Abstract
Centrosomes play a crucial role in the directed migration of developing neurons. However, the underlying mechanism is poorly understood. This study has identified a novel disrupted in schizophrenia 1 (DISC1)-interacting protein, named CAMDI after coiled-coil protein associated with myosin II and DISC1, which translocates to the centrosome in a DISC1-dependent manner. Knockdown of CAMDI by shRNA revealed severely impaired radial migration with disoriented centrosomes. A yeast two-hybrid screen identified myosin II as a binding protein of CAMDI. CAMDI interacts preferentially with phosphomyosin II and induces an accumulation of phosphomyosin II at the centrosome in a DISC1-dependent manner. Interestingly, one single nucleotide polymorphism of the CAMDI gene (R828W) is identified, and its gene product was found to reduce the binding ability to phosphomyosin II. Furthermore, mice with overexpression of R828W in neurons exhibit an impaired radial migration. Our findings indicate that CAMDI is required for radial migration probably through DISC1 and myosin II-mediated centrosome positioning during neuronal development.
Keywords: Cell Migration, Development, Myosin, Neurobiology, Neurodevelopment
Introduction
Neuronal migration is a critical phase in nervous system development, and the regulation of the centrosome underlies the directed migration of neurons (1–7). Centrosomes require not only a radial array of cytoplasmic microtubules and the activity of the microtubule motor dynein, but also actomyosin-driven forces (8). Recent studies have demonstrated that myosin II is critical for centrosome separation and positioning during mitotic spindle assembly (9, 10) and coordinated movement of the centrosome during neuronal migration (11); however, little is known about the molecular basis for myosin II-dependent centrosome positioning.
Understanding the molecular mechanisms underlying neuronal migration has advanced in recent years due to the identification of genes involved in human neuronal migration disorders (12–14). DISC13 is identified as a risk factor for major mental illness, originally discovered in a large Scottish pedigree linked to major depression and schizophrenia (15, 16). DISC1 localizes to the centrosome by interacting with centrosomal proteins (17–19). Thereby it has been proposed to, in part, control centrosome function and microtubule dynamics, notably interacting with NUDEL, LIS1, and dynein (20–23). A C-terminal truncated DISC1 protein, encoded from a mutation in the DISC1 gene, failed to localize to the centrosome and disrupted the dynein-microtubule network in vitro (18, 24). In vivo analyses of DISC1 mutant mice and DISC1 down-regulation by shRNA showed that both give rise to phenotypes related to major mental illness, suggesting that DISC1 is involved in cortical architecture, most likely through the regulation of centrosome-microtubule dynamics (24–31).
Here, we identified a novel DISC1-interacting protein, CAMDI, which controls centrosome positioning by regulation probably via the myosin II pathway. CAMDI may represent a missing link between the actomyosin and the DISC1-centrosome-microtubule complex. Our findings might provide a clue to understand the molecular pathology for DISC1-related mental diseases including schizophrenia and autism.
EXPERIMENTAL PROCEDURES
In Situ Hybridization and Immunohistochemistry
For in situ hybridization and immunohistochemical analysis, embryos were fixed by immersion in 4% paraformaldehyde in 0.1 m PBST (phosphate-buffered saline containing 0.1% Tween 20), cryoprotected in 20% sucrose, frozen in OCT compound, and stored at −80 °C until sectioning. Coronal and sagittal sections (20 μm) were cut with a cryostat and stored at −80 °C prior to used. For in situ hybridization, antisense riboprobes were labeled with digoxigenin-11-d-UTP (Roche Applied Science) according to the directions of the supplier. Tissue sections were hybridized with digoxigenin-labeled riboprobes. For immunohistochemical analysis, tissue sections were blocked for 1 h at room temperature in PBS containing 5% horse serum and then incubated overnight at 4 °C with primary antibody. For analysis of neuronal morphology and dendrite number in vivo, three-dimensional reconstructions of each EGFP-positive neuron were produced by Z-series stacks of confocal images. The projection images were automatically traced with ImageJ software (National Institutes of Health).
In Utero Electroporation
Briefly, in utero electroporation to dorsal neocortex was performed by injecting the DNA plasmid solution (5 mg/ml) plus 1% Fast Green using a glass capillary into the E14.5 ICR mouse ventricle. DNA mixture was 2–3-fold higher than that of the EGFP plasmid, which was the electroporation marker. Electroporation was performed using a CUY-21 electroporator (NEPA GENE) and the following parameters: four 50-ms-long pulse separated by 950-ms-long intervals at 33 V.
Antibodies
Anti-CAMDI antibody was produced by immunizing a rabbit with synthetic peptide corresponding to amino acid sequence 1356–1364 of the mouse CAMDI. Rabbit anti-DISC1 antibody was obtained from Novus Biologicals. Anti-FLAG M2 monoclonal and anti-γ-tubulin antibodies were obtained from Sigma-Aldrich. Anti-HA high affinity antibody was obtained from Roche Applied Science. Anti-GFP rabbit polyclonal antibody and secondary antibodies conjugated with Alexa Fluor 350, 488, and 594 were obtained from Invitrogen. Anti-GFP mouse monoclonal and polyclonal antibodies were purchased from Clontech. Hoechst 33258 was obtained from Nacalai Tesque. Anti-pericentrin and anti-Ki67 antibodies were obtained from BD Transduction Laboratories. Anti-phosphohistone H3 and anti-active caspase3 antibodies were obtained from Millipore. Anti-p-MRLC antibody was purchased from Cell Signaling.
