Abstract
Chromosomal translocations targeting the mixed lineage leukemia (MLL) gene result in MLL fusion proteins that are found in aggressive human acute leukemias. Disruption of MLL by such translocations leads to overexpression of Hox genes, resulting in a blockage of hematopoietic differentiation that ultimately leads to leukemia. Menin, which directly binds MLL, has been identified as an essential oncogenic co-factor required for the leukemogenic activity of MLL fusion proteins. Here, we characterize the molecular basis of the MLL-menin interaction. Using 13C-detected NMR experiments, we have mapped the residues within the intrinsically unstructured fragment of MLL that are required for binding to menin. Interestingly, we found that MLL interacts with menin with a nanomolar affinity (Kd ∼ 10 nm) through two motifs, MBM1 and MBM2 (menin binding motifs 1 and 2). These motifs are located within the N-terminal 43-amino acid fragment of MLL, and the MBM1 represents a high affinity binding motif. Using alanine scanning mutagenesis of MBM1, we found that the hydrophobic residues Phe9, Pro10, and Pro13 are most critical for binding. Furthermore, based on exchange-transferred nuclear Overhauser effect measurements, we established that MBM1 binds to menin in an extended conformation. In a series of competition experiments we showed that a peptide corresponding to MBM1 efficiently dissociates the menin-MLL complex. Altogether, our work establishes the molecular basis of the menin interaction with MLL and MLL fusion proteins and provides the necessary foundation for development of small molecule inhibitors targeting this interaction in leukemias with MLL translocations.
Keywords: Biophysics, Calorimetry, NMR, Peptide Conformation, Protein-Protein Interactions, Menin, Mixed Lineage Leukemia
Introduction
Chromosomal translocations involving the mixed lineage leukemia (MLL)3 gene result in human acute myeloid and lymphoid leukemias, affecting both children and adults (1, 2). Fusion of MLL with one of 60 partner genes forms chimeric oncogenes encoding MLL fusion proteins, which results in enhanced proliferation and blockage of blood cell differentiation ultimately leading to the development of acute leukemia (3). Translocations of MLL are particularly prevalent in infants with acute myeloid leukemia and acute lymphoblastic leukemia and constitute up to 80% of all infant acute leukemia cases (4). Patients with leukemias harboring MLL translocations have a very poor prognosis using available therapies (20% event-free survival at 3 years), and it is clear that novel targeted therapies are urgently needed to treat these leukemias (3, 5).
MLL belongs to the evolutionary conserved family of TRX (Drosophila Trithorax) proteins that positively regulate gene expression during development (6–8). MLL has been shown to associate with promoters of >5000 human genes, suggesting that it might play a global role in transcription (9). MLL is an important regulator of Hox gene expression, which is required for normal hematopoiesis (10). Disruption of MLL by chromosomal translocations up-regulates expression of Hox genes, including Hoxa7, Hoxa9, and the Hox cofactor Meis1, resulting in blockage of hematopoietic differentiation that leads to leukemia (11). MLL is involved in a complex network of interactions with multiple proteins, including menin (12, 13). Importantly, the direct interaction with menin is critical for the oncogenic function of MLL fusion proteins (13–15).
Menin is a highly specific partner for MLL proteins and is an essential component of the MLL SET1-like histone methyltransferase complex (12, 16). The MLL-menin interaction is required to regulate expression of MLL target genes, including Hoxa7, Hoxa9, Hoxa10, Hoxc7, and Meis1 (12–15). Menin is a 67-kDa tumor suppressor protein encoded by the Men1 (multiple endocrine neoplasia I) gene (17), which directly controls cell growth in selected organs, including parathyroid, pancreatic islets, and the pituitary gland (18). In leukemias, menin functions as an essential oncogenic co-factor of MLL fusion proteins (13). Because menin binds to the N terminus of MLL, this interaction is preserved among wild-type MLL and all MLL fusion proteins (13, 14). Mutations within the N terminus of MLL fusions disrupt its association with menin and abolish its oncogenic properties in vitro and in vivo (13–15). Furthermore, the expression of a dominant negative polypeptide corresponding to the N-terminal MLL sequence inhibits growth of MLL-AF9-transformed bone marrow cells (14). Overall, the MLL interaction with menin is critical for the oncogenic activity of MLL fusions, validating the MLL-menin interaction as a potential target for molecular therapy (13, 14). Recent findings strongly suggest that MLL fusion proteins require the co-expression of wild-type MLL to induce leukemia (19). Therefore, inhibition of the association of menin with both MLL and MLL fusions by small molecule inhibitors might result in new therapeutic agents for MLL-associated leukemias.
To date, the physical interaction of menin with MLL has been demonstrated by co-immunoprecipitation and in vivo experiments (13, 14). However, the detailed characterization of this interaction is currently missing, and the exact MLL fragment involved in the interaction with menin has not been identified. According to Yokoyama et al. menin recognizes only a short, five-amino acid RWRFP motif located at the very N terminus of MLL (13). In contrast, more recent work by Caslini et al. emphasized the requirement of a much longer, nearly 40-amino acid-long fragment of MLL for high affinity binding to menin (14). In this work, we have characterized in detail the MLL-menin interaction using a collection of biophysical methods. We found that menin forms a very stable complex with MLL (Kd = 10 nm) and recognizes two fragments of MLL, MBM1 and MBM2 (menin binding motifs 1 and 2). We have extensively characterized the interaction of MBM1 with menin, and by employing alanine scanning mutagenesis of MBM1 we found that hydrophobic residues have the most significant contribution to the binding affinity. Based on the exchange-transferred NOE NMR experiments we found that MBM1 binds to menin in an extended conformation. Importantly, we have also shown that targeting the MBM1 binding site is sufficient to dissociate the MLL-menin interaction fully. Therefore, the MBM1-menin interaction represents an attractive target for development of small molecule inhibitors as potential drugs for leukemias with MLL translocations.
