Abstract
Pseudallescheria boydii (Scedosporium apiospermum) is a saprophytic fungus widespread in the environment, and has recently emerged as an agent of localized as well as disseminated infections, particularly mycetoma, in immunocompromised and immunocompetent hosts. We have previously shown that highly purified α-glucan from P. boydii activates macrophages through Toll-like receptor TLR2, however, the mechanism of P. boydii recognition by macrophage is largely unknown. In this work, we investigated the role of innate immune receptors in the recognition of P. boydii. Macrophages responded to P. boydii conidia and hyphae with secretion of proinflammatory cytokines. The activation of macrophages by P. boydii conidia required functional MyD88, TLR4, and CD14, whereas stimulation by hyphae was independent of TLR4 and TLR2 signaling. Removal of peptidorhamnomannans from P. boydii conidia abolished induction of cytokines by macrophages. A fraction highly enriched in rhamnomannans was obtained and characterized by NMR, high performance TLC, and GC-MS. Preparation of rhamnomannans derived from P. boydii triggered cytokine release by macrophages, as well as MAPKs phosphorylation and IκBα degradation. Cytokine release induced by P. boydii-derived rhamnomannans was dependent on TLR4 recognition and required the presence of non-reducing end units of rhamnose of the rhamnomannan, but not O-linked oligosaccharides from the peptidorhamnomannan. These results imply that TLR4 recognizes P. boydii conidia and this recognition is at least in part due to rhamnomannans expressed on the surface of P. boydii.
Keywords: Carbohydrate, Cytokine, Fungi, Inflammation, Innate Immunity, Macrophage, MyD88, Pathogen-associated Molecular Pattern (PAMP), Toll-like Receptors (TLR), Tumor Necrosis Factor (TNF)
Introduction
Fungal infections are escalating recently, especially as a consequence of growing incidence in the population of immunocompromised individuals (1). Strong risk factors for the development of invasive fungal infections are therapy with corticoids, cytotoxic chemotherapy, transplant followed by immunosuppressive therapy, and TNF neutralization (2–4). These conditions strongly delineate the essential role of immunity, mainly innate immunity mediated by phagocytosis and recruitment of polymorphonuclear leukocytes, in the control of fungal infections (5, 6). Toll-like receptors (TLRs)3 are pattern recognition receptors homologues to the Toll receptor of Drosophila melanogaster (7). The Drosophila Toll receptor was initially characterized as a molecule involved in the immunity during the infection with the filamentous fungus Aspergillus fumigatus, leading to the induction of an antifungal peptide, drosomycin, and resistance against this pathogen (8). Mammalian TLRs recognize pathogen-associated molecular patterns, for example, lipopolysaccharides from Gram-negative bacteria, bacterial lipoproteins, flagellin, and viral and bacterial non-methylated CpG motifs are recognized, respectively, by TLR4, TLR2, TLR5, and TLR9 (7). TLRs also have been implicated in the recognition and triggering of immunity during fungal infections in mammals (9, 10). TLR2 and TLR4 mediate cytokine release and NFκB activation in response to different developmental stages of A. fumigatus (11–13). Leukocyte activation induced by Candida albicans also involves TLR2 and TLR4 triggering (14–16). The relevance of TLR2 and TLR4 for the recognition of these important fungal pathogens is demonstrated by the observations that Tlr2−/− and Tlr4−/− mice show a higher susceptibility to C. albicans and A. fumigatus infections (14–18).
Pseudallescheria boydii is a saprophytic fungus, extremely widespread in the environment, that presents different developmental stages (19, 20). Hyphal forms of P. boydii grow as branching septated structures producing structures of dispersion, the conidia. P. boydii infections present a large spectrum of manifestations varying from localized mycetomas, sinusitis, and pulmonary infections to disseminated infections, especially in immunodeficient patients. P. boydii is one of the most common pathogenic fungi that cause mycetoma, whose incidence extends from subtropical to temperate areas. This infection is a major cause of morbidity, particularly in rural areas, where treatment and diagnosis of infections are extremely difficult (19, 20).
In the absence of an adequate clearance by phagocytic cells, P. boydii conidia that have reached deep tissues can differentiate in hyphal forms and promote tissue dissemination. Although innate immunity clearly plays an essential role in resistance against P. boydii infection, the mechanisms of recognition of this pathogen by the innate immune cells are largely uncharacterized (21). We have recently observed that highly purified α-glucan from P. boydii activates macrophages and dendritic cells through TLR2, thus indicating a role for TLRs on P. boydii recognition (22). In this work we investigated the role of innate immune receptors on recognition of P. boydii developmental forms. We provide evidence that P. boydii conidia are recognized by TLR4, our results also suggest that rhamnomannans isolated from this fungus induce macrophage activation through TLR4 signaling.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 (wild-type) mice were obtained from the Universidade Federal do Rio de Janeiro Breeding Unit (Rio de Janeiro, Brazil). Tlr4−/−, Tlr2−/−, Cd14−/−, and Myd88−/− on a C57BL/6 background were provided by Drs. Shizuo Akira (Osaka University, Japan), Douglas Golenbock (University of Massachusetts), and Ricardo Gazzinelli (UFMG, Brazil). The animals were kept at constant temperature (25 °C) with free access to chow and water in a room with a 12-h light/dark cycle. The experiments were approved by the Institutional Animal Welfare Committee.
