Abstract
Smac mimetic compounds (SMCs) potentiate TNFα-mediated cancer cell death by targeting the inhibitor of apoptosis (IAP) proteins. In addition to TNFα, the tumor microenvironment is exposed to a number of pro-inflammatory cytokines, including IL-1β. Here, we investigated the potential impact of IL-1β on SMC-mediated death of cancer cells. Synergy was seen in a subset of a diverse panel of 21 cancer cell lines to the combination of SMC and IL-1β treatment, which required IL-1β-induced activation of the NF-κB pathway. Elevated NF-κB activity resulted in the production of TNFα, which led to apoptosis dependent on caspase-8 and RIP1. In addition, concurrent silencing of cIAP1, cIAP2, and X-linked IAP by siRNA was most effective for triggering IL-1β-mediated cell death. Importantly, SMC-resistant cells that produced TNFα in response to IL-1β treatment were converted to an SMC-sensitive phenotype by c-FLIP knockdown. Reciprocally, ectopic expression of c-FLIP blocked cell death caused by combined SMC and IL-1β treatment in sensitive cancer cells. Together, our study indicates that a positive feed-forward loop by pro-inflammatory cytokines can be exploited by SMCs to induce apoptosis in cancer cells.
Keywords: Anticancer Drug, Apoptosis, Cytokine Action, Drug Resistance, NF-kappa B, Tumor Necrosis Factor (TNF), IAP, IL-1beta, Smac Mimetic Compounds, c-FLIP
Introduction
Inflammation is intricately linked to various aspects of tumorigenesis. An inflammatory microenvironment contributes to cancer progression by driving the proliferation of tumor cells as well as by promoting survival, angiogenesis, and metastasis (1–3). Furthermore, the pro-inflammatory condition within tumors represses adaptive immune response and rewires signaling pathways that respond to hormones and chemotherapeutics (1–3). Cytokines and chemokines are the principal mediators of pro-inflammatory responses that shape the progression of malignancy. In particular, tumor cells or infiltrating cells such as macrophages produce abundant amounts of tumor necrosis factor α (TNFα) and interleukin-1β (IL-1β) at tumor sites (1–3) where these molecules have an important role in activating the inflammatory process that promotes angiogenesis, invasiveness, and repression of anti-tumor immunity (4, 5). TNFα and IL-1β further contribute to the pro-inflammatory response through the activation of transcription factor NF-κB. Recent evidence implicates that the cellular inhibitor of apoptosis 1 and 2 (cIAP1 and cIAP2) proteins are critical modulators for TNFα-induced activation of NF-κB (6, 7). Targeting of cIAP1 and cIAP2 by siRNA or small molecule inhibitor blunts TNFα-induced NF-κB activation, switching the cellular response from a pro-inflammatory to a pro-apoptotic signal (6, 7).
Smac mimetic compounds (SMCs)2 are a novel class of experimental small molecule cancer therapeutics that target the cIAPs and X-linked IAP (XIAP) (8, 9). The mode of action for SMCs in sensitive cancer cells involves binding of SMCs to the BIR motifs of the cIAPs, which induces the rapid activation of the E3 ligase domain of the cIAPs, leading to their autoubiquitination and subsequent proteasomal degradation (10–14). The loss of cIAPs results in the accumulation of their substrate, NF-κB-inducing kinase (NIK), an activator of the alternative NF-κB pathway (13, 15). In addition, the absence of cIAP1 and cIAP2 facilitates the association of receptor interacting protein 1 (RIP1) to the TNF receptor 1 (TNF-R1) complex, promoting activation of the classical NF-κB pathway (14). The induction of these NF-κB pathways leads to the production and release of TNFα, which acts in an autocrine fashion. With the loss of the cIAPs after SMC treatment, TNFα can no longer promote ubiquitination of RIP1 (6, 7, 11). As a result, the downstream NF-κB response is delayed or blunted (6, 7). Furthermore, nonubiquitinated RIP1 is released from the TNF-R1 complex to associate with caspase-8 and FADD that results in the formation of the death-inducing complex II, activating the extrinsic apoptotic pathway (16, 17).
The investigation into the SMC mechanism has revealed some remarkable roles for the IAPs in the regulation of signals initiated from cell surface cytokine receptors. In addition to sensitizing cells to TNFα-mediated apoptosis, down-regulation of the cIAPs also promotes cell death in response to TNF-related apoptosis-inducing ligand (TRAIL), DR5/TRAILR2 agonist antibody, and FasL treatment. TRAIL mediates its apoptotic effects through DR4/TRAILR1 and DR5/TRAILR2, and the combination of SMC and TRAIL treatment synergistically induces cell death in a wide array of cancer cell lines (17–20). In an MDA-MB-231 tumor xenograft model, co-treatment of SMC and DR5 agonist antibody was found to promote tumor regression in a TNFα-independent manner (21). The loss of cIAPs induced by SMC treatment can also promote sensitization of cancer cells to FasL-mediated cell death through the recruitment of RIP1 to death-inducing signaling complex and death-inducing complex II (22). Additionally, the combination of SMC and FasL treatment causes regression of MDA-MB-231 tumor xenograft (21). Interestingly, TNF-like weak inducer of apoptosis can also down-regulate cIAP1 in a lysosome-dependent fashion (23). Down-regulation of cIAP1 by TNF-like weak inducer of apoptosis is sufficient to sensitize cells to TNFα-induced cell death (23). Hence, the IAPs are central in determining cellular fate in response to ligands that engage the extrinsic apoptotic pathway, and SMCs can convert a pro-inflammatory TNFα response into an apoptotic signal. The abundance of IL-1β in the tumor microenvironment raises the possibility that this cytokine might impact on SMC-mediated cell death.
To investigate whether the IAPs are biologically relevant to IL-1β signaling, we combined IL-1β and SMC treatment with siRNA knockdown and determined the effects of IAP antagonism on cancer cells. We demonstrate that IL-1β treatment induces NF-κB-dependent production of TNFα, which triggers RIP1- and caspase 8-dependent apoptosis when the IAPs are down-regulated by SMC treatment. Furthermore, in those cancer cells that are resistant to SMC and IL-1β treatment, cellular resistance can be overcome by the down-regulation of the caspase-8 inhibitor c-FLIP.
EXPERIMENTAL PROCEDURES
Reagents
Smac mimetic compound SM-164 was synthesized as described previously (24). Smac mimetic compound AEG40730, as described previously (11), was synthesized by Vibrant Pharma Inc. (Brantford, Ontario, Canada). Human IL-1β recombinant protein was obtained from eBioscience Inc. (San Diego). Lipopolysaccharide (LPS) was obtained from Sigma.
Cell Culture
Tumor cell lines were maintained at 37 °C and 5% CO2 in DMEM supplemented with 10% heat-inactivated fetal calf serum, penicillin, and streptomycin and 1% nonessential amino acids (all from Invitrogen). All cell lines were obtained from ATCC with the following exceptions: SF295 (Brain Tumor Research Center, University of California, San Francisco), SNB75 (Dr. D. Stojdl, Children's Hospital of Eastern Ontario Research Institute, Ottawa, Canada), and ovarian cancer cell lines (Dr. B. Vanderhyden, Ottawa Health Research Institute, Ottawa, Canada).
