Abstract
Mutations in amelogenin sequence result in defective enamel, and the diverse group of genetically altered conditions is collectively known as amelogenesis imperfecta (AI). Despite numerous studies, the detailed molecular mechanism of defective enamel formation is still unknown. In this study, we have examined the biophysical properties of a recombinant murine amelogenin (rM180) and two point mutations identified from human DNA sequences in two cases of AI (T21I and P41T). At pH 5.8 and 25 °C, wild type (WT) rM180 and mutant P41T existed as monomers, and mutant T21I formed lower order oligomers. CD, dynamic light scattering, and fluorescence studies indicated that rM180 and P41T can be classified as a premolten globule-like subclass protein at 25 °C. Thermal denaturation and refolding monitored by CD ellipticity at 224 nm indicated the presence of a strong hysteresis in mutants compared with WT. Variable temperature tryptophan fluorescence and dynamic light scattering studies showed that WT transformed to a partially folded conformation upon heating and remained stable. The partially folded conformation formed by P41T, however, readily converted into a heterogeneous population of aggregates. T21I existed in an oligomeric state at room temperature and, upon heating, rapidly formed large aggregates over a very narrow temperature range. Thermal denaturation and refolding studies indicated that the mutants are less stable and exhibit poor refolding ability compared with WT rM180. Our results suggest that alterations in self-assembly of amelogenin are a consequence of destabilization of the intrinsic disorder. Therefore, we propose that, like a number of other human diseases, AI appears to be due to the destabilization of the secondary structure as a result of amelogenin mutations.
Keywords: Biophysics, Protein Chemistry, Protein Conformation, Protein Denaturation, Protein Self-assembly, Amelogenesis Imperfecta, Amelogenin, Circular Dichroism, Enamel Biomineralization, Protein Thermal Stability
Introduction
Tooth enamel is one of the hardest and most heavily mineralized vertebrate tissues that can withstand wear without catastrophic failure during the entire life span of an organism (1, 2). The formation of enamel takes place in an extracellular environment, including an array of complex proteins and proteases in three main stages, namely the secretory, transition, and maturation stages (3). As the enamel development progresses, proteases such as enamelysin (MMP-20) and later kallikrein 4 (KLK-4) cleave the proteins, which are removed from the mineralization site in the extracellular matrix enabling the growth of enamel crystals and resulting in enamel hardness. Thus, during amelogenesis, the soft mineralized tissue formed during the early stages becomes hard and tough and almost devoid of any proteins at the maturation stage. Amelogenin is the major constituent (∼90%) of the protein matrix during the secretory stage and, together with other proteins in enamel, is responsible for the hierarchical structure observed in the enamel prisms (4–6).
In vitro amelogenin self-assembles into spherical structures, and this property depends on pH, ionic strength, and protein concentration (7). Using CD spectropolarimetry and NMR, we have reported that recombinant porcine amelogenin, under acidic conditions, can be classified as an intrinsically disordered or natively unfolded protein (IDP)4 (8, 9). In addition, we used various computational and experimental approaches to identify unfolded regions in amelogenin, further supporting the notion that it is a member of the IDP family (10). Unlike folded proteins, IDPs lack regular secondary or tertiary structure and instead exist in an ensemble of conformations (9). The resultant extended structure is thought to provide an increased surface area of interaction, the flexibility to interact with different partners, induced folding upon binding to partners, and accessibility to post-translational modifications (11–13). Notably, amelogenin exhibits properties that are hallmarks of IDPs, such as susceptibility to proteolysis, a high abundance of proline, and the ability to interact with other matrix components (8, 14). A number of acidic extracellular matrix proteins (ECM) associated with bone and tooth formation have also been characterized as being disordered (15, 16). The amino acid composition of amelogenin, however, differs from that of other ECM proteins. It lacks an appreciable amount of charged residues but contains higher amounts of hydrophobic residues (supplemental Fig. S1). Thus, in a charge-hydrophobicity plot amelogenin is placed at the border between the completely disordered and natively folded proteins (8, 9), whereas other ECM proteins completely fall in the disordered region.5 Based on double wavelength CD plot spectra, we have defined porcine amelogenin in the subgroup of a “pre-molten globule” (PMG) state, meaning that it is not completely disordered but has residual secondary structure. Although a number of ECM proteins associated with bone and dentin have strong affinity for calcium, amelogenin is a relatively weak calcium binder but has a strong tendency to self-assemble (5, 7, 9).
Mutations observed in the AMELX, ENAM, KLK-4, and MMP-20 gene sequences lead to abnormal enamel formation and the diverse groups of genetically altered conditions known as amelogenesis imperfecta (18–22). Of the 23 mutations reported to date, 14 of the mutations are identified in the AMELX gene. These coding errors can disturb the secretion of amelogenin, produce mutations in the tyrosine-rich amelogenin polypeptide (TRAP) region, and cause truncation of amelogenin at the C terminus. All of these changes can contribute to defective enamel formation. Clinical manifestations of AI are heterogeneous and vary depending on the affected gene and location of the mutation. The enamel structure of the affected individuals is defined as either hypoplasia or hypomineralization or a combination of these (19). Three missense mutations at the N terminus of human amelogenin involve substitution of threonine 21 to isoleucine (T21I), proline 40 to threonine (P40T), and histidine 47 to isoleucine (H47I). The T21I substitution in human amelogenin results in a phenotype described as hypomineralized/hypomatured enamel with brown discoloration (20). The P40T mutation results in hypomaturation with a discolored phenotype (21). The regular hierarchical prism structure observed in enamel is lacking the hypomineralized enamel, and the overall enamel shows porous and disoriented carbonated apatite crystals (22). It has been shown that the hypomineralized enamel exhibits poor hardness and elastic modulus compared with the unaffected enamel (23, 24). As a result, the defective enamel fractures readily.
The amino acid sequences of amelogenin across different vertebrates are highly conserved (25). Although the T21I point mutation from the human sequence occurs in an identical position in mouse amelogenin, the human P40T mutation occurs as Pro-41 in the mouse due to an additional methionine at residue 29 in the mouse (supplemental Fig. S1). Using recombinant mouse amelogenin and the AI mutants, Moradian-Oldak et al. (26) have shown that the mutant amelogenins generated more heterogeneous assemblies than the wild type. It has been further reported that the interactions between wild type and AI variants are reduced by about 43% for the T21I mutant and 26% for the P41T mutants (27). Tanimoto et al. (28) have shown that the P41T mutation reduced the interaction of amelogenin with MMP-20, and the observed reduction in processing was purported to play an important role in the defective enamel formation.
Despite these studies, details on the etiology of AI and the associated phenotypes remain largely unknown. Alterations in the self-assembly properties of amelogenin as a result of mutations are a clear manifestation of differences in enamel structure, and considerable insights can be gained by examining their physical and chemical properties.
