Abstract
The proteins from the thioredoxin family are crucial actors in redox signaling and the cellular response to oxidative stress. The major intracellular source for oxygen radicals are the components of the respiratory chain in mitochondria. Here, we show that the mitochondrial 2-Cys peroxiredoxin (Prx3) is not only substrate for thioredoxin 2 (Trx2), but can also be reduced by glutaredoxin 2 (Grx2) via the dithiol reaction mechanism. Grx2 reduces Prx3 exhibiting catalytic constants (Km, 23.8 μmol·liter−1; Vmax, 1.2 μmol·(mg·min)−1) similar to Trx2 (Km, 11.2 μmol·liter−1; Vmax, 1.1 μmol·(mg·min)−1). The reduction of the catalytic disulfide of the atypical 2-Cys Prx5 is limited to the Trx system. Silencing the expression of either Trx2 or Grx2 in HeLa cells using specific siRNAs did not change the monomer:dimer ratio of Prx3 detected by a specific 2-Cys Prx redox blot. Only combined silencing of the expression of both proteins led to an accumulation of oxidized protein. We further demonstrate that the distribution of Prx3 in different mouse tissues is either linked to the distribution of Trx2 or Grx2. These results introduce Grx2 as a novel electron donor for Prx3, providing further insights into pivotal cellular redox signaling mechanisms.
Keywords: Glutathione, Mitochondria, Oxidative Stress, Peroxidase, Thiol, Glutaredoxin, Peroxiredoxin, Redox Signaling, Thioredoxin
Introduction
Most cellular processes are regulated by redox signaling, including cell proliferation, differentiation, apoptosis, and gene expression. Essentially every protein containing surface-exposed cysteinyl residues constitutes a possible target for redox-based modifications. Reversible modifications include thiols being oxidized to disulfides or sulfenic acid and glutathionylation, displaying a fast and specific way of regulating the activity and function of proteins. Sustained oxidative conditions may lead to irreversible modifications, such as oxidation of thiol groups to sulfinic or sulfonic acid. These modifications may render proteins inactive and thereby inhibit signaling and metabolic pathways. Like other signal transduction pathways, redox signaling needs to be tightly controlled.
The thioredoxin family of proteins is crucial for regulating the cellular redox homeostasis (1–3). This protein family includes thioredoxins (Trxs),5 glutaredoxins (Grxs), and peroxiredoxins (Prxs), whose isoforms are ubiquitously expressed in mammals (4–6). Trx family proteins share the thioredoxin fold and an active site motif with one or two cysteine residues, which are essential for their catalytic activity. The Trx system consists of Trx, the selenoprotein thioredoxin reductase (TrxR), and the electron donor NADPH. Reduced Trx binds to a protein disulfide and reduces the target protein. The N-terminal active site cysteine, characterized by a low pKa, starts the nucleophilic attack on the target disulfide. A covalently bound mixed disulfide intermediate with the attacked sulfur group is formed, which can be reduced by the C-terminal active site cysteine. Via this so-called dithiol mechanism a disulfide is formed in the active site of the redoxin, and the target disulfide is reduced to two thiol groups (7). The disulfide in the Trx active site is reduced by TrxR at the expense of NADPH. Compared with Trxs, Grxs cannot only reduce protein disulfides, but can also catalyze the formation or reduction of GSH-mixed disulfides via the monothiol mechanism (8, 9). This mechanism only requires the N-terminal active site cysteine forming a GSH-mixed disulfide with a glutathionylated protein. The GSH-mixed disulfide of Grx is subsequently reduced by a second GSH molecule generating GSSG and the reduced protein. GSSG is kept reduced by glutathione reductase (GR). Mammalian cells contain two Trx and Grx systems, that are localized in the cytosol (GR, Grx1, Trx1, TrxR1) and in mitochondria (GR, Grx2, Trx2, TrxR2).
Prxs are unique among the peroxidases because they utilize cysteine(s) at their active site receiving electrons from the Trx system (10). Depending on their catalytic mechanism they can be divided into three groups: 2-Cys Prxs, atypical 2-Cys Prxs and 1-Cys Prxs (11, 12). The typical 2-Cys Prxs (in mammals Prx1-Prx4) constitute the largest class of Prxs. They exist as homodimers, often within oligomeric torin structures, possessing conserved N-terminal and C-terminal cysteines. The N-terminal cysteine is oxidized by peroxides to sulfenic acid, which then forms an intermolecular disulfide with the C-terminal cysteine of the other subunit (12). Both atypical 2-Cys Prxs (Prx5) and 1-Cys Prxs (Prx6) only contain the conserved N-terminal cysteine. Atypical 2-Cys Prxs require an additional, nonconserved cysteine for their catalytic activity generating an intramolecular disulfide during the reaction cycle. 1-Cys Prxs require additional thiol-containing molecules, for instance, GSH. Nowadays it is becoming apparent that Prxs function as signaling molecules in cells reducing not only peroxides but also peroxynitrite, giving them an outstanding role in cell physiology and pathophysiology (13, 14).