Plasmids
The mouse CAMDI was derived from Image clone 6831397. The rat full-length CAMDI was derived from the Image clone 7104862. The human CAMDI was derived from the FLJ26337 clone (National Institute of Technology Evaluation (NITE)). The human DISC1 was derived from Image clone 5756143. PCR was performed using these Image clones as templates. To construct a plasmid for FLAG-CAMDI, the PCR product of CAMDI was inserted into the NotI/XbaI sites of pFLAG-CMV-2a. To construct a plasmid for EGFP-DISC1, the PCR product of DISC1 was inserted into the EcoRI/SalI sites of pEGFP-C1. Although the PCR product of DISC1 contained one nucleotide deletion, the error was corrected using the QuikChange Site-directed Mutagenesis kit (Stratagene). Several shRNA-targeted sequences are shown below (with an order of sense, loop (underlined), and antisense): control-sh (5′-ACTACCGTTGTATAGGTGTTCAAGAGACACCTATAACAACGGTAGT-3′), CAMDI-sh (5′-GGGTAGCCTATAATGACAAGCTTCAAGAGAGCTTGTCATTATAGGCTACCC-3′), DISC1-sh (5′-GGCAAACACTGTGAAGTGCTTCAAGAGAGCACTTCACAGTGTTTGCC-3′). The knockdown efficiencies of DISC1-sh and control-sh were established previously (24). To construct a plasmid for control-sh, CAMDI-sh, and DISC1-sh, two primers were annealed, and the product was inserted into BamHI/HindIII sites of pSilencer 3.1-H1. To construct a plasmid for the CAMDI mutant resistant to CAMDI-sh, silent mutations were introduced using the QuikChange kit and primers 5′-GACATGGTGGTCGCGTACAATGACAAGC-3′ and 5′-GCTTGTCATTGTACGCGACCCACCATGTC-3′. To construct a plasmid for CAMDI-R828W, the mutation was introduced using QuikChange and primers 5′-GCAGATTCACCTCTGGTGCTCTCAGGAAAAGC-3′ and 5′-GCTTTTCCTGAGAGCACCAGAGGTGAATCTGC-3′. To construct a plasmid for MRLC-DD and -AA mutants (Thr18 and Ser19), mutations were introduced using QuikChange. Mutagenesis near the phosphorylatable serine, AA-MRLC and DD-MRLC (Thr18 and Ser19), has been reported to mimic the unphosphorylated and phosphorylated forms of MRLC, respectively (32, 33).
Cell Culture and Transfections
SH-SY5Y and HEK293 cells were obtained from the American Type Culture Collection, and the COS-7 cell line was provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. These cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C, in 5% CO2, in a humidified chamber. Transfection was carried out using Lipofectamine 2000 (Invitrogen). Primary cortical neurons were prepared from ICR mice (embryonic day 16) and plated on a poly-l-lysine-coated slide glass in Minimum Eagle's Medium containing 2% FBS and N2 supplement. Transfection was carried out using the calcium phosphate method (Clontech).
Immunoprecipitation and Western Blotting
Culture cells were lysed in Nonidet P-40 lysis buffer (20 mm Tris-HCl, pH 7.2, 2 mm EDTA, 0.5% Nonidet P-40, 8% sucrose, 80 mm dithiothreitol). The lysate was clarified by centrifugation at 15,000 × g for 10 min and immunoprecipitated with the appropriate antibody. Immunoprecipitates were washed three times with lysis buffer. After boiling for 3 min, equal protein amounts of the lysates were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore). Membranes were blocked for 1 h at room temperature in 5% skim milk in PBST with gentle shaking and incubated with primary antibodies overnight at 4 °C. After washing the membranes three times with PBST, they were incubated with secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature. The blotted membranes were developed using the Immobilon Western chemiluminescent HRP substrate (Millipore) according to the manufacturer's instructions.
Immunofluorescence
Cells were fixed for 20 min in PBS containing 4% paraformaldehyde or cold methanol and permeabilized with 0.2% Triton X-100. After incubation in PBS containing 1% bovine serum albumin for 30 min, the cells were reacted with first antibody for overnight at 4 °C, followed by incubation with secondary antibody and/or TRITC-conjugated phalloidin. The staining was analyzed by confocal microscope (Olympus FV1000).
Yeast Two-hybrid Screening
The Matchmaker two-hybrid system kit (Clontech) was used for detecting specific proteins interacting with DISC1 and CAMDI as described by the manufacturer.
Centrosome Isolation
Centrosome was prepared from COS-7 cells according to the methods described previously (34). COS-7 cells were treated with 50 ng/ml nocodazole for 2 h. After centrifugation of cell lysates by a discontinuous gradient consisting sucrose solutions, fractions were collected. The centrosome fractions were determined by immunoblotting with anti-γ-tubulin.
Ethics Statement
All animals were maintained under the university guidelines for the care and use of animals. The experiments were performed after securing Tokyo University of Pharmacy and Life Sciences Animal Use Committee Protocol approval.
