Abstract
O6-Methylguanine (O6-methylG) is highly mutagenic and is commonly found in DNA exposed to methylating agents, even physiological ones (e.g. S-adenosylmethionine). The efficiency of a truncated, catalytic DNA polymerase ι core enzyme was determined for nucleoside triphosphate incorporation opposite O6-methylG, using steady-state kinetic analyses. The results presented here corroborate previous work from this laboratory using full-length pol ι, which showed that dTTP incorporation occurs with high efficiency opposite O6-methylG. Misincorporation of dTTP opposite O6-methylG occurred with ∼6-fold higher efficiency than incorporation of dCTP. Crystal structures of the truncated form of pol ι with O6-methylG as the template base and incoming dCTP or dTTP were solved and showed that O6-methylG is rotated into the syn conformation in the pol ι active site and that dTTP misincorporation by pol ι is the result of Hoogsteen base pairing with the adduct. Both dCTP and dTTP base paired with the Hoogsteen edge of O6-methylG. A single, short hydrogen bond formed between the N3 atom of dTTP and the N7 atom of O6-methylG. Protonation of the N3 atom of dCTP and bifurcation of the N3 hydrogen between the N7 and O6 atoms of O6-methylG allow base pairing of the lesion with dCTP. We conclude that differences in the Hoogsteen hydrogen bonding between nucleotides is the main factor in the preferential selectivity of dTTP opposite O6-methylG by human pol ι, in contrast to the mispairing modes observed previously for O6-methylG in the structures of the model DNA polymerases Sulfolobus solfataricus Dpo4 and Bacillus stearothermophilus DNA polymerase I.
Keywords: DNA Damage, DNA Structure, Enzyme Kinetics, Enzyme Mechanisms, Kinetics, Nucleic Acid Enzymology, X-ray Crystallography, Misincorporation
Introduction
Alkylating agents damage DNA by reacting with the nitrogen and oxygen atoms in DNA bases. Human exposure to alkylating agents arises from endogenous (e.g. food-derived nitrosamines) and exogenous (e.g. tobacco-specific nitrosoamines and chemotherapeutic agents including temozolomide and streptozotocin) sources (1). The endogenously produced compound S-adenosylmethionine also methylates DNA (2). Many alkylating agents that react with DNA form the mutagenic and cytotoxic DNA lesion O6-alkylguanine (3), along with other methylated bases. The mutagenicity of O6-alkylated DNA is a factor in human diseases such as cancer, teratogenic defects, and premature aging (1). Work focused on the adduct O6-methylG2 has shown that it causes G:C→A:T transition mutations (4).
The mutagenic potential of O6-methylG arises from the ability of DNA polymerases to incorporate dTTP opposite the lesion efficiently (5–8). This incorporation occurs, although H1 NMR and Tm studies have demonstrated that the O6-methylG:T base pair is less stable than the O6-methylG:C base pair in duplex DNA (9–12). Two possible base-pairing mechanisms, isosteric Watson-Crick pairing and wobble pairing, have been proposed for the O6-methylG:C base pair from results obtained in NMR experiments with oligonucleotides and crystal structures of DNA polymerase ternary complexes (12–15). A crystal structure of the Bacillus stearothermophilus DNA polymerase I showed cytosine paired opposite O6-methylG in an isosteric Watson-Crick geometry (14). The crystal structure of the Y-family DNA polymerase Sulfolobus solfataricus Dpo4 revealed wobble base pairing of cytosine opposite O6-methylG (13). The NMR results and crystal structures demonstrate that the O6-methylG:T base pair forms a pseudo Watson-Crick pair, with a single hydrogen bond formed between the O2 atom of T and the N2 atom of O6-methylG (11, 13, 14, 16). The shape of the Watson-Crick-like O6-methylG:T base pair fits in the active site of the DNA polymerase without distorting the DNA, thereby contributing to the incorporation of dTTP opposite the lesion (17). Also, the sequence context of O6-methylG has been shown to influence the extent of dTTP incorporation by DNA polymerases opposite the lesion (18).
Replicative DNA polymerases catalyze the misincorporation of dTTP opposite O6-alkylG with similar or even higher efficiency than incorporation of the correct dCTP (4, 7, 14, 18). Similarly, the Y-family DNA polymerases η, ι, and κ all show poor nucleotide discrimination when bypassing O6-alkylG (5, 8). Pols η and κ have similar efficiencies for dTTP and dCTP incorporation opposite O6-methylG; however, pol ι incorporates dTTP with ∼10-fold higher efficiency than dCTP opposite the lesion (5). Differences in the catalytic mechanisms of the Y-family DNA polymerases, inferred from kinetic and structural analyses, offer insights and possible explanations for the different abilities of the Y-family DNA polymerases to bypass O6-alkylG DNA lesions, in particular O6-methylG. Pols κ and η mainly use Watson-Crick base pairing during nucleotide incorporation (19–23). Pol ι, however, has been shown to use a Hoogsteen base-pairing mechanism for efficient nucleotide incorporation opposite unadducted and some adducted template purines (24–27).