Hitherto, the mammalian 2-Cys Prxs have been reported to rely exclusively on Trxs for their reduction (see for instance Refs. 11, 15). These initial studies have used the cytosolic Grx1 to test the potential role of Grxs as electron donors, even when the mitochondrial 2-Cys Prxs were assayed. However, the mammalian mitochondrial Grx2 is unique because it can receive electrons from both GSH and TrxR, thus combining the activity profile of both Grxs and Trxs (16). These facts prompted us to hypothesize that Grx2 might be able to catalyze the reduction of the mammalian mitochondrial 2-Cys Prxs in vitro and that it might contribute to their redox state in vivo. The results presented here provide strong evidence that both mitochondrial redox systems, Grx2 and Trx2, contribute to the redox state of the typical 2-Cys Prx3 in vivo, but not to the reduction of the atypical 2-Cys Prx5, which appears to rely on the Trx system for the reduction of the disulfide formed during its reaction cycle.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Chemicals and enzymes (GR) were purchased from Sigma-Aldrich unless otherwise stated and were of analytical grade or better. SDS-PAGE was run using the Novex Mini-Cell and precast NuPAGE gels (12% acrylamide, BisTris; Invitrogen) or 4–20% Precise Gels (Pierce) according to the manufacturers' instructions.
Protein Purification and Expression
The cloning, recombinant expression, and purification of Grx2, Prx3, Prx5, Trx1, and Trx2 were described in Ref. 4. The Grx2 mutant C35S was introduced in Ref. 16. TrxRs of human and rat origin were a kind gift of Elias Arnér, Karolinska Institutet, Stockholm, Sweden.
Enzyme Kinetics
The activity of purified Prx3 and Prx5 was measured in optical assays in 96-well plates (200 μl/well) containing 100 mm Tris-HCl, pH 7.0, 1 mm EDTA, 120 μm H2O2, ≥1 μg TrxR, 200 μm NADPH, and Grx2 or Trx2 (1–80 μm). In separate experiments TrxR was replaced by 1 mm GSH and yeast GR as described previously for other GSH-dependent Grx reactions (17). The reaction was started by the addition of the Prx (0.05–1 μm) and followed as a decrease in absorbance at 340 nm. The linear part of the reaction was used to determine the rates of NADPH consumption. Kinetic constants were determined by nonlinear curve fitting of the primary [S] versus v plots against the Michaelis-Menten equation using Grace.
Cell Culture and Transfection
HeLa cells were cultivated in low glucose DMEM (PAA, Cölbe, Germany) supplemented with 10% heat-inactivated fetal calf serum and 100 units/ml penicillin and streptomycin at 37 °C in a 90% humidified atmosphere containing 5% CO2. Cells were transiently transfected with siRNA against hGrx2 (sense, GGU GCA ACU GAC ACU CAU; antisense, UAU GAG UGU CAG UUG CAC) and hTrx2 (sense, GGA UCU CCU UGA CAA CCU; antisense, AAG GUU GUC AAG GAG AUC), as well as unspecific “scrambled” siRNA as control (sense, CAU UCA CUC AGG UCA UCA; antisense, CUG AUG ACC UGA GUG AAU). Briefly, 3.5 million HeLa cells were resuspended in electroporation buffer (21 mm HEPES, 137 mm NaCl, 5 mm KCl, 0.7 mm Na2HPO4, 6 mm d-glucose, pH 7.15), mixed with 15 μg of siRNA and were electroporated in a total volume of 600 μl at 250 mV and 1500 microfarads. FCS was directly added to the cells before seeding them out in fresh medium. Sufficient knockdown of Trx2 was observed after 3 days. To knock down Grx2, cells were transfected a second time after 3 days.
Antibodies
The generation of the antibodies, Western blotting procedures, and immunohistochemistry methods have been described in Refs. 4 and 18.