RESULTS
Identification of CAMDI as a Novel DISC1-interacting Protein
To understand the molecular basis underlying DISC1 function in the centrosome, we performed a yeast two-hybrid screen of a mouse whole brain cDNA library using the coiled-coil domains of DISC1 as bait. We obtained several DISC1-binding candidates, including a previously uncharacterized gene, CCDC141 (hereafter referred to as CAMDI). The mouse CAMDI gene encodes an open frame of 1,451 amino acids containing spectrin-like repeats in the N terminus, a tandem repeat of three coiled-coil domains in the middle, and I-set domains in the C terminus (Fig. 1A and supplemental Fig. 1A). Western blot analyses indicated the expression of CAMDI to be mainly in the developing brain (supplemental Fig. 1B). CAMDI orthologs exist in vertebrates (supplemental Fig. 1C) but not in lower organisms such as Drosophila and Caenorhabditis elegans. In the brain, CAMDI mRNA is detected in various regions, including the cerebrum, eye, cerebellum, and hippocampus in developing and adult mice (supplemental Fig. 1D). Interactions of CAMDI and DISC1 in HEK293 cells and mouse brain lysates were confirmed by immunoprecipitation followed by an immunoblotting (IP-IB) assay (Fig. 1B). A yeast two-hybrid screen identified a CAMDI fragment containing coiled-coil domains, suggesting the coiled-coil domain-mediated interaction. A pulldown assay using three GST-fused coiled-coil domains of CAMDI indicated that three coiled-coil domains of CAMDI interact with DISC1 (Fig. 1C, M1–M3). Furthermore, a pulldown assay using each GST-fused coiled-coil domain of CAMDI suggested that the second coiled-coil domain of CAMDI is mainly responsible for the interaction with DISC1 (Fig. 1C, M2). On the other hand, a pulldown assay using each GST-fused coiled-coil domain of DISC1 suggested that the second coiled-coil domain of DISC1 is mainly responsible for the interaction with CAMDI (Fig. 1D, CC2). Thus, the second coiled-coil domains of CAMDI and DISC1 appear to mediate their specific interaction as illustrated in Fig. 1A. Taken together, these results demonstrate that CAMDI is a novel DISC1-interacting protein expressed in the developing brain.
FIGURE 1.
Identification of CAMDI as a novel DISC1-interacting protein. A, schematic illustration of interaction between CAMDI and DISC1. Breakpoint of the truncated DISC1 is shown by a vertical line. B, interactions of CAMDI with DISC1 both in vitro and in vivo. Immunoprecipitation followed by immunoblot (IP-IB) assays were performed with the indicated antibodies in HEK293 cells and E18 mouse brain. Equal amounts of protein expression in total lysates are shown below. C, CAMDI interacting with DISC1 through the second coiled-coil domain. HEK293 cells were co-transfected with EGFP-DISC1 and each HA-CAMDI fragment (M1–M3, 576–1106; C, 1106–1499 amino acids; M1, 576–706; M2, 707–906; M3, 907–1106), followed by IP with anti-HA antibody and IB with anti-EGFP antibody. The position of each HA-CAMDI fragment is indicated by an asterisk. D, second coiled-coil domain of DISC1 mainly responsible for interaction with CAMDI. Three purified DISC1 coiled-coil domains fused to GST (CC1, 340–400 amino acids; CC2, 446–507; CC3, 533–590) were subjected to pulldown assay using FLAG-CAMDI-expressing cell lysates, followed by IB with anti-FLAG antibody. Coomassie Brilliant Blue (CBB) staining shows the amounts of GST or GST-fused proteins. An equal amount of FLAG-CAMDI in total lysate is shown below. Densitometric analysis of each interaction indicates a specific interaction between FLAG-CAMDI and GST-CC2. The intensity of each FLAG-CAMDI band was normalized to corresponding Coomassie Brilliant Blue-stained bands indicated by asterisks.
Translocation of CAMDI to the Centrosome in a DISC1-dependent Manner
Co-localization of CAMDI with DISC1 was observed at/around the centrosome (Fig. 2A). Expression of CAMDI alone revealed a diffuse cytoplasmic distribution without centrosomal localization, whereas co-expression with DISC1 induced CAMDI translocation to the centrosome (Fig. 2B, top and middle). Translocation did not occur, however, with the C terminus-truncated DISC1 mutant, which retains the binding ability to CAMDI (data not shown) but fails to localize to the centrosome (Fig. 2B, top). Biochemical analysis, using sucrose density gradients, also demonstrated a DISC1-dependent shift of CAMDI to the centrosome fraction (supplemental Fig. 2). To determine whether DISC1 is required for centrosomal localization of endogenous CAMDI in cortical neurons, the effect of DISC1-knockdown by shRNA (DISC1-sh) on CAMDI localization was examined. Accumulation of endogenous CAMDI around the centrosome is indicated by an arrow (Fig. 2C, top). The centrosomal CAMDI decreased in EGFP-positive DISC1-knockdown neurons by DISC1-sh, but not in control-sh or EGFP-negative DISC1-sh neurons (Fig. 2C, middle and bottom). The diffuse staining pattern of γ-tubulin is due to the cell fixation by paraformaldehyde, which is suitable for detection of change in the amount of γ-tubulin (supplemental Fig. 3). These results demonstrated that CAMDI localizes to the centrosome, at least in part, in a DISC1-dependent manner both in vitro and in vivo.