The high potential for mutation from misincorporation of nucleotides opposite O6-methylG and the potential roles of Y-family DNA polymerases in bypassing the adduct in vivo motivated our structural analyses of pol ι and the O6-methylG lesion. The results provide an understanding of the molecular mechanism facilitating the high efficiency of misincorporation by DNA pol ι opposite O6-methylG. The high efficiency of dTTP incorporation arising from aberrant Watson-Crick-like base pairing with O6-methylG is well established (4, 5, 7, 8, 14, 18). The increasing evidence that pol ι utilizes rotation of the purine template into a syn conformation led to a hypothesis that the increased efficiency of dTTP over dCTP incorporation opposite O6-methylG by pol ι arises from a more stable Hoogsteen base pairing of T opposite O6-methylG than C in the pol ι active site (24–27). Here, we report that the crystal structure of pol ι, in complex with a template O6-methylG, demonstrates that the lesion adopts a syn conformation in the pol ι active site. Important differences in the hydrogen bonding and base positioning of dCTP or dTTP opposite O6-methylG contribute to the increased efficiency of dTTP misincorporation opposite O6-methylG by pol ι.
EXPERIMENTAL PROCEDURES
Oligonucleotides
The self-complementary 18-mer DNA oligonucleotides 5′-TCTXGGGTCCTAGGACC(ddC)-3′ (X: O6-methylG and ddC: dideoxy CMP), 5′-TCTXGGGTCCTAGGACCC-3′, and 5′-TCTGGGGTCCTAGGACCC-3′ were purchased from Midland Certified Reagent Co. (Midland, TX) and purified using reversed-phase HPLC by the manufacturer.
Expression and Purification of Human Pol ι
The recombinant catalytic fragment of pol ι (amino acids 1–420) was expressed as an GST:pol ι or MBP:pol ι (gift from F. W. Perrino, Wake Forest University, Winston-Salem, NC) fusion protein (GST or MBP positioned at the N terminus of pol ι) with a PreScission Protease (GE Healthcare) cut site (LEVLFQG) four (GST) or seven (MBP) residues from the pol ι N-terminal methionine. The PreScission Protease recognition sequence and pol ι coding sequence were verified by nucleotide sequence analysis. The plasmid constructs were transfected into Escherichia coli BL21 Gold (DE3) cells (Stratagene, La Jolla, CA) for overexpression. Cells were grown at 37 °C to O.D.600 0.5. After induction with 1 mm isopropyl β-d-thiogalactopyranoside, the cells were allowed to grow for 15 h at 17 °C. Cell extracts were prepared, and the GST-pol ι fusion protein was bound to a 1-ml GSTrap HP column (GE Healthcare) in 50 mm Tris-HCl buffer (pH 7.5) containing 1 mm EDTA, 150 mm NaCl, and 1 mm dithiothreitol. The fusion protein was cleaved directly on the column by the addition of PreScission Protease (2 units/ml) and incubation at 4 °C for 12–48 h. An eluted fraction containing human pol ι was collected and concentrated using a Centriprep® centrifugal concentrator (Amicon, Beverly, MA; supplemental Fig. S1). A280 measurements (ϵ280 = 16,390 m−1 cm−1) (28) indicated a yield of ∼10–30 mg of pol ι per 4-liter preparation.
Primer Extension and Steady-state Kinetic Assays
For primer extension and kinetic assays, the G- and O6-methylG-containing DNA oligonucleotides were 5′ [γ-32P]ATP end-labeled (PerkinElmer Life Sciences) and annealed before being added to reactions containing 20 mm Tris-HCl buffer (pH 7.5) and 2 mm dithiothreitol, 0.1 mg/ml BSA, 200 μm dNTPs, and the amount of pol ι indicated (in the figure legends). Incubations were done for 10 min at 37 °C, and reactions were quenched with 30 μl of C2H5OH. Samples were dried and resuspended in 6 μl of a 95% formamide/0.02% (w/v) bromphenol blue/0.02% (w/v) xylene cyanol dye solution. Extension products were separated on 8 m urea/23% (w/v) polyacrylamide gels, visualized using a phosphorimaging system, and quantified using Quantity OneTM software (Bio-Rad). For steady-state kinetic assays, the site-specific insertion procedure of Boosalis et al. (29) was used. The reaction conditions were the same as for the primer extension assays except that the concentration of pol ι was 5 nm, and only single nucleoside triphosphates were used, either dCTP or dTTP. The amount of pol ι in the reactions yielded ≤20% extended product. Incubations were for 10 min at 37 °C, and reactions were processed as described above. All extended product bands were used to determine kinetic parameters (Km and kcat) by nonlinear regression using GraphPad Prism 5.0 software (GraphPad, San Diego, CA). Relative insertion frequencies were calculated as (kcat/Km)incorrect/(kcat/Km)correct.