Grx2 ELISA
A specific sandwich ELISA was used to quantify cellular levels of Grx2 as described in reference (18). 96-well plates were coated with 0.5 μg/ml affinity-purified antibodies against Grx2 overnight at 4 °C. After blocking for 2 h with 10 mg/ml bovine serum albumin diluted cell extracts were added and incubated overnight at 4 °C, as well as standards in the range of 0–32 ng/ml. Grx2 was detected by incubation for 2 h with 0.5 μg/ml biotinylated secondary antibody and 1 h with alkaline phosphatase-conjugated streptavidin, before adding the substrate p-nitrophenyl phosphate (Sigma) and measuring the absorbance at 405 nm in a 96-well plate reader (Tecan, Männedorf, Switzerland).
Prx3 Redox Blot
Cells were incubated in N-ethylmaleimide-containing buffer (40 mm HEPES, pH 7.4, 50 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1× protease inhibitors, and 100 mm N-ethylmaleimide) for 15 min at room temperature and lysed by the addition of 2% CHAPS (19) and were flash frozen in liquid nitrogen. Cell extracts were centrifuged at 13,000 rpm for 15 min; protein levels of the supernatant were determined using Bradford reagent (Bio-Rad).
Samples (5–20 μg of total protein) were diluted and combined with sample loading buffer (0.3 m Tris-HCl, pH 7, 50% glycerol, 5% SDS, 0.1% bromphenol blue). Proteins were separated using nonreducing SDS-PAGE and transferred to PVDF membranes, which were blocked with 5% milk powder and 1% bovine serum albumin in Tris-buffered saline containing 0.05% Tween 20. Prx3 was detected using specific primary antibodies, a horseradish peroxidase-coupled secondary antibody and the enhanced chemoluminescence method.
RESULTS
Reduction of Prx3 and Prx5 by Trxs and Grx2 in Vitro
The catalytic activity of recombinantly expressed and purified proteins, i.e. Grx2, Prx3, Prx5, Trx1, Trx2, and TrxRs, was measured in optical assays using hydrogen peroxide as the substrate for the Prxs. The reaction and data analysis were optimized so that the substrate concentration exceeded the enzyme concentration at least 20-fold and that in neither case did NADPH, TrxR or H2O2 become the limiting factor for the reaction.
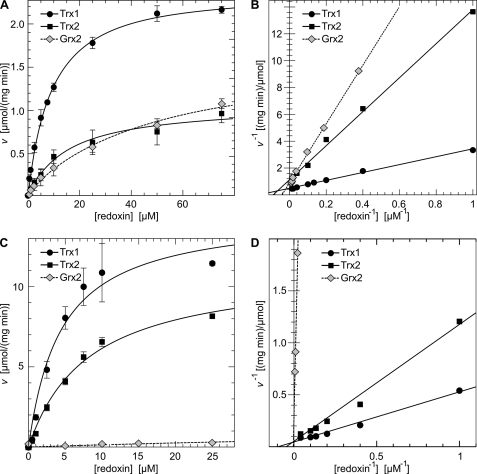
As described before, the mitochondrial thioredoxin system (NADPH, TrxR2, and Trx2) efficiently regenerates oxidized Prx3 (Fig. 1, A and B) and Prx5 (Fig. 1, C and D). Surprisingly, cytosolic Trx1 was 6.3-fold more efficient in the reduction of Prx3 and 2.4-fold more efficient as an electron donor for Prx5 (Table 1). Grx2, in the absence of GSH and the presence of TrxR, reduced Prx3 with kinetic parameters (apparent Km, 23.8 μmol·liter−1, apparent Vmax, 1.2 μmol·(mg·min)−1) similar to those of Trx2 (apparent Km, 11.2 μmol·liter−1, apparent Vmax, 1.1 μmol·(mg·min)−1), resulting in a catalytic efficiency of Grx2 of 55% compared with Trx2 (Table 1). The Grx2 C35S mutant, which only supports the monothiol reaction mechanism (16), did not support the Prx3-catalyzed reduction of H2O2 (data not shown). In contrast to Prx3, Grx2 was not able to support the reduction of catalytically oxidized Prx5 efficiently (Fig. 1, C and D, and Table 1). These data prove that, in vitro, both mitochondrial redoxins can serve as reductants for the typical 2-Cys Prx 3 in a dithiol reaction mechanism, but that the reduction of the catalytic disulfide in the atypical 2-Cys Prx 5 is restricted to the Trx system.
FIGURE 1.
Reduction of Prx3 and Prx5 by Grx2, Trx1, and Trx2 in vitro. The catalytic activity of Prx3 and Prx5 was measured in an optical assay including NADPH, TrxR, and hydrogen peroxide by following the decrease in absorbance at 340 nm. A and C, Michaelis-Menten plot for the activity of Prx3 and Prx5, respectively, using Trx1, Trx2, and Grx2 as electron donors. B and D, Lineweaver-Burk plot of A and C.