FIGURE 2.
CAMDI translocates to the centrosome in a DISC1-dependent manner. A, co-localization of CAMDI with DISC1. COS-7 cells co-expressing FLAG-CAMDI and EGFP-DISC1 were double immunostained with anti-EGFP (green) and anti-FLAG antibodies (red). Merged images indicate the co-localization of CAMDI with DISC1 at perinuclear in both cells (arrows). Scale bar, 5 μm. B, DISC1-dependent translocation of CAMDI to the centrosome in vitro. COS-7 cells transfected with indicated vectors were analyzed by immunostaining. Centrosomal CAMDI indicated by arrows are magnified in the insets. −, control vector; WT, DISC1-wild type; mut, C-terminal truncated DISC1 mutant. Scale bar, 5 μm. C, DISC1-dependent centrosomal localization of endogenous CAMDI in cortical neurons. Endogenous CAMDI is co-localized with γ-tubulin (arrow; upper). Note that the centrosomal CAMDI disappeared in EGFP-positive DISC1-knockdown neurons by DISC1-sh (arrowhead), but not in control-sh or EGFP-negative DISC1-sh neurons (arrows). Scale bar, 10 μm.
CAMDI Knockdown Impairs Cortical Migration and Centrosomal Positioning
A strong signal of CAMDI mRNA was detected in the intermediate zone (IZ) at embryonic day (E) 16.5, and in the cortical plate (CP) at E18.5, in the mouse frontal cortex by in situ hybridization analysis (Fig. 3A). Immunohistochemical analysis using anti-CAMDI antibody also showed that the expression pattern of CAMDI protein was similar to that of CAMDI mRNA (Fig. 3B). These results suggest the possible involvement of CAMDI in cortical migration because the IZ at E16.5 is a critical point in development: neurons migrate out, and polarity formation commences.
FIGURE 3.
CAMDI is required for radial migration and centrosome positioning. A and B, in situ hybridization (A) and immunohistochemical (B) analyses of CAMDI in the sagittal sections of mouse frontal cortex at E16.5 and E18.5. Scale bar, 100 μm. C, impaired cortical migration by CAMDI knockdown. Sagittal sections of mice brains electroporated at E14 in utero with indicated vectors plus EGFP were analyzed at P2. Statistic data are shown in the bottom. The numbers of EGFP-positive neurons (>600 cells) in the VZ, IZ, and CP are measured from three independent samples, and the average ratio (%) in each area is calculated in the bottom. Results represent the means ± S.E. (error bars). Asterisks indicate significant differences between control-sh versus CAMDI-sh and CAMDI-sh versus CAMDI-resi; **, p < 0.01; *, p < 0.05, Student's t test. Scale bar, 100 μm. D, aberrant leading processes with disoriented centrosomes in CAMDI-knockdown neurons. Morphology between typical EGFP-positive control-sh- and CAMDI-sh-transfected cells in the CP as shown in Fig. 3C are compared by a high magnification analysis (top). The direction of VZ-CP axis is shown on the right. Arrow indicates an abnormal leading process extending to opposite direction. Computer-based reconstructions of these typical EGFP-positive cells are shown below. Centrosomes in the typical EGFP-positive control-sh- and CAMDI-sh-transfected cells as described above were labeled with anti-pericentrin antibody (arrowheads). The positioning of centrosomes in these cells (200 cells each) is measured by angles relative to the VZ-CP axis and compared by a bar graph (bottom). Scale bar, 10 μm. ***, p < 0.001, Student's t test.
To assess the role of CAMDI in radial migration, a pSilencer-based RNAi plasmid (shRNA) against the mouse CAMDI gene (CAMDI-sh) and a CAMDI construct resistant to this shRNA (CAMDI-resi) were designed in supplemental Fig. 4A and “Experimental Procedures.” Expression of CAMDI wild-type, but not CAMDI-resi, was reduced by >90% in CAMDI-sh-transfected HEK293 cells (supplemental Fig. 4A, left). Endogenous CAMDI in cultured cortical neurons was also reduced by 80% by CAMDI-sh compared with control-sh. Furthermore, immunohistochemical analysis demonstrated the in vivo knockdown of endogenous CAMDI by CAMDI-sh, but not by control-sh, in migrating cortical neurons (supplemental Fig. 4A, right). Thus, these constructs are available and applied to the following experiments. These constructs, with plasmid encoding EGFP to mark transfected cells, were injected into the mouse embryonic cerebral ventricle followed by in utero electroporation at E14; the mice were analyzed at postnatal day (P) 2. In this period, a massive number of cortical neurons underwent radial migration from the ventricular zone (VZ)/sub-VZ toward the CP and through the IZ. These constructs, with plasmid encoding EGFP to mark transfected cells, were injected into the mouse embryonic cerebral ventricle followed by in utero electroporation at E14; the mice were analyzed at P2. In this period, a massive number of cortical neurons underwent radial migration from the VZ/sub-VZ toward the CP through the IZ. Therefore, it is suitable to detect whether there are any defect in radial migration. In contrast to the normal CP localization of cortical neurons by control-sh, CAMDI-sh caused an abnormal accumulation of cortical neurons at the IZ (Fig. 3C). This effect of CAMDI-sh was completely restored by a co-transfection of the CAMDI-resi, indicating the specific phenomenon induced by CAMDI knockdown. Statistical analysis indicated that compared with control-sh or CAMDI-sh plus CAMDI-resi, CAMDI-sh caused a significant increase at the IZ and decrease at the CP in the number of cortical neurons. Similar accumulations at the IZ of cortical neurons were also observed when embryos were harvested at early stages, E17 and P0 (supplemental Fig. 4B). The IZ is a critical point for neurons to decide the direction and time to migrate out along the radial glia toward the CP. Therefore, in many cases, the abnormal accumulation of neurons at the IZ means an impaired migration. However, there is a small possibility that this is due to the cell death or cell cycle inhibition during early neural proliferation at the VZ. To deny this possibility, we examined the effect of CAMDI knockdown on cell survival and cell cycle in proliferative neural cells. Immunohistochemical analysis using an anti-active caspase3 antibody demonstrated that apoptosis is not elevated by CAMDI silencing in the VZ (supplemental Fig. 5A). In addition, a mitotic index stained with anti-phosphohistone H3 (G2M marker) and anti-Ki67 (proliferation marker) antibodies is not changed in CAMDI-silenced cortical progenitors (supplemental Fig. 5B), suggesting that CAMDI knockdown does not affect cell survival and cell cycle in proliferative neural cells. These results indicated that CAMDI silencing specifically interfered with radial migration.