Crystallization of Pol ι:O6-MethylG Ternary Complexes
The purified pol ι catalytic core was concentrated to ∼11 mg of protein/ml using a Centriprep® centrifugal concentrator. The pol ι was mixed at a 1:1.2 molar ratio with the 3′ dideoxyC-terminated O6-methylG-containing oligonucleotide. To study ternary complexes, MgCl2 and dCTP (or dTTP) were added to final concentrations of 10 and 20 mm, respectively. Crystals grew from solutions prepared as described by Nair et al. (25), i.e. containing 0.2–0.4 m (NH4)2SO4, 12.5–15% PEG 5000 monomethyl ether (w/v), and 0.1 m MES (pH 6.5). Crystal trays were incubated at 4 °C, and diffraction quality crystals appeared in 1–3 days. The crystals belonged to space group P6522 and had cell dimensions of a = b = 97.9 Å, c = 202.5 Å for dCTP-containing crystals and a = b = 97.9 Å, c = 202.7 Å for dTTP-containing crystals, with α = β = 90°, γ = 120°. For data collection, crystals were step-soaked for 5 min in mother liquor solutions containing 0–25% glycerol (w/v) and flash frozen in liquid nitrogen.
Structure Determination and Refinement
X-ray diffraction data were collected at a wavelength of 1.54 Å using a Bruker Microstar microfocus rotating-anode x-ray generator with a Bruker Proteum PT135 CCD detector (Bruker AXS, Madison, WI) in the Vanderbilt Center for Structural Biology or on the 21-ID-D beam line at the Advanced Photon Source (APS) at Argonne National Laboratory (Argonne, IL). The data were indexed, integrated, and scaled using HKL2000 (30). Phases were calculated using molecular replacement with Phaser software, which generated a unique solution when pol ι (Protein Data Bank code 2ALZ) minus DNA was used as a search model (31). Electron density maps (calculated to 2.8 Å (dCTP) and 1.9 Å (dTTP)) showed clear density around the O6-methylG lesion and both incoming dCTP and dTTP. Models were built using COOT software and refined using REFMAC5 software using TLS refinement (32–34). The refined model converged to an Rcryst = 21.5% and Rfree = 28.3% for the dCTP-containing complex and an Rcryst = 20.8% and Rfree = 24.1% for the dTTP-containing complex. Ramachandran plots for the refined models show good stereochemistry, with 90.0 (dCTP-containing) and 92.5% (dTTP-containing) of residues in the favored regions and 0.0 (dCTP-containing) and 0.0% (dTTP-containing) in the disallowed regions. Figures were prepared using PyMOL (35).
RESULTS
Extension of G- and O6-MethylG-containing Oligonucleotide Primers by Human Pol ι
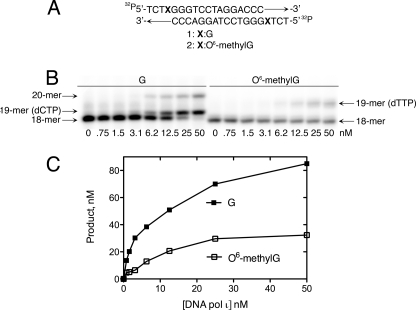
Human pol ι incorporated (the correct) dCTP opposite template G but preferentially misincorporated dTTP opposite template O6-methylG. The ability of the pol ι catalytic core to extend a primer annealed to a G- or O6-methylG-containing DNA template was determined in a primer extension experiment with a self-complementary 18-mer oligonucleotide used as both the primer and template in the reaction (self-annealing of the oligonucleotide generated 4-base overhangs at both ends of the duplex DNA) (Fig. 1A). The results of the primer extension reaction demonstrated that pol ι incorporated nucleotides opposite template G and extended the primer by one nucleotide using this substrate (Fig. 1B). The lack of further extension is consistent with the previously demonstrated low processivity of pol ι (5, 24, 36–38). Pol ι incorporated a single nucleotide opposite O6-methylG but was unable to extend from the nascent O6-methylG base pair (Fig. 1B). The 19-mer product bands ran faster in the gel when G was the template base compared with the 19-mer product bands formed using O6-methylG. This result suggests that a different incoming nucleotide was incorporated opposite G than O6-methylG. Based on primer extension experiments performed in the presence of single nucleotides, it was determined that the 19-mer product bands had dCTP incorporated opposite G and dTTP misincorporated opposite O6-methylG (data not shown). Quantification of the primer extension data demonstrated that O6-methylG inhibited extension catalyzed by pol ι compared with that using template G (Fig. 1C). The results of the primer extension experiments using the pol ι catalytic fragment (amino acids 1–420) are consistent with previous results of primer extension opposite template G and O6-methylG using full-length pol ι (amino acids 1–715) (5).
FIGURE 1.
Primer extension of pol ι opposite G- and O6-methylG-containing DNA templates. A, the 18-mer oligonucleotide was self-complementary and generated two extendable 3′ termini at both ends of the duplex. X was either G or O6-methylG. B, a primer extension experiment was performed using G- and O6-methylG-containing DNA templates (50 nm) and increasing pol ι concentrations in the presence of MgCl2 (2 mm). C, quantified primer extension data is shown.