Viability Assay
Tumor cells were plated onto 96-well flat bottom plates at various cell densities depending on the cell type. Cells were incubated overnight and then treated with combinations of SM-164 and IL-1β for 48 h. Cell viability was determined by resazurin/Alamar Blue vital dye (Sigma) that measures the activity of the mitochondrial respiratory chain (25).
Clonogenic Assay
SF295, SNB75, and U2OS cells were seeded overnight at equal densities on 6-well plates and treated with vehicle, SM-164 or SMC-AEG40730, in the absence or presence of IL-1β. At 24 h after treatment, cells were trypsinized and re-seeded. Cells were cultured for 7 days and fixed with methanol, and colonies were stained with crystal violet.
Apoptosis Assay
Drug-treated cells were used to determine the translocation of phosphatidylserine from the inner to the outer surface of the plasma membrane during apoptosis by using the human phospholipid-binding protein, annexin V, conjugated to fluorescein isothiocyanate (FITC), according to the manufacturer's instructions (BD Biosciences). The percentage of apoptotic (annexin V-positive and propidium iodide-negative) cells was determined by flow cytometric analysis (BD Biosciences Immunocytometry Systems, BD FACSDiva, and De Novo FCS Express V3).
Generation of Adenoviral Vector Expressing Dominant Negative IKKβ
Plasmid vector expressing the kinase-dead (D145N) dominant negative form of IKKβ (IKKβ-DN) (26) was kindly provided by Dr. Bernd Baumann (University of Ulm, Ulm, Germany). The coding sequence of IKKβ-DN was subcloned into pAdTrack-CMV, and adenovirus particles were generated using the AdEasy system (27). Adenovirus expressing c-FLIP was obtained from Vector Biolabs (Philadelphia).
ELISA
Cells were cultured on 96-well plates overnight and then stimulated with 1 ng/ml IL-1β or 100 nm SM-164. After 24 h of drug treatment, cell culture supernatants were collected, clarified by centrifugation, and processed for ELISA for TNFα according to the manufacturer's instructions (R&D Systems catalog no. D4210 or StressGen catalog no. 900-099). In some experiments, cells were transduced with an adenoviral vector that expresses GFP or co-expresses GFP and IKKβ-DN prior to drug treatment.
Transfection of Small Interfering RNA
siRNAs targeting caspase-8, caspase-9, RIP1, NIK, cIAP1, cIAP2, XIAP, TNF-R1, c-FLIP (ON-TARGETplus SMARTpool) and a nontargeting siRNA control (Accell) were obtained from Dharmacon. Cells were reverse-transfected using DharmaFECT I Reagent (Dharmacon) according to the manufacturer's protocol.
Western Immunoblotting
For immunoblotting, equal amounts of SDS-solubilized samples were separated on polyacrylamide gels and transferred to nitrocellulose as described previously (28). Following protein transfer, individual proteins were detected by Western immunoblotting using the following antibodies g: β-actin (clone AC-15) from Sigma; cleaved caspase-3 (9661), caspase-9 (9502), IκBα (4812), IKKβ (2678), NIK (4994), p100/p52 (4882), p65 (clone C22B4), phospho(S536)-p65 (clone 93H1), and TNF-R1 (3736) from Cell Signaling Technology; TRAF6 (1660-1) from Epitomics; caspase-8 (AHZ0502) from Invitrogen; c-FLIP (clone NF6) from Alexis Biochemicals; IL-1R2 (sc-52678) and MyD88 (sc-74532) from Santa Cruz Biotechnology; and RIP1 (clone 38) from BD Biosciences. Our rabbit anti-rat IAP1 and IAP3 (RIAP1 and -3) polyclonal antibodies were used to detect cIAP1/2 and XIAP, respectively (6). Bound primary antibodies were reacted with secondary antibodies conjugated with Alexa Fluor® 680 (Molecular Probes) or with IRDyeTM 800 (Rockland) and the infrared fluorescent signals were detected using the Odyssey® infrared imaging system (LI-COR) (29, 30).
Protein Preparation and Immunoprecipitation
Protein samples were prepared as described before (30). Protein content was determined by the protein assay (Bio-Rad) using bovine serum albumin as a standard. For endogenous caspase-8 immunoprecipitation, ∼1 × 107 cells were washed twice with PBS and lysed in a buffer (50 mm Tris-HCl, pH 8.0, containing 10% glycerol, 1% Triton X-100, 150 mm NaCl, 1 mm NaF, 0.1 mm phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin A, and 10 μg/ml each of leupeptin and aprotinin) on ice for 15 min. Endogenous caspase-8 complex was immunoprecipitated overnight at 4 °C with 4 μg of anti-caspase-8 (Santa Cruz Biotechnology, SC6136), and complexes were recovered with 40 μl of protein G-agarose. Protein complexes were separated and solubilized by heating at 100 °C for 5 min in SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, containing 2% SDS, 1% β-mercaptoethanol, and 5% glycerol).
RESULTS
SMC Synergizes with IL-1β to Induce Apoptosis in Cancer Cells
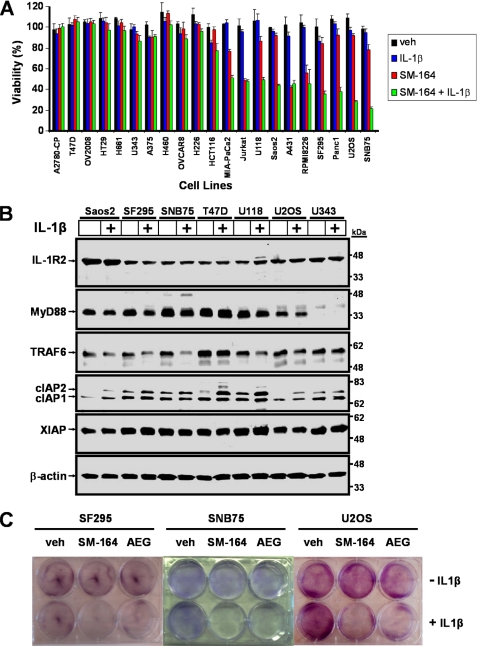
IL-1β is abundant in the tumor microenvironment (4, 31) and therefore could impact on SMC-mediated cell death. As a first step, we screened a diverse panel of cancer cells originating from the bone, breast, central nervous system, colon, lung, lymphatic system, ovaries, pancreas, and skin and measured cytotoxicity in response to the combinatorial treatment of IL-1β and SMC SM-164 (Fig. 1A). Of the 21 cancer cell lines examined, 10 were responsive to SMC treatment alone or in combination with IL-1β (Fig. 1A). Notably, none of the cell lines examined was sensitive to IL-1β treatment alone (Fig. 1A). To examine the possibility that IL-1β failed to synergize with SMC treatment in some cancer cells because they lack the corresponding receptor for IL-1β, we analyzed the expression of IL-1R2 by Western immunoblotting. All cancer cell lines examined express IL-1R2 (Fig. 1B and supplemental Fig. 1), suggesting that IL-1β could at least bind to IL-1R2 and possibly engage in signal transduction. To examine the possibility that defects exist in IL-1R-mediated signal transduction in nonresponsive cells, we also analyzed for the expression levels of MyD88 and TRAF6. The absence of MyD88 expression in U343 and A2780-CP cells and the absence of TRAF6 expression in A431 and RPMI-8226 cells correlated with the lack of synergism of these cells to combined IL-1β and SMC treatment (Fig. 1, A and B, and supplemental Fig. 1). Therefore, in some cancer cell lines, IL-1β may not synergize with SMC to cause cell death because they are missing components that are necessary for IL-1R-mediated signal transduction.