A high magnification microscopic analysis of CAMDI-knockdown neurons revealed aberrant leading processes showing random direction, which correlates with a disoriented centrosome (Fig. 3D). This phenotype was previously observed in DISC1-knockdown neurons, although the result was milder than that seen for CAMDI-knockdown neurons (24). Taken together, these results demonstrated that CAMDI plays a critical role in radial migration and centrosomal function.
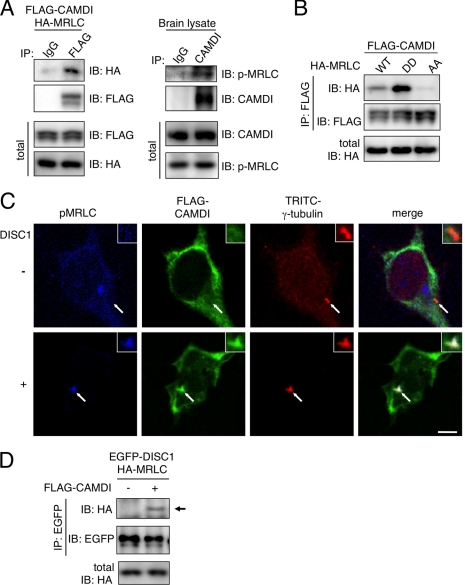
CAMDI Mediates Cross-linking between Myosin II and DISC1-centrosome
To understand CAMDI-dependent centrosomal regulation, we searched for target molecule(s) of CAMDI through a yeast two-hybrid screen of the mouse whole brain cDNA library using the CAMDI coiled-coil domain as bait. We obtained several CAMDI-binding candidates, including a MRLC 2a. We focused on MRLC in the CAMDI-DISC1 signaling pathway because myosin II is required for centrosome positioning and neuronal migration (10, 11, 35, 36). An IP-IB assay demonstrated a specific interaction of CAMDI with MRLC both in vitro and in vivo (Fig. 4A). MRLC is activated by phosphorylation, and the mutagenesis near the phosphorylatable serine, AA-MRLC and DD-MRLC (Thr18 and Ser19), has been shown to mimic the unphosphorylated and phosphorylated forms of MRLC, respectively. To understand the functional relationship between CAMDI and MRLC, we examined the binding abilities of CAMDI with active and inactive forms of MRLC. CAMDI was found to bind preferentially to phosphomimetic MRLC (MRLC-DD) but not to nonphosphorylation form of MRLC (MRLC-AA) (Fig. 4B). It is conceivable that p-MRLC exposes an interactive domain with the CAMDI coiled-coil region via conformational change, although this mechanism is currently unknown. Importantly, co-expression of CAMDI with DISC1 induced p-MRLC accumulation at/around the centrosome (Fig. 4C). Furthermore, DISC1 was found to interact with MRLC in a CAMDI-dependent manner (Fig. 4D). Taken together, CAMDI may function as a cross-linker between MRLC and the DISC1-centrosome complex.
FIGURE 4.
Identification of MRLC as a target of CAMDI. A, interactions of CAMDI with MRLC both in vitro and in vivo. IP-IB assays were performed in SH-SY5Y cells and E18 mouse brain. Equal amounts of protein expression in total lysates are shown below. B, binding of CAMDI specifically to phosphomimetic MRLC (MRLC-DD) but not to nonphosphorylation form (MRLC-AA). Equal amounts of HA-MRLC in total lysates are shown below. C, accumulation of p-MRLC to the centrosome by co-expression of CAMDI and DISC1 in COS-7 cells. Arrow indicates the position of centrosome. Scale bar, 3 μm. D, association of DISC1 with MRLC in a CAMDI-dependent manner. An arrow indicates the position of DISC1.