EXPERIMENTAL PROCEDURES
Protein Expression
A cDNA encoding the full-length human menin was cloned into the pET32a vector (Promega) containing an N-terminal thioredoxin-His6 tag. Protein was expressed in RosettaTM (DE3) cells (EMD) and purified using affinity chromatography employing nickel-agarose (GE Healthcare) followed by the ion exchange using Q-Sepharose (GE Healthcare). To remove the fusion tag, the protein was cleaved by thrombin and applied onto nickel-agarose to extract the thioredoxin-His6 tag. In the final step, menin was dialyzed to 50 mm Tris buffer, pH 7.5, 50 mm NaCl, 1 mm DTT and frozen at −80 °C for further experiments.
The synthetic gene encoding N terminus of MLL was ordered from GenScript. To avoid oxidation and dimerization of MLL160 and MLL46 constructs, a single cysteine residue in the N terminus of MLL (Cys2) was mutated to an alanine. As shown in Table 1, this mutation does not affect binding to menin. The synthetic construct encoding the 160-residue-long fragment of human MLL (MLL160) was cloned into pET32a expression vector. The thioredoxin-MLL160 construct was expressed in RosettaTM (DE3) cells as an insoluble fraction. The inclusion bodies were purified from the cell pellet and solubilized in 6 m guanidine hydrochloride, and the protein was refolded using dialysis to 50 mm Tris buffer, pH 7.5, 50 mm NaCl, 1 mm DTT. The fusion protein was subsequently digested with enterokinase, and MLL160 was purified by an ion exchange using SP-Sepharose (GE Healthcare).
TABLE 1.
Binding affinities of various MLL fragments toward menin
The binding constants (Kd) were determined by employing ITC and/or FP. All FP measurements were done for synthetic peptides labeled with the N-terminal fluorescein. The ITC experiments were carried out for unlabeled peptides.
| MLL fragment | Kd | Method |
|---|---|---|
| nm | ||
| MLL15 | 90 ± 27 | ITC |
| MLL15 C2A (MBM1) | 49 ± 2.8 | ITC |
| MLL4–15 | 72 ± 22 | ITC |
| 53 ± 4.2 | FP | |
| MLL23–40 (MBM2) | 1,400 ± 424 | FP |
| MLL4–15 F9A | ∼80,000 | FP |
| MLL46 | 6.8 ± 1.7 | ITC |
| MLL160 | 9.8 ± 2.5 | ITC |
| MLL160 F9A | 1,550 ± 353 | ITC |
To obtain MLL46 we introduced stop codon mutation into MLL160 construct. The thioredoxin-MLL46 fusion expressed as soluble protein and was purified using affinity chromatography (nickel-agarose) followed by enterokinase cleavage and ion-exchange chromatography using SP-Sepharose. The 15N or 13C,15N-labeled proteins for NMR were obtained from bacterial cells cultured in minimal medium and purified using the same protocols as for unlabeled proteins.
Peptides
All peptides (>95% purity) were ordered from GenScript.
Detection of MLL-Menin Interaction by NMR
To examine interaction of MLL160 with menin we measured 1H-15N HSQC spectra. Two samples were prepared for these experiments: 30 μm 15N-MLL160 and 30 μm 15N-MLL160 with 30 μm menin in 50 mm phosphate buffer, pH 6.8, 50 mm NaCl, and 1 mm DTT. The experiments were recorded at 25 °C employing Bruker Avance III 600-MHz spectrometer equipped with cryogenic probe.
Assignment and Binding Studies of MLL46 with Menin
To assign chemical shifts of MLL46 we employed 13C-detected experiments measured for the sample containing 0.7 mm 13C,15N-labeled MLL46 in 50 mm Tris, pH 7.5, 50 mm NaCl, 1 mm DTT. The assignment was based on a series of two-dimensional CACO, CANCO, CBCACO, NCO spectra (20). To increase the sensitivity of these experiments, we employed experiments originating with 1H excitation (21). Mapping of the MLL46-menin interaction was carried out based on CACO and NCO spectra measured for 50 μm 13C,15N-MLL46 in the absence and presence of 40 μm menin. The spectra for the complex were collected with three times as many transients to account for lower signal-to-noise due to global broadening of resonances. To map the MLL residues involved in the binding to menin we analyzed peak broadening of MLL46 in the complex by calculating the ratio of peak heights derived from CACO experiments for MLL46-menin and free MLL46. All 13C-detected experiments were carried out at 200 MHz 13C frequency using Bruker Avance III 800-MHz spectrometer equipped with cryogenic probe. The spectra were measured at 25 °C.
Measurement of Tr-NOESY Spectra and Structure Calculation
To measure exchange-transferred Overhauser effect (Tr-NOEs) we prepared two samples: 3 mm MLL15 C2A mutant and 3 mm MLL15 C2A with 40 μm menin. Samples were prepared in 50 mm phosphate buffer, pH 6.8, 50 mm NaCl, and 1 mm DTT. All NMR measurements were carried out at 20 °C. Two-dimensional NOESY spectra were collected with 150-ms and 250-ms mixing times, 6000-Hz sweep width, and 300 complex points in indirect dimension. An additional two-dimensional TOCSY experiment with 80-ms mixing time was collected to complete assignment of the MLL peptide. All NMR spectra with 1H detection were measured using Bruker Avance III 600-MHz spectrometer equipped with cryogenic probe. Processing and analysis of NMR spectra were carried out employing NMRDraw (22) and Sparky, respectively (T. D. Goddard and J. M. Kneller, University of California, San Francisco).