FIGURE 1.
Responsiveness of a panel of cancer cell lines to treatment with IL-1β and Smac mimetic compound SM-164. A, diverse panel of 21 cell lines representing cancer cells originating from the bone (Saos2 and U2OS), breast (T47D), central nervous system (U343, U118, SF295, and SNB75), colon (HT29 and HCT116), lung (H661, H460, and H226), lymphatic system (Jurkat and RPMI8226), ovaries (A2780-CP, OV2008, and OVCAR8), pancreas (MIA-PaCa2 and Panc1), skin (A375), and vulvar (A431) were tested for their sensitivity to combinations of 1 ng/ml IL-1β and 100 nm SM-164 or vehicle (veh, 0.1% DMSO) alone. Cells were treated for 48 h, and cell viability was measured with Alamar Blue. Percentage viability relative to nontreated ± S.D. (n = 4) was plotted. B, subset of the cancer cell line panel from A was treated with 1 ng/ml IL-1β for 24 h. Proteins were extracted and Western immunoblotted with the indicated antibodies. β-Actin was used as a loading control. C, SF295, SNB75, and U2OS cells were treated with combinations of 1 ng/ml IL-1β with 500 nm SM-164 or 500 nm SMC-AEG40730 for 24 h. Cells were washed and trypsinized, and equal numbers of cells were re-seeded. After 7 days, colonies were stained with crystal violet. The combination of SMC and IL-1β treatment repressed clonogenic survival of cancer cells.
To further investigate the synergism between SMC and IL-1β, we selected U2OS osteosarcoma cells and SF295 and SNB75 glioblastoma cells for more in-depth analysis. Dose-response curves showed that the IC50 values of SM-164 for IL-1β synergism were 83, 1.6, and 50 nm for SF295, SNB75, and U2OS cells, respectively (supplemental Fig. 2, A, C, and E), whereas the IC50 values of IL-1β for SM-164 synergism were 36 pg/ml, 340 fg/ml, and 40 pg/ml for SF295, SNB75, and U2OS cells, respectively (supplemental Fig. 2, B, D, and F). Furthermore, only combined treatment of IL-1β and SM-164 or with an alternative SMC AEG40730 prevented clonogenic survival of these three cell lines (Fig. 1C). Together, these results demonstrate that SMC and IL-1β synergize to induce cancer cell death.
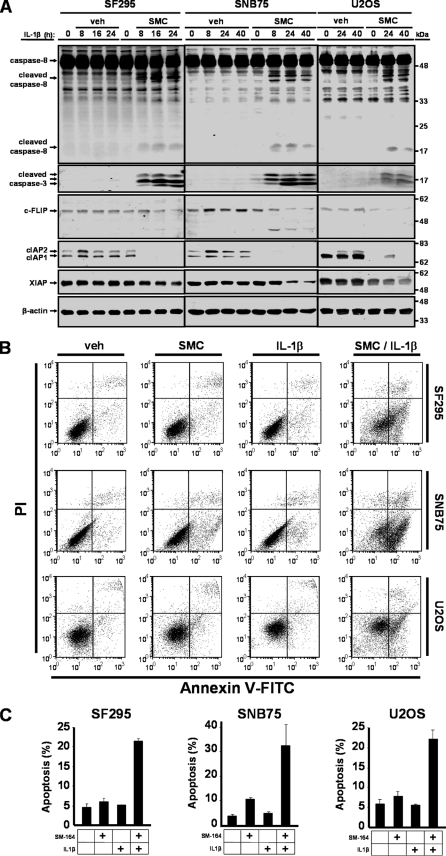
To decipher the underlying mechanism of cell death in response to SMC and IL-1β treatment, we assessed caspase activity in sensitive cancer cells. In SF295, SNB75, and U2OS cells, we detected processing and activation of caspase-3 and caspase-8 following combined SMC and IL-1β treatment (Fig. 2A). Down-regulation of cIAP1 and cIAP2 by SMC treatment was also confirmed (Fig. 2A). As activation of the caspase cascade is integral to the execution of apoptosis, we quantified the percentage of cells that were undergoing apoptosis by identifying cells that were stained with annexin V-FITC without propidium iodide uptake using flow cytometry. In SF295, SNB75, and U2OS cells, the combined treatment of SMC and IL-1β resulted in marked apoptosis (Fig. 2, B and C). Single treatment alone was not sufficient to induce apoptosis in these cell lines, with the exception of a modest induction by SMC treatment alone in SNB75 cells (Fig. 2C). These results demonstrate that apoptosis is the underlying mechanism for IL-1β-induced SMC-mediated cell death.
FIGURE 2.
SMC synergizes with IL-1β to induce apoptosis in cancer cells. A, SF295 cells were treated with 500 nm SM-164 (SMC) and 1 ng/ml IL-1β for the indicated times. SNB75 and U2OS cells were treated with vehicle (veh) or 100 nm SM-164 (SMC) for 24 h and then treated with 1 ng/ml IL-1β. At the indicated times, cells were harvested, and protein extracts were subjected to Western immunoblotting with antibodies recognizing caspase-8, cleaved caspase-3, c-FLIP, cIAP1, cIAP2, and XIAP. β-Actin was used as a loading control. B, SF295, SNB75 and U2OS cells were treated with combinations of 500 nm SM-164 (SMC) and 1 ng/ml IL-1β for 24 h, and cells were stained with annexin V and propidium iodide to measure the rate of apoptosis by flow cytometry. C, plotted numbers represent the average ± S.D. of three independent assays. The co-treatment of SMC and IL-1β induces apoptosis in cancer cells.
SMC-mediated IL-1β-induced Cell Death Depends on Caspase-8 and RIP1
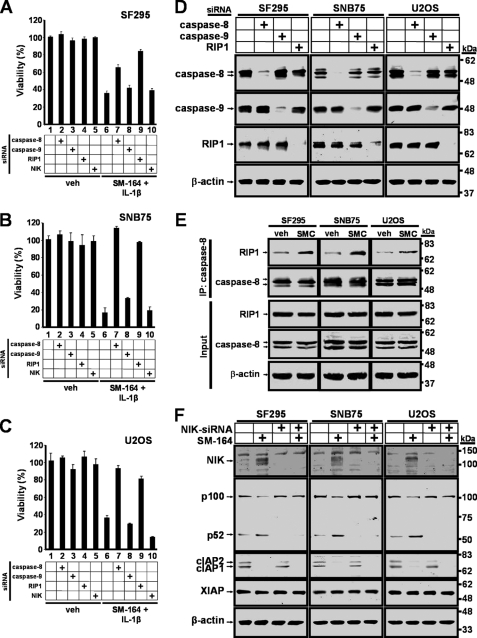
As a stand-alone agent, SMC exerts its effects in responsive cells through the accumulation of NIK that facilitates the production of TNFα, thereby killing cells in an autocrine fashion (13, 14, 32). SMC-mediated cell death induced by TNFα requires the formation of the death-inducing complex II containing RIP1 and caspase-8 (16, 17). Furthermore, caspase-9 does not appear to play a role in the execution of apoptosis in response to SMC treatment (13). To assess for the functional roles of these molecules in the context of SMC-mediated IL-1β-induced cell death, we individually silenced these molecules by siRNA prior to drug treatment. Down-regulation of caspase-8 prevented SMC-mediated IL-1β-induced cell death in SNB75 and U2OS cells, and to a lesser extent in SF295 cells (Fig. 3, A–C, lane 7). By contrast, down-regulation of caspase-9 failed to rescue cell death (Fig. 3, A–C, lane 8). Similar to caspase-8 silencing, down-regulation of RIP1 also protected these cells against SMC-mediated IL-1β-induced cell death (Fig. 3, A–C, lane 9). Down-regulation of each molecule by siRNA was confirmed (Fig. 3D).