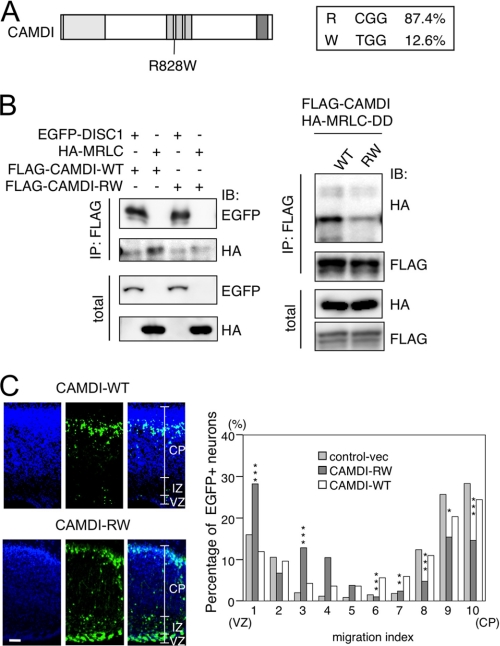
Overexpression of CAMDI-R828W Impairs Cortical Migration
On the International HapMap data base, one single-nucleotide polymorphism (SNP) rs12988301 (C/T) of the CAMDI gene is found in all the human races in a constant ratio, ∼5–19% (Fig. 5A). This SNP is an arginine to tryptophan substitution at residue 828 (R828W). As R828W is located in the coiled-coil region of CAMDI, which is responsible for interaction with MRLC and DISC1, we examined whether the R828W polymorphism affects these interactions. An IP-IB assay indicated that the interaction of R828W with MRLC or MRLC-DD was significantly attenuated, whereas the interaction of R828W with DISC1 was not changed (Fig. 5B). We next examined the effect of R828W overexpression on radial migration. In utero electroporation assay indicated an abnormal accumulation of R828W-overexpressing neurons at the VZ and IZ (Fig. 5C, left). Statistical analysis from three independent experimental results demonstrated that compared with CAMDI-WT or -R828W caused a significant increase at the VZ and IZ and decrease at the CP in the number of cortical neurons (Fig. 5C, right). A high magnification microscopic analysis of R828W-overexpressing neurons revealed a disoriented centrosome similar to CAMDI-sh phenotype (data not shown). Thus, an interaction of CAMDI with MRLC may be critical for radial migration probably via the centrosome positioning.
FIGURE 5.
Interaction of CAMDI with MRLC is required for radial migration. A, identification of SNP (R828W) in human CAMDI. B, reduced interaction of human CAMDI-R828W with MRLC and phosphomimetic MRLC. IP-IB assays indicated the reduced interaction of human CAMDI-R828W with MRLC-DD in SH-SY5Y cells (left). Note that the interaction of CAMDI-R828W with DISC1 is not changed. Amounts of protein expression in total lysates are shown below. C, CAMDI-R828W overexpression impairs radial migration. In utero electroporation experiments were performed with EGFP/control vector, EGFP/CAMDI-WT, or EGFP/CAMDI-R828W. The numbers of EGFP-positive neurons (CAMDI-WT, n = 950; CAMDI- R828W, n = 507) in the VZ, IZ, and CP are measured from three independent samples, and the average ratio (%) in each area is calculated in the right panel. Results represent the means ± S.E. Asterisks indicate significant difference between CAMDI-WT versus CAMDI R828W; ***, p < 0.001; **, p < 0.01; *, p < 0.05, Student's t test. Scale bar, 100 μm.
DISCUSSION
In this study we identified a novel DISC1-interacting protein CAMDI, which directly binds with DISC1 through the second coiled-coil domain. Both CAMDI and DISC1 genes are evolutionally conserved from the vertebrates, and the expression pattern of CAMDI in the developing mouse cortical neurons is very similar to that of DISC1. In addition, knockdown of CAMDI or DISC1 showed a similar phenotype, an impaired migration of cortical neurons with disorientated centrosomes. These lines of evidence indicate a close functional relationship between CAMDI and DISC1. A C-terminal truncated DISC1 protein, encoded from a mutation in the DISC1 gene, failed to localize to the centrosome, indicating that DISC1 plays a key role in centrosome function. Therefore, the DISC1-dependent centrosomal translocation of CAMDI suggests that CAMDI controls the cortical migration, in part, through the regulation of centrosome in concert with DISC1.
CAMDI was found to bind preferentially to p-MRLC, and co-expression with DISC1 induces an accumulation of p-MRLC at the centrosome. In migrating cortical neurons, centrosomes move continuously and often far in advance of nuclei toward the CP, followed by nuclear translocation with contraction of the soma. A previous study demonstrated that inhibition of myosin II specifically blocked the nuclear translocation (36). Thus, p-MRLC may play a pivotal role in nuclear translocation via centrosome regulation. Induction of p-MRLC may be induced by the integrin-mediated RhoA-ROCK pathway or gap junction-mediated calcium mobilization that leads to the LKB1-AMPK pathway (37–40). Myosin II has been proposed to play a part in squeezing the cell soma and forcing the nucleus forward (36). We argue that not only pushing the nucleus forward, the CAMDI-myosin II signaling pathway regulates the centrosome positioning and maturation, including microtubule dynamics associated with dynein pulling forces that enable nuclear movement. Indeed, a recent study suggested that myosin II and F-actin dynamics may function to pull the centrosome and soma forward during glial-guided migration (11).