Steady-state Kinetics of Nucleotide Incorporation Opposite G and O6-MethylG by Pol ι
To quantify more precisely the results of the primer extension experiment, steady-state kinetic analyses were performed using the pol ι catalytic fragment (Table 1). Pol ι was more efficient at misincorporation of dTTP opposite O6-methylG than correct incorporation of dCTP. Steady-state kinetic analyses using full-length enzyme show that pol ι has a ∼10-fold higher efficiency for dTTP incorporation compared with dCTP incorporation opposite O6-methylG (5). The catalytic core of pol ι was similar to the full-length enzyme and maintained a high efficiency of dTTP misincorporation opposite O6-methylG; dTTP misincorporation efficiency was ∼6-fold higher than dCTP incorporation opposite O6-methylG (Table 1). The increased efficiency of dTTP misincorporation opposite O6-methylG is mainly due to a Km effect; no changes were observed in the kcat values for dCTP or dTTP incorporation (Table 1). Thus, the different efficiencies of dCTP or dTTP incorporation opposite O6-methylG might result from binding and positioning of the nucleotide differently in the pol ι active site.
TABLE 1.
Steady-state kinetics of nucleotide incorporation opposite G and O6-methylG by pol ι
Km and kcat values were determined by quantifying gel band intensities using QuantityOne software (Bio-Rad) and nonlinear regression analysis of product versus [dNTP] curves using GraphPad Prism 5.0.
| Template | dNTP | Km | kcat | kcat/Km | finca |
|---|---|---|---|---|---|
| μm | s−1 | s−1 mm−1 | |||
| O6-MethylG | C | 1200 ± 700 | 0.03 ± 0.007 | 2.5 × 10−2 | 1 |
| T | 140 ± 60 | 0.02 ± 0.002 | 1.4 × 10−1 | 5.6 | |
| G | C | 90 ± 10 | 0.04 ± 0.002 | 4.4 × 10−1 | 1 |
| T | 860 ± 360 | 0.01 ± 0.003 | 1.2 × 10−2 | 0.27 |
a finc was calculated as (kcat/Km)incorrect/(kcat/Km)correct.
Crystal Structures of Human Pol ι:O6-MethylG Ternary Complexes
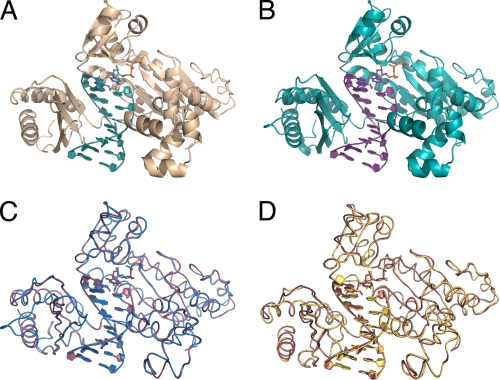
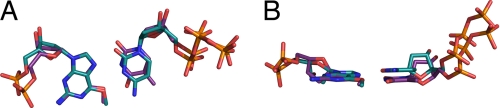
Several crystal structures of pol ι ternary complexes with normal and adducted DNA templates have demonstrated the ability of pol ι to rotate template purines to the syn conformation (24–27). Thus, the high efficiency of pol ι for dTTP misincorporation opposite O6-methylG may be due to dCTP and dTTP base pairing in different modes with the Hoogsteen edge of O6-methylG. To test this hypothesis, the crystal structures of pol ι:O6-methylG ternary complexes were solved with incoming dTTP or dCTP (Fig. 2, A and B, and Table 2). The structural data indicate that pol ι bound O6-methylG in a ternary complex with incoming dCTP or dTTP without major changes in the protein fold or the position of specific amino acid side chains around the active site. The structures of the pol ι:O6-methylG complex were similar, regardless of whether dCTP or dTTP was the incoming nucleotide and superimpose with a root mean square deviation value of 0.34 Å (Fig. 2C). Also, the structure of the pol ι:O6-methylG complex is similar to the pol ι:G ternary complex (2ALZ), and the structures superimpose with a root mean square deviation value of 0.42 Å (Fig. 2D). These data indicate that the preference of pol ι for dTTP incorporation opposite O6-methylG does not result from major conformational changes in the protein, therefore suggesting that preferential dTTP incorporation occurs as a result of differences in the base pairing of dTTP and dCTP with O6-methylG.
FIGURE 2.
Crystal structures of pol ι in complex with O6-methylG and incoming dCTP and dTTP. A, pol ι with template O6-methylG and incoming dCTP. B, pol ι with template O6-methylG and incoming dTTP. C, comparison of the pol ι complexes containing template O6-methylG and incoming dCTP (pink) and dTTP (blue). D, comparison of the pol ι with template G (2ALZ; yellow) and pol ι with template O6-methylG and incoming dCTP (red).
TABLE 2.
Data collection and refinement statistics
| Data collection | Pol ι:O6-methylG with dCTP | Pol ι:O6-methylG with dTTP |
| Resolution (Å)a | 2.8 (2.85-2.80) | 1.9 (1.97-1.90) |
| No. of measured reflections | 255,620 | 723,837 |
| No. of unique reflections | 14,304 | 46,192 |
| Completeness (%) | 96.8 (86.7) | 99.8 (98.9) |
| Redundancy | 17.9 (3.8) | 15.7 (8.8) |
| Rmergeb (%) | 17.3 (87.2) | 6.8 (52.5) |
| Mean I/σ | 11.2 (1.1) | 37.9 (3.2) |
| Refinement | ||
| Resolution range (Å) | 80–2.8 | 85–1.9 |
| Reflections | 13,705 | 43,636 |
| Rcrystc (%) | 21.5 | 20.8 |
| Rfreed (%) | 28.3 | 24.1 |
| Root mean square deviation bond lengths (Å) | 0.014 | 0.026 |
| Root mean square deviation bond angles (°) | 1.7 | 2.2 |
| Mean B-factor (Å2) | ||
| Protein | 13.8 | 16.8 |
| DNA | 19.1 | 18.6 |
| H2O | 10.9 | 24.6 |
a Values for outermost shells are given in parentheses.
