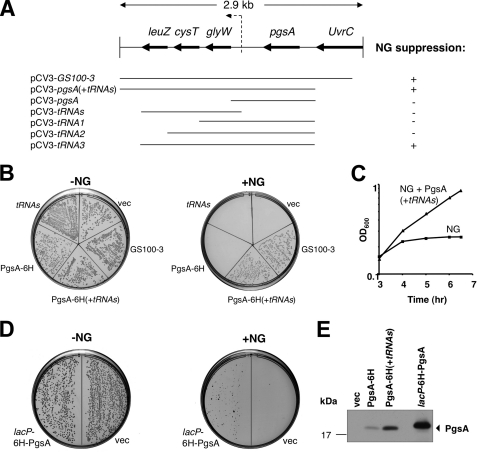
FIGURE 2.
Deletion and functional analysis of clones harboring the pgsA gene. A, schematic representation of several deletion constructs that were characterized. B, plasmids harboring the deletion constructs shown in A, containing 6 histidine tags at the C terminus of PgsA, were transformed into E. coli Top10/pT7-5-tacP-NG and examined for their ability to suppress the NG toxicity. C, overnight cultures of cells co-expressing NG and either plain vector (NG) or PgsA (NG+pgsA(+tRNAs)) were diluted and grown in LB broth. At A600 (OD600) of 0.03, the cultures were induced with 0.2 mm IPTG and grown for additional 5 h. Growth was followed by measuring the optical density of the cultures. D, Aska clone JW1897 (25) harboring the His6-pgsA gene under the lac promoter (lacP) (without the tRNAs operon) was transformed into E. coli Top10/pCV-5-araP-NG and plated on LB agar plates with 0.5% arabinose and without IPTG, the PgsA inducer. E, E. coli cells co-expressing NG and either plain vector (vec), PgsA-His6, PgsA-His6 (+tRNAs), or lacP-His6-PgsA (Aska clone JW1897) were grown in LB broth. Membrane fractions were isolated, and protein expression was analyzed by Western blotting using India HisProbe-HRP (for His6-tagged PgsA identification). His6-PgsA expressed under the regulation of lacP is larger than PgsA-His6 expressed under its endogenous promoter due to addition of 15 amino acids derived from insertion of SfiI restriction sites (25).
Characterization of the Effect of PgsA Expression on NG Toxicity
The increase in anionic phospholipids in cells overexpressing PgsA raised the possibility that the NG toxicity is alleviated by an excess of acidic lipids. As shown in Fig. 2, B and C, induction of NG expression alone abolished growth, whereas cells co-expressing NG and various constructs harboring pgsA grew relatively well. It is interesting that cells tolerated PgsA expression only when the pgsA gene was followed by the flanking 3′ region (Fig. 2, pCV3-pgsA(+tRNAs)). A similar observation was made in the past where cells tolerated plasmid-borne pgsA only with an unrelated insert at its 3′ end that differs from the tRNAs sequences found here (20, 23). This suggests a nonspecific polar effect of 3′ sequences on the expression of the pgsA gene. To examine further if the 3′ tRNA sequences are functionally essential, we repeated the experiments with a plasmid encoding His6-PgsA under the regulation of the lac promoter (lacP) (lacP His6-PgsA) (Aska clone JW1897) (17). This clone does not contain the 3′ tRNA region. Fig. 2D shows that NG-overexpressing cells that simultaneously express His6-PgsA under the regulation of lacP did restore growth, although not as well as the chromosomal clone (compare the right plates in Fig. 2, B and D). To investigate why the expression of His6-PgsA from the lac promoter seems to impose toxicity (Fig. 2D, right plate) we examined the PgsA expression level in cells transformed with plasmids encoding either of the three tested constructs as follows: PgsA-His6 under its endogenous promoter, with and without the flanking tRNAs 3′ region, and PgsA-His6 under the lac promoter (Fig. 2E). The results suggest that only moderate expression of PgsA (from pgsA-His6+tRNAs) can fully restore the NG toxicity, whereas low expression (from pgsA-His6) could not suppress NG toxicity, and very high expression (from lacP His6-pgsA) seems to be toxic, as observed in the past with other PgsA-encoding high copy number plasmids (see below) (23). Notably, the PgsA-His6 protein migrated slowly on the gel (Fig. 2E) because it harbors additional 15 amino acids encoded by the insertion of SfiI restriction sites (17). These results demonstrate that PgsA alone is sufficient for suppressing the NG toxicity. The 3′ gene sequences in our studies (tRNAs) and others (20, 23) probably affect the level of expression by an uncharacterized mechanism (such as mRNA stability or translation regulation). For consistency, we used the PgsA clone with the flanking 3′ tRNAs region throughout most of the experiments.