Reagents
LPS O111:B4 from Escherichia coli was obtained from Sigma. Bacterial lipoprotein, Pam3Cys-Ser-(Lys)4 (Pam3Cys), was obtained from EMC collections. Polymixin B was purchased from Bedford Laboratories. RPMI medium for macrophage culture was obtained from Sigma and was supplemented with fetal calf serum (FCS) and penicillin-streptomycin (Invitrogen).
P. boydii Growth and Isolation of Conidial and Hyphal Forms for in Vitro Stimulation Assays
P. boydii strain HLPB, isolated from eumycotic mycetoma, was kindly supplied by Bodo Wanke from Evandro Chagas Hospital, Instituto Oswaldo Cruz, Rio de Janeiro, Brazil. The P. boydii identity was confirmed by sequencing performed by Dr. Kathrin Tintelnot (Robert Koch-Institut, Berlin, Germany). The sequencing of the ITS regions revealed that this strain belongs to clade 4 (Scedosporium apiospermum sensu stricto) according to the taxonomy proposed by Gilgado et al. (23). Cells were grown on Sabouraud solid slants, inoculated in liquid culture medium, and incubated for 7 days at 25 °C with shaking. Cultures were then transferred to the same medium and incubated for 7 days at the same temperature with shaking; the mycelium was filtered, washed with distilled water, and stored at −20 °C. Conidial forms of P. boydii were grown on agar-Sabouraud for 7 days. The culture plates were washed with phosphate-buffered saline and filtered through sterile gauze to remove hyphae fragments and debris. Conidial suspensions were counted in a hemocytometer, washed three times with apyrogenic saline, and heat killed at 115 °C for 15 min. For the extraction of peptidopolysaccharides, P. boydii conidia were extracted with 0.05 m phosphate buffer, pH 7.2, at 100 °C for 2 h. Conidia were recovered by centrifugation at 1160 × g for 5 min, washed three times with apyrogenic saline, and counted in a hemocytometer. P. boydii mycelia was washed three times with saline apyrogenic, and then hyphal fragments were prepared by mechanical disruption and sonication of mycelia for 10 min, amounts of hyphae employed in the experiments were normalized by wet weight. For inactivating the hyphae, stock preparations of hyphae were heat killed at 115 °C for 15 min. Because hyphal preparations constitute extremely heterogeneous suspensions with filaments varying greatly in morphology and length, the experiments of macrophage stimulation were performed taking into account the wet weight of hyphal suspensions.
Purification of Rhamnomannans and Chemical Treatments
P. boydii mycelia (120 g) were submitted to an alkaline extraction (KOH 2% w/v, 2 h, 100 °C), then neutralized with glacial acetic acid and centrifuged, polysaccharides in supernatant were precipitated with 3 volumes of ethanol, suspended in distilled water, dialyzed, and lyophilized. Polysaccharides were then fractionated by gel filtration in a Superdex 200 column (30 cm x 10 cm), previously equilibrated, and fractions were eluted in a sodium phosphate buffer (0.01 m, pH 7.0) with 0.15 m NaCl, at a flow rate of 0.5 ml/min using a FPLC system with a ÄKTA device (GE Healthcare). Eluted fractions were monitored by A280 for protein and colorimetrically (A490) for carbohydrate (24). Fractions containing the polysaccharide were pooled, dialyzed against distilled water, and lyophilized. Neutral carbohydrates were determined by the phenol/sulfuric acid method (24), protein was determined by the Lowry method (25), phosphate by the procedure of Ames (26), and hexosamines by the method of Belcher et al. (27). Partial acid hydrolysis was performed by the treatment of rhamnomannans (2 mg) with trifluoroacetic acid (0.1 m TFA at 100 °C for 20 min), degraded rhamnomannans were then dialyzed against distilled water and then lyophilized. Peptidorhamnomannans (5 mg) was chemically de-O-glycosylated by mild reductive alkaline treatment under reducing conditions, and the liberated O-linked oligosaccharide alditol fraction was recovered on dialysis (28).
Monosaccharide Analysis
For qualitative analysis of carbohydrates, fractions were hydrolyzed with TFA (3 m, 100 °C, 3 h), and samples were analyzed by high performance TLC, in comparison to a standard mix of known sugars (25 μg/μl). High performance TLC plates were treated with 0.3 m KH2PO4, dried at room temperature, developed two times in 1-butanol/acetone/H2O (4:5:1, v/v/v) and stained with orcinol/sulfuric acid. To quantify the carbohydrates, the hydrolyzed polysaccharide was reduced with NaBH4 for 1 h, neutralized with glacial acetic acid, and acetylated with acetic anhydride/pyridine (1:1, v/v) for 1 h at 100 °C. The resulting alditol acetates were examined by GC using a capillary column of DB-225 (25 m × 0.22 mm) at a temperature of 170–210 °C with a variation of 20 °C/min.