b Rmerge = Σ|I − 〈I〉|ΣI, where I is the integrated intensity of a given reflection.
c Rcryst = Σ‖Fobserved − Fcalculated‖/Σ|Fobserved|.
d Rfree was calculated using 5% random data omitted from the refinement.
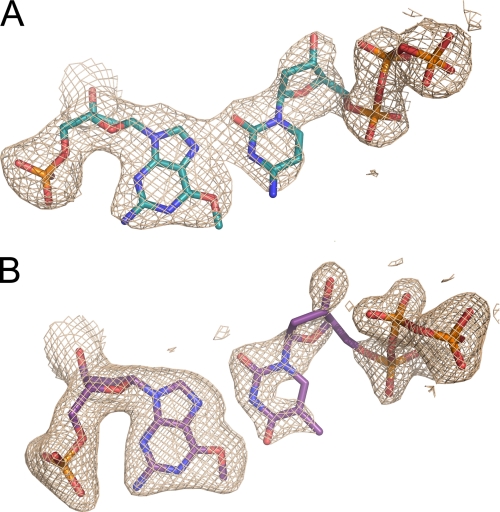
The electron density around the O6-methylG lesion indicates that the base is rotated from the anti to the syn conformation in the pol ι active site. The ability of pol ι to rotate template purine bases into a syn conformation was shown previously in crystal structures of pol ι complexed with a normal G template (2ALZ), a 1,N6-ethenodeoxyadenosine lesion (2DPJ), and an N2-ethylG lesion (3EPG) (24–26). These crystal structures have demonstrated a Hoogsteen base pairing mechanism for pol ι (24–26). The Hoogsteen edge of the adducted template base forms hydrogen bonds with the incoming dCTP and dTTP, and clear electron density is seen around O6-methylG and each incoming dCTP or dTTP (Fig. 3, A and B). Interestingly, for minor groove DNA adducts, e.g. at the N2 position of G, the ability of pol ι to form Hoogsteen base pairs results in high efficiency of correct nucleotide incorporation (24, 39). However, this may not be the case with O6-methylG, where an increased efficiency of misincorporation is observed (Table 1) (5).
FIGURE 3.
Pol ι active site. A, electron density map showing the template O6-methylG in the syn conformation paired with incoming dCTP. B, electron density map showing the template O6-methylG in the syn conformation paired with incoming dTTP.
Hydrogen Bonding Between dCTP and (syn) O6-MethylG
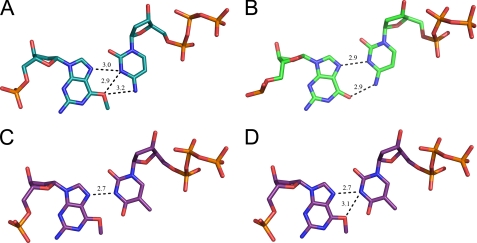
Preferential incorporation of dTTP opposite O6-methylG is the result of a more energetically favored base pair formed with the Hoogsteen edge of O6-methylG compared with dCTP. Different hydrogen bonds were formed whether dCTP or dTTP pair with O6-methylG in the pol ι active site (Fig. 4). The N3 atom of cytosine must be protonated to function as a hydrogen donor to the N7 and O6 atoms of O6-methylG. A positively charged cytosine has been hypothesized to bind in the active site of pol ι, as a hydrogen atom at the N3 position is needed to form a base pair with the syn G- and N2-ethylG-template bases (24, 25). The geometry of cytosine, bound in the active site of pol ι, suggests that the N3 atom opposite O6-methylG is protonated and donates its hydrogen to the N7 and O6 acceptor atoms of O6-methylG in a bifurcated hydrogen bond (Fig. 4A). The distances between the N3 atom of dCTP and the two hydrogen acceptors of O6-methylG are 2.9 Å and 3.0 Å, respectively. A second hydrogen bond is formed between the N4 atom of dCTP and the O6 atom of O6-methylG, with a distance of 3.2 Å (Fig. 4A). The hydrogen bond network between dCTP and O6-methylG is different from the hydrogen bonds formed between dCTP and the unadducted G. The structure of pol ι with unmodified G (2ALZ) showed two hydrogen bonds formed between the N7·N3 and O6·N4 atoms of G and dCTP, respectively (Fig. 4B).
FIGURE 4.
Hoogsteen edge hydrogen bonding in the pol ι active site. A, template O6-methylG forms a bifurcated hydrogen bond with the protonated dCTP. B, template G forms two hydrogen bonds with the protonated dCTP (2ALZ). C, single hydrogen bond forms between template O6-methylG and incoming dTTP. D, bifurcated hydrogen bond does not form between O6-methylG and dTTP.