TABLE 1.
Kinetic parameters of the Prx3- and Prx5-catalyzed reduction of hydrogen peroxide using Trx1, Trx2, or Grx2 as electron donor
The efficiencies (Kcat·Km−1) were normalized to the parameters of mitochondrial Trx2.
| Km | Vmax | Efficiency | |
|---|---|---|---|
| μmol·liter−1 | μmol·(mg·min)−1 | Kcat·Km−1 (%) | |
| Prx3 | |||
| Trx1 | 4.0 | 2.4 | 630 |
| Trx2 | 11.2 | 1.1 | 100 |
| Grx2 | 23.8 | 1.2 | 55 |
| Prx5 | |||
| Trx1 | 4.5 | 14.6 | 240 |
| Trx2 | 8.1 | 11.2 | 100 |
| Grx2 | 121 | 1.8 | 1 |
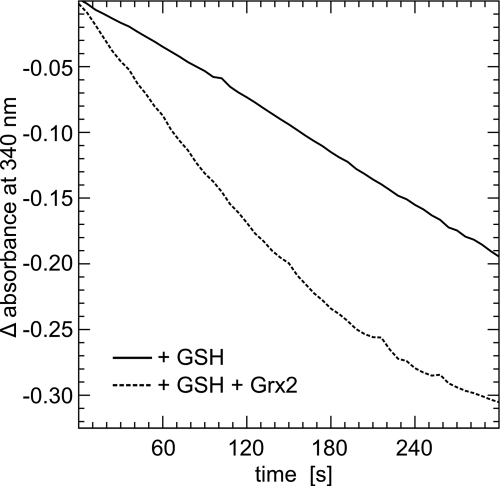
Grx2 can use electrons from both TrxR and GSH (16). We have thus analyzed the reduction of H2O2 by Prx3 with NADPH, GR, GSH, and Grx2 as the reducing system. As depicted in Fig. 2, Grx2 reduced by GSH does support the reduction of Prx3. Because of the high background of GR/GSH in the reduction of H2O2, however, it was not possible to determine kinetic constants of this reaction.
FIGURE 2.
Reduction of Prx3 by Grx2 using electrons from GSH. The catalytic activity of Prx3 in the presence of NADPH, GR, GSH, and/or Grx2 was followed as decrease in absorbance at 340 nm.
Contribution of Grx2 and Trx2 to the Redox State of Prx3 in Vivo
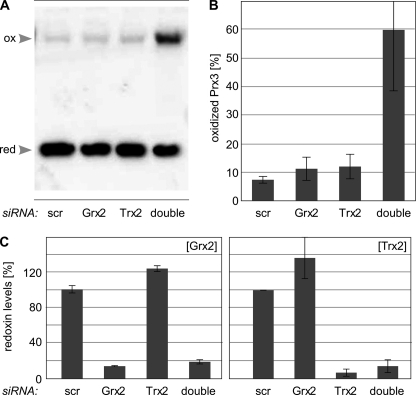
We have employed siRNAs to silence the expression of Trx2 and Grx2 to evaluate their individual contribution to the redox state of Prx3 in vivo. Both individual and combined silencing of Grx2 and Trx2 expression reduced the levels of the redoxins in HeLa cells to 5–20% compared with control siRNAs (Fig. 3C). Neither the individual silencing of Grx2, nor Trx2, significantly affected the levels of the other redoxin (Fig. 3C). The redox state of Prx3 was assayed by Western blotting following nonreducing SDS-PAGE of cell extracts with blocked thiol redox chemistry. Because of the intermolecular disulfide formed in the reaction cycle of 2-Cys Prxs, catalytically oxidized Prx3 can be identified as dimeric band in this assay. Reduced Prx3 corresponds to the monomeric band. As seen in Fig. 3, A and B, silencing of Grx2 or Trx2 alone did not significantly affect the redox state of Prx3 in HeLa cells. Only when the expression of both redoxins was silenced did catalytically oxidized Prx3 start to accumulate (Fig. 3, A and B). To examine whether this accumulation could be the result of a general increase in oxidative stress in these cells, we have analyzed protein carbonylation and glutathionylation but could not record any differences between the differently treated cell cultures (data not shown).
FIGURE 3.