FIGURE 3.
SMC-mediated IL-1β-induced cell death depends on caspase-8 and RIP1. SF295 (A), SNB75 (B), and U2OS (C) cells were transfected with siRNA targeting caspase-8, caspase-9, RIP1, NIK, or nontargeting siRNA as a control. At 48 h transfection, cells were treated with vehicle (veh) or 100 nm SM-164 and 1 ng/ml IL-1β for 48 h. Cell viability was determined by Alamar Blue. The percentage viability relative to nontargeting siRNA and vehicle ± S.D. (n = 4) was plotted. D, SF295, SNB75, and U2OS cells were transfected with siRNA targeting caspase-8, caspase-9, RIP1, or a nontargeting siRNA as a control. At 48 h transfection, cells were harvested, and protein expression levels of targeted genes were analyzed by Western immunoblotting. β-Actin was used as a loading control. E, SF295, SNB75, and U2OS cells were treated with vehicle (veh) or 100 nm SM-164 (SMC) for 24 h, and endogenous caspase-8-associated complexes were isolated by immunoprecipitation, resolved by SDS-PAGE, and probed for the presence of RIP1. Input levels of RIP1, caspase-8, and β-actin were assessed and are shown in the lower panel. F, SF295, SNB75, and U2OS cells were transfected with siRNA targeting NIK or nontargeting siRNA for 48 h, and cells were treated with vehicle (veh) or 100 nm SM-164 (SMC) for an additional 8 h. Cells were harvested, and protein expression levels of NIK, p100, cIAP1, cIAP2, and XIAP were analyzed by Western immunoblotting. β-Actin was used as a loading control. Down-regulation of caspase-8 or RIP1 rescues cells from SMC-mediated IL-1β-induced cell death.
The requirement of RIP1 and caspase-8 in SMC-mediated IL-1β-induced cell death suggests that the death-inducing complex II might have been formed in response to SMC treatment (16, 17). To assess for the formation of this complex, we determined the level of RIP1 recruitment by immunoprecipitating caspase-8 following SMC treatment. We found that in response to SMC treatment, RIP1 increased its association with caspase-8 in all three cell lines tested (Fig. 3E). These results demonstrate that cell death induced by SMC and IL-1β treatment is dependent on caspase-8 and RIP1.
In the absence of the cIAPs, NIK accumulates and thereby activates the alternative NF-κB pathway (13, 15). Indeed, SMC treatment led to the accumulation of NIK, leading to increased processing of p100 to p52, indicating activation of the alternative NF-κB pathway (Fig. 3F). As expected, silencing of NIK by siRNA prevented the processing of p100 to p52 in response to SMC treatment. To explore for a potential role for NIK, we knocked down NIK and assessed for cell viability in SMC and IL-1β-treated cells. Down-regulation of NIK did not alter cell viability in SF295 and SNB75 cells and further promoted SMC-mediated IL-1β-induced cell death in U2OS cells (Fig. 3, A–C, lane 10). These results suggest that the alternative NF-κB pathway does not partake in promoting cell death in response to SMC and IL-1β treatment.
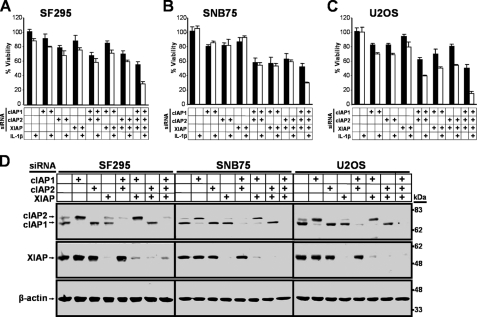
cIAP1, cIAP2, and XIAP Cooperatively Protect against IL-1β-induced Cell Death
The ability of SMCs to antagonize the cIAPs, and possibly XIAP, is critical for sensitizing cells to TNFα-mediated apoptosis (12–14, 32–34). To analyze the potential contribution of each of these IAPs in blocking IL-1β-induced cell death, we individually and jointly silenced cIAP1, cIAP2, and XIAP prior to IL-1β treatment. In SF295 and SNB75 cells, sensitization to IL-1β-mediated cell death required the combined down-regulation of cIAP1, cIAP2, and XIAP (Fig. 4, A and B), whereas the combined down-regulation of any two of the three IAPs was sufficient to sensitize U2OS cells to IL-1β-mediated cell death (Fig. 4C). The down-regulation of IAPs in the absence of exogenous IL-1β resulted in varying degrees of cell death, with the lowest cell viability observed with the combined down-regulation of all three IAPs (Fig. 4, A–C). The efficiency of cIAP1, cIAP2, and XIAP knockdown by siRNA was confirmed (Fig. 4D). These results demonstrate that the combined targeting of cIAP1, cIAP2, and XIAP is the most effective approach to trigger IL-1β-induced cell death.
FIGURE 4.
cIAP1, cIAP2, and XIAP cooperatively protect against IL-1β-induced cancer cell death. SF295 (A), SNB75 (B), and U2OS (C), cells were transfected with siRNA targeting cIAP1, cIAP2, XIAP, or nontargeting siRNA as a control. 48 h after siRNA-mediated silencing, cells were treated with vehicle (black bars) or with 1 ng/ml IL-1β (white bars) for an additional 48 h. Cell viability was determined by Alamar Blue. The percentage viability relative to nontargeting siRNA and vehicle ± S.D. (n = 4) was plotted. D, cells were transfected with siRNA as in A–C for 48 h and harvested, and protein expression levels of cIAP1, cIAP2, and XIAP were analyzed by Western immunoblotting. β-Actin was used as a loading control. The combined silencing of cIAP1, cIAP2, and XIAP by siRNA results in the highest sensitization to IL-1β-mediated cancer cell death.