Hydrogen Bonding Between dTTP and (syn) O6-MethylG
A single hydrogen bond, with a distance of 2.7 Å, forms between the N7 and N3 atoms of O6-methylG and dTTP, respectively (Fig. 4C). The length of the bond provides some indication of its strength, and the short distance of the hydrogen bond formed between dTTP and O6-methylG suggests a strong hydrogen bonding interaction that is potentially favored over the relatively weak bifurcated hydrogen bond formed with dCTP. The possibility that dTTP may also form a bifurcated hydrogen bond with O6-methylG is unlikely given the position of the base and the asymmetrical distances of the bond lengths (the bond between the O6 atom and the N3 atoms is 3.1 Å; Fig. 4D).
Positioning of dCTP and dTTP Opposite O6-MethylG in the Pol ι Active Site
Different hydrogen bond networks form depending on whether the dCTP or dTTP base is paired with O6-methylG in the pol ι active site. These differences affect how the incoming dCTP or dTTP binds in the active site of pol ι when paired with O6-methylG. Superposition of the O6-methylG bases demonstrates how the incoming dCTP and dTTP are accommodated and positioned differently opposite the lesion. Comparison of the positions of the incoming nucleotides relative to the position of O6-methylG revealed that the dCTP:O6-methylG base pair is sheared relative to the dTTP:O6-methylG base pair by ∼1.0 Å (Fig. 5A). The N3 atom of dTTP is positioned to be directly in line with the N7 atom of O6-methylG, whereas the N3 atom of dCTP is positioned equidistant between the N7 and O6 atoms of O6-methylG (Fig. 5A). The comparison showed that the dCTP base has a different rise relative to the dTTP base when paired opposite O6-methylG where the dTTP base lies in a plane ∼1.2 Å below the base of the incoming dCTP (Fig. 5B).
FIGURE 5.
Positions of dCTP and dTTP bases paired opposite O6-methylG. A, template O6-methylG aligned to show how dCTP and dTTP position differently opposite the adduct in the pol ι active site. B, rotated view showing differences in the rise between the O6-methylG:dCTP and dTTP base pairs.
DISCUSSION
Utilization of the Hoogsteen edge of adducted and unadducted template purines, when base pairing with incoming nucleotides, provides the basis for the higher efficiency of dTTP misincorporation opposite O6-methylG observed with pol ι compared with other Y-family DNA polymerases (5). The Hoogsteen edge of O6-methylG presents a unique hydrogen bond platform for the C and T nucleoside triphosphates. Differences in the hydrogen bonds that form (observed in the crystal structures) between dTTP or dCTP paired opposite O6-methylG appear to contribute to the ∼10-fold higher steady-state efficiency of dTTP misincorporation by human pol ι (Table 1) (5). To form a hydrogen bond between dCTP and (syn) O6-methylG, the N3 atom of dCTP must be protonated (40). The pKa of C in solution is 4.5, thus the N3 atom of C is not protonated at physiological pH (41). However, the pKa of C has been reported to increase to 7.5 when paired in a hemiprotonated C:C(H+) base pair (42). Also, the possibility that C has an elevated pKa when bound in the active site of pol ι has been suggested (25). The crystal structure of pol ι and O6-methylG indicates that the protonated N3 atom of dCTP acts as a hydrogen donor to both the N7 and O6 acceptor atoms of O6-methylG to form a bifurcated hydrogen bond. Splitting the proton between two acceptor atoms lowers the energy of the bifurcated hydrogen bond compared with the simple hydrogen bond formed between a single donor and acceptor pair (43). Thus, the protonation of the N3 atom of dCTP and formation of a bifurcated hydrogen bond between O6-methylG and dCTP may factor together in lowering the efficiency of DNA pol ι-catalyzed dCTP incorporation opposite O6-methylG. Moreover, it is likely that this bifurcated hydrogen bond is only formed in a fraction of the encounters with dCTP. In many cases cytosine is not protonated, and the resulting repulsion will then prevent formation of a productive complex. The crystal structure of pol ι and O6-methylG:dTTP indicates that a single hydrogen bond forms between the N3 and N7 atoms of dTTP and O6-methylG, respectively. The short length of the hydrogen bond (2.7 Å) suggests that the strength of the O6-methylG:dTTP bond may be greater than typical N–H–N hydrogen bonds (2.9–3.0 Å) that normally form (with a bond energy of ∼5 kcal/mol). The increased strength of the O6-methylG:dTTP hydrogen bond may stabilize the bound nucleoside triphosphate in the pol ι active site and contribute to the high efficiency of dTTP misincorporation opposite the lesion.