Reduction of Prx3 by Trx2 and Grx2 in vivo. A, monomer:dimer ratio of Prx3 detected by a specific 2-Cys peroxiredoxin redox blot in HeLa cells with siRNA-decreased expression levels of Grx2, Trx2, or both. Cells treated with unspecific siRNA (scr) were used as control. B, levels of oxidized Prx3 in transfected HeLa cells, constituted in percent. C, protein levels of Grx2 and Trx2 in HeLa cells treated with specific siRNA constructs, analyzed by a specific sandwich ELISA (Grx2) or Western blotting (Trx2), respectively. The average of ≥three independent experiments is shown.
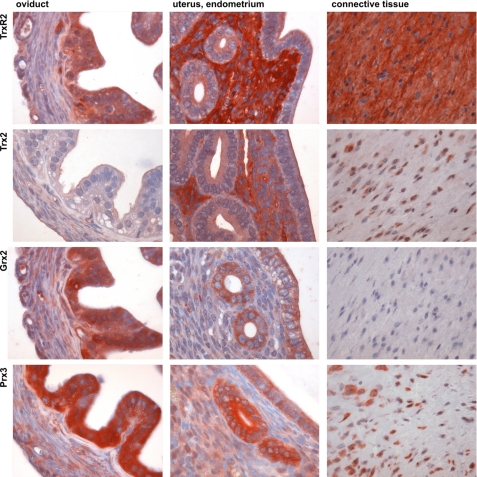
Additional evidence for a contribution of both mitochondrial redox systems to the reduction of Prx3 comes from our recently presented systematic identification of Grx, Prx, and Trx family proteins in various tissues of the mouse (4). Fig. 4 depicts the distribution of Grx2, Prx3, Trx2, and TrxR2 in three exemplary tissues, oviduct, uterus/endometrium, and connective tissue. These three tissues clearly show a different correlation between the different redoxins. In oviduct, TrxR2, Grx2, and Prx3 show a strong correlation, whereas Trx2 is basically absent from the epithelial cells. In the endometrium of the uterus TrxR2 and Trx2 on one side and Grx2 and Prx3 on the other side strongly correlate, but the distribution pattern for both pairs is almost inverse. Cells of the connective tissue show a good correlation between Prx3 and Trx2, but Grx2 antigenicity could not be recognized. Although these data have to be taken with care (for a discussion, see Ref. 4), they further support the idea that both mitochondrial redox systems, Grx2 and Trx2, contribute to the redox state of Prx3 in vivo and that this contribution may vary between different tissues and cell types.
FIGURE 4.
Distribution of Prx3, Trx2, TrxR2, and Grx2 immunoreactivity in mouse tissues, oviduct, uterus, and connective tissue. The oviduct (magnification, ×500) showed strong staining for TrxR2, Grx2, and Prx3, whereas Trx2 was basically absent. The endometrium of the uterus (×500) displayed a correlative staining for TrxR2 and Trx2 as well as for Grx2 and Prx3, with an apparent inverse staining pattern. Cells of the connective tissue (×250) were strongly stained for Prx3 and Trx2, whereas no staining could be detected for Grx2.
DISCUSSION
In this study we have analyzed the potential contribution of Grx2 to the reduction of the catalytic disulfide of Prxs in mitochondria. Our results suggest that both mitochondrial thiol-disulfide reductase systems, Grx2 and Trx2, contribute to the reduction of the catalytic disulfide in the typical 2-Cys Prx3 in vivo. The reaction cycle of the atypical 2-Cys Prx5, on the other hand, relies on the Trx system as electron donor. With some exceptions, for instance in plants (20), 2-Cys Prxs are exclusively reduced by Trxs, and this has also been reported for mammalian Prx3 and Prx5 before (11, 15). The mammalian mitochondrial Grx2 is a unique member of the Trx family of proteins because it combines the activity profile of both Grxs and Trxs. This protein efficiently catalyzes reactions employing both the monothiol and the dithiol reaction mechanisms, and, unlike other Grxs such as mammalian Grx1, it can receive electrons from both GSH and TrxR (16). It is thus not surprising that Grx2 is able to reduce the catalytic disulfide in Prx3, although earlier studies excluded Grxs as electron donors. These studies investigated Grx1 as electron donor (15), Grx2 and its unique properties were only identified later (16, 21, 22). Fig. 5 summarizes the experimentally confirmed electron pathways in the mitochondrial matrix. The data from the “Redox Atlas of the Mouse” (4) (Fig. 4) indicate that in many tissues, without being mutually exclusive, the expression of Prx3 is linked to the expression of either GSH/Grx2, TrxR2/Grx2 or TrxR2/Trx2. These data suggest that the individual contribution of the different redox systems in the regulation of ROS signals varies between cell types.