PgsA Overexpression Changes the Phospholipid Composition
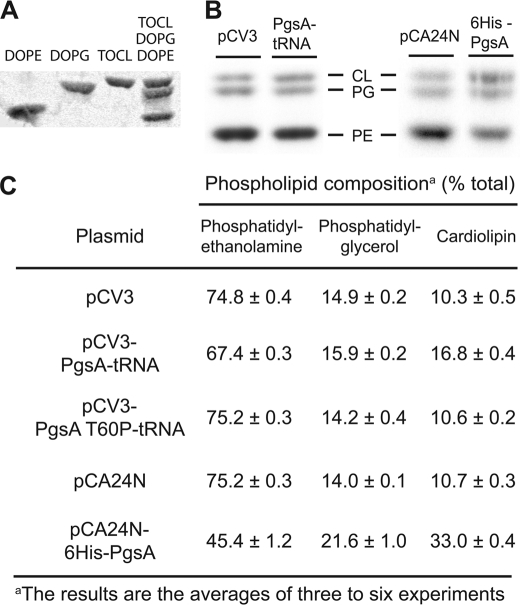
Of special relevance to our studies are previous reports on the importance of acidic lipids that participate in the regulation of pathways that involve peripheral membrane proteins such as MinD (16, 24). It was shown that overexpression of PgsA alters cellular phospholipid composition, as manifested by an increase of about 10% in the concentrations of the acidic phospholipids phosphatidylglycerol and cardiolipin (23, 25). To verify that overexpression of PgsA indeed increased the concentration of acidic phospholipids in our expression systems, we examined the phospholipid compositions of E. coli harboring empty vectors or plasmids encoding various PgsA constructs. Fig. 3C clearly shows that all the tested active PgsA constructs increased the amount of acidic phospholipids in the cells compared with cells harboring empty vectors or the inactive mutant PgsA(T60P) (see below). It is likely that the toxicity observed with His6-PgsA (Fig. 2D) is caused by the dramatic increase in the content of anionic phospholipids, in addition to problems associated with PgsA overexpression in cells harboring pCA24N-lacP-His6-pgsA (Fig. 3C). Hence, this construct could only partially restore the growth of NG-overexpressing cells.
FIGURE 3.
Effect of PgsA overexpression on lipid composition in vivo. A, thin layer chromatography separation of phospholipid standards. B, E. coli Top10 harboring plasmid-encoded variants of PgsA or empty vectors were labeled with [32P]phosphoric acid at a final concentration of 1 μCi/ml and grown for 3 h at 37 °C. Lipids were extracted and separated by thin layer chromatography. C, quantitation of the phospholipid composition (moi % of lipid phosphorus calculated from radioactivity of each spot, see “Experimental Procedures”). The data shown are mean values, and the standard deviations are calculated from two or three different batches of cultures.