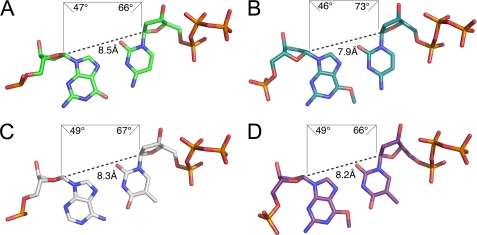
The thermodynamic and energetic properties of the hydrogen bonds formed with the Hoogsteen edge of O6-methylG appear to be more important to the aberrant incorporation of dTTP opposite O6-methylG than the geometry of the base pair fit into the active site of pol ι. Comparison of the pol ι:O6-methylG structures with structures of pol ι and template G paired with dCTP (2ALZ) (25) or template A paired with dTTP (2FLL) (44) reveal a relatively undistorted geometry for all of the base pairs (Fig. 6). An exception is observed in the glycosidic bond angle (73°) of dCTP when paired opposite O6-methylG, which may contribute to the lower efficiency of dCTP incorporation opposite O6-methylG (Fig. 6B). Yet, the similarity of the geometry and fit of the unadducted and adducted Hoogsteen base pairs in the pol ι active site suggests an important role for the hydrogen bond in the base selectivity for this Y-family DNA polymerase. Similarly in the case of S. solfataricus Dpo4, hydrogen bonding is more important than base pair geometry for nucleotide selectivity opposite O6-methylG. The crystal structures of Dpo4 with O6-methylG and O6-benzylG have shown that a wobble base pair forms when dCTP pairs opposite the lesion (13, 16). However, unlike pol ι, Dpo4 utilizes wobble base pairing to preferentially incorporate dCTP opposite O6-methylG, thus maintaining high fidelity during lesion bypass (13). When incorporating nucleotides opposite O6-methylG, the Y-family DNA polymerases differ from replicative DNA polymerases, which rely on Watson-Crick geometry for nucleotide selectivity and which have been proposed to tolerate the fit of the less thermally stable Watson-Crick-like O6-methylG:dTTP base pair better than the more thermally stable O6-methylG:dCTP wobble base pair with distorted Watson-Crick geometry (10). The structure of the B. stearothermophilus DNA polymerase I (BF) revealed that a O6-methylG:dTTP base pair with pseudo-Watson-Crick geometry fits in the active site of the DNA polymerase (14). Interestingly, the structure showed that an isosteric O6-methylG:dCTP base pair was also accommodated in the active site. The authors suggested that, upon binding the adducted base pair, the DNA polymerase introduced changes to the shape of the DNA so that Watson-Crick geometry was maintained despite energetic barriers (14).
FIGURE 6.
Geometric properties of Hoogsteen base pairs G:dCTP, O6-methylG:dCTP, A:dTTP, and O6-methylG:dTTP in the active site of pol ι. The C1′–C1′ distances (Å) and glycosidic bond angles (°) are shown for the G:dCTP (2ALZ) (A), O6-methylG:dCTP (B), A:dTTP (2FLL) (C), and O6-methylG:dTTP (D) base pairs.
The results of the structural analysis with human pol ι provide evidence that the differences in the hydrogen bonds formed between the incoming nucleotides dCTP or dTTP paired opposite O6-methylG are important to facilitating the increased misincorporation of dTTP opposite the lesion. Formation of a Hoogsteen base pair during nucleotide incorporation is a unique property of pol ι, allowing it to bypass minor groove DNA adducts efficiently, e.g. the N2 position of G. The N2-ethylG lesion, for example, is rotated out of the active site of pol ι and does not interfere with hydrogen bond formation between the template base and incoming nucleotide (24). In contrast, the major groove O6-methylG adduct, when rotated to the syn conformation, remains on the hydrogen bonding face of the template base and influences the hydrogen bonding properties of the adduct. The modified hydrogen bond pattern formed between O6-methylG and dCTP lowers the potential for binding of dCTP in the pol ι active site and results in decreased efficiency of correct nucleotide incorporation. The ability of DNA polymerases to misincorporate dTTP opposite O6-methylG results in a high number of G:C→A:T transition mutations caused by O6-alkylG DNA adducts. The reason for such low discrimination by DNA polymerases when replicating O6-methylG is not well understood despite substantial biochemical and structural work done to address this problem. The structure of pol ι (in complex with O6-methylG) shows that the adduct does not change the shape of the active site, suggesting that hydrogen bonding has an important role in the selection of incorporated nucleotides during pol ι-catalyzed bypass of O6-methylG. Differences in the hydrogen bonds formed between O6-methylG and dTTP or dCTP demonstrate how small changes to the geometry and strength of the hydrogen bond can affect base pairing and thereby influence the efficiency and fidelity of nucleotide incorporation by DNA polymerases.
Supplementary Material
Acknowledgments
We thank F. W. Perrino for the purified MBP-pol ι and S. Anderson for assistance with x-ray data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES010375 (to F. P. G.), P01 ES005355 (to M. E.), P30 ES000267 (to F. P. G. and M. E.), and T32 ES007028 (to F. P. G. and M. G. P.). Vanderbilt University is a member institution of LS-CAT at the Advanced Photon Source (APS), Argonne, Illinois. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-06CH11357.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 3NGD and 3OSN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- O6-methylG
- O6-methylguanine
- MBP
- maltose-binding protein
- pol ι
- DNA polymerase ι.