In this study, to gain more insight into the detailed secondary structures, degree of intrinsic disorder, and the changes in physico-chemical properties, we assessed the impact of T21I and P41T point mutations on recombinant full-length wild type (WT) mouse amelogenin (rM180) by circular dichroism, dynamic light scattering, and intrinsic tryptophan fluorescence techniques. We justify the use of recombinant proteins for the following reasons. 1) The differences in sequence between recombinant and native amelogenin are minimal and do not have a significant effect on protein self-assembly (26, 29). 2) Recombinant amelogenin has been demonstrated to be an excellent model for the study of amelogenin self-assembly (5, 7, 30). 3) Purification of native mouse amelogenin in substantial quantities free of contamination is difficult.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Preparation of the recombinant His-tagged rM180 and the mutants have been described elsewhere (26). The proteins were further purified on a Jupiter C4 semi-preparative reversed phase column (10 × 250 mm, 5 μm) Varian Prostar HPLC system (ProStar/Dynamics 6, version 6.41, Varian, Palo Alto, CA). A linear gradient of 60% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 2 ml/min was used. The amino acid sequence of mouse and human amelogenins shares ∼86% identity, and the two mutational spots are highly conserved (supplemental Fig. S1). Compared with the native mouse amelogenin sequence, all the proteins contain “RGHHHHHHGS” residues at the N terminus and lack a phosphorylation on Ser-16 (supplemental Fig. S2) (26, 30).
CD Spectropolarimetry
CD spectra were obtained with a Jasco J-810 spectropolarimeter (A J-815 was used to perform the experiments displayed in supplemental Fig. S3) at a protein concentration of 0.4 mg/ml in 25 mm sodium acetate buffer, pH 5.8. Unless otherwise stated, the same concentration and buffer were used for all other experiments. The far UV-CD spectra were recorded using a 0.1-cm path length cell under constant nitrogen flush with a step size of 0.1 nm, bandwidth of 2 nm, and an averaging time of 3 s. The final spectra reported were an average of 16 scans. All the spectra were background-subtracted and smoothed by the Savitzky-Golay method using a window size of 5 nm (no smoothing was performed for any of the scans performed by the J-815). For the variable temperature CD spectra, an external thermostat was used that controlled the temperature to within 0.1 °C. The thermal denaturation of proteins was determined by monitoring the changes in CD intensity at 200 and 224 nm for all the proteins. The protein solutions were heated from 10 to 70 °C and then cooled back to 10 °C at a rate of 5 °C/min. For the thermal stability measurements, the data obtained from CD intensity at 200 nm were fit into a two-state model using the Origin 7.5 software. The thermodynamic parameters were determined by the procedure reported by Pace and Scholtz (31). For the refolding kinetics, protein solutions (0.4 mg/ml) were heated to 70 °C and kept at that temperature for 5 min before being cooled to 25 °C. Changes in CD intensity at 200 nm as a function of time were then monitored. We noted that rM180 and P41T rapidly returned to equilibrium. We therefore monitored the changes from the denaturation temperature onward. The samples reached 25 °C in 70 s, and the data were fit into a single exponential function using Origin 7.5 software.
Fluorescence Spectroscopy
Intrinsic tryptophan fluorescence spectra were recorded on a QuantaMaster QM-4SE spectrofluorometer (Photon Technology International). The protein solution (0.4 mg/ml, pH 5.8, sodium acetate buffer (25 mm), 25 °C) was excited at 290 nm, and the emission spectra were monitored between 300 and 400 nm with a step size of 1 nm, using a 10-mm path length cell. For variable temperature measurements, the Peltier controller was used. The temperature scan was performed at 5 °C intervals from 25 to 70 °C. Measurements were made at each temperature after 5 min to allow the sample to equilibrate in the cell. 8-Anilinonaphthalene sulfonate (ANS) fluorescence was recorded after excitation of the dye or dye-protein complexes at 350 nm. The emission spectra were monitored between 400 and 600 nm.
Dynamic Light Scattering
Measurements of the hydrodynamic radii of the various amelogenin proteins (0.4 mg/ml, pH 5.8, 25 mm sodium acetate buffer) were performed using a Wyatt DynaPro Nanostar dynamic light scattering instrument (Wyatt Technology, Santa Barbara, CA). For room temperature experiments, the temperature was set to 25 °C, although variable temperature experiments were performed between 20 and 60 °C at a ramp rate of 0.25 °C/min. The data were analyzed using Dynamics 7.0 software. The dynamic light scattering data were produced by the program performing a regularization fit using the Dynals algorithm on the resultant autocorrelation functions. A Rayleigh sphere model was used for the analysis, meaning that the hydrodynamic radii calculated were sphere-equivalent radii. By measuring the fluctuations in the laser light intensity scattered by the sample, the instrument was able to detect the speed (diffusion coefficient) at which the particles were moving through the medium. This value is converted to hydrodynamic radius using the Stokes-Einstein relation shown in Equation 1,
 |
where D is the is the diffusion coefficient; k is the Boltzmann constant; T is the absolute temperature; η is the viscosity, and RH is the sphere-equivalent hydrodynamic radius (32).
RESULTS
Conformation and Hydrodynamic Radii of WT and Mutant Amelogenins
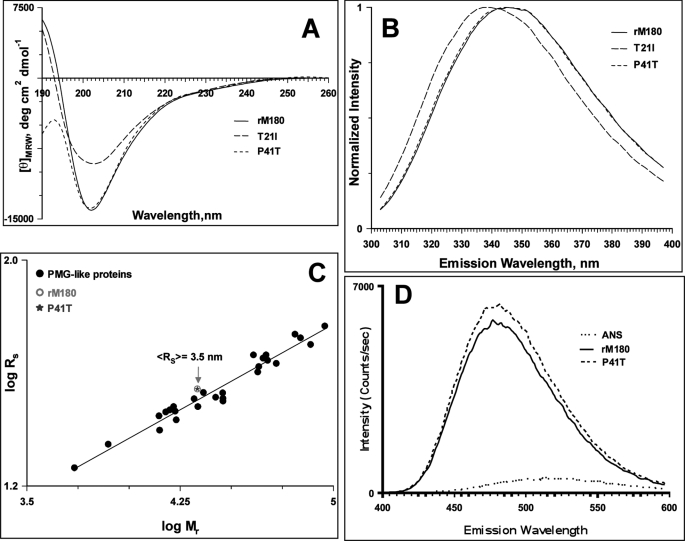
The secondary structures of WT rM180 and mutant amelogenins were recorded in 25 mm sodium acetate buffer, pH 5.8. At 25 °C, WT and mutant amelogenins all exhibited a strong negative minimum around 202 nm, which is characteristic of intrinsically disordered proteins (Fig. 1A). For the T21I mutant, CD spectra showed a similar minimum around 202 nm, but the CD intensity was much lower than the other two proteins.
FIGURE 1.