Enzymatic Activity of PgsA Is Required for Suppressing NG Toxicity
PgsA may affect the toxic phenotype of NG through its enzymatic activity by elevating the level of acidic lipids (Fig. 3) or by direct or indirect physical interaction with NG. To distinguish between these possibilities, we mutated PgsA and examined how the inactive PgsA mutant (PgsA(T60P)) (26) affects the NG toxicity. As shown in Fig. 4A, cells expressing NG did not grow when co-transformed with a plasmid expressing PgsA(T60P). By utilizing a 6-histidine tag, we confirmed that the mutant is expressed (Fig. 4B), suggesting that the enzymatic activity of PgsA is required for the rescue of NG-expressing cells. Taken together, the results strongly support the notion that NG toxicity can be relieved by increasing the abundance of acidic lipids.
FIGURE 4.
Inactive PgsA mutant does not suppress NG toxicity. A, inactive PgsA mutant (T60P) was examined for its ability to suppress the NG toxic effect as in Fig. 2. vec, vector. B, E. coli co-expressing NG and either plain vector, PgsA-His6 (+tRNAs) or PgsA(T60P)-His6(+tRNAs) were grown in LB broth. Membrane fractions were isolated, and protein expression was analyzed by Western blotting using antibodies against FtsY for NG identification (left panel) or India HisProbe-HRP for PgsA identification (right panel).
PgsA Expression Restores the Biological Activity of NG
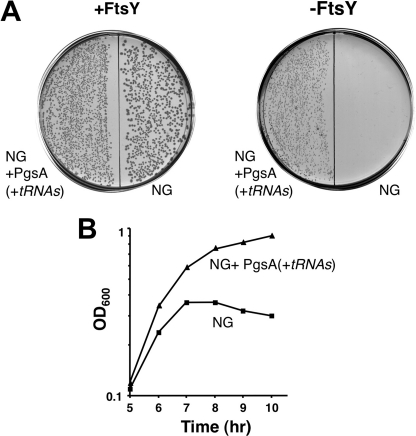
How do elevated levels of anionic phospholipids suppress the toxicity of NG? One possible explanation is that acidic lipids bind and sequester NG, leaving a crucial membrane-attachment site free to accommodate the functional wild-type FtsY. Alternatively, it is also possible that an excess of acidic lipids might restore the function of the lipid-binding element at the N terminus of NG and thereby revive its biological activity. To examine the latter, we used cells depleted of FtsY, which co-expressed NG and PgsA. In the absence of FtsY, such cells would grow only if NG functions properly as an SRP receptor. Fig. 5A shows that in the presence of the FtsY inducer (arabinose), all the transformants formed colonies on agar plates. However, in the absence of arabinose, only cells co-expressing NG and PgsA were able to grow. This phenomenon was also observed when cells were grown in liquid media (Fig. 5B). The results indicate that the nonfunctional NG is activated by elevated amounts of anionic phospholipids.
FIGURE 5.
NG activation by PgsA. A, E. coli IY28-pT75-tacP-NG cells were transformed either with a plain vector (NG) or PgsA (NG+pgsA(+tRNAs)) and plated with (as a control) or without arabinose, the FtsY inducer. B, overnight cultures were grown with arabinose, washed four times, and diluted in LB broth without arabinose. Growth was followed by measuring the optical density of the cultures.
PgsA Expression Affects NG Stability
What is the mechanism underlying the restoration effect of acidic lipids on NG activity? It was previously shown that the addition of one amino acid to NG at its N terminus abolished its toxicity and restored its function (18). The crystal structure of the active mutant (NG+1) (14) revealed that it has an ordered N terminus that folds as an amphipathic helix. In contrast, the N terminus of NG is disordered (27). The N-terminal amphipathic helix of NG+1 was found to be essential for the lipid-stimulated GTPase activity of the receptor in the context of its complex with the SRP (13, 14). Interestingly, analysis of the purified NG protein revealed that it is further processed in vivo and consequently lacks its first methionine (Met-195) (14). Therefore, the so-called toxic NG protein actually lacks two amino acids at its N terminus compared with NG+1 and is consequently unable to form an amphipathic helix. We hypothesized that if acidic lipids protect the N-terminal Met-195 residue of NG, the protein would be able to partially re-establish the formation of a helix in its N terminus and consequently its biological activity. To test this, NG was expressed alone or in the presence of PgsA and purified from the cytoplasmic and membrane fractions (data not shown). The N-terminal amino acid sequences of the purified NG proteins were determined. The results of this experiment rule out the suggestion that the NG N terminus is protected by access of acidic lipids because even when NG was expressed in the presence of PgsA, it lacks Met-195, indicating that Met-195 processing was not prevented by anionic phospholipids (data not shown).