MLL15 structures were calculated solely based on distance restraints from Tr-NOESY experiment recorded at 250-ms mixing time. Distance restraints were derived from the peak intensities and were categorized into two broad distance ranges: 1.8–4.5Å and 1.8–6.0 Å. We assigned a total of 138 distance restraints including: 61 intraresidual distances, 58 sequential, and 19 short range distances. For structure calculations we generated the peptide in extended conformation and calculated 300 structures employing simulated annealing protocol in the CNS program (23). We selected 51 of the lowest energy conformers for further analysis. Root mean square deviations values for all 51 structures (residues 7–13) are 1.3 ± 0.5 Å and 2.7 ± 0.8 Å for backbone and heavy atoms, respectively. The structures were superimposed and clustered in MolMol (24).
Isothermal Titration Calorimetry (ITC)
Menin and MLL fragments were dialyzed extensively at 4 °C against 50 mm phosphate buffer, pH 7.5, 50 mm NaCl, 1 mm β-mercaptoethanol, and degassed prior to measurement. The titrations were performed using the VP-ITC system (MicroCal) at 27 °C. The calorimetric cell, which contained menin (concentrations in the range 5–10 μm), was titrated with the MLL160-, MLL46-, and MLL-derived peptides (50–100 μm) injected in 10-μl aliquots. Data were analyzed using Origin 7.0 (OriginLab) to obtain Kd and stoichiometry.
Fluorescence Polarization Experiments
Fluorescein-labeled MLL-derived peptides, FITC-MLL4–43 at 1 nm, FITC-MBM1 at 15 nm, and FITC-MBM2 at 0.2 μm, were titrated with a range of menin concentrations in the FP buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 1 mm DTT) used for binding experiments (Kd measurements). In the competition experiments, fluorescein-labeled MLL peptides, menin, and varying concentrations of the unlabeled MLL peptides in FP buffer were used for IC50 determination (the concentrations of fluorescein-labeled peptides and menin used for IC50 determination are listed in the legend of Fig. 3). After a 1-h incubation of the protein-peptide complexes, changes in fluorescence polarization and anisotropy were monitored at 525 nm after excitations at 495 nm using PHERAstar microplate reader (BMG). Results were used to assess binding or inhibition for MLL-derived peptides with the Origin 7.0 program.
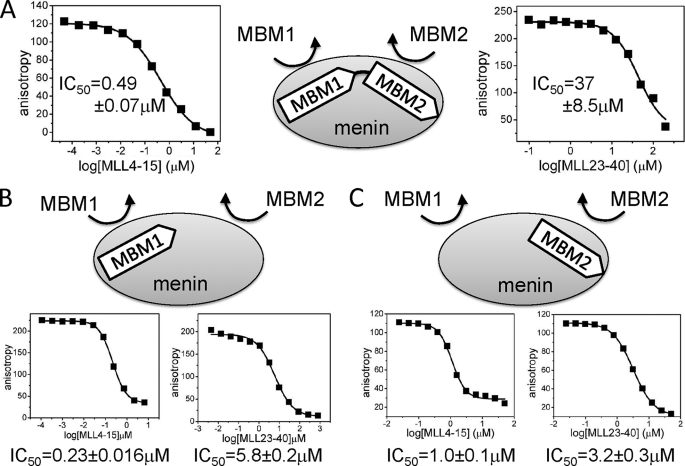
FIGURE 3.
Competition between different fragments of MLL for binding to menin. A series of FP experiments was measured to quantify the dissociation of the complex of menin bound to the fluorescein-labeled MLL-derived peptides by unlabeled MLL fragments. A, IC50 values for the displacement of FITC-MLL4–43 from menin by MBM1 and MBM2. FITC-labeled MLL4–43 peptide and menin were at 1 nm and 4 nm, respectively. B, displacement of FITC-MBM1 (15 nm) from menin (150 nm) by MBM1 and MBM2. C, displacement of FITC-MBM2 (0.2 μm) from menin (2.0 μm) by MBM1 and MBM2.
RESULTS
Menin Interacts with the N Terminus of MLL with High Affinity
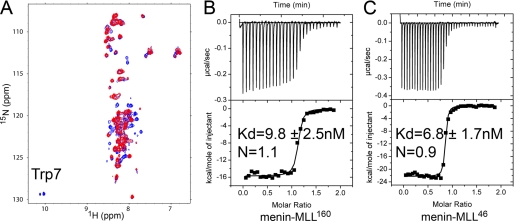
The interaction of menin with the N-terminal fragment of MLL has been demonstrated previously by co-immunoprecipitation and in vivo experiments (13, 14). However, detailed biophysical characterization of this interaction including binding affinity determination, stoichiometry of binding, and structural studies is currently missing. Furthermore, there is a disparity in the reported length of the MLL amino acid sequence required for the interaction with menin (13, 14). To map the exact MLL motif required for binding to menin, we expressed the 160-residue-long N terminus of MLL (MLL160) and then tested this protein for binding to menin using NMR spectroscopy and isothermal titration calorimetry. The 1H-15N HSQC spectrum of 15N-labeled MLL160 protein alone shows poor signal dispersion, which is indicative of unstructured protein (Fig. 1). To test whether MLL160 interacts with menin, we recorded a 1H-15N HSQC experiment for MLL160 mixed with menin (1:1 ratio). The addition of menin to MLL160 resulted in strong broadening of a subset of signals on the MLL160 spectrum while leaving the remaining peaks unperturbed (Fig. 1A). This demonstrates direct interaction between menin and MLL160 and also indicates that a relatively short fragment of MLL160 is involved in this interaction. Interestingly, the indole peak of the sole tryptophan (Trp7) was among the broadened signals, demonstrating that the very N-terminal part of MLL interacts with menin.