Characterization of wild type rM180 and mutant amelogenins. A, CD spectra of amelogenins in 25 mm sodium acetate buffer, pH 5.8, at 25 °C. B, normalized fluorescence emission spectra of amelogenins at 25 °C. C, dependence of the hydrodynamic radii on the relative molecular mass for PMG-like subclass of IDPs taken from Ref. 36. An open circle and a star represent the hydrodynamic radii of rM180 and P41T mutant determined by DLS, respectively. D, ANS fluorescence in the presence of rM180 and P41T at 25 °C.
Steady state tryptophan fluorescence is a useful technique to probe the environment around the aromatic amino acid residues (33, 34). Illuminating proteins with 290-nm excitation wavelength selectively excites tryptophan. Fig. 1B shows the plot of normalized emission intensity versus emission wavelength for all three amelogenins at 25 °C. Both the WT and the P41T mutant had an emission maximum around 346 nm, a characteristic feature of “class III” proteins that have tryptophan residues exposed to mobile water molecules (34). However, for the T21I mutant, the maximum was blue-shifted to 338 nm (characteristic of “class II” proteins) indicating partially buried nature of the fluorophores.
DLS allows the measurement of hydrodynamic radii of proteins. DLS studies revealed that WT rM180 and the P41T mutant had hydrodynamic radii of 3.4 and 3.5 nm, respectively (Table 1), which are smaller than expected for a completely extended conformation (4.2 nm) and larger than for a globular conformation (2.2 nm) of proteins with the same amino acid chain lengths (35). Based on the analysis of CD spectra and hydrodynamic dimensions, Uversky classified IDPs into intrinsic coil-like and intrinsic premolten globule-like (PMG) proteins (36). The two subclasses can be distinguished by the relationship between increases in hydrodynamic radii and molecular weight (36). Fig. 1C compares the hydrodynamic radii of PMG-like proteins with that of rM180 and P41T mutant. The hydrodynamic radii for the two proteins have an excellent agreement with the dimensions of PMG-like proteins. To further confirm that rM180 and P41T belong to the PMG-like subclass, ANS binding assays were recorded (Fig. 1D). Excitation of ANS at 350 nm had an emission maximum at 505 nm in acetate buffer, pH 5.8. However, when added to rM180 or P41T mutant, the fluorescence maximum was blue-shifted to 475 nm, and an 18–20-fold increase in intensity of ANS was observed in comparison with the emission in the absence of rM180 or P41T (Fig. 1D). Thus, ANS fluorescence and DLS measurements confirm that at pH 5.8 and 25 °C, rM180 and the P41T mutant are monomeric and more compact (PMG-like) than completely extended IDPs. Interestingly, at the same pH and protein concentration, the T21I mutant existed as an oligomer with a hydrodynamic radius of 7 nm (Table 1).
TABLE 1.
Hydrodynamic radii and polydispersity values (% PD) of rM180 and the mutants at 25 °C, pH 5.8, as determined by DLS analysis
| Protein | RH | Mass | PD |
|---|---|---|---|
| nm | % | % | |
| rM180 | 3.41 ± 0.15 | 97.04 ± 1.09 | 24.29 ± 10.97 |
| T21I | 7.07 ± 0.23 | 98.81 ± 1.52 | 34.6 ± 9.81 |
| P41T | 3.52 ± 0.11 | 98.98 ± 1.06 | 41.97 ± 12.19 |
Denaturation and Refolding Hysteresis in the WT rM180 and Mutant Amelogenins
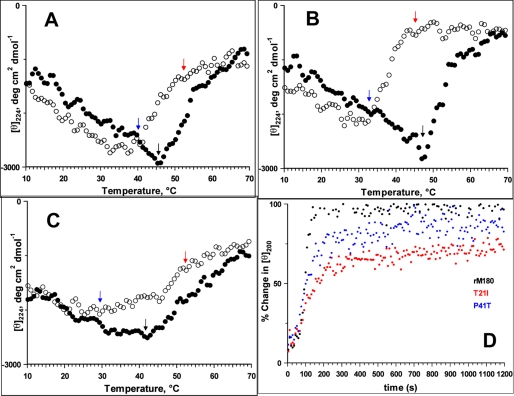
A hallmark of IDPs is that the heat-induced conformational transition is reversible (37). To infer the effect of temperature on WT rM180 and the two AI mutants, the thermal denaturation/refolding behavior of amelogenins was examined by monitoring the CD intensity at 224 nm as a function of temperature. Fig. 2, A–C, show the changes in CD intensity at 224 nm as a function of temperature for the three proteins. Unlike the linear thermal transition observed for recombinant porcine amelogenin and other IDPs, rM180 exhibited a biphasic behavior (Fig. 2A) (8, 9). At low temperatures, the CD intensity increased with temperature indicating a noncooperative unfolded-to-folded transition. Above 45 °C, denaturation began with a concomitant reduction in CD intensity (indicated by black arrow in Fig. 2A), as has been observed for folded proteins (38). On cooling (open circles), the CD intensity retraced back slowly, and refolding began at 50 °C (indicated by red arrow in Fig. 2A). Complete refolding occurred at around 41 °C (indicated by blue arrow in Fig. 2A), and the CD intensity followed the same path as the heating cycle upon further cooling. Thus, WT rM180 exhibited a weak hysteresis upon denaturation (heating) and refolding (cooling). While heating the T21I mutant, the CD intensity increased at a similar rate, but it decreased much more sharply above 45 °C and reached a plateau at higher temperatures (Fig. 2B). On cooling, the intensity retraced more slowly than rM180 and refolding began below 42 °C (indicated by red arrow in Fig. 2B), and the process was completed at 33 °C (indicated by blue arrow in Fig. 2B). Thus, the T21I mutant exhibited a larger hysteresis deflection than was observed for rM180. The denaturation of the P41T mutant followed the same path as rM180 (Fig. 2C). The onset of refolding, however, occurred around 45 °C (indicated by red arrow in Fig. 2C), but complete refolding was achieved only below 30 °C (indicated by blue arrow in Fig. 2B). The loss in CD intensity at elevated temperatures suggests that rM180 and the mutants denature to an aggregated state. To further understand the thermal behavior, all three proteins were subjected to multiple heating/cooling cycles and monitored as before (supplemental Fig. S3). For rM180, the CD intensity patterns derived from various cycles could be superimposed. However, both mutants showed considerable lag after the 1st heating cycle (supplemental Fig. S3).
FIGURE 2.
Effects of mutations on the thermal properties of rM180. Thermal denaturation (solid circles)/refolding (open circles) and refolding kinetics of amelogenins monitored by CD spectra are shown: rM180 (A), T21I (B), and P41T (C). The arrows in the figures indicate the temperatures of onset of denaturation (black arrow), onset of refolding (red arrow), and complete refolding (blue arrow). The proteins were denatured by heating from 10 to 70 °C at 5 °C/min. Refolding was monitored between 70 and 10 °C at 5 °C/min. D, refolding kinetics of amelogenin monitored by CD ellipticity at 200 nm and 25 °C. The heat-denatured proteins were cooled to 25 °C within ∼70 s, and the change in ellipticity was monitored over time. The time-dependent changes in the CD intensity at 200 nm are normalized to the initial CD intensity (at 25 °C) before denaturation of the proteins.