SMC-mediated IL-1β-induced Cell Death Requires TNF-R1 Signaling
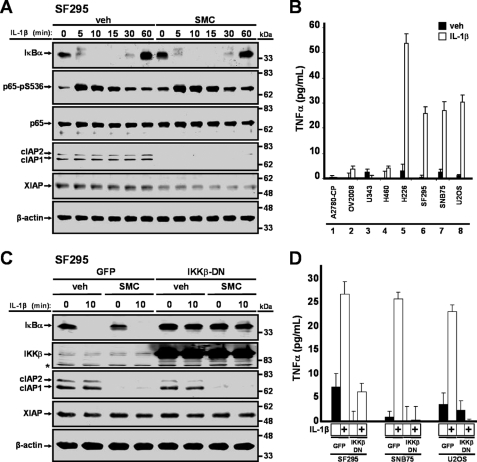
SMC treatment either blunts or delays activation of the TNFα-induced classical NF-κB pathway (7, 17). Inactivation of the classical NF-κB pathway can in part sensitize cells to TNFα-mediated apoptosis (6, 7). As NF-κB pathway activation is also modulated by IL-1β (35), and because SMC treatment can sensitize cells to IL-1β-induced apoptosis, we asked whether SMC treatment might also blunt NF-κB activation mediated by IL-1β. To measure activation of NF-κB, we analyzed the degradation of IκBα and phosphorylation of p65 following IL-1β treatment. As expected, IL-1β activated the NF-κB pathway in SF295, SNB75, and U2OS cells, and similar kinetics were seen with prior treatment of SMC (Fig. 5A and supplemental Fig. 3, A and B). These results indicate that SMC treatment does not interfere with activation of the NF-κB pathway in response to IL-1β stimulation.
FIGURE 5.
IL-1β induces TNFα production via the NF-κB pathway. A, SF295 cells were treated with vehicle (veh) or 100 nm SM-164 (SMC) for 2 h and then treated with 1 ng/ml IL-1β for the indicated times, and protein lysates were immunoblotted for IκBα, p65-pS536, p65, cIAP1, cIAP2, and XIAP. β-Actin was used as a loading control. B, cells were treated with 1 ng/ml IL-1β for 24 h, and cell culture supernatants were processed for the presence of TNFα through ELISA. C, SF295 cells were transduced with adenoviral vectors expressing GFP or co-expressing GFP and a dominant negative form of IKKβ at a multiplicity of infection of 100 for 24 h. Cells were treated with vehicle (veh) or 100 nm SM-164 (SMC) for 2 h and then treated with 1 ng/ml IL-1β for 10 min. Cells were harvested, and protein expression levels of IκBα, IKKβ, cIAP1, cIAP2, and XIAP were analyzed by Western immunoblotting. β-Actin was used as a loading control. Asterisk denotes a nonspecific band. D, cells were transduced with an adenoviral vector that expresses GFP or co-expressing GFP and a dominant negative form of IKKβ for 24 h and then treated with 1 ng/ml IL-1β for an additional 24 h. Cell culture supernatants were collected for ELISA analysis for the presence of TNFα. Inhibition of the NF-κB pathway prevented IL-1β-mediated production of TNFα.
Because SMC treatment does not alter activation of the NF-κB pathway in response to IL-1β treatment and TNFα is an NF-κB-targeted gene (36), we next explored the possibility that TNFα might be a mediator for IL-1β-induced cell death. We detected production of TNFα in sensitive cells SF295, SNB75, and U2OS in response to IL-1β treatment (Fig. 5B, lanes 6–8) but not to SMC treatment (data not shown). In contrast, TNFα was not detected in A2780-CP, OV2008, U343, and H460 cells (Fig. 5B, lanes 1–4) which were resistant to cell death by SMC and IL-1β treatment. To confirm that the NF-κB pathway is required for TNFα production in response to IL-1β treatment, we blocked the NF-κB pathway by expressing a dominant negative form of IKKβ (IKKβ-DN) (26). Expression of IKKβ-DN prevented IL-1β-mediated degradation of IκBα and hence activation of the NF-κB pathway (Fig. 5C and supplemental Fig. 4). As expected, inhibition of the IL-1β-mediated NF-κB pathway repressed the production of TNFα in response to IL-1β treatment (Fig. 5D).
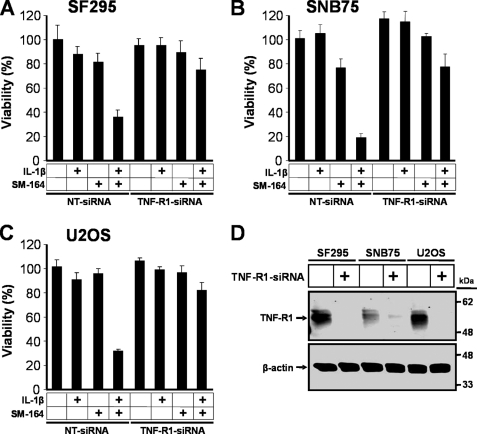
Next, we investigated whether TNFα signaling has a functional role in SMC-mediated IL-1β-induced cell death by down-regulating TNF-R1 using siRNA. In SF295, SNB75, and U2OS cells, silencing of TNF-R1 blocked cell death induced by SMC and IL-1β treatment (Fig. 6, A–C). Down-regulation of TNF-R1 in each of the cell lines was also confirmed (Fig. 6D). Together, these results indicate that cancer cell apoptosis induced by SMC and IL-1β co-treatment requires TNFα signaling.
FIGURE 6.
SMC-mediated IL-1β-induced cell death requires TNF-R1 signaling. SF295 (A), SNB75 (B), and U2OS (C) were transfected with nontargeting siRNA or TNF-R1-siRNA. At 48 h transfection, cells were treated with combinations of 1 ng/ml IL-1β and 100 nm SM-164 for 48 h, and then cell viability was determined by Alamar Blue. The percentage viability relative to nontargeting siRNA and vehicle ± S.D. (n = 4) was plotted. D, cells were transfected with siRNA as in A–C. At 48 h transfection, cells were harvested, and protein expression levels of TNF-R1 were analyzed by Western immunoblotting. β-Actin was used as a loading control. The silencing of TNF-R1 prevented SMC-mediated IL-1β-induced cell death.