NMR Spectroscopy
For NMR experiments, the samples were deuterium exchanged by repeated dissolution in D2O and freeze drying. Spectra were obtained from solutions in D2O at 30 °C, using sodium-3-trimethylsilyl propionate as standard (δ = 0). All spectra were obtained with a Bruker 400 MHz AVANCE III NMR spectrometer with a 5-mm inverse gradient probe. Signal assignments in the one-dimensional 1H (zgpr) and 13C NMR (zgpg decoupled) spectra were carried out using edited HSQC (hsqcedetgp), COSY (cosygpprqf), and TOCSY (mlevphpr.2) programs. The two-dimensional experiments were recorded for quadrature detection in the indirect dimension, COSY spectra were acquired using 8 scans per series of 2 K × 256 W data points, and two-dimensional TOCSY spectra were acquired using 16 scans per series of 2 × 512 W data points, edited HSQC, COSY, and TOCSY spectra were acquired using 8, 4, and 8 scans, respectively, per series of 2 K × 512 W data points with zero filling in F1 (4 K) prior to Fourier transformation.
Macrophage Culture and Stimulation
Elicited peritoneal macrophages were obtained by intraperitoneal injection of 2 ml of 3% sterile thioglycollate (Sigma). After 4 days, mice were sacrificed; peritoneal cells were harvested with chilled Hanks' balanced salt solution and plated at a density of 2 × 105 cells/well, in 96-well plates. Non-adherent cells were washed and macrophages were cultured in RPMI medium for the stimulation. For the culture of bone marrow-derived macrophages, bone marrow cells were harvested from murine femur and tibia, and cultured in RPMI, FCS (20%), L929 supernatant (30%), antibiotics, and β-marcaptoethanol. After 3 days, medium was exchanged, and on the sixth day, cells were plated at 2 × 105 cells/well in 96-well plates. The following day, differentiated macrophages were stimulated. Macrophages were stimulated with live conidia, inactivated hyphae, or heat-killed conidia, as indicated in the figure legends. In some experiments, LPS and Pam3Cys were included in the stimuli as positive controls for TLR4 and TLR2 activation, respectively. Polymixin B (10 μg/ml) was included in the stimuli with conidia and hyphae to exclude possible endotoxin contamination. Stimulations were also performed in the absence of polymixin B with similar results.
Cytokine Quantification by ELISA
IL-6, IP-10/CXCL10, IL-12p40, and IL-10 ELISA were obtained from R&D Systems and performed according to the manufacturer's instructions, the TNF ELISA was obtained from Peprotech and performed following the manufacturer's instructions.
Western Blot Analysis for MAPKs Phosphorylation and IκBα Degradation
To evaluate ERK1/2 and p38 phosphorylation, elicited peritoneal macrophages were plated in 6-well plates at a density of 2.5 × 106 cells/well, non-adherent cells were removed by washing with medium, and adherent cells were stimulated as indicated in the figure legends. After 15, 30, or 60 min, cells were lysed in a buffer consisting of Tris-HCl (50 mm), NaCl (150 mm), Nonidet P-40 (1%), sodium deoxycholate (0.25%), EDTA (1 mm), aprotinin (5 μg/ml), leupeptin (5 μg/ml), pepstatin (5 μg/ml), PMSF (1 mm), sodium orthovanadate (1 mm), and NaF (1 mm), pH 7.5. Cell lysates were centrifuged and the supernatants were boiled and subjected to electrophoresis in SDS-polyacrylamide gel (12%) in reducing conditions. The proteins were transferred to a nitrocellulose membrane at 4 °C for 2 h. Then the membranes were blocked with Tris-buffered saline solution with 0.05% Tween 20 (TBS-T) and 5% fat-free milk. The membranes were incubated overnight with anti-phospho-ERK1/2 (1/1000) (Santa Cruz Biotechnology) or anti-phospho-p38 (1/1000) (Cell Signaling), diluted in blocking solution. The membranes were washed in TBS-T and incubated for 2 h with horseradish peroxidase-conjugated goat anti-rabbit (1/5000) or goat anti-mouse (1/5000) IgG polyclonal antibodies (Santa Cruz Biotechnology). The bands were revealed by chemiluminescence, using ECL substrate. The normalization of MAPK phosphorylation was performed by stripping membranes during 30 min at 50 °C in stripping buffer (β-mercaptoethanol 100 mm, SDS 2%, 62.5 mm Tris-HCl, pH 6.7). After stripping, membranes were washed with TBS-T, blocked with 5% fat-free milk TBS-T, incubated with rabbit anti-ERK2 (1/1000) overnight, and detected as described above. The degradation of IκB protein was analyzed by Western blot with polyclonal rabbit anti-IκB (Sigma) diluted (1/1000) in block solution.