The presence of a hysteresis during thermal denaturation implies that unfolding/refolding takes a different path and varies for the two mutants compared with WT rM180. For example, during refolding hardly any original conformation was detectable at 37 °C for the two mutants, whereas rM180 reaches the original conformation below 42 °C. Therefore, to assess the influence of mutations on the refolding ability, the proteins were completely denatured by heating to 70 °C and cooled rapidly to 25 °C. The change in ellipticity values at 200 nm was monitored over time. Fig. 2D shows the kinetics of refolding for WT and the mutant proteins. For all the proteins, a large change in signal occurred within the first few seconds. This burst phase was accompanied by a slower observable phase. For rM180, an almost complete conversion from an aggregated state to the monomeric conformation was reached within seconds (time required to reach 50% monomer conformation, t50% = ∼93 s) implying a rapid refolding ability. For the T21I mutant, the initial burst phase was followed by a very slow refolding (t50% = ∼155 s). The refolding was considerably slower than rM180 and even after 2 h managed to achieve only ∼78% of the initial oligomeric conformation. The P41T mutation followed the same path as rM180 during the initial phase of the refolding. However, its refolding rate was slower than rM180, and ∼95% monomeric conformation was achieved after 2 h (t50% = ∼101 s).
The time courses for the wild type rM180 and P41T mutants were fit into a double exponential function to determine the intrinsic rate constants for refolding (supplemental Fig. S4). For rM180, the rate constant for the faster reaction (k1) was 15.3 ± 3.1 × 10−3 s−1 and for the slower reaction (k2) was 6.5 ± 0.93 s−1. The k1 and k2 values for P41T mutant were 9.2 ± 0.2 × 10−3 and 0.73 ± 0.05 × 10−3 s−1, respectively. These results indicate about a 9-fold decrease in k2 as a result of P41T substitution, although k1 was not affected significantly. However, for the T21I mutant only ∼78% of the initial intensity was achieved after 2 h. When extrapolated to the CD intensity at 100% initial intensity, the k1 and k2 values were 6.3 ± 0.08 × 10−3 and 33.82 ± 0.67 × 10−6 s−1, respectively.
Variable Temperature Fluorescence Spectra of Amelogenins
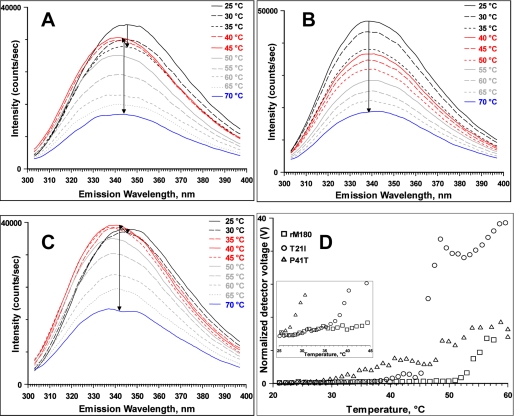
To gain further insight into the nature of the intermediates observed in the thermal transition, the fluorescence emission spectrum of each of the proteins was recorded from 25 to 70 °C at 5 °C intervals. For rM180, the emission intensity decreased with increasing temperature from 25 to 35 °C without apparent change in the emission maximum (Fig. 3A). After reaching the transition region (40 °C), the intensity increased, and the emission maximum was blue-shifted to 340 nm indicating partial burial of the tryptophan fluorophores. As the temperature was increased further, the intensity of the emission spectra decreased with simultaneous broadening and red shifting to 343 nm. For the T21I mutant, the emission intensity decreased continuously with the increase in temperature, and no intermediate shift in the emission maximum was observed (Fig. 3B). Above 50 °C, the emission maximum was red-shifted from 338 to 342 nm indicating heat-induced aggregation of the protein. The emission maximum for the P41T mutant began to shift as soon as the sample was heated to 30 °C (Fig. 3C). Raising the temperature to 35 °C resulted in a significant increase in the emission intensity and blue shift of the emission maximum to 338 nm. The intensity increased further without any change in emission maxima at 40 °C. A further rise in temperature resulted in a decrease in the intensity of emission maximum at 338 nm with a concomitant red shift of the emission maximum to 342 nm at higher temperatures. The blue shift in the emission maximum and a strong reduction in fluorescence intensity at elevated temperature are indicative of temperature-induced aggregation (39).
FIGURE 3.
Effects of mutation on structural properties of rM180. Thermal denaturation of amelogenins monitored by tryptophan fluorescence rM180 (A), T21I (B), and P41T (C). The arrows inside the panels indicate the progression of the transition. D, heat-induced changes in scattering intensity of rM180 and the mutants monitored by DLS. The change in scattering intensity with temperature between 25 and 45 °C is expanded in the inset to indicate significant scattering observed for the mutants.
Hydrodynamic Properties of the Intermediates
To understand the nature of intermediates, we performed variable temperature light scattering measurements (Fig. 3D). These measurements reflect the overall intensity of light scattered by the sample. As particles with hydrodynamic radii over 100 nm scatter light far more intensely than protein monomers and small oligomers, Fig. 3D effectively represents the concentration of aggregates within the samples by reporting a voltage value for the intensity of light detected. Each value was normalized to show the voltage that would have been detected if the laser power was 100% and the attenuation level was 0%.
For rM180, no apparent increase in scattering intensity and hence no aggregation were observed until 50 °C. When the T21I mutant was heated, no apparent increase in scattering was observed until 37 °C, although the most significant increase in scattering occurred very rapidly between 45 and 46 °C. The P41T mutant gradually increased in scattering intensity from 28 °C upward, implying that aggregation was occurring slowly.
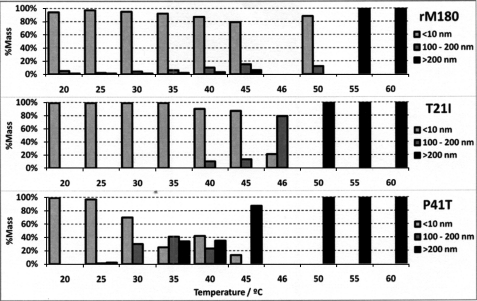
The data in Fig. 4 were derived from the same measurements in Fig. 3D and represent the dynamic light scattering analysis of the samples at temperature intervals of 5 °C. The sample composition is divided into “monomers and oligomers” (species with RH <10 nm), “small aggregates” (RH = 100–200 nm), and “large aggregates” (RH >200 nm). Particles with RH between 10 and 100 nm were not observed in significant quantities (>1% by mass) in any of the measurements and were therefore not included in the graphic.
FIGURE 4.