Resistance of Cancer Cells to SMC and IL-1β Treatment Is Overcome by c-FLIP Knockdown
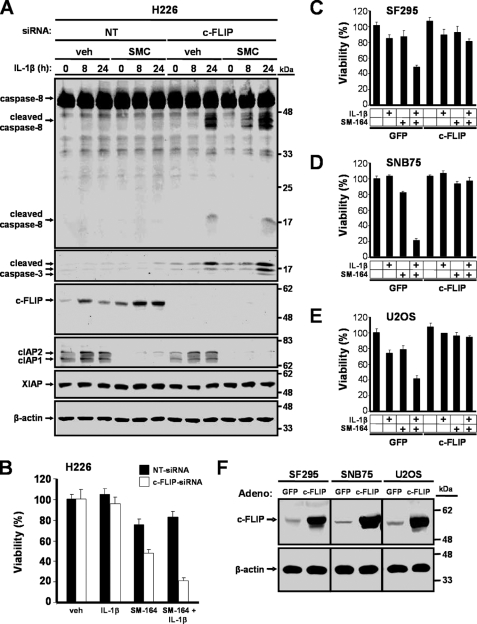
IL-1β-mediated release of TNFα promotes cell death in response to SMC treatment. H226 cells are resistant to SMC and IL-1β treatment (Fig. 1A) despite the fact that TNFα is secreted following IL-1β addition (Fig. 5B). These results indicate that TNFα signaling is necessary but is not sufficient to induce cell death. In SMC-resistant cells, we previously reported that c-FLIP blocks SMC-mediated TNFα-induced apoptosis (17). In this study, we found that c-FLIP levels decreased in response to SMC and IL-1β treatment in the sensitive cell lines SF295, SNB75, and U2OS (Fig. 2A). We reasoned that the same principle might apply to explain the cellular resistance to SMC and IL-1β treatment, given that TNFα signaling is a critical intermediate. We therefore examined whether c-FLIP is responsible for resistance to SMC-mediated IL-1β-induced cell death by silencing c-FLIP with siRNA in H226 cells prior to treatment with SMC and IL-1β. Depletion of c-FLIP led to the processing and activation of caspase-8 and caspase-3 (Fig. 7A) and reversed the resistance of SMC-mediated IL-1β-induced cell death (Fig. 7B). To further analyze the contribution of c-FLIP, we ectopically expressed c-FLIP by adenoviral transduction prior to exposure to combined IL-1β and SMC treatment. We observed that ectopic expression of c-FLIP blocked cell death induced by the combined IL-1β and SMC treatment in SF295, SNB75, and U2OS cells (Fig. 7, C–E). Expression of c-FLIP was also confirmed (Fig. 7F). These results demonstrate that c-FLIP prevents the induction of cell death by SMC and IL-1β treatment.
FIGURE 7.
Resistance of cancer cells to SMC and IL-1β treatment is circumvented by c-FLIP knockdown. A, H226 cells were transfected with siRNA targeting c-FLIP or nontargeting (NT) siRNA as a control. At 24 h after siRNA-mediated silencing, these cells were treated with vehicle (veh) or 100 nm SM-164 (SMC) for 2 h and then treated with 1 ng/ml IL-1β for the indicated times. Cells were harvested and immunoblotted for caspase-8, cleaved caspase-3, c-FLIP, cIAP1, cIAP2, and XIAP. β-Actin was used as a loading control. B, H226 cells were transfected with siRNA against c-FLIP or nontargeting (NT) siRNA for 24 h and then treated with combinations of 1 ng/ml IL-1β and 100 nm SM-164 for 48 h. Cell viability was determined by Alamar Blue. The percentage viability relative to nontargeting siRNA and vehicle ± S.D. (n = 4) was plotted. SF295 (C), SNB75 (D), and U2OS (E) cells were transduced with adenoviral vectors expressing GFP or c-FLIP for 24 h at a multiplicity of infection of 100 for SF295 and SNB75, and a multiplicity of infection of 40 for U2OS cells. Cells were then treated with combinations of 1 ng/ml IL-1β and 100 nm SM-164 (SMC) for 48 h. Cell viability was determined by Alamar Blue. The percentage viability relative to cells transduced with adeno-GFP and treated with vehicle ± S.D. (n = 3) was plotted. F, cells were transduced with an adenoviral vector that expresses GFP or c-FLIP for 24 h and harvested, and protein expression levels of c-FLIP were analyzed by Western immunoblotting. β-Actin was used as a loading control.
LPS Potentiates SMC-mediated Cell Death
To extend our observations on the ability of an inflammatory cytokine to potentiate SMC-mediated cell death, we analyzed for the potential of endotoxin LPS to similarly cause cancer cell death. We selected SF295, SNB75, and U2OS cells for further analysis because of their robust response to combined IL-1β and SMC treatment, and the LPS receptor TLR4 was expressed in these cells (data not shown). We found that LPS potentiated SMC-mediated cell death in the selected cell lines (supplemental Fig. 5). In sum, these results indicate that inflammatory signaling can synergize with SMC treatment to induce cancer cell death.
DISCUSSION
SMC-mediated IL-1β-induced Apoptosis Depends on TNFα Signaling Involving Caspase-8 and RIP1
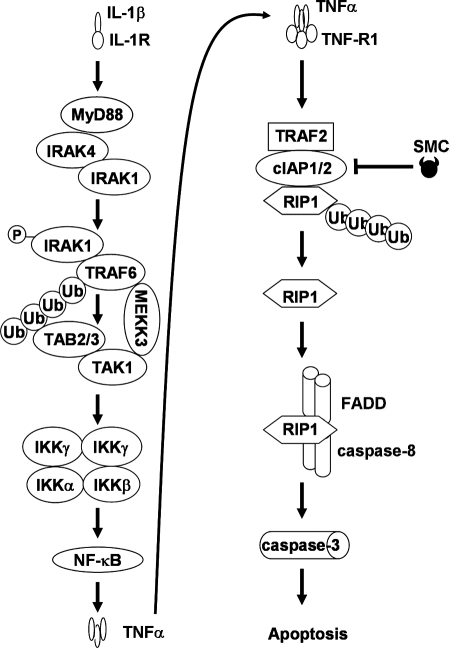
Smac mimetic compounds are IAP antagonists that are under clinical development as cancer therapeutics. SMCs function either by activating the NF-κB pathways to produce TNFα for autocrine signaling (13, 14, 32) or by potentiating cytokines TNFα, TRAIL, or Fas ligand within the tumors to induce cancer cell death (17–22, 37–39). The abundance of the cytokine IL-1β, a known inducer of the NF-κB pathway, within the tumor microenvironment (1–3, 35) suggests that IL-1β could impact on the efficacy of SMC treatment. Here, we investigated the potential synergism between SMC and IL-1β for the induction of apoptosis in cancer cells. We found that SMC synergizes with IL-1β to induce cytotoxicity in a diverse subset of cancer cell lines that include glioblastoma, osteosarcoma, and pancreatic cancer. We found that the IC50 values of IL-1β for SM-164 synergism were 36 pg/ml, 340 fg/ml, and 40 pg/ml for SF295, SNB75, and U2OS cells, respectively. Because cultured alveolar macrophages from patients of non-small cell lung cancer produce 238 pg/ml IL-1β (40), our IC50 values for IL-1β-induced death in SMC-treated cells suggest that the endogenous level of IL-1β might be sufficient to synergize with SMC treatment. The synergism in sensitive cell lines requires activation of the NF-κB pathway by IL-1β treatment, which in turn leads to the production of TNFα. SMC-mediated depletion of cIAP1 and cIAP2 has two consequences with respect to TNFα signaling. First, the absence of cIAP1 and cIAP2 can blunt TNFα-mediated activation of the classical NF-κB pathway (6, 7). Second, depletion of cIAP1 and cIAP2 prevents TNFα-mediated RIP1 ubiquitination (6, 7), resulting in the release of RIP1 from the TNF-R1 complex. Released nonubiquitinated RIP1 is free to form the death-inducing complex II with FADD and caspase-8, thereby activating the extrinsic apoptotic pathway (16, 17). However, it should be noted that most cells that are resistant to SMC and IL-1β treatment do not produce TNFα in response to IL-1β. However, if resistant cells do produce TNFα following IL-1β treatment, resistance to SMC treatment can be circumvented by c-FLIP knockdown. Thus, our data demonstrate that a pathway in cancer cells exists by which IL-1β-mediated cell death is potentiated by SMC treatment (Fig. 8).