FIGURE 1.
Menin interacts with the N terminus of MLL with high affinity. A, comparison of 1H-15N HSQC spectra for 30 μm MLL160 (blue) and 30 μm MLL160 with 30 μm menin (red). Spectra were recorded in 50 mm phosphate buffer, pH 6.8, 50 mm NaCl, and 1 mm DTT at 25 °C. B, ITC experiment showing the titration of menin with MLL160. C, ITC for binding of menin with MLL46.
To quantify the MLL-menin interaction we employed ITC and obtained a nanomolar binding affinity (Kd = 9.8 nm) and 1:1 stoichiometry (Fig. 1B) for MLL160. Based on the previous work of Caslini et al. (14) we designed a shorter MLL construct encoding the 46-amino acid-long N terminus of MLL (MLL46) and tested its binding to menin by ITC. Indeed, using MLL46 we obtained a very similar binding affinity (Kd = 6.8 nm) as for MLL160 (Fig. 1, B and C), demonstrating that the menin binding motif is located within the N-terminal 46-amino acid fragment of MLL.
MLL Comprises Two Menin Binding Motifs
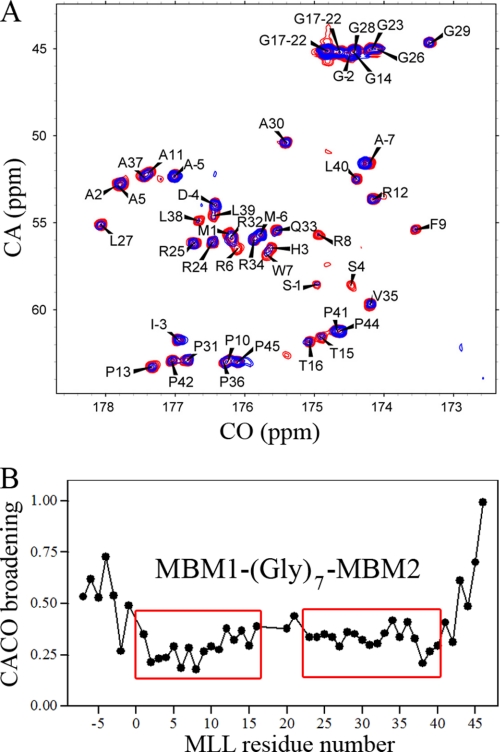
To map the residues of MLL46 involved in the binding to menin, we employed NMR spectroscopy. As judged from 1H-15N HSQC spectrum and 15N{1H} NOE experiment (supplementary Fig. S1), the N terminus of MLL is unstructured in solution. Application of proton-detected NMR experiments commonly used for assignment and binding studies is limited for unstructured polypeptides due to unfavorable H-D exchange properties of backbone amides and lack of sufficient chemical shift dispersion. For example, the 1H-15N HSQC recorded for MLL46 (pH 7.5, 25 °C) shows the presence of only about 10 peaks for backbone amides (supplementary Fig. S2). On the contrary, 13C-detected CACO recorded at the same conditions shows all expected resonances and excellent peak dispersion (Fig. 2A). Therefore, we chose to employ 13C-detected experiments (20) to map the residues of MLL46 required for binding to menin. We obtained nearly complete assignment of MLL46 based on combination of two-dimensional CACO, CANCO, CBCACO, and NCO experiments (Fig. 2A).
FIGURE 2.
Mapping of the MLL residues involved in binding to menin by employing 13C-detected experiments. A, superposition of two-dimensional CACO spectra measured for 50 μm 13C,15N MLL46 (red), and 50 μm 13C,15N MLL46 in the presence of 40 μm menin (blue). B, plot showing quantitative analysis of the signal broadening derived from CACO spectra for MLL46 upon binding to menin. The broadening values have been normalized to Val46 resonance, which shows the smallest effect. The two regions with strongest broadening are labeled, respectively, MBM1 and MBM2 (see “Results”). Residues with negative numbers correspond to 7 residues (AMADIGS) from cloning into pET32a vector.
To map the MLL residues involved in the interaction with menin, we measured CACO spectra for MLL46 in the presence of a substoichiometric concentration of menin. Binding of the highly flexible unstructured MLL46 to menin should result in the ordering of a set of MLL46 residues in the complex. For these residues, a very strong broadening is expected as a result of formation of a high molecular weight complex with a low nanomolar binding affinity. Indeed, we observed such an effect upon binding of MLL46 to menin (Fig. 2A). Interestingly, quantitative analysis of signal broadening revealed the presence of two MLL fragments: 2–15 and 23–40, which experienced a strong decrease in peak intensities upon binding to menin (Fig. 2B). We have labeled these MLL fragments, respectively, as MBM1 and MBM2. MBM1 and MBM2 are separated by a 7-amino acid polyglycine linker, which remains flexible in the complex and most likely is dispensable for binding to menin. These results were further corroborated by analysis of NCO spectra (see supplementary Fig. S3).