Variable temperature DLS data showing the particle size distribution of rM180 and the mutants. For clarity we divided the distribution into three size categories (less than 10 nm, 100–200 nm, and larger than 200 nm). Note that the size distribution is little affected by temperature for rM180, whereas P41T showed a significant population of larger sized particles (100–200 nm) at 30 °C.
The mass percentage of monomers and oligomer present in the rM180 sample was maintained above 80% until the 45 °C interval (Fig. 4). At 50 °C and above, large aggregates were the dominant species. As the particle size did not change significantly between 35 and 40 °C for rM180, it implied that the intermediate observed in the fluorescence spectra was a partially folded monomer. The T21I mutant remained primarily in its oligomeric form (RH ≈7 nm) until 45 °C (Fig. 4). After a 1 °C interval, the composition of the sample suddenly switched from 80% oligomers and 20% small aggregates to 20% oligomers and 80% small aggregates. From 50 °C upward, the mutant protein existed as large aggregates. For the P41T mutant, no significant change in scattering intensity was observed below 28 °C (Fig. 4). However, a mixture of monomers and small aggregates was observed at 30 °C. Between 30 and 45 °C, the size distribution was heterogeneous, consisting of monomers, small aggregates, and large aggregates. Above 45 °C, only large aggregates were observed. These results confirm that, for the P41T mutant, the partially folded monomer observed below 30 °C is readily converted to small aggregates above 30 °C, whereas the partially folded intermediate formed by wild type rM180 remained stable up to 50 °C.
Conformational Stability of rM180 and Mutant Amelogenins
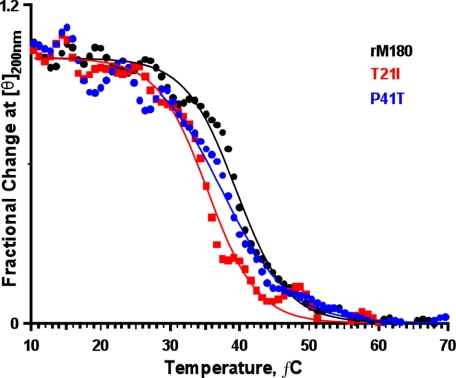
Numerous studies have shown that subtle alterations in the chemical structure of proteins can cause substantial changes in the conformational stability (40–43). To determine the conformational stability and the corresponding enthalpy and entropy changes, we studied the thermal denaturation of amelogenins by monitoring the change in CD ellipticity at 200 nm from 10 to 70 °C (44). Fig. 5 illustrates the thermal denaturation curves for rM180 and the mutants. Both of the mutants had a lower temperature of denaturation (Tm, indicated by the mid-point of denaturation/aggregation) compared with wild type rM180. The data were fitted to a two-state model and used to estimate the difference in stability (ΔΔG) between rM180 and the mutants (Table 2). A negative ΔΔG value indicates the mutant protein is less stable than rM180. The results showed that Tm and the conformational stability of the mutants were decreased relative to rM180; the decrease was greater for T21I mutant. Comparing these results with fluorescence and DLS studies, at 37 °C, the P41T mutant exists in an intermediate heterogeneous large oligomeric state, whereas rM180 exists in a partially folded conformation. On the other hand, the T21I mutant exists predominantly in the small oligomeric state at 37 °C.
FIGURE 5.
Effects of mutations on the conformational stability of rM180. Proteins were heated from 10 to 70 °C, and changes in CD intensity at 200 nm were monitored. Note that the mid-point of the transition (Tm) is lowered for the mutants compared with rM180.
TABLE 2.
Thermodynamic parameters obtained from thermal denaturation of rM180 and mutant amelogenins
Data were analyzed based on Ref. 24.
| Protein | ΔHma | ΔSmb | Tm | ΔTmc | ΔΔGd |
|---|---|---|---|---|---|
| kcal/mol | cal/mol/K | °C | °C | kcal/mol | |
| rM180 | 176 ± 7.1 | 562.9 ± 22.6 | 40.6 ± 0.1 | ||
| T21I | 102.9 ± 5.7 | 333.5 ± 18.5 | 35.3 ± 0.1 | −5.3 | −2.98 |
| P41T | 144.7 ± 5.8 | 466.1 ± 23.3 | 37.5 ± 0.2 | −3.1 | −1.75 |
a ΔHm indicates enthalpy of denaturation at Tm.
b ΔSm = ΔHm/Tm.
c ΔTm = Tm(mutant) − Tm(rM180).
d ΔΔG = 0.563 (ΔSm for rM180 in kcal/mol) × ΔTm. A negative value indicates a decrease in stability.
DISCUSSION
The physiological function of amelogenin protein in controlling enamel mineralization has been well established. Although in vivo and in vitro studies support the view that amelogenin self-assembly is critical for its structural role, detailed information on its structure and the mechanisms of its self-assembly is still lacking. Here, we investigated protein folding and refolding paradigms to understand the phenotype-genotype correlations of the amelogenin mutations occurring in patients with AI. The structural properties of two mutations observed at the N terminus of human amelogenin were studied using recombinant mouse amelogenin and characterized by variable temperature CD, light scattering, and intrinsic tryptophan fluorescence spectroscopy. We demonstrated that the mutations perturbed the secondary structure and decreased the thermal stability of rM180. The thermal properties of the two mutants were different, corroborating the phenotype variations observed in the defective enamel.
Effect of Mutations on the Secondary Structure of rM180
At 25 °C, the CD spectrum of the P41T mutant was similar to wild type rM180 with a strong minimum around 202 nm characteristic of intrinsic disorder proteins. Although the CD spectra of the T21I mutant showed a negative minimum around 202 nm, the ellipticity value was reduced significantly indicating this mutant amelogenin was more folded compared with wild type rM180 and the P41T mutant. Our data suggest that threonine to isoleucine substitution increases the secondary structure in amelogenin, whereas proline to threonine substitution does not alter the secondary structure at 25 °C. This increase in folding may have been caused by the oligomerization of the T21I mutant protein, as observed by DLS (Fig. 4).
In steady state fluorescence spectroscopy, the shape and wavelength of emission maxima are sensitive to the environment around tryptophan and can vary from 308 to 358 nm (45). rM180 and the mutants contain three tryptophan residues, two of them are located in the TRAP region (proximal to the mutation sites) and the other is located at the C terminus (supplemental Fig. S1). For proteins containing multiple tryptophan residues, the observed emission spectrum is composed of contributions from one of the five spectral classes (34). At 25 °C, the emission spectrum of rM180 and the P41T mutant were similar to the class III component that is contributed by solvent-exposed fluorophores. The similarity between rM180 and P41T further suggests that the conformation is little affected by this substitution at 25 °C. The emission maximum in T21I, on the other hand, was significantly blue-shifted to 338 nm and similar to a class II component that represents fluorophores exposed to bonded water molecules. The CD and fluorescence spectra of rM180 and the P41T mutant at 25 °C suggested that the two proteins are more compact than fully extended coil-like proteins. Comparing the hydrodynamic radii with PMG-like proteins further confirmed that rM180 and the P41T mutant are monomeric at pH 5.8 and more compact compared with extended coil-like proteins. The blue shift and large increase in ANS fluorescence intensity indicated the presence of exposed hydrophobic patches in rM180 and P41T, further substantiating the PMG-like conformation.