CAMDI may regulate myosin II-dependent centrosome function in concert with DISC1. In our preliminary results, CAMDI overexpression in the hippocampus in vivo resulted in multiple dendritic spine morphology. Myosin II is reported to play a critical role in dendritic spine morphology and synaptic function (41, 42). We identified several CAMDI-interacting proteins in a yeast two-hybrid screen using coiled-coil, spectrin, and I-set domains as baits. These CAMDI-binding proteins clearly indicate CAMDI function in myosin II-dependent regulation, including vesicle trafficking to control the expression level of neurotransmitter receptors. Thus, it may be stated that myosin II is a principal target of CAMDI. Furthermore, the deregulation of the CAMDI-myosin II pathway may contribute to the pathogenesis of neurological and psychiatric disorders. Indeed, we found one SNP (R828W) in the CAMDI gene, which shows a reduced affinity for p-MRLC and impairs cortical migration (Fig. 5C). Therefore, it raises the possibility that this SNP may be a risk factor for major mental illnesses. Association studies and array-CGH analysis indicate that a candidate region for autism or related disease is mapped on chromosome 2q31 (43, 44) where the CAMDI gene is located. The DISC1 gene was also reported to be associated with autism and Asperger syndrome (45). Compared with schizophrenia, autism is a pervasive developmental disorder diagnosed in early childhood. CAMDI down-regulation caused a more severe migration defect than DISC1 down-regulation. Therefore, it is urgent to analyze the relationship between autism and the CAMDI gene including R828W. Further studies are needed to address the physiological and pathological function of CAMDI in neuronal network and developmental disorder.
Supplementary Material
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology and the Japan Society for the Promotion of Science (to T. F., R. I., and S. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) AB540232.
- DISC1
- disrupted in schizophrenia 1
- CAMDI
- coiled-coil protein associated with myosin II and DISC1
- CP
- cortical plate
- E
- embryonic day
- IP-IB
- immunoprecipitation followed by immunoblotting
- IZ
- intermediate zone
- MRLC
- myosin II regulatory light chain
- P
- postnatal day
- SNP
- single-nucleotide polymorphism
- VZ
- ventricular zone.
REFERENCES
- 1.Rakic P. (1972) J. Comp. Neurol. 145, 61–83 [DOI] [PubMed] [Google Scholar]
- 2.Kriegstein A. R., Noctor S. C. (2004) Trends Neurosci. 27, 392–399 [DOI] [PubMed] [Google Scholar]
- 3.Tsai L. H., Gleeson J. G. (2005) Neuron 46, 383–388 [DOI] [PubMed] [Google Scholar]
- 4.Schaar B. T., McConnell S. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13652–13657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marín O., Valdeolmillos M., Moya F. (2006) Trends Neurosci. 29, 655–661 [DOI] [PubMed] [Google Scholar]
- 6.Higginbotham H. R., Gleeson J. G. (2007) Trends Neurosci. 30, 276–283 [DOI] [PubMed] [Google Scholar]
- 7.Métin C., Vallee R. B., Rakic P., Bhide P. G. (2008) J. Neurosci. 28, 11746–11752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burakov A., Nadezhdina E., Slepchenko B., Rodionov V. (2003) J. Cell Biol. 162, 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulding M. B., Canman J. C., Senning E. N., Marcus A. H., Bowerman B. (2007) J. Cell Biol. 178, 1177–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenblatt J., Cramer L. P., Baum B., McGee K. M. (2004) Cell 117, 361–372 [DOI] [PubMed] [Google Scholar]
- 11.Solecki D. J., Trivedi N., Govek E. E., Kerekes R. A., Gleason S. S., Hatten M. E. (2009) Neuron 63, 63–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatten M. E. (2005) J. Cell Biol. 170, 867–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LoTurco J. J., Bai J. (2006) Trends Neurosci. 29, 407–413 [DOI] [PubMed] [Google Scholar]
- 14.Gressens P. (2006) Curr. Opin. Neurol. 19, 135–140 [DOI] [PubMed] [Google Scholar]
- 15.Millar J. K., Wilson-Annan J. C., Anderson S., Christie S., Taylor M. S., Semple C. A., Devon R. S., St. Clair D. M., Muir W. J., Blackwood D. H., Porteous D. J. (2000) Hum. Mol. Genet. 9, 1415–1423 [DOI] [PubMed] [Google Scholar]
- 16.Chubb J. E., Bradshaw N. J., Soares D. C., Porteous D. J., Millar J. K. (2008) Mol. Psychiatry 13, 36–64 [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi K., Asanuma M., Miyazaki I., Diaz-Corrales F. J., Katayama T., Tohyama M., Ogawa N. (2004) Biochem. Biophys. Res. Commun. 317, 1195–1199 [DOI] [PubMed] [Google Scholar]
- 18.Morris J. A., Kandpal G., Ma L., Austin C. P. (2003) Hum. Mol. Genet. 12, 1591–1608 [DOI] [PubMed] [Google Scholar]