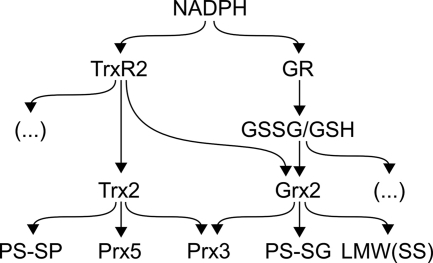
FIGURE 5.
Overview of the electron flow of mitochondrial Trx family proteins. NADPH as the ultimate electron donor reduces thioredoxin reductase 2 (TrxR2), which among other substrates ( … ), reduces Trx2. Trx2 reduces protein disulfides, the 2-Cys Prx3, and the atypical 2-Cys Prx5. Grx2 cannot only use electrons from NADPH/GR/GSH but also from NADPH/TrxR2 for the reduction of low molecular weight disulfides (LMW(SS)), GSH-mixed disulfides (PS-SG), and the 2-Cys Prx3.
Over the past few years reactive oxygen species, including hydrogen peroxide and peroxynitrite, have been recognized as important signaling molecules, see for instance (14, 23). Hydrogen peroxide can oxidize critical cysteine residues in numerous proteins thereby regulating their activity. Prxs with their very high affinity for peroxide substrates seem to possess a central role in these redox circuits by regulating the levels of the signaling molecules in both temporal and spatial manner (24–26). For instance, a recent study demonstrated that the inactivation of Prx1 associated with membranes by transient and local phosphorylation allows for the limited accumulation of H2O2 in defined areas, while preventing the toxic accumulation of H2O2 elsewhere in the cell (27). Such defined mechanisms of regulation have not been defined for the mitochondrial Prxs; however, their importance for the regulation of mitochondrial peroxide concentrations is undisputed. Prx3 knock-out animals displayed increased levels of reactive oxygen species and were more sensitive to oxidative stress, for instance to lipopolysaccharide-induced lung injury (28). Prx3 plays a crucial role in the protection of mitochondria in the cardiovascular system and hippocampal neurons (29–31). Unlike Prx3, Prx5 is not localized exclusively in mitochondria; it has also been detected in the cytosol, peroxisomes, and the nucleus. Its role in the mitochondrial redox networks is not clear; overexpression of Prx5 in mitochondria decreased mtDNA damages caused by exogenously added hydrogen peroxide (32).
The active site cysteines of Prx cannot only form the catalytic sulfenic acid and disulfide, they can also be “over”-oxidized to sulfinic and sulfonic acids, for instance overoxidized Prx3 accumulates in aged rat liver mitochondria (33). The formation of protein sulfinic acids is usually irreversible; however, in the case of Prxs this oxidation can be reduced by specialized enzymes, the sulfiredoxins (34, 35). Prxs have also been reported to undergo reversible S-glutathionylation. Glutathionylation of Prx5, for instance, has been demonstrated in both resting and menadione-treated cells (36). Grxs are highly efficient catalysts of protein (de-)glutathionylation; however, in the case of human Prxs it seems that this reaction is also the privilege of the sulfiredoxins (37).
Plant type II Prxs belong to the atypical 2-Cys Prx subfamily. In contrast to human Prx5, some of these, for instance chloroplastic PrxIIB and PrxIIE, prefer the GSH/Grx system over the Trx system as electron donor (38, 39), whereas others, such as PrxIIF, are able to utilize both the Trx and GSH/Grx systems (40). No obvious reasons for these differences in electron donor preference could be detected by comparing the sequences and, when available, structures of these proteins (data not shown). The reduction of plant type II Prxs required the deglutathionylation of the catalytic cysteine by Grx (Gama et al.; a reaction we could not detect for human Prx5 and Grx2). Thus, the ability of the atypical 2-Cys Prxs to interact directly with GSH may be the discriminating factor.