FIGURE 8.
Schematic of IL-1β-mediated SMC-induced apoptosis. Upon IL-1β occupancy, IL-1R transduces a signal by recruiting adaptor protein MyD88, which in turn recruits IRAK1 and IRAK4 (51). IRAK1 is likely phosphorylated and activated by IRAK4, allowing IRAK1 to interact with TRAF6. Autoubiquitination of TRAF6 recruits TAB2 and TAB3, and the binding of MEKK3 to TAK1 may stabilize TRAF6-associated signaling complex, allowing the cascade involving MEKK3, TAK1, and IKK complex to proceed, leading to NF-κB activation. In turn, activation of NF-κB results in production of TNFα, which is secreted to engage the TNF-R1 signaling complex in an autocrine fashion. In the presence of TRAF2, cIAP1, and cIAP2, cells respond to TNFα stimulation by promoting RIP1 ubiquitination. However, as a result of SMC treatment, cIAP1 and cIAP2 are down-regulated, thereby preventing ubiquitination of nonubiquitinated RIP1 and allowing RIP1 to bind and activate the death-inducing complex II that includes caspase-8 and FADD. Death-inducing complex II in turn activates caspase-3, promoting apoptosis.
Improving the Efficacy of SMC-based Cancer Therapies
Effective targeting of the IAPs is crucial for the induction of cell death by SMC treatment. This study and others have shown that the concurrent down-regulation of cIAP1, cIAP2, and XIAP by SMC or siRNA treatment, either in the absence or presence of an extrinsic apoptotic trigger, can induce cytotoxicity in responsive cancer cells (12, 33, 34). Although the level of XIAP in response to SMC treatment appears to somewhat fluctuate over the early and intermediate time points (10 min to 8 h), prolonged treatment results in noticeable XIAP down-regulation. Fluctuation of XIAP levels at earlier times may be due to the result of rapid cIAP1 degradation by SMC treatment, as the ubiquitin E3 ligase RING domain of cIAP1 can mediate the degradation of other IAPs, including XIAP (30, 41, 42). Therefore, the loss of cIAP1 might allow XIAP levels to undergo transient recovery. Nonetheless, in addition to changes in the structural design of SMCs to improve specificity, either prolonged exposure or increased dosage of SMC should also lead to increased targeting of XIAP.
Interestingly, resistance to SMC-induced death in cancer cells has also been reported, but the mechanism for this resistance appears complex and is dependent on the molecular signature of the particular cancer cell. However, attempts have been made to increase our understanding of the basis of SMC resistance, resulting in some emerging themes. First, persistent c-FLIP levels can block cancer cell death triggered by SMC treatment when combined with TNFα or TRAIL (17). Silencing of c-FLIP restored sensitivity toward combined SMC and TNFα, TRAIL (17), or IL-1β treatment in previously resistant cancer cells. Current chemotherapeutics such as paclitaxel and bortezomib are known to down-regulate c-FLIP (43, 44), raising the possibility that these agents can be used with SMC treatment for an improved therapeutic outcome. Second, some cancer cells are resistant to SMC-induced apoptosis because the cells become refractory to SMC-mediated degradation of cIAP2 (45). As protein expression level of cIAP2 is also modulated by the phosphatidylinositol 3-kinase (PI3K) pathway, exposure to a PI3K inhibitor can repress cIAP2 expression and re-sensitize cancer cells to SMC and TNFα treatment (45). Third, inactivation of caspase-8 renders certain cancer cells to be nonresponsive to SMC and TNFα treatment (17). Because interferon has been reported to reactivate caspase-8 expression (46, 47), interferon treatment can potentially restore sensitivity to SMC and TNFα treatment in cancer cells deficient in caspase-8 expression. Therefore, the concurrent targeting of caspase inhibitors, IAPs and c-FLIP, may provide the broadest success in causing cancer cell death triggered by an intact extrinsic apoptotic pathway.
Tumor microenvironments are frequently subjected to inflammatory stress, including exposure to the pro-inflammatory cytokines TNFα and IL-1β that are produced either by the tumor cells or infiltrating cells such as macrophages (1–3). An emerging approach for cancer therapy is to harness this existing inflammatory condition (48). The ability of SMCs to promote TNFα-induced apoptosis suggests that SMCs may provide anti-tumor activity in situ. Indeed, SMCs have demonstrated in vivo efficacy in preclinical xenograft models, either as a stand-alone therapy or in combination with additional agents (32, 33, 49, 50). From our current data, we propose that IL-1β contributes to SMC synergism by acting as a TNFα inducer in cancer cells through the NF-κB pathway, providing a positive feed-forward loop of pro-inflammatory cytokines that activate SMC-mediated cancer cell death.
Supplementary Material
This work was supported by Canadian Institutes of Health Research, Ottawa Neuroblastoma Fund, Ottawa Regional Cancer Foundation, James Birrell Neuroblastoma Research Fund, and joint funding from Ontario Institute for Cancer Research (OICR) and Terry Fox Research Institute. Funding for OICR is provided by the Ministry of Research and Innovation from the province of Ontario. R. G. K. is a founder and shareholder of Aegera Therapeutics Inc., which designed SMC-AEG40730. S.W. is a shareholder and consultant for Ascenta Therapeutics Inc., which has licensed technologies related to Smac mimetic compounds.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- SMC
- Smac mimetic compound
- IAP
- inhibitor of apoptosis
- cIAP
- cellular inhibitor of apoptosis
- NIK
- NF-κB-inducing kinase
- TRAIL
- TNF-related apoptosis-inducing ligand
- XIAP
- X-linked IAP.