Statistical Analysis
Data are presented as mean ± S.E. Results were analyzed using a statistical software package (GraphPad Prism 4). Statistical differences among the experimental groups were evaluated by means of Student's t test. Values of p < 0.05 were regarded as significant.
RESULTS
P. boydii Conidia Induced Cytokine Secretion by Macrophages
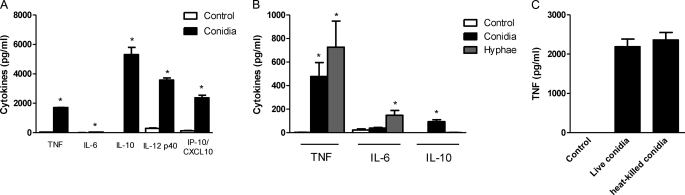
The mechanisms by which the innate immune system recognizes P. boydii, as well the induction of cytokines by the morphological stages of this fungus, are unknown. Thus to evaluate macrophage activation by P. boydii, macrophages were stimulated with live conidia, the developmental form responsible for initiating host colonization (19–21). P. boydii live conidia induced the secretion of substantial amounts of TNF, IL-12, IP-10/CXCL10, and IL-10, but low amounts of IL-6 (Fig. 1A). Filamentous fungal pathogens present different developmental phases, such as conidia and hyphae, and the morphological transition promotes a distinct recognition of these fungal structures, which induce different leukocyte responses (13, 29–32). During stimulation of macrophages with P. boydii live conidia, these fungal structures differentiated into hyphae. Thus, to investigate the differential recognition of P. boydii conidia and hyphae by macrophages, resting heat-inactivated conidia and hyphae were used. Stimulation of macrophages with hyphae and heat-killed conidia resulted in cytokine induction, with hyphae inducing a maximal TNF and IL-6 release at 5 mg/ml (no IL-10 release), whereas heat-killed conidia were more effective at 5 conidia per macrophage and induced TNF, IL-10, IL-6, and IP-10/CXCL10 release (Fig. 1B and data not shown). To evaluate if conidia viability affected macrophage activation, peritoneal macrophages were stimulated with these two conidial preparations. Induction of TNF secretion by heat-killed conidia was similar to that of live conidia (Fig. 1C). Furthermore, the same pattern of cytokine induction was obtained by conidia inactivated with thimerosal and heat-killed conidia, which also stimulated TNF production from mouse bone marrow-derived macrophages and human macrophages (data not shown). These results indicate that recognition of P. boydii conidia, and hyphae, induce a strong activation of macrophages.
FIGURE 1.
P. boydii conidia induced cytokine release by macrophages. Macrophages were stimulated with P. boydii live conidia at a ratio of 5 conidia/macrophage, after 24 h the supernatant was recovered, centrifuged, and TNF, IL-6, IL-12, IP-10/CXCL10, and IL-10 were evaluated by ELISA, results represent mean ± S.E. of two experiments *, p ≤ 0.05 (A). Macrophages were stimulated with heat-killed P. boydii conidia (5 conidia/macrophage) or heat-killed hyphae (5 mg/ml), after 24 h the supernatant was recovered and TNF, IL-6, and IL-10 were evaluated by ELISA, results represent mean ± S.E. of two different experiments *, p ≤ 0.05 (B). Macrophages were stimulated with live P. boydii conidia (5 conidia/macrophage) or heat-killed P. boydii conidia (5 conidia/macrophage), after 24 h supernatants were recovered and TNF was evaluated by ELISA, results represent mean ± S.E. of two experiments (C).
P. boydii Conidia-induced IκBα Degradation and MAPKs Phosphorylation
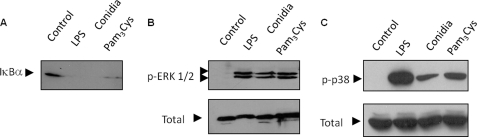
Activation of NFκB and MAP kinases is essential for TNF production induced by pathogen-associated molecular patterns (7). To determine involvement of the NFκB pathway in macrophage activation by P. boydii conidia, we characterized the content of IκBα by Western blot. Heat-killed conidia, LPS (a TLR4 ligand), and Pam3Cys (a TLR2 ligand) induced degradation of IκBα, indicating activation of the NFκB signaling pathway (Fig. 2A). Activation of macrophages by pathogen-associated molecular patterns also cause the phosphorylation of ERK1/2 and p38 (1). The stimulation of macrophages by P. boydii, LPS, and Pam3Cys caused the phosphorylation of these MAPKs (Fig. 2, B and C). These results indicate that P. boydii conidia induced the activation of MAP kinases and NFκB signaling pathways.
FIGURE 2.