This study was specifically designed to analyze the reduction of the protein disulfide formed during the Prx reaction cycle, which strictly requires the dithiol reaction mechanism. The similar catalytic activity of Prx3 with either Grx2 or Trx2 as electron donor and the ability of Prx3 in vivo to still form the disulfide-bound dimer excludes the formation of higher amounts of over-oxidized Prx3 during the reaction. Moreover, Grx2 donated electrons to Prx3 even in the absence of GSH. We can thus exclude that our results have been obscured by, for instance the reduction of sulfinic acids or glutathionylation/deglutathionylation reactions. In addition to the ubiquitous mitochondrial Grx2a, humans and mice possess additional cytosolic/nuclear Grx2 isoforms (41, 42). Worth mentioning, while screening for potential dithiol mechanism substrates of these isoforms of Grx2, we have also identified 2-Cys Prxs as potential Grx2 interaction partners.6
In conclusion, mitochondrial 2-Cys Prx3 is not only substrate for Trx2, but can also be reduced by Grx2 with similar catalytic efficiency via the dithiol reaction mechanism. The reduction of the catalytic disulfide of the atypical 2-Cys Prx5 is limited to the Trx system. In HeLa cells, only the combined silencing of Grx2 and Trx2 expression led to a significant accumulation of catalytically oxidized protein. The expression of Prx3 in different mouse tissues is, in many cases, linked to the expression of either Grx2 or Trx2. This study introduces Grx2 as a novel electron donor for Prx3, yielding further insights into essential redox signaling mechanisms in the compartment with the highest prevalence for reactive oxygen species, mitochondria.
Acknowledgments
We thank Elias Arnér, Karolinska Institutet, Stockholm, Sweden, for the kind gift of TrxRs, and Sabrina Oesteritz for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB593-N01 and by a grant from the von Behring-Röntgen Foundation.
L. D. Schütte and C. H. Lillig, unpublished results.
- Trx
- thioredoxin
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- GR
- glutathione reductase
- Grx
- glutaredoxin
- Prx
- peroxiredoxin
- TrxR
- thioredoxin reductase.
REFERENCES
- 1.Lillig C. H., Holmgren A. (2007) Antiox. Redox Signal. 9, 25–47 [DOI] [PubMed] [Google Scholar]
- 2.Lillig C. H., Berndt C., Holmgren A. (2008) Biochim. Biophys. Acta 1780, 1304–1317 [DOI] [PubMed] [Google Scholar]
- 3.Rhee S. G., Chae H. Z., Kim K. (2005) Free Rad. Biol. Med. 38, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 4.Godoy J. R., Funke M., Ackermann W., Haunhorst P., Oesteritz S., Capani F., Elsässer H., Lillig C. H. (2010) Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]
- 5.Aon-Bertolino M. L., Romero J. I., Galeano P., Holubiec M., Badorrey M. S., Saraceno G. E., Hanschmann E. M., Lillig C. H., Capani F. (2010) Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]
- 6.Dammeyer P., Arnér E. S. J. (2010) Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]
- 7.Kallis G. B., Holmgren A. (1980) J. Biol. Chem. 255, 10261–10265 [PubMed] [Google Scholar]
- 8.Holmgren A. (1978) J. Biol. Chem. 253, 7424–7430 [PubMed] [Google Scholar]
- 9.Gravina S. A., Mieyal J. J. (1993) Biochemistry 32, 3368–3376 [DOI] [PubMed] [Google Scholar]
- 10.Chae H. Z., Robison K., Poole L. B., Church G., Storz G., Rhee S. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7017–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo M. S., Kang S. W., Kim K., Baines I. C., Lee T. H., Rhee S. G. (2000) J. Biol. Chem. 275, 20346–20354 [DOI] [PubMed] [Google Scholar]
- 12.Rhee S. G., Kang S. W., Chang T. S., Jeong W., Kim K. (2001) IUBMB Life 52, 35–41 [DOI] [PubMed] [Google Scholar]
- 13.Essler S., Dehne N., Brüne B. (2009) FEBS Lett. 583, 3531–3535 [DOI] [PubMed] [Google Scholar]
- 14.Forman H. J., Maiorino M., Ursini F. (2010) Biochemistry 49, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chae H. Z., Kim H. J., Kang S. W., Rhee S. G. (1999) Diabetes Res. Clin. Pract. 45, 101–112 [DOI] [PubMed] [Google Scholar]