During our studies, we noted that NG is usually better expressed in cells co-expressing PgsA (Fig. 6A). Similarly, previous studies showed that when expressed under identical conditions, the amount of NG was substantially lower than that of the active mutant NG+1, suggesting that, in addition to its functional role, the N-terminal helix of NG+1 may also have a stabilizing effect. To test the possibility that an excess of acidic lipids (in cells expressing PgsA) stabilizes NG, we performed the following experiment. E. coli cultures harboring plasmid-borne NG, alone or together with a constitutively expressed PgsA, were induced for NG expression, rinsed several times to remove the inducer, and grown again. Samples were taken at different time points, and cells were fractionated. Fig. 6, B and C, shows that the level of NG fell in time when expressed alone. However, when NG was co-expressed with PgsA, its amount remained constant throughout the experiment. Importantly, in the absence of PgsA, NG is unable to complement FtsY-depletion even when highly induced.6
Because in the presence of PgsA, the amount of NG at the membrane remained constant, we tested the ability of NG and NG+1 to associate with membranes containing different amounts of anionic phospholipids in an in vitro binding assay (Fig. 6D, upper panel) (14). The association of NG with large unilamellar vesicles increases with increasing amounts of phosphatidylglycerol. Interaction was not observed with zwitterionic phospholipids. Therefore, our data support a crucial role of anionic phospholipids in functional membrane localization of NG. Importantly, when NG+1 containing an intact lipid-binding α-helix was tested in this experiment (Fig. 6D, lower panel), the efficiency of lipid association was significantly higher, as described previously (14). Taken together, the results presented here imply that anionic phospholipid-enriched membranes can induce structural changes in NG that not only restore its biological function at the membrane but also stabilize the protein. In light of these results, it is surprising that pgsA null mutants of E. coli are viable in the absence of phosphatidylglycerol and cardiolipin (28). Other studies, however, offered a likely explanation as they showed that other acidic species accumulate in membranes of pgsA-deleted cells (29, 30). These observations indirectly suggest that the functional interaction of FtsY with acidic lipids is probably not specific for a certain lipid.
In summary, our studies offer strong genetic support for the notion that acidic lipids play a crucial role in the function of the E. coli SRP receptor in vivo, probably at a defined step during the SRP pathway that requires GTP hydrolysis by the SR-SRP complex, when ribosomes translating membrane proteins should be transferred to and assembled on the translocon.
This work was supported by the German-Israeli Foundation for Scientific Research and Development (to I. S. and E. B.), by the Israel Science Foundation (to E .B.), and by Deutsche Forschungsgemeinschaft Collaborative Research Grant GRK 1188 (to I. S.).
I. Yosef and E. Bibi, unpublished data.
A. M. Zelazny and E. Bibi, unpublished data.
E. Erez and E. Bibi, unpublished data.
- SRP
- signal recognition particle
- IPTG
- isopropyl β-thiogalactoside.