To examine whether MBM1 and MBM2 represent two independent binding motifs, we designed two fluorescein-labeled peptides: MLL4–15 (MBM1; MLL residues 1–3 are not involved in binding to menin as shown by ITC experiments, see below and Table 1) and MLL23–40 (MBM2). Binding of these peptides to menin was determined by fluorescence polarization (FP). We found that both peptides bind to menin, albeit with different affinities, suggesting that there are two distinct sites on menin for MLL binding. The dissociation constants obtained for MBM1 and MBM2 binding to menin are, respectively, 53 ± 4.2 nm and 1.4 ± 0.42 μm (Table 1). Hence, MBM1, which binds menin with 20 times higher affinity compared with MBM2, represents the high affinity binding motif. Very similar binding affinity was also obtained for MLL15, which comprises the entire 15 amino acid N-terminal fragment of MLL, in an independent ITC experiments (Kd = 90 ± 27 nm; see Table 1).
MBM1 Represents a Key Target Site for Inhibition of MLL-Menin Complex
The MLL-menin interaction has been validated to be a potential drug target for leukemias with MLL translocations (13–15). The presence of two binding sites on menin for MLL raises the question as to which site represents a more relevant target for inhibition by small molecules. To answer this question, we carried out a series of competition experiments and examined whether MBM1 and MBM2 are capable of causing dissociation of the MLL-menin complex. For these experiments we used fluorescein-labeled MLL3–42 (FITC-MLL3–42), which comprises both menin binding motifs, and unlabeled MBM1 and MBM2 peptides as competitors. We found that MBM1 can efficiently displace FITC-MLL4–43 from menin with IC50 = 0.49 ± 0.07 μm. MBM2, which binds to menin with much weaker affinity, was able to disrupt the MLL-menin complex but only at much higher concentrations, with an IC50 value of 37 ± 8.5 μm (Fig. 3).
Interestingly, the complete dissociation of MLL4–43 from menin by MBM1 suggests that MBM1 is capable of causing dissociation of not only MBM1 but also the MBM2 peptide from menin. To verify this, we carried out a series of competition experiments and tested whether binding of MBM1 to menin affects the binding of MBM2 and vice versa. We found that unlabeled MBM1 efficiently dissociates MBM2 with an IC50 value of 1.0 ± 0.1 μm (Figs. 3 and supplemental Fig. S4). The MBM2 peptide is also capable of causing dissociation of MBM1 from menin, although it is a weaker competitor with an IC50 of 5.8 ± 0.2 μm (Fig. 3). Overall, these experiments strongly suggest an allosteric regulation between binding of MBM1 and MBM2 peptides to menin. MBM1, which binds to menin with a higher affinity, can dissociate peptides from both sites on menin, ultimately resulting in disruption of the entire MLL-menin complex. This demonstrates that MBM1 represents the essential MLL fragment required for high affinity binding to menin. Importantly, our results provide strong evidence that the MBM1 site on menin is the key target site that could be utilized to develop small molecule inhibitors targeting the MLL-menin interaction.
Characterization of the High Affinity Menin Binding Motif
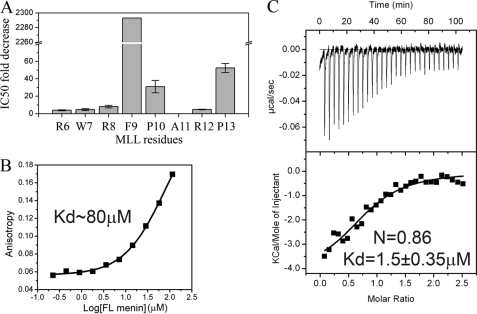
To determine the contribution of the individual MLL MBM1 residues to the binding to menin, we carried out alanine scanning mutagenesis. First, we found that three N-terminal MLL residues are not required for menin binding and a shorter peptide corresponding to the MLL fragment 4–15 (MLL4–15) binds to menin with the same affinity as MLL15 (Table 1). Subsequently, we introduced a set of point mutations into MLL4–15 to replace all residues within the Arg6 to Pro13 fragment to alanines (Fig. 4 and supplemental Table S1) and tested these peptides in the competition experiments for their ability to displace fluorescein-labeled wild-type MLL4–15 from menin. Comparison of relative IC50 values for all peptides is shown in Fig. 4A (see also supplemental Table S1). We found that three hydrophobic residues of MLL, Phe9, Pro10, and Pro13, are the most critical for binding to menin, with Phe9 showing the most pronounced effect (Fig. 4). Mutations of the other residues have significantly weaker effect (up to an 8-fold increase in IC50 value for R8A; Fig. 4 and supplemental Table S1). Overall, our competition experiments clearly demonstrate that menin recognizes a patch of hydrophobic residues within the MLL MBM1 motif.
FIGURE 4.
Mapping the MLL residues essential for the high affinity interaction of MBM1 with menin. A, comparison of IC50 for a series of alanine mutants of MBM1. The IC50 values were obtained for MLL4–15 peptides with single alanine mutations in competition experiments with FITC-MLL4–15 peptide (see supplemental Table S1 for IC50 values). B, dissociation constant for binding of MLL4–15 F9A and menin determined in FP experiment for fluorescein-labeled peptide. C, ITC experiment showing the binding of MLL160 F9A to menin.
The most significant decrease in the binding affinity resulted upon mutation of Phe9 to Ala, which reduced the IC50 by more than 2,000-fold. To verify this observation further, we tested the direct binding of fluorescein-labeled MLL4–15 F9A peptide in the FP experiment and found only a very weak interaction, with an estimated Kd of 80 μm (Fig. 4B and Table 1). Subsequently, we also introduced the F9A mutation into MLL160 and employed ITC to characterize the interaction with menin. We found that the MLL160 F9A mutant interacts with menin with Kd = 1.5 ± 0.35 μm, which indicates a nearly complete lack of interaction via MBM1 and is consistent with the affinity observed for MBM2 alone (Fig. 4C and Table 1). Overall, these experiments clearly indicate that MLL residue Phe9 is critical for high affinity interaction with menin.