- 16.Johansson C., Lillig C. H., Holmgren A. (2004) J. Biol. Chem. 279, 7537–7543 [DOI] [PubMed] [Google Scholar]
- 17.Holmgren A. (1979) J. Biol. Chem. 254, 3672–3678 [PubMed] [Google Scholar]
- 18.Lundberg M., Fernandes A. P., Kumar S., Holmgren A. (2004) Biochem. Biophys. Res. Commun. 319, 801–809 [DOI] [PubMed] [Google Scholar]
- 19.Cox A. G., Pullar J. M., Hughes G., Ledgerwood E. C., Hampton M. B. (2008) Free Rad. Biol. Med. 44, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 20.Rouhier N., Gelhaye E., Jacquot J. P. (2002) J. Biol. Chem. 277, 13609–13614 [DOI] [PubMed] [Google Scholar]
- 21.Lundberg M., Johansson C., Chandra J., Enoksson M., Jacobsson G., Ljung J., Johansson M., Holmgren A. (2001) J. Biol. Chem. 276, 26269–26275 [DOI] [PubMed] [Google Scholar]
- 22.Gladyshev V. N., Liu A., Novoselov S. V., Krysan K., Sun Q. A., Kryukov V. M., Kryukov G. V., Lou M. F. (2001) J. Biol. Chem. 276, 30374–30380 [DOI] [PubMed] [Google Scholar]
- 23.Groeger G., Quiney C., Cotter T. G. (2009) Antiox. Redox Signal. 11, 2655–2671 [DOI] [PubMed] [Google Scholar]
- 24.Neumann C. A., Cao J., Manevich Y. (2009) Cell Cycle 8, 4072–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flohé L. (2010) Methods Enzymol. 473, 1–39 [DOI] [PubMed] [Google Scholar]
- 26.Cox A. G., Winterbourn C. C., Hampton M. B. (2010) Biochem. J. 425, 313–325 [DOI] [PubMed] [Google Scholar]
- 27.Woo H. A., Yim S. H., Shin D. H., Kang D., Yu D. Y., Rhee S. G. (2010) Cell 140, 517–528 [DOI] [PubMed] [Google Scholar]
- 28.Li L., Shoji W., Takano H., Nishimura N., Aoki Y., Takahashi R., Goto S., Kaifu T., Takai T., Obinata M. (2007) Biochem. Biophys. Res. Commun. 355, 715–721 [DOI] [PubMed] [Google Scholar]
- 29.Araki M., Nanri H., Ejima K., Murasato Y., Fujiwara T., Nakashima Y., Ikeda M. (1999) J. Biol. Chem. 274, 2271–2278 [DOI] [PubMed] [Google Scholar]
- 30.Matsushima S., Ide T., Yamato M., Matsusaka H., Hattori F., Ikeuchi M., Kubota T., Sunagawa K., Hasegawa Y., Kurihara T., Oikawa S., Kinugawa S., Tsutsui H. (2006) Circulation 113, 1779–1786 [DOI] [PubMed] [Google Scholar]
- 31.Hattori F., Murayama N., Noshita T., Oikawa S. (2003) J. Neurochem. 86, 860–868 [DOI] [PubMed] [Google Scholar]
- 32.Banmeyer I., Marchand C., Clippe A., Knoops B. (2005) FEBS Lett. 579, 2327–2333 [DOI] [PubMed] [Google Scholar]
- 33.Musicco C., Capelli V., Pesce V., Timperio A. M., Calvani M., Mosconi L., Zolla L., Cantatore P., Gadaleta M. N. (2009) Biochim. Biophys. Acta 1787, 890–896 [DOI] [PubMed] [Google Scholar]
- 34.Rhee S. G., Jeong W., Chang T. S., Woo H. A. (2007) Kidney Int. Suppl. 106, S3–S8 [DOI] [PubMed] [Google Scholar]
- 35.Jönsson T. J., Lowther W. T. (2007) Subcell. Biochem. 44, 115–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fratelli M., Demol H., Puype M., Casagrande S., Villa P., Eberini I., Vandekerckhove J., Gianazza E., Ghezzi P. (2003) Proteomics 3, 1154–1161 [DOI] [PubMed] [Google Scholar]
- 37.Park J. W., Mieyal J. J., Rhee S. G., Chock P. B. (2009) J. Biol. Chem. 284, 23364–23374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bréhélin C., Meyer E. H., de Souris J. P., Bonnard G., Meyer Y. (2003) Plant Physiol. 132, 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gama F., Bréhélin C., Gelhaye E., Meyer Y., Jacquot J. P., Rey P., Rouhier N. (2008) Physiol. Plant. 133, 599–610 [DOI] [PubMed] [Google Scholar]
- 40.Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L. J., Dietz K. J. (2005) J. Biol. Chem. 280, 12168–12180 [DOI] [PubMed] [Google Scholar]
- 41.Hudemann C., Lönn M. E., Godoy J. R., Zahedi Avval F., Capani F., Holmgren A., Lillig C. H. (2009) Antiox. Redox Signal. 11, 1–14 [DOI] [PubMed] [Google Scholar]
- 42.Lönn M. E., Hudemann C., Berndt C., Cherkasov V., Capani F., Holmgren A., Lillig C. H. (2008) Antiox. Redox Signal. 10, 547–558 [DOI] [PubMed] [Google Scholar]