REFERENCES
- 1.Luirink J., von Heijne G., Houben E., de Gier J. W. (2005) Annu. Rev. Microbiol. 59, 329–355 [DOI] [PubMed] [Google Scholar]
- 2.Pool M. R. (2005) Mol. Membr. Biol. 22, 3–15 [DOI] [PubMed] [Google Scholar]
- 3.Herskovits A. A., Bibi E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zelazny A., Seluanov A., Cooper A., Bibi E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6025–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibi E., Herskovits A. A., Bochkareva E. S., Zelazny A. (2001) Trends Biochem. Sci. 26, 15–16 [DOI] [PubMed] [Google Scholar]
- 6.Herskovits A. A., Shimoni E., Minsky A., Bibi E. (2002) J. Cell Biol. 159, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelini S., Deitermann S., Koch H. G. (2005) EMBO Rep. 6, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mircheva M., Boy D., Weiche B., Hucke F., Graumann P., Koch H. G. (2009) BMC Biol. 7, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millman J. S., Andrews D. W. (1999) J. Biol. Chem. 274, 33227–33234 [DOI] [PubMed] [Google Scholar]
- 10.de Leeuw E., Poland D., Mol O., Sinning I., ten Hagen-Jongman C. M., Oudega B., Luirink J. (1997) FEBS Lett. 416, 225–229 [DOI] [PubMed] [Google Scholar]
- 11.de Leeuw E., te Kaat K., Moser C., Menestrina G., Demel R., de Kruijff B., Oudega B., Luirink J., Sinning I. (2000) EMBO J. 19, 531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braig D., Bär C., Thumfart J. O., Koch H. G. (2009) J. Mol. Biol. 390, 401–413 [DOI] [PubMed] [Google Scholar]
- 13.Bahari L., Parlitz R., Eitan A., Stjepanovic G., Bochkareva E. S., Sinning I., Bibi E. (2007) J. Biol. Chem. 282, 32168–32175 [DOI] [PubMed] [Google Scholar]
- 14.Parlitz R., Eitan A., Stjepanovic G., Bahari L., Bange G., Bibi E., Sinning I. (2007) J. Biol. Chem. 282, 32176–32184 [DOI] [PubMed] [Google Scholar]
- 15.Lam V. Q., Akopian D., Rome M., Henningsen D., Shan S. O. (2010) J. Cell Biol. 190, 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szeto T. H., Rowland S. L., Rothfield L. I., King G. F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15693–15698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. (2005) DNA Res. 12, 291–299 [DOI] [PubMed] [Google Scholar]
- 18.Eitan A., Bibi E. (2004) J. Bacteriol. 186, 2492–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohn C., Bouloc P. (1998) J. Bacteriol. 180, 6072–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishnan A. S., Chen Y. C., Temkin M., Dowhan W. (1986) J. Biol. Chem. 261, 1329–1338 [PubMed] [Google Scholar]
- 21.Raetz C. R., Dowhan W. (1990) J. Biol. Chem. 265, 1235–1238 [PubMed] [Google Scholar]
- 22.Mileykovskaya E., Dowhan W. (2005) Curr. Opin. Microbiol. 8, 135–142 [DOI] [PubMed] [Google Scholar]
- 23.Ohta A., Waggoner K., Radominska-Pyrek A., Dowhan W. (1981) J. Bacteriol. 147, 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mileykovskaya E., Dowhan W. (2009) Biochim. Biophys. Acta 1788, 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H., Nishiyama K., Tokuda H. (1999) J. Biol. Chem. 274, 31020–31024 [DOI] [PubMed] [Google Scholar]
- 26.Usui M., Sembongi H., Matsuzaki H., Matsumoto K., Shibuya I. (1994) J. Bacteriol. 176, 3389–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montoya G., Svensson C., Luirink J., Sinning I. (1997) Nature 385, 365–368 [DOI] [PubMed] [Google Scholar]
- 28.Shiba Y., Yokoyama Y., Aono Y., Kiuchi T., Kusaka J., Matsumoto K., Hara H. (2004) J. Bacteriol. 186, 6526–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi S., Shibuya I., Matsumoto K. (2000) J. Bacteriol. 182, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mileykovskaya E., Ryan A. C., Mo X., Lin C. C., Khalaf K. I., Dowhan W., Garrett T. A. (2009) J. Biol. Chem. 284, 2990–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]