P. boydii conidia-induced IκBα degradation of MAPKs phosphorylation. Macrophages were either non-stimulated or stimulated with conidia (5 conidia/macrophage), LPS, or Pan3Cys (both at 100 ng/ml) during the indicated periods of time. Cell extracts were prepared and submitted to electrophoresis. Detection of non-phosphorylated ERK1/2 was performed to normalize the amount of protein run on the lanes. A, IκBα degradation; B, ERK1/2; and C, p38 phosphorylation were detected by immunoblotting using anti-IκBα, anti-phospho-ERK1/2, or anti-phospho-p38 polyclonal antibodies, respectively. The figures are representative of two experiments with similar results.
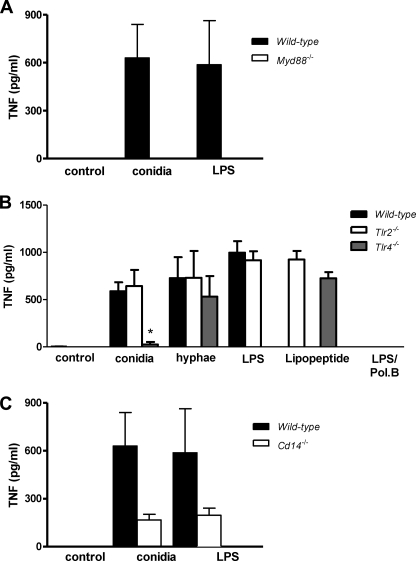
Induction of Cytokine Secretion by Macrophages in Response to P. boydii Conidia Was Dependent of MyD88, TLR4, and CD14
TLRs perform pathogen recognition and are essential to the induction of pro-inflammatory mediators and the transition from innate to adaptive immunity. TLRs signal through a conserved pathway that employs TIR domain containing adaptor molecules. MyD88 is an adaptor protein that contains a TIR domain and is essential to the signaling of TLRs, with the exception of TLR3 (7). Thus, to evaluate a possible role of TLRs in the recognition of P. boydii conidia by macrophages, we stimulated peritoneal macrophages obtained from wild-type (WT) and Myd88−/− mice with heat-killed conidia. Macrophages from Myd88−/− mice were unable to release TNF in response to the stimuli with heat-killed conidia or LPS (Fig. 3A). These results indicate a role for one or more TLRs in the recognition of P. boydii conidia. TLR2 and TLR4 were pointed out as receptors involved in the recognition of fungal pathogens (11–18). TNF secretion in response to heat-killed P. boydii conidia was abolished on Tlr4−/− macrophages (Fig. 3B). P. boydii conidia-induced IL-10 and IP-10/CXCL10 secretion were also dependent on TLR4 signaling (data not shown). In contrast, Tlr2−/− macrophages showed a similar TNF release in response to activation with heat-killed conidia, when compared with WT macrophages (Fig. 3B). The secretion of TNF and IL-6 induced by P. boydii hyphae was independent of both TLR4 and TLR2 signaling (Fig. 3B). As positive controls to TLR4 and TLR2 activation, we employed LPS and Pam3Cys, respectively. Stimulations with heat-killed P. boydii conidia and hyphae were performed in the presence of polymyxin B, in a concentration able to completely neutralize LPS in a concentration as high as 100 ng/ml. Stimulations with conidia performed in the absence or presence of polymyxin B gave similar results (Fig. 3B and data not shown). CD14 is a co-receptor important for cell signaling of several TLR2 and TLR4 ligands, promoting a higher sensitivity to small concentrations of agonists (33). Moreover, CD14 has been shown to participate in the recognition of fungal molecules (34–36). Thus we investigated the role of CD14 in the recognition of conidia by macrophages. These experiments were conducted in the absence of serum to avoid any exogenous source of soluble CD14. Macrophages from Cd14−/− mice stimulated with conidia presented an impaired production of TNF as compared with WT macrophages (Fig. 3C). As expected, at the concentration tested (100 ng/ml), LPS was unable to induce an optimal TNF release from Cd14−/− macrophages (Fig. 3C).
FIGURE 3.
The P. boydii conidia-induced TNF release by macrophages requires functional MyD88, TLR4, and CD14. Peritoneal macrophages were obtained from WT, Myd88−/− (A), Tlr2−/−, Tlr4−/− (B), and Cd14−/− (C) mice and stimulated with heat-killed P. boydii conidia (5 conidia/cell). As controls, LPS and Pam3Cys (both at 100 ng/ml) were included in the experimental settings. Polymyxin B (10 μg/ml) was added during stimulation of macrophages with conidia. Supernatants were collected and TNF was evaluated by ELISA. Results represent mean ± S.E. and are representative of two or three experiments. * p ≤ 0.05.