MLL High Affinity Binding Motif Interacts with Menin in an Extended Conformation
The structures of menin and the MLL-menin complex are currently unknown. Here, we employed NMR to determine the structure of the MLL high affinity menin binding motif (MBM1) in the menin-bound conformation. MLL46 is natively unstructured in solution (see supplementary Fig. S1). However, binding of MLL to menin should result in the formation of a well defined conformation. One of the approaches that can be used to probe such a conformation is based on detection of Tr-NOE (25). Observation of Tr-NOEs requires fast exchange between bound and free ligands, typically corresponding to Kd > 1 μm (25). Because of the high affinity of MLL46 toward menin (Kd = 6.8 nm), such an approach would be most likely ineffective. Instead, we assumed that MBM1 might be amenable to such studies. To detect Tr-NOEs we measured two-dimensional NOESY spectra for free MLL15 C2A peptide and for its complex with menin (Fig. 5, A and B). Indeed, we observed a significant number of cross-peaks arising from transferred NOE in the spectrum of MLL15 C2A peptide in complex with menin.
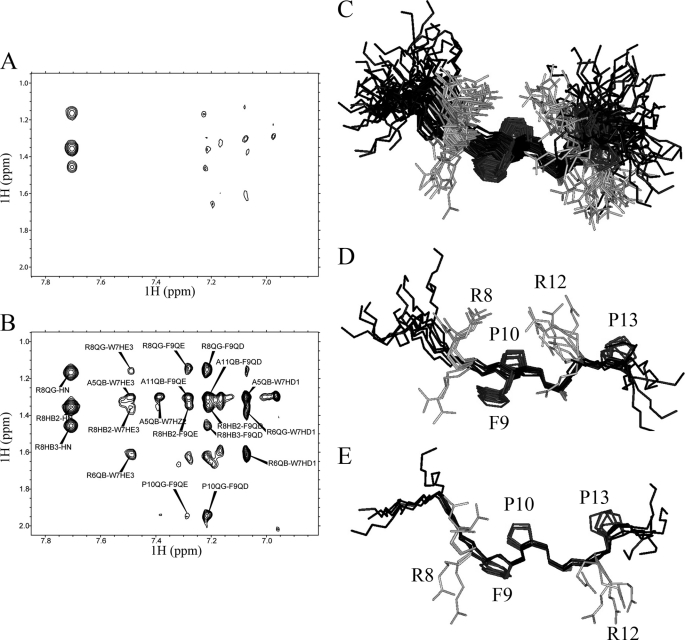
FIGURE 5.
MBM1 binds to menin in extended conformation. A, a fragment of two-dimensional NOESY spectrum with 250-ms mixing time recorded for 3 mm MBM1 peptide (MLL15 C2A). B, a fragment of assigned two-dimensional NOESY spectrum recorded for 3 mm MBM1 peptide with 40 μm menin. The experimental conditions were identical to those for the peptide alone. C, a set of 51 lowest energy conformers (MLL fragment 3–15; backbone in black) for MLL15 C2A based on Tr-NOEs. The side chains of hydrophobic residues (Phe9, Pro10, Pro13) which have the most significant contribution for binding to menin are shown in dark gray. D and E, superposition of the conformers for two most populated clusters.
To determine the conformation of MLL15 C2A bound to menin, we assigned proton chemical shifts for the free peptide based on two-dimensional NOESY and TOCSY spectra. Subsequently, the assignment was used for quantitative interpretation of Tr-NOEs observed on the spectra of MLL15 C2A in the presence of menin. The vast majority of observed Tr-NOEs arise from sequential residues (see “Experimental Procedures”), and only a small set of peaks corresponds to medium range (predominantly i; i+2) interactions. We then calculated a set of structures for MLL15 C2A employing standard simulated annealing simulation using distance restraints derived from Tr-NOEs as the sole structural information. As expected from the pattern of NOEs, all calculated structures show that MBM1 binds to menin in an extended conformation (Fig. 5). The backbone root mean square deviation for central residues of MBM1 motif (MLL residues 7–13) within a set of 51 lowest energy structures is 1.3 ± 0.5Å. Because of the presence of two proline residues (Pro10 and Pro13) in MBM1 we observe a minor population of the peptide which results from cis-trans isomerization. Analysis of NOEs indicates that the predominant form of MBM1 (>80%) exists in an all-trans conformation. Importantly, this is also the conformation that gives rise to the Tr-NOEs used for structure determination.
The detailed analysis of the calculated structures of MLL15 C2A indicates some conformational heterogeneity that we have attributed to the limited accuracy of distance restraints. Most likely the sensitivity of Tr-NOESY experiments is affected by the slow dissociation rate of the MLL15-menin C2A complex. We analyzed calculated structures by clustering them into several well defined conformations. Interestingly, in the most populated clusters the side chains of hydrophobic residues Phe9, Pro10, and Pro13 are clearly facing the same direction (Fig. 5, D and E), suggesting that they interact with the same binding pocket on menin. Such a conformation agrees very well with the alanine mutagenesis data which demonstrate that these hydrophobic residues are the most critical for binding to menin (Fig. 4A). Based on these studies we concluded that menin binds MBM1 in an extended conformation and recognizes the hydrophobic patch formed by the MLL residues Phe9, Pro10, and Pro13.