- 19.Kamiya A., Tan P. L., Kubo K., Engelhard C., Ishizuka K., Kubo A., Tsukita S., Pulver A. E., Nakajima K., Cascella N. G., Katsanis N., Sawa A. (2008) Arch. Gen. Psychiatry 65, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka T., Serneo F. F., Higgins C., Gambello M. J., Wynshaw-Boris A., Gleeson J. G. (2004) J. Cell Biol. 165, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu T., Ayala R., Nguyen M. D., Xie Z., Gleeson J. G., Tsai L. H. (2004) Neuron 44, 263–277 [DOI] [PubMed] [Google Scholar]
- 22.Brandon N. J., Millar J. K., Korth C., Sive H., Singh K. K., Sawa A. (2009) J. Neurosci. 29, 12768–12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korth C. (2009) Rev. Neurosci. 20, 321–330 [DOI] [PubMed] [Google Scholar]
- 24.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., Ross C. A., Hatten M. E., Nakajima K., Sawa A. (2005) Nat. Cell Biol. 7, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 25.Clapcote S. J., Lipina T. V., Millar J. K., Mackie S., Christie S., Ogawa F., Lerch J. P., Trimble K., Uchiyama M., Sakuraba Y., Kaneda H., Shiroishi T., Houslay M. D., Henkelman R. M., Sled J. G., Gondo Y., Porteous D. J., Roder J. C. (2007) Neuron 54, 387–402 [DOI] [PubMed] [Google Scholar]
- 26.Hikida T., Jaaro-Peled H., Seshadri S., Oishi K., Hookway C., Kong S., Wu D., Xue R., Andradé M., Tankou S., Mori S., Gallagher M., Ishizuka K., Pletnikov M., Kida S., Sawa A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14501–14506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pletnikov M. V., Ayhan Y., Xu Y., Nikolskaia O., Ovanesov M., Huang H., Mori S., Moran T. H., Ross C. A. (2008) Mol. Psychiatry 13, 173–186 [DOI] [PubMed] [Google Scholar]
- 28.Li W., Zhou Y., Jentsch J. D., Brown R. A., Tian X., Ehninger D., Hennah W., Peltonen L., Lonnqvist J., Huttunen M. O., Kaprio J., Trachtenberg J. T., Silva A. J., Cannon T. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18280–18285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kvajo M., McKellar H., Arguello P. A., Drew L. J., Moore H., MacDermott A. B., Karayiorgou M., Gogos J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen S., Lang B., Nakamoto C., Zhang F., Pu J., Kuan S. L., Chatzi C., He S., Mackie I., Brandon N. J., Marquis K. L., Day M., Hurko O., McCaig C. D., Riedel G., St. Clair D. (2008) J. Neurosci. 28, 10893–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niwa M., Kamiya A., Murai R., Kubo K., Gruber A. J., Tomita K., Lu L., Tomisato S., Jaaro-Peled H., Seshadri S., Hiyama H., Huang B., Kohda K., Noda Y., O'Donnell P., Nakajima K., Sawa A., Nabeshima T. (2010) Neuron 65, 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweeney H. L., Yang Z., Zhi G., Stull J. T., Trybus K. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamisoyama H., Araki Y., Ikebe M. (1994) Biochemistry 33, 840–847 [DOI] [PubMed] [Google Scholar]
- 34.Mitchison T. J., Kirschner M. W. (1986) Methods Enzymol. 134, 261–268 [DOI] [PubMed] [Google Scholar]
- 35.Bellion A., Baudoin J. P., Alvarez C., Bornens M., Métin C. (2005) J. Neurosci. 25, 5691–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai J. W., Bremner K. H., Vallee R. B. (2007) Nat. Neurosci. 10, 970–979 [DOI] [PubMed] [Google Scholar]
- 37.Elias L. A., Wang D. D., Kriegstein A. R. (2007) Nature 448, 901–907 [DOI] [PubMed] [Google Scholar]
- 38.Asada N., Sanada K., Fukada Y. (2007) J. Neurosci. 27, 11769–11775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams T., Brenman J. E. (2008) Trends Cell Biol. 18, 193–198 [DOI] [PubMed] [Google Scholar]
- 40.Wei C., Wang X., Chen M., Ouyang K., Song L. S., Cheng H. (2009) Nature 457, 901–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H., Webb D. J., Asmussen H., Niu S., Horwitz A. F. (2005) J. Neurosci. 25, 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryu J., Liu L., Wong T. P., Wu D. C., Burette A., Weinberg R., Wang Y. T., Sheng M. (2006) Neuron 49, 175–182 [DOI] [PubMed] [Google Scholar]
- 43.Mencarelli M. A., Caselli R., Pescucci C., Hayek G., Zappella M., Renieri A., Mari F. (2007) Am. J. Med. Genet. A 143, 858–865 [DOI] [PubMed] [Google Scholar]
- 44.Conroy J., Cochrane L., Anney R. J., Sutcliffe J. S., Carthy P., Dunlop A., Mullarkey M., O'hici B., Green A. J., Ennis S., Gill M., Gallagher L. (2009) Am. J. Med. Genet. B Neuropsychiatr. Genet. 150B, 535–544 [DOI] [PubMed] [Google Scholar]
- 45.Kilpinen H., Ylisaukko-Oja T., Hennah W., Palo O. M., Varilo T., Vanhala R., Nieminen-von Wendt T., von Wendt L., Paunio T., Peltonen L. (2008) Mol. Psychiatry 13, 187–196 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.