Interestingly, DLS studies indicated that T21I existed predominantly as an oligomer with a 7-nm hydrodynamic radius under the same conditions. The observed blue shift in the tryptophan fluorescence emission maxima and increased order in the CD spectra of T21I were attributed to the presence of oligomers and suggested that this substitution causes an increase in both intra- and intermolecular interactions in the otherwise unstructured rM180.
Thermal Properties of rM180 and the Mutant Amelogenins
The absence of any significant structural difference between wild type rM180 and the P41T mutant at 25 °C raises the question of how the mutation influences the differences observed in the enamel structure and proteolytic processing by MMP-20. Thermal denaturation and refolding studies clearly provided further evidence of intrinsic differences at a molecular level between wild type and mutant proteins. All of the amelogenins studied in this work underwent biphasic thermal transition on heating, contrary to the linear thermal transition observed for porcine amelogenin and other IDPs (8, 46–50). The differences observed between the porcine and mouse amelogenin sequences could explain this behavior (8).
The T21I mutant exhibited considerable delay in refolding causing pronounced hysteresis. The P41T mutant, however, refolded in a similar manner to rM180 during the initial stages but reached the monomeric conformation at a much lower temperature. Accordingly, k2 for P41T was about nine times lower than rM180. The effect of the T21I mutation on the refolding kinetics was so pronounced that even after 2 h only ∼78% had refolded to the initial oligomeric conformation, and there was about a 200-fold reduction in k2. These results suggest a significant kinetic barrier intervening between the denatured and the initial monomeric/oligomeric states for the mutants. Similar hysteresis has been reported for soluble NSF attachment protein receptor (SNARE) assembly, transthyretin, and bacterial luciferase during denaturation and refolding by urea and was attributed to the presence of stable intermediates (51–53). In accordance with the DLS data, it is likely that the intermediate aggregates/oligomers formed after the burst phase require more time to form the monomer, thus reducing the k2 for the two mutants. However, for rM180 this transition could occur rapidly because the intermediate is a partially folded monomer.
Thermal Properties of the Intermediates
Variable temperature intrinsic tryptophan fluorescence spectra and DLS provided further insight into the nature of the intermediates observed during thermal denaturation of rM180 and the mutant amelogenins. Upon heating, a considerable blue shift (∼5 nm) in the emission maxima was observed for rM180 followed by an increase in fluorescence intensity at 45 °C. As there was no apparent change in the scattering intensity observed for rM180 in the DLS experiments between 25 and 40 °C, it is reasonable to conclude that the buried nature of the fluorophore is due to partial folding of the disordered rM180. P41T exhibited a blue shift immediately after heating to 30 °C, concomitant with the DLS observation that heterogeneous aggregation began to occur at around 28 °C. This suggests that the P41T partially folded intermediate is far less stable than the intermediate formed by rM180. CD, light scattering, and fluorescence studies demonstrated that both rM180 and P41T transformed into a partially folded conformation on raising the temperature (the mutant transforms at lower temperature than rM180) and then to an aggregated state. The transformation of the partially folded intermediate to the aggregates occurs above 40 °C for rM180. However, substitution of proline 41 to threonine facilitates the transition to aggregates, as shown by a large scattering observed in the DLS experiments, above 28 °C. We suggest that Pro-41 plays a key role in maintaining equilibrium between the intrinsically disordered and the partially folded intermediate state, and the equilibrium is destabilized when substituted with threonine. In contrast, a greater destabilization occurs in the T21I mutant resulting in soluble oligomeric state even at 25 °C.
Mutants Populate Oligomers by Destabilizing the Disordered Conformation
The conversion of monomers into soluble oligomers and then to a fibrillar assembly is a pathogenic hallmark of unstructured polypeptides associated with neurodegenerative disease (54, 55). Mutations that populate the oligomeric states have an increased propensity to form higher order oligomers (56). Using bioinformatics, we have shown that the TRAP domain of amelogenin has a high propensity to form an ordered structure, although the remaining portions are highly disordered (9, 10). The conversion of monomers to oligomers seen with the T21I mutation suggests that the bulky hydrophobic substitution in the TRAP domain promotes intermolecular interactions and stabilizes the oligomer. However, the heterogeneous population of small aggregates seen with the P41T mutation (above 30 °C) is associated with the destabilization of the partially folded conformation and suggests that proline functions as “gatekeeper” inhibiting the access to oligomeric states that trigger an aggregation cascade. Alternatively, proline 41 can function to preserve some structural features in the monomer that favors formation of a particular oligomeric state structure required for the formation of the basic “native” nanospheres existing in the forming enamel (5). A high conservation of proline at this position in diverse vertebrate amelogenins further confirms the important role played by this residue in preventing formation of oligomers from partially folded conformations (57).
In addition, CD studies indicated that both of the mutants exhibited a moderate decrease in stability and reduced refolding rate. Our results are in complete agreement with a number of studies that showed an inverse correlation between conformational stability and propensity to form aggregates (58–60). It has been observed that substitution by proline or glycine residues at the β-turn position increases the thermal stability of proteins by decreasing the conformational entropy of the denatured state. The extent of increase in stability varies from 0.7 to 1.3 kcal/mol (61, 62). The NMR structure of porcine amelogenin revealed that Pro-41 is in a cis-configuration and assumes a β-turn conformation (8). Substitution of Pro at position 41 by Thr thus destabilizes the protein by ∼1.2 kcal/mol, in agreement with the above observations. Therefore, we suggest that Pro-41 plays an important role in conferring rigidity to the otherwise disordered amelogenin; this local rigidity is lost upon substitution of threonine at this position in the mutant condition and is therefore a concomitant destabilization. Several studies have shown that hydrogen bonding contributes significantly to conformational stability through intramolecular and intermolecular hydrogen bonding (63–65). Mutations that remove hydrogen bonding between protein and water molecules contribute ∼2.2 kcal/mol per hydrogen bond (63). The observed decrease of 2.98 kcal/mol indicates that Thr-21 may be involved in other stabilizing interactions apart from hydrogen bonding. Substitution by hydrophobic isoleucine results in the removal of this interaction, promoting oligomerization and decreasing thermal stability. Thus, the two mutations exert different influences on the secondary structure and on rM180.