DISCUSSION
Interaction of menin with MLL fusion proteins has been shown to be critical to MLL-mediated leukemogenesis (13–15). Therefore, inhibition of this interaction by small molecules might represent an attractive therapeutic strategy for leukemia patients with MLL translocations. Several studies have emphasized that development of such compounds could result in novel targeted drugs for MLL-related leukemias (13–15). The first step toward development of small molecule inhibitors targeting MLL-menin requires detailed characterization of this interaction. In this work we found that (i) menin interacts with MLL with a low nanomolar affinity (Kd ∼ 10 nm); (ii) two short fragments of MLL, MBM1 (MLL4–15), and MBM2 (MLL23–40), are involved in the interaction with menin; (iii) MBM1 represents the high affinity menin binding motif; (iv) in competition experiments, the MBM1 peptide is capable of causing efficient displacement of MLL comprising MBM1 and MBM2 from menin; (v) MBM1 binds to menin in an extended conformation, and (vi) a set of hydrophobic residues (Phe9, Pro10, and Pro13) has the most significant contribution to the binding affinity.
MLL is a large, >400-kDa multidomain protein that might interact with menin in a complex manner. Intriguingly, based on previous studies, the menin binding motif has been mapped to involve either a 5-amino acid-long (13) or much longer 40-residue-long motif (14). To map this interaction accurately, we started with measuring the affinity of a significantly longer fragment of MLL, which consists of 160 amino acids. Subsequently, we found that the shortest MLL fragment that retains intact menin binding affinity requires 46 residues from the N terminus of MLL. Interestingly, more detailed analysis revealed that this fragment contains two separate menin binding motifs. These results emphasize that mapping protein-protein interactions involving natively unstructured proteins requires very careful analysis.
MBM1 and MBM2 Bind to Menin with Negative Cooperativity
The N terminus of MLL is natively unstructured in solution, and we found that two short fragments, which we labeled MBM1 and MBM2, are required for high affinity binding to menin. Interestingly, isolated peptides corresponding to MBM1 and MBM2 are capable of independent binding to menin with 53 ± 4.2 nm and 1.4 ± 0.42 μm affinities, respectively. Surprisingly, the binding affinity of a longer MLL fragment (MLL46) comprising both MBM1 and MBM2 is increased only by 8-fold compared with MBM1 alone. Such a modest increase in the binding affinity suggests that MBM1 and MBM2 interfere with each other when they bind to menin. The cooperativity of the binding of polyvalent ligands can be characterized by the cooperativity factor α (26). We calculated α using the experimentally determined binding affinities of MBM1, MBM2, and MLL46 to menin and found that α = 0.6, which indicates a strong negative cooperativity for the binding of MBM1 and MBM2. Indeed, we have verified this experimentally and found a significant competition between MBM1 and MBM2 for binding to menin (Fig. 3). One of the most significant consequences of such a negative cooperativity between MBM1 and MBM2 is the possibility of disrupting the MLL-menin complex by targeting only one of the binding sites on menin. For example, we found that the MBM1 peptide can fully dissociate the MLL4–43-menin complex.
13C-detected Experiments Are Well Suited to Study Interactions of Intrinsically Unstructured Proteins
NMR is a key experimental method for obtaining site-specific information for intrinsically unstructured proteins. The 13C-detected experiments are particularly well suited for this purpose due to high chemical shift dispersion and narrow line widths of peaks for highly flexible unfolded proteins (20, 27). Contrary to standard HN-detected triple resonance experiments, the 13C-detected experiments are independent of H-D exchange of amide protons (see supplementary Fig. S2). Here, we have demonstrated that 13C-detected experiments are also highly suitable for characterization of the interactions of natively unstructured proteins with globular domains. Based on CACA and NCO experiments we were able to map MLL residues involved in the interaction with menin accurately. Such an approach, in general, represents a powerful strategy to characterize the interactions of unstructured proteins.
Implications for Targeting Menin-MLL
Results from our studies will have a strong impact on development of small molecule inhibitors of the MLL-menin interaction. Disruption of this interaction should have the potential to reverse the oncogenic activity of MLL fusion proteins. It has been demonstrated that excision of a short, 5-amino acid-long (RWRFP) fragment from the N terminus of the MLL-GAS7 fusion, resulted in a protein that lost oncogenic potential in vivo (13). Based on our experiments, such a deletion would inevitably disrupt the interaction of the MBM1 motif with menin. The remaining MBM2 is therefore not sufficient for maintaining high affinity association of MLL fusions with menin and for their oncogenic activity. Therefore, physiologically relevant inhibition of the MLL-menin interaction could be achieved by targeting the MBM1 binding site on menin. Based on the series of alanine mutations in MBM1, we identified three hydrophobic MLL residues (Phe9, Pro10, Pro13) as the most relevant for high affinity binding to menin. The MBM1 binds in an extended conformation exposing these hydrophobic residues toward menin. Introduction of a point mutation F9A into MLL160 significantly impairs the interaction of MLL with menin to the level observed for MBM2 alone. Currently, the structure of menin and the MLL-menin complex is not known. Nevertheless, based on our biophysical characterization of the MLL interaction with menin we predict that the MBM1 binding site is predominantly hydrophobic. Accommodation of three bulky side chains of Phe9, Pro10, and Pro13 further defines a large hydrophobic pocket on menin. Overall, our data suggest that development of small molecule inhibitors targeting the menin interaction with MLL fusions is achievable and might ultimately lead to development of novel classes of anti-leukemia drugs.
Supplementary Material
Acknowledgments
We thank Dr. John Bushweller (University of Virginia) for providing the research environment for carrying out part of these studies and Dr. Michael L. Cleary (Stanford University) for the clone encoding human menin.