Isolation and Characterization of P. boydii Rhamnomannans
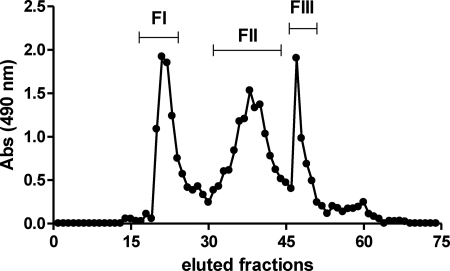
Mannans are molecular patterns expressed by pathogenic fungi like C. albicans, and triggers TLR4 activation, as well as Dectin-2 and mannose receptor, promoting cytokine secretion by macrophages (14, 37). Because our results pointed to a role for TLR4 in P. boydii conidia recognition, we hypothesized that polysaccharides similar to mannans could be the molecular patterns expressed in P. boydii conidia involved in TLR4 activation. Thus, we isolated and characterized rhamnomannans from P. boydii using a hot alkaline extraction. Polysaccharides were then fractioned by gel filtration, and further analyses were carried out using fraction II that consisted predominantly of rhamnomannans with a low amount of protein (Fig. 4, Tables 1 and 2). This fraction showed the presence of rhamnose, mannose, glucose, and traces of galactose, whereas fractions I and III presented only glucose (data not shown). To precisely determine the composition of monosaccharides in the rhamnomannan fraction purified from P. boydii, we analyzed the alditol acetates by GC-MS. As indicated in Table 1, fraction II present rhamnose (23.5%), mannose (45.5%), glucose (31.0%), and traces of galactose. In contrast, fractions I and III were constituted essentially by glucose and minor traces of mannose (data not shown). We also performed quantification of total sugars, protein, phosphate, and hexosamine, and all fractions were free of phosphate and hexosamine (Table 2).
FIGURE 4.
Purification of rhamnomannans from P. boydii. Polysaccharides from P. boydii were extracted by hot alkaline treatment and successive precipitation with ethanol, polysaccharides were submitted to gel filtration in a Superdex 200 column with a phosphate/NaCl buffer. Carbohydrates were quantified by reaction in the presence of 5% phenol and sulfuric acid and reading at 490 nm. The result represents the pattern of elution of different fractions in relationship to their carbohydrate contents.
TABLE 1.
Chemical composition (%) of rhamnomannan fraction of P. boydii
TABLE 2.
Quantitative composition of monosaccharides present in the polysaccharides obtained from P. boydii by hot alkaline extraction as evaluated by gas chromatography
Monosaccharides were analyzed by detection of their alditol-acetate derivatives using a capillary column DB-225 (25 × 0.22 mm) programmed for a temperature of 170–210 °C with a variation of 20 °C/min.
| Percentual of monosaccharides |
||
|---|---|---|
| Unfractionated polysaccharides | Fraction II | |
| Rhamnose | 7.9 | 26.5 |
| Mannose | 16.5 | 38.5 |
| Glucose | 72.4 | 28.2 |
| Galactose | 3.2 | 7.7 |
RMN Analysis of Preparations Containing Rhamnomannans
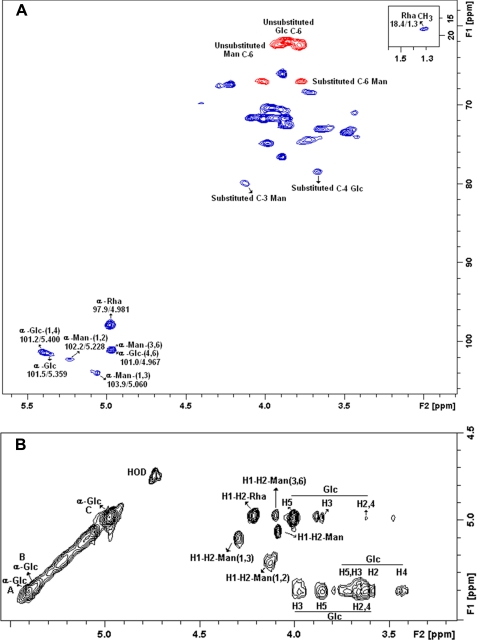
One dimensional and two-dimensional NMR analysis confirmed the structures of the polysaccharides present in Fraction II, suggesting that it contains typical signals of α-glucan and rhamnomannan (22, 38). Edited HSQC spectrum showed substituted C-1 and C-4 signals at δ 101.2/5.400, 101.5/5.359, and 78.8/3.667 of α-d-Glcp units (Fig. 5A). The α-glucan was confirmed by a TOCSY experiment using a mixing time of 120 ms, which allows observing the glucopyranosyl connectivity (Fig. 5B). Total 1H-1H axial correlations were observed for the three α-d-Glcp units at δ 5.400-A, 5.359-B, and 4.976-C, corroborating a glycogen-like structure. These features are shown in the partial TOCSY spectrum in the diagonal and its cross-peaks (Fig. 5B). Rhamnomannan identification was determined by one-dimensional (1H and 13C) and two-dimensional COSY, TOCSY, and HSQC experiments. The NMR data of fraction II showed at C-1 signals at δ 97.9/4.981, 101.0/4.967, 102.2/5.228, and 103.9/5.060, typical of terminal α-rhamnose units, O-3,6-substituted-α-mannopyranose (α-d-Manp-(1 → 3,6)), O-2-substituted-α-mannopyranose (α-d-Manp-(1 → 2)), and α-Manp-3-O-substituted units, respectively (38, 39). The signal at δ 79.9/4.127 confirms the 3-O-substituted α-Manp units (Fig. 5A). The phase-sensitive edited HSQC gave inverted signals of CH2 at δ 62.0/3878, 62.4/3.785, and 67.0/4.013; 3.771, which correspond to non-substituted C-6 units of Glcp and Manp and O-substituted C-6 of Manp units. These signals were observed on negative phase (red), and the C-6 units of Rhap were observed at δ 18.4/1.300 on positive phase (blue) (see Fig. 5A). COSY and TOCSY complemented the identification of the rhamnomannan and showed the characteristic low connectivity of the Rhap and Manp units, easily visualized when compared with higher connectivity of the Glcp units (Fig. 5B).