REFERENCES
- 1.Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A., Romero P., Palucka A. K., Marincola F. M. (2008) Lancet 371, 771–783 [DOI] [PubMed] [Google Scholar]
- 3.Grivennikov S. I., Greten F. R., Karin M. (2010) Cell 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apte R. N., Dotan S., Elkabets M., White M. R., Reich E., Carmi Y., Song X., Dvozkin T., Krelin Y., Voronov E. (2006) Cancer Metastasis Rev. 25, 387–408 [DOI] [PubMed] [Google Scholar]
- 5.Wu Y., Zhou B. P. (2010) Br. J. Cancer 102, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahoney D. J., Cheung H. H., Mrad R. L., Plenchette S., Simard C., Enwere E., Arora V., Mak T. W., Lacasse E. C., Waring J., Korneluk R. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varfolomeev E., Goncharov T., Fedorova A. V., Dynek J. N., Zobel K., Deshayes K., Fairbrother W. J., Vucic D. (2008) J. Biol. Chem. 283, 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaCasse E. C., Mahoney D. J., Cheung H. H., Plenchette S., Baird S., Korneluk R. G. (2008) Oncogene 27, 6252–6275 [DOI] [PubMed] [Google Scholar]
- 9.Flygare J. A., Fairbrother W. J. (2010) Expert Opin. Ther. Pat. 20, 251–267 [DOI] [PubMed] [Google Scholar]
- 10.Gaither A., Porter D., Yao Y., Borawski J., Yang G., Donovan J., Sage D., Slisz J., Tran M., Straub C., Ramsey T., Iourgenko V., Huang A., Chen Y., Schlegel R., Labow M., Fawell S., Sellers W. R., Zawel L. (2007) Cancer Res. 67, 11493–11498 [DOI] [PubMed] [Google Scholar]
- 11.Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 12.Lu J., Bai L., Sun H., Nikolovska-Coleska Z., McEachern D., Qiu S., Miller R. S., Yi H., Shangary S., Sun Y., Meagher J. L., Stuckey J. A., Wang S. (2008) Cancer Res. 68, 9384–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 14.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 15.Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W., Korneluk R. G., Cheng G. (2008) Nat. Immunol. 9, 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., Du F., Wang X. (2008) Cell 133, 693–703 [DOI] [PubMed] [Google Scholar]
- 17.Cheung H. H., Mahoney D. J., Lacasse E. C., Korneluk R. G. (2009) Cancer Res. 69, 7729–7738 [DOI] [PubMed] [Google Scholar]
- 18.Li L., Thomas R. M., Suzuki H., De Brabander J. K., Wang X., Harran P. G. (2004) Science 305, 1471–1474 [DOI] [PubMed] [Google Scholar]
- 19.Dai Y., Liu M., Tang W., Li Y., Lian J., Lawrence T. S., Xu L. (2009) BMC Cancer 9, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecis D., Drago C., Manzoni L., Seneci P., Scolastico C., Mastrangelo E., Bolognesi M., Anichini A., Kashkar H., Walczak H., Delia D. (2010) Br. J. Cancer 102, 1707–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varfolomeev E., Alicke B., Elliott J. M., Zobel K., West K., Wong H., Scheer J. M., Ashkenazi A., Gould S. E., Fairbrother W. J., Vucic D. (2009) J. Biol. Chem. 284, 34553–34560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geserick P., Hupe M., Moulin M., Wong W. W., Feoktistova M., Kellert B., Gollnick H., Silke J., Leverkus M. (2009) J. Cell Biol. 187, 1037–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., Yeoh G., Brink R., Vaux D. L., Silke J. (2008) J. Cell Biol. 182, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H., Nikolovska-Coleska Z., Lu J., Meagher J. L., Yang C. Y., Qiu S., Tomita Y., Ueda Y., Jiang S., Krajewski K., Roller P. P., Stuckey J. A., Wang S. (2007) J. Am. Chem. Soc. 129, 15279–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed S. A., Gogal R. M., Jr., Walsh J. E. (1994) J. Immunol. Methods 170, 211–224 [DOI] [PubMed] [Google Scholar]
- 26.Herrmann O., Baumann B., de Lorenzi R., Muhammad S., Zhang W., Kleesiek J., Malfertheiner M., Köhrmann M., Potrovita I., Maegele I., Beyer C., Burke J. R., Hasan M. T., Bujard H., Wirth T., Pasparakis M., Schwaninger M. (2005) Nat. Med. 11, 1322–1329 [DOI] [PubMed] [Google Scholar]
- 27.Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., He T. C. (2007) Nat. Protoc. 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 28.Cheung H. H., Gurd J. W. (2001) J. Neurochem. 78, 524–534 [DOI] [PubMed] [Google Scholar]
- 29.Cheung H. H., Lynn Kelly N., Liston P., Korneluk R. G. (2006) Exp. Cell Res. 312, 2347–2357 [DOI] [PubMed] [Google Scholar]
- 30.Cheung H. H., Plenchette S., Kern C. J., Mahoney D. J., Korneluk R. G. (2008) Mol. Biol. Cell 19, 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apte R. N., Voronov E. (2008) Immunol. Rev. 222, 222–241 [DOI] [PubMed] [Google Scholar]
- 32.Petersen S. L., Wang L., Yalcin-Chin A., Li L., Peyton M., Minna J., Harran P., Wang X. (2007) Cancer Cell 12, 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Probst B. L., Liu L., Ramesh V., Li L., Sun H., Minna J. D., Wang L. (2010) Cell Death Differ. 17, 1645–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ndubaku C., Varfolomeev E., Wang L., Zobel K., Lau K., Elliott L. O., Maurer B., Fedorova A. V., Dynek J. N., Koehler M., Hymowitz S. G., Tsui V., Deshayes K., Fairbrother W. J., Flygare J. A., Vucic D. (2009) ACS Chem. Biol. 4, 557–566 [DOI] [PubMed] [Google Scholar]
- 35.Arend W. P. (2002) Cytokine Growth Factor Rev. 13, 323–340 [DOI] [PubMed] [Google Scholar]
- 36.Kasibhatla S., Brunner T., Genestier L., Echeverri F., Mahboubi A., Green D. R. (1998) Mol. Cell 1, 543–551 [DOI] [PubMed] [Google Scholar]
- 37.Foster F. M., Owens T. W., Tanianis-Hughes J., Clarke R. B., Brennan K., Bundred N. J., Streuli C. H. (2009) Breast Cancer Res. 11, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai Y., Lawrence T. S., Xu L. (2009) Am. J. Transl. Res. 1, 1–15 [PMC free article] [PubMed] [Google Scholar]
- 39.Servida F., Lecis D., Scavullo C., Drago C., Seneci P., Carlo-Stella C., Manzoni L., Polli E., Lambertenghi Deliliers G., Delia D., Onida F. (2010) Invest. New Drugs, in press [DOI] [PubMed] [Google Scholar]
- 40.Siziopikou K. P., Harris J. E., Casey L., Nawas Y., Braun D. P. (1991) Cancer 68, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 41.Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mace P. D., Shirley S., Day C. L. (2010) Cell Death Differ. 17, 46–53 [DOI] [PubMed] [Google Scholar]
- 43.Sayers T. J., Brooks A. D., Koh C. Y., Ma W., Seki N., Raziuddin A., Blazar B. R., Zhang X., Elliott P. J., Murphy W. J. (2003) Blood 102, 303–310 [DOI] [PubMed] [Google Scholar]
- 44.Safa A. R., Day T. W., Wu C. H. (2008) Curr. Cancer Drug Targets 8, 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen S. L., Peyton M., Minna J. D., Wang X. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11936–11941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S., Kang J., Evers B. M., Chung D. H. (2004) J. Pediatr. Surg. 39, 509–515 [DOI] [PubMed] [Google Scholar]
- 47.Casciano I., De Ambrosis A., Croce M., Pagnan G., Di Vinci A., Allemanni G., Banelli B., Ponzoni M., Romani M., Ferrini S. (2004) Cell Death Differ. 11, 131–134 [DOI] [PubMed] [Google Scholar]
- 48.Gyrd-Hansen M., Meier P. (2010) Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 49.Dineen S. P., Roland C. L., Greer R., Carbon J. G., Toombs J. E., Gupta P., Bardeesy N., Sun H., Williams N., Minna J. D., Brekken R. A. (2010) Cancer Res. 70, 2852–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huerta S., Gao X., Livingston E. H., Kapur P., Sun H., Anthony T. (2010) Surgery 148, 346–353 [DOI] [PubMed] [Google Scholar]
- 51.Yamazaki K., Gohda J., Kanayama A., Miyamoto Y., Sakurai H., Yamamoto M., Akira S., Hayashi H., Su B., Inoue J. (2009) Sci. Signal. 2, ra66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.