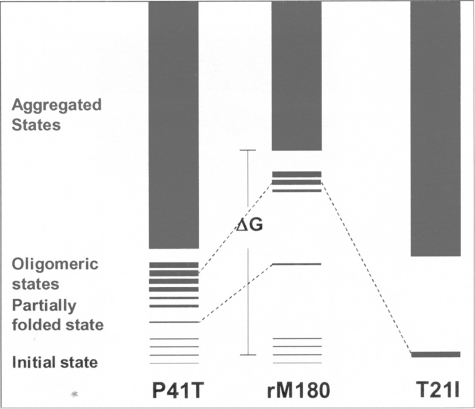
In Fig. 6, we offer a schematic representation of the structural effects of the two mutations on amelogenin secondary structure. The T21I mutation destabilizes the disordered structure, and in turn, the relative population of partially folded soluble oligomers is increased. On the other hand, the P41T mutation destabilizes the disordered structure by populating a heterogeneous distribution of small aggregates, which may be the molecular basis for reduced proteolytic processing by MMP-20 (28).
FIGURE 6.
Schematic diagram representing hypothesized energy levels for rM180 and the mutants. The initial state of rM180 is represented as a collection of isoenergetic states to indicate the presence of a conformation ensemble (i.e. intrinsic disorder). The partially folded state is represented as a single state to distinguish it from the disordered state. The intermediate oligomeric state is represented as several isoenergetic states of varying thickness to show the presence of aggregates of various sizes. Note that P41T denatures by reducing the energy difference between the disordered and partially folded state, whereas T21I denatures by reducing the energy difference between disordered and oligomeric state.
The observed differences in thermal behavior of the two mutants may be responsible for the alterations observed in the enamel phenotypes, presumably by interfering with amelogenin nanosphere assembly and proteolytic processing of amelogenin. The data produced in this study reinforce these observations and shed light on the more detailed mechanism of defective enamel formation. Recent studies have shown that, in some protein deposition diseases, small oligomers are thought to be primary pathogenic species (66). In the case of abnormal enamel formation in AI, the presence of misfolded mutant amelogenin may disturb the assembly and function of the extracellular matrix in controlling mineralization. Future study of the interaction of mutant proteins with other matrix components, minerals, and with the ameloblasts may provide a better understanding of the etiology of AI.
Supplementary Material
Acknowledgments
We thank Prof. Ralf Langen, Department of Biochemistry and Molecular Biology, Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, for use of CD facilities. We are grateful to Dr. Nickolas Chelyapov at the University of Southern California NanoBiophysics Core Facility for the use of fluorescence facilities and valuable suggestions on thermal hysteresis.
This work was supported, in whole or in part, by National Institutes of Health Grants DE-013414, DE-02009, and DE-15644 (to J. M. O.) and DE-13045 and DE-06988 (to M. L. S.) from NIDCR.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
R. Lakshminarayanan and J. Moradian-Oldak, unpublished observations.
- IDP
- intrinsically disordered protein
- DLS
- dynamic light scattering
- IDP
- intrinsically disordered protein
- AI
- amelogenesis imperfecta
- ANS
- 8-anilinonaphthalene sulfonate
- rM180
- recombinant mouse amelogenin
- PMG
- premolten globule-like
- ECM
- extracellular matrix protein
- TRAP
- tyrosine-rich amelogenin polypeptide.
REFERENCES
- 1.Robinson C., Connell S., Kirkham J., Shore R., Smith A. (2004) J. Mater. Chem. 14, 2242–2248 [Google Scholar]
- 2.Chai H., Lee J. J., Constantino P. J., Lucas P. W., Lawn B. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7289–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmer J. P., Fincham A. G. (1995) Crit. Rev. Oral Biol. Med. 6, 84–108 [DOI] [PubMed] [Google Scholar]
- 4.Snead M. L., Zeichner-David M., Chandra T., Robson K. J., Woo S. L., Slavkin H. C. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7254–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moradian-Oldak J. (2001) Matrix Biol. 20, 293–305 [DOI] [PubMed] [Google Scholar]
- 6.Gibson C. W., Yuan Z. A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J. T., Decker S., Piddington R., Harrison G., Kulkarni A. B. (2001) J. Biol. Chem. 276, 31871–31875 [DOI] [PubMed] [Google Scholar]
- 7.Wiedemann-Bidlack F. B., Beniash E., Yamakoshi Y., Simmer J. P., Margolis H. C. (2007) J. Struct. Biol. 160, 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delak K., Harcup C., Lakshminarayanan R., Sun Z., Fan Y., Moradian-Oldak J., Evans J. S. (2009) Biochemistry 48, 2272–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakshminarayanan R., Il Y., Hegde B. G., Du C., Fan D., Moradian-Oldak J. (2009) Proteins 76, 560–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moradian-Oldak J., Lakshminarayanan R. (2010) Amelogenins: Multifaceted Proteins for Dental and Bone Formation (Goldberg M. ed) p. 106, Bentham Science Publishers Ltd., Dubai, U.A.E. [Google Scholar]
- 11.Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradoviæ Z. (2002) Biochemistry 41, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 12.Dunker A. K., Silman I., Uversky V. N., Sussman J. L. (2008) Curr. Opin. Struct. Biol. 18, 756–764 [DOI] [PubMed] [Google Scholar]
- 13.Mészáros B., Tompa P., Simon I., Dosztányi Z. (2007) J. Mol. Biol. 372, 549–561 [DOI] [PubMed] [Google Scholar]
- 14.Fan D., Du C., Sun Z., Lakshminarayanan R., Moradian-Oldak J. (2009) J. Struct. Biol. 166, 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huq N. L., Cross K. J., Ung M., Reynolds E. C. (2005) Arch. Oral. Biol. 50, 599–609 [DOI] [PubMed] [Google Scholar]
- 16.Fisher L. W., Torchia D. A., Fohr B., Young M. F., Fedarko N. S. (2001) Biochem. Biophys. Res. Commun. 280, 460–465 [DOI] [PubMed] [Google Scholar]
- 17.Le T. Q., Gochin M., Featherstone J. D., Li W., DenBesten P. K. (2006) Eur. J. Oral. Sci. 114, Suppl. 1, 320–326 [DOI] [PubMed] [Google Scholar]
- 18.Wright J. T. (2006) Am. J. Med. Genet. A 140, 2547–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright J. T., Hart P. S., Aldred M. J., Seow K., Crawford P. J., Hong S. P., Gibson C. W., Hart T. C. (2003) Connect. Tissue Res. 44, Suppl. 1, 72–78 [PubMed] [Google Scholar]