FIGURE 5.
Partial two-dimensional NMR spectra (edited HSQC and TOCSY) of fraction II. A, partial edited HSQC spectrum, assignment of the main signals from the anomeric region and carbohydrate linkages, the positive phase (blue) correspond to CH and CH3 carbons, and the negative phase (red) correspond to CH2 carbons. B, partial TOCSY from the anomeric region showing the main cross-peaks of the rhamnomannan and α-glucan.
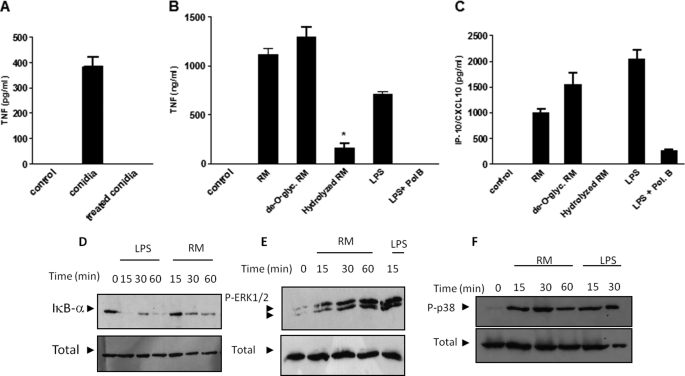
Rhamnomannan Preparations from P. boydii-induced Cytokine Release, IκBα Degradation, and MAPK Phosphorylation
Peptidopolysaccharides are abundant antigens that are expressed in P. boydii conidia. To investigate if these molecules are involved on cytokine induction by macrophages stimulated with conidia, these fungal structures were treated for 2 h at 100 °C in phosphate buffer (50 mm, pH 7.2), a protocol that was previously demonstrated to remove peptidopolysaccharides from P. boydii (39). Removal of peptidopolysaccharides in conidia abolished TNF release by macrophages stimulated with P. boydii conidia (Fig. 6A). Then we evaluated if rhamnomannans were able to induce cytokine release and activation of signaling pathways involved on macrophage activation. We assessed two prototypical cytokines induced by TLR4 signaling, TNF and IP-10/CXCL10. Purified rhamnomannans induced TNF and IP-10/CXCL10 release by macrophages (Fig. 6, B and C). To define the structural determinants of rhamnomannans involved in cytokine inducing, we evaluated cytokine induction by de-O-glycosylated peptidorhamnomannans, obtained by removal of the O-linked oligosaccharides from the peptidorhamnomannan by a mild reductive alkaline treatment under reducing conditions, and rhamnomannans submitted to partial acid hydrolysis that removes rhamnopyranosyl non-reducing end units (28). The de-O-glycosylated peptidorhamnomannans induced TNF and IP-10/CXCL10 release by macrophages in the same amounts of that induced by rhamnomannans (supplemental Fig. S1, A and B). In contrast, removal of rhamnopyranosyl units by partial acid hydrolysis eliminates TNF and IP-10/CXCL10 induction by rhamnomannans, but had no effect on LPS-induced cytokine secretion (Fig. 6, B and C, and data not shown). Peptidorhamnomannans and de-O-glycosylated peptidorhamnomannans induced TNF secretion in a dose-response fashion, in this condition 10 μg/ml was the minimum dose able to cause TNF secretion and the concentration of 50 μg/ml reached the maximum stimulation (supplemental Fig. 1, A and B). Then we set up to investigate the activation of signaling pathways involved on cytokine induction by fractions of rhamnomannans. For this purpose, we evaluated IκBα degradation and phosphorylation of ERK1/2 and p38 in macrophages stimulated with preparations of P. boydii-derived rhamnomannans. Preparations of rhamnomannans were able to induce JNK and ERK1/2 phosphorylation, as well as IκBα degradation, which are essential components of signaling pathways triggered by TLRs. LPS, a TLR4 activator and a positive control to our experimental conditions, was able to induce a similar pattern of signal transduction, with JNK, ERK1/2 phosphorylation, and IκB degradation (Fig. 6D).