REFERENCES
- 1.Drabløs F., Feyzi E., Aas P. A., Vaagbø C. B., Kavli B., Bratlie M. S., Peña-Diaz J., Otterlei M., Slupphaug G., Krokan H. E. (2004) DNA Repair 3, 1389–1407 [DOI] [PubMed] [Google Scholar]
- 2.Shank R. C. (1987) Arch. Toxicol. Suppl. 10, 204–216 [DOI] [PubMed] [Google Scholar]
- 3.Loveless A. (1969) Nature 223, 206–207 [DOI] [PubMed] [Google Scholar]
- 4.Loechler E. L., Green C. L., Essigmann J. M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 6271–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J. Y., Chowdhury G., Zang H., Angel K. C., Vu C. C., Peterson L. A., Guengerich F. P. (2006) J. Biol. Chem. 281, 38244–38256 [DOI] [PubMed] [Google Scholar]
- 6.Snow E. T., Foote R. S., Mitra S. (1984) J. Biol. Chem. 259, 8095–8100 [PubMed] [Google Scholar]
- 7.Woodside A. M., Guengerich F. P. (2002) Biochemistry 41, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 8.Haracska L., Prakash S., Prakash L. (2000) Mol. Cell. Biol. 20, 8001–8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaffney B. L., Marky L. A., Jones R. A. (1984) Biochemistry 23, 5686–5691 [DOI] [PubMed] [Google Scholar]
- 10.Gaffney B. L., Jones R. A. (1989) Biochemistry 28, 5881–5889 [DOI] [PubMed] [Google Scholar]
- 11.Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. (1986) Biochemistry 25, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 12.Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. (1986) Biochemistry 25, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 13.Eoff R. L., Irimia A., Egli M., Guengerich F. P. (2007) J. Biol. Chem. 282, 1456–1467 [DOI] [PubMed] [Google Scholar]
- 14.Warren J. J., Forsberg L. J., Beese L. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19701–19706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginell S. L., Kuzmich S., Jones R. A., Berman H. M. (1990) Biochemistry 29, 10461–10465 [DOI] [PubMed] [Google Scholar]
- 16.Eoff R. L., Angel K. C., Egli M., Guengerich F. P. (2007) J. Biol. Chem. 282, 13573–13584 [DOI] [PubMed] [Google Scholar]
- 17.Tan H. B., Swann P. F., Chance E. M. (1994) Biochemistry 33, 5335–5346 [DOI] [PubMed] [Google Scholar]
- 18.Singer B., Chavez F., Goodman M. F., Essigmann J. M., Dosanjh M. K. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 8271–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasquez-Del Carpio R., Silverstein T. D., Lone S., Swan M. K., Choudhury J. R., Johnson R. E., Prakash S., Prakash L., Aggarwal A. K. (2009) PLoS One 4, e5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lone S., Townson S. A., Uljon S. N., Johnson R. E., Brahma A., Nair D. T., Prakash S., Prakash L., Aggarwal A. K. (2007) Mol. Cell 25, 601–614 [DOI] [PubMed] [Google Scholar]
- 21.Irimia A., Eoff R. L., Guengerich F. P., Egli M. (2009) J. Biol. Chem. 284, 22467–22480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang H., Taylor J. S. (2005) Biochemistry 44, 4850–4860 [DOI] [PubMed] [Google Scholar]
- 23.Wolfle W. T., Washington M. T., Kool E. T., Spratt T. E., Helquist S. A., Prakash L., Prakash S. (2005) Mol. Cell. Biol. 25, 7137–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pence M. G., Blans P., Zink C. N., Hollis T., Fishbein J. C., Perrino F. W. (2009) J. Biol. Chem. 284, 1732–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2005) Structure 13, 1569–1577 [DOI] [PubMed] [Google Scholar]
- 26.Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2006) Nat. Struct. Mol. Biol. 13, 619–625 [DOI] [PubMed] [Google Scholar]
- 27.Nair D. T., Johnson R. E., Prakash S., Prakash L., Aggarwal A. K. (2004) Nature 430, 377–380 [DOI] [PubMed] [Google Scholar]
- 28.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boosalis M. S., Petruska J., Goodman M. F. (1987) J. Biol. Chem. 262, 14689–14696 [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34.Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 35.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 36.Tissier A., McDonald J. P., Frank E. G., Woodgate R. (2000) Genes Dev. 14, 1642–1650 [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Yuan F., Wu X., Wang Z. (2000) Mol. Cell. Biol. 20, 7099–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson R. E., Washington M. T., Haracska L., Prakash S., Prakash L. (2000) Nature 406, 1015–1019 [DOI] [PubMed] [Google Scholar]
- 39.Washington M. T., Minko I. G., Johnson R. E., Wolfle W. T., Harris T. M., Lloyd R. S., Prakash S., Prakash L. (2004) Mol. Cell. Biol. 24, 5687–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams L. D., Shaw B. R. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 1779–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawson R. M. C., Elliott D. C., Elliott W. H., Jones K. M. (eds) (1986) Data for Biochemical Research, 3rd Ed., pp. 80–85, Clarendon Press, Oxford [Google Scholar]
- 42.Gray D. M., Cui T., Ratliff R. L. (1984) Nucleic Acids Res. 12, 7565–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonin A. V., Vashchenko A. V. (2010) Magn. Reson. Chem. 48, 309–317 [DOI] [PubMed] [Google Scholar]
- 44.Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2006) Structure 14, 749–755 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.