- 20.Lench N. J., Winter G. B. (1995) Hum. Mutat. 5, 251–259 [DOI] [PubMed] [Google Scholar]
- 21.Hart P. S., Aldred M. J., Crawford P. J., Wright N. J., Hart T. C., Wright J. T. (2002) Arch. Oral Biol. 47, 261–265 [DOI] [PubMed] [Google Scholar]
- 22.Mahoney E. K., Rohanizadeh R., Ismail F. S., Kilpatrick N. M., Swain M. V. (2004) Biomaterials 25, 5091–5100 [DOI] [PubMed] [Google Scholar]
- 23.Xie Z., Swain M., Munroe P., Hoffman M. (2008) Biomaterials 29, 2697–2703 [DOI] [PubMed] [Google Scholar]
- 24.Xie Z. H., Mahoney E. K., Kilpatrick N. M., Swain M. V., Hoffman M. (2007) Acta Biomater. 3, 865–872 [DOI] [PubMed] [Google Scholar]
- 25.Toyosawa S., O'hUigin C., Figueroa F., Tichy H., Klein J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moradian-Oldak J., Paine M. L., Lei Y. P., Fincham A. G., Snead M. L. (2000) J. Struct. Biol. 131, 27–37 [DOI] [PubMed] [Google Scholar]
- 27.Paine M. L., Lei Y. P., Dickerson K., Snead M. L. (2002) J. Biol. Chem. 277, 17112–17116 [DOI] [PubMed] [Google Scholar]
- 28.Tanimoto K., Le T., Zhu L., Witkowska H. E., Robinson S., Hall S., Hwang P., Denbesten P., Li W. (2008) J. Dent. Res. 87, 451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmer J. P., Lau E. C., Hu C. C., Aoba T., Lacey M., Nelson D., Zeichner-David M., Snead M. L., Slavkin H. C., Fincham A. G. (1994) Calcif. Tissue Int. 54, 312–319 [DOI] [PubMed] [Google Scholar]
- 30.Fincham A. G., Moradian-Oldak J., Sarte P. E. (1994) Calcif. Tissue Int. 55, 398–400 [DOI] [PubMed] [Google Scholar]
- 31.Pace C. N., Scholtz J. M. (1997) in Protein Structure: A Practical Approach (Creighton T. E. ed) pp. 299–321, IRL Press, Oxford, UK [Google Scholar]
- 32.Schärtl W. (2007) Light Scattering from Polymer Solutions and Nanoparticle Dispersions, 1st Ed., Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 33.Lakowicz J. R. (1999) Principles of Fluorescence Spectroscopy, Kluwer Academic/Plenum Press, New York [Google Scholar]
- 34.Shen C., Menon R., Das D., Bansal N., Nahar N., Guduru N., Jaegle S., Peckham J., Reshetnyak Y. K. (2008) Proteins 71, 1744–1754 [DOI] [PubMed] [Google Scholar]
- 35.Uversky V. N. (2002) Eur. J. Biochem. 269, 2–12 [DOI] [PubMed] [Google Scholar]
- 36.Uversky V. N. (2002) Protein Sci. 11, 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uversky V. N. (2009) Protein J. 28, 305–325 [DOI] [PubMed] [Google Scholar]
- 38.Hill R. B., DeGrado W. F. (2000) Structure 8, 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duy C., Fitter J. (2006) Biophys. J. 90, 3704–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almstedt K., Mårtensson L. G., Carlsson U., Hammarström P. (2008) Biochemistry 47, 1288–1298 [DOI] [PubMed] [Google Scholar]
- 41.Matthews B. W. (1995) Adv. Protein Chem. 46, 249–278 [DOI] [PubMed] [Google Scholar]
- 42.Shortle D. (1992) Q. Rev. Biophys. 25, 205–250 [DOI] [PubMed] [Google Scholar]
- 43.Harada A., Yagi H., Saito A., Azakami H., Kato A. (2007) Biosci. Biotechnol. Biochem. 71, 2952–2961 [DOI] [PubMed] [Google Scholar]
- 44.Nicolini C., Ravindra R., Ludolph B., Winter R. (2004) Biophys. J. 86, 1385–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pain R. H. (2004) Current Protocols in Protein Science, pp. 7.7.1–7.7.20, John Wiley & Sons, Inc., New York [Google Scholar]
- 46.Lim K. H., Collver H. H., Le Y. T., Nagchowdhuri P., Kenney J. M. (2007) Biochem. Biophys. Res. Commun. 353, 443–449 [DOI] [PubMed] [Google Scholar]
- 47.Ishijima J., Nagasaki N., Maeshima M., Miyano M. (2007) J. Biochem. 142, 201–211 [DOI] [PubMed] [Google Scholar]
- 48.Danielsson J., Jarvet J., Damberg P., Gräslund A. (2005) FEBS J. 272, 3938–3949 [DOI] [PubMed] [Google Scholar]
- 49.Lam S. L., Hsu V. L. (2003) Biopolymers 69, 270–281 [DOI] [PubMed] [Google Scholar]
- 50.Soulages J. L., Kim K., Arrese E. L., Walters C., Cushman J. C. (2003) Plant Physiol. 131, 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fasshauer D., Antonin W., Subramaniam V., Jahn R. (2002) Nat. Struct. Biol. 9, 144–151 [DOI] [PubMed] [Google Scholar]
- 52.Sinclair J. F., Ziegler M. M., Baldwin T. O. (1994) Nat. Struct. Biol. 1, 320–326 [DOI] [PubMed] [Google Scholar]
- 53.Lai Z., McCulloch J., Lashuel H. A., Kelly J. W. (1997) Biochemistry 36, 10230–10239 [DOI] [PubMed] [Google Scholar]
- 54.Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 55.Uversky V. N., Lee H. J., Li J., Fink A. L., Lee S. J. (2001) J. Biol. Chem. 276, 43495–43498 [DOI] [PubMed] [Google Scholar]
- 56.Bitan G., Vollers S. S., Teplow D. B. (2003) J. Biol. Chem. 278, 34882–34889 [DOI] [PubMed] [Google Scholar]
- 57.Steward A., Adhya S., Clarke J. (2002) J. Mol. Biol. 318, 935–940 [DOI] [PubMed] [Google Scholar]
- 58.Ramirez-Alvarado M., Merkel J. S., Regan L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8979–8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramírez-Alvarado M., Regan L. (2002) J. Mol. Biol. 323, 17–22 [DOI] [PubMed] [Google Scholar]
- 60.Stathopulos P. B., Rumfeldt J. A., Scholz G. A., Irani R. A., Frey H. E., Hallewell R. A., Lepock J. R., Meiering E. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7021–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trevino S. R., Schaefer S., Scholtz J. M., Pace C. N. (2007) J. Mol. Biol. 373, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prajapati R. S., Das M., Sreeramulu S., Sirajuddin M., Srinivasan S., Krishnamurthy V., Ranjani R., Ramakrishnan C., Varadarajan R. (2007) Proteins 66, 480–491 [DOI] [PubMed] [Google Scholar]
- 63.Pace C. N., Shirley B. A., McNutt M., Gajiwala K. (1996) FASEB J. 10, 75–83 [DOI] [PubMed] [Google Scholar]
- 64.Bulaj G., Goldenberg D. P. (2001) J. Mol. Biol. 313, 639–656 [DOI] [PubMed] [Google Scholar]
- 65.Myers J. K., Pace C. N. (1996) Biophys. J. 71, 2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campioni S., Mannini B., Zampagni M., Pensalfini A., Parrini C., Evangelisti E., Relini A., Stefani M., Dobson C. M., Cecchi C., Chiti F. (2010) Nat. Chem. Biol. 6, 140–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.