This work was supported by Leukemia and Lymphoma Society TRP Grant 6070-09 (to J. G.), a University of Virginia Cancer Center grant (to J. G. and T. C.), a Children's Leukemia Research Foundation grant (to J. G.), and startup funds provided by the Department of Pathology, University of Michigan (to J. G. and T. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- MLL
- mixed lineage leukemia
- FP
- fluorescence polarization
- HSQC
- heteronuclear single quantum coherence
- ITC
- isothermal titration calorimetry
- MBM1
- menin binding motif 1
- MBM2
- menin binding motif 2
- TOCSY
- total correlation spectroscopy
- Tr-NOE
- exchange-transferred nuclear Overhauser effect.
REFERENCES
- 1.Sorensen P. H., Chen C. S., Smith F. O., Arthur D. C., Domer P. H., Bernstein I. D., Korsmeyer S. J., Hammond G. D., Kersey J. H. (1994) J. Clin. Invest. 93, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox M. C., Panetta P., Lo-Coco F., Del Poeta G., Venditti A., Maurillo L., Del Principe M. I., Mauriello A., Anemona L., Bruno A., Mazzone C., Palombo P., Amadori S. (2004) Am. J. Clin. Pathol. 122, 298–306 [DOI] [PubMed] [Google Scholar]
- 3.Eguchi M., Eguchi-Ishimae M., Greaves M. (2003) Int. J. Hematol. 78, 390–401 [DOI] [PubMed] [Google Scholar]
- 4.Greaves M. F. (1996) Leukemia 10, 372–377 [PubMed] [Google Scholar]
- 5.Slany R. K. (2005) Hematol. Oncol. 23, 1–9 [DOI] [PubMed] [Google Scholar]
- 6.Tkachuk D. C., Kohler S., Cleary M. L. (1992) Cell 71, 691–700 [DOI] [PubMed] [Google Scholar]
- 7.Sedkov Y., Tillib S., Mizrokhi L., Mazo A. (1994) Development 120, 1907–1917 [DOI] [PubMed] [Google Scholar]
- 8.Yu B. D., Hess J. L., Horning S. E., Brown G. A., Korsmeyer S. J. (1995) Nature 378, 505–508 [DOI] [PubMed] [Google Scholar]
- 9.Guenther M. G., Jenner R. G., Chevalier B., Nakamura T., Croce C. M., Canaani E., Young R. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8603–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hess J. L. (2004) Crit. Rev. Eukaryot. Gene Expr. 14, 235–254 [DOI] [PubMed] [Google Scholar]
- 11.Zeisig B. B., Milne T., García-Cuéllar M. P., Schreiner S., Martin M. E., Fuchs U., Borkhardt A., Chanda S. K., Walker J., Soden R., Hess J. L., Slany R. K. (2004) Mol. Cell. Biol. 24, 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes C. M., Rozenblatt-Rosen O., Milne T. A., Copeland T. D., Levine S. S., Lee J. C., Hayes D. N., Shanmugam K. S., Bhattacharjee A., Biondi C. A., Kay G. F., Hayward N. K., Hess J. L., Meyerson M. (2004) Mol. Cell 13, 587–597 [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama A., Somervaille T. C., Smith K. S., Rozenblatt-Rosen O., Meyerson M., Cleary M. L. (2005) Cell 123, 207–218 [DOI] [PubMed] [Google Scholar]
- 14.Caslini C., Yang Z., El-Osta M., Milne T. A., Slany R. K., Hess J. L. (2007) Cancer Res. 67, 7275–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y. X., Yan J., Keeshan K., Tubbs A. T., Wang H., Silva A., Brown E. J., Hess J. L., Pear W. S., Hua X. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1018–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D. J., Kitabayashi I., Herr W., Cleary M. L. (2004) Mol. Cell. Biol. 24, 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrasekharappa S. C., Guru S. C., Manickam P., Olufemi S. E., Collins F. S., Emmert-Buck M. R., Debelenko L. V., Zhuang Z., Lubensky I. A., Liotta L. A., Crabtree J. S., Wang Y., Roe B. A., Weisemann J., Boguski M. S., Agarwal S. K., Kester M. B., Kim Y. S., Heppner C., Dong Q., Spiegel A. M., Burns A. L., Marx S. J. (1997) Science 276, 404–407 [DOI] [PubMed] [Google Scholar]
- 18.Marx S. J. (2005) Nat. Rev. Cancer 5, 367–375 [DOI] [PubMed] [Google Scholar]
- 19.Thiel A. T., Blessington P., Zou T., Feather D., Wu X., Yan J., Zhang H., Liu Z., Ernst P., Koretzky G. A., Hua X. (2010) Cancer Cell 17, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bermel W., Bertini I., Duma L., Felli I. C., Emsley L., Pierattelli R., Vasos P. R. (2005) Angew Chem. Int. Ed. Engl. 44, 3089–3092 [DOI] [PubMed] [Google Scholar]
- 21.Bermel W., Bertini I., Csizmok V., Felli I. C., Pierattelli R., Tompa P. (2009) J. Magn. Reson. 198, 275–281 [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 23.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 24.Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph 14, 29–32, 51–55 [DOI] [PubMed] [Google Scholar]
- 25.Post C. B. (2003) Curr. Opin. Struct. Biol. 13, 581–588 [DOI] [PubMed] [Google Scholar]
- 26.Mammen M., Choi S. K., Whitesides G. M. (1998) Angew Chem. Int. Ed. Engl. 2755–2794 [DOI] [PubMed] [Google Scholar]
- 27.Bermel W., Bertini I., Felli I. C., Lee Y. M., Luchinat C., Pierattelli R. (2006) J. Am. Chem. Soc. 128, 3918–3919 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.