FIGURE 6.
Rhamnomannans isolated from P. boydii induced TNF release, MAPKs activation, and IκBα degradation. A, P. boydii heat-inactivated conidia or P. boydii conidia treated with hot phosphate buffer and extensive washing were used to stimulate mouse macrophages, after 24 h the supernatant was recovered and TNF was analyzed by ELISA. Results represent mean ± S.E. and are representative of two different experiments with similar results. Macrophages were stimulated with different concentrations of preparations of P. boydii-derived rhamnomannans (RM), de-O-glycosylated peptidorhamnomannans (de-O-glyc RM), or acid-hydrolyzed rhamnomannans (hydrolyzed RM), TNF (B), and IP-10/CXCL10 (C) were measured from supernatants by ELISA. Results represent mean ± S.E. and are representative of two or three different experiments with similar results. D, macrophages were stimulated with preparations of P. boydii-derived rhamnomannans (RM, 50 μg/ml) or LPS (100 ng/ml). Cell extracts were prepared and submitted to electrophoresis. Detection of non-phosphorylated ERK1/2 was performed to normalize the amount of protein run on the lanes. D, IκBα degradation; E, ERK1/2; and F, p38 phosphorylation were detected by immunoblotting using anti-IκBα, anti-phopho-ERK1/2, or anti-phospho-p38 polyclonal antibodies, respectively. The figures are representative of two experiments with similar results.
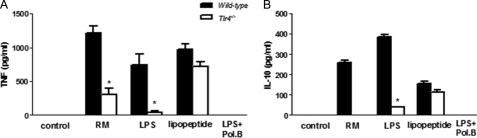
Cytokine Release Induced by Rhamnomannans Derived from P. boydii Required TLR4 Recognition
Cytokine release by macrophages in response to P. boydii conidia is dependent on TLR4 signaling, and removal of peptidopolysaccharides from conidia reduces cytokine release by macrophages. Based on these results, the role of TLR4 on macrophage activation by rhamnomannan was investigated. P. boydii-derived rhamnomannans induced TNF, IL-6, IL-10, and IP-10/CXCL10 production by WT macrophages, but not by macrophages from Tlr4−/− mice (Fig. 7, A and B, and data not shown). Activation of bone marrow-derived macrophages by P. boydii-derived rhamnomannans was also dependent on TLR4 signaling, as observed by the reduced TNF and IL-10 release by Tlr4−/− macrophages (data not shown). All experiments were performed in the presence of polymyxin B (10 μg/ml), used in a concentration able to completely neutralize LPS at 100 ng/ml (data not shown). Induction of TNF and IP-10/CXCL10 release by de-O-glycosylated rhamnomannans also required TLR4 signaling, as demonstrated by the impaired TNF release by Tlr4−/− macrophages in comparison to the WT macrophages (data not shown).
FIGURE 7.
Preparations of rhamnomannans derived from P. boydii-induced cytokine release by macrophages through TLR4 signaling. Macrophages were obtained from WT or Tlr4−/− mice and stimulated with preparations of P. boydii-derived rhamnomannans (RM, 50 μg/ml), LPS (100 ng/ml), Pam3Cys (100 ng/ml), and polymyxin B (Pol. B) (10 μg/ml) were added during stimuli with preparations of rhamnomannans and LPS in some wells. Supernatants were recovered and TNF (A) and IL-10 (B) were quantified by ELISA, results represent mean ± S.E. and are representative of two or three different experiments with similar results. *, p ≤ 0.05.
DISCUSSION
Although P. boydii represents an emergent pathogen with a ubiquitous distribution in the environment, there is a great gap in the knowledge about the mechanisms of resistance triggered by the immune recognition of this pathogen. In this work we investigated the activation of innate immunity by different developmental stages of P. boydii. Our results indicate that P. boydii conidia and hyphae induce macrophage activation, as observed by cytokine release, but use distinct mechanisms with conidia inducing TLR4 but not TLR2 signaling, whereas hyphae recognition is independent of both TLR2 and TLR4.
Macrophages responded to live conidia stimulation secreting TNF, IL-12, IP-10/CXCL10, and IL-10. Because the differentiation of live conidia to hyphae occurred during stimulation, recognition of P. boydii by macrophages could involve one or both of these two different developmental forms. Both P. boydii conidia and hyphae induced TNF secretion by macrophages, but heat-killed conidia promoted IL-10 secretion by macrophages, whereas hyphae did not induce IL-10 release. The mechanisms involved in the differences of IL-10 secretion by conidia and hyphae are not clear. It is possible that a different expression of molecules could result in triggering of distinct receptors. C-type lectin receptors like mannose receptor or DC-SIGN and its mouse counterpart SIGNR1 are involved in the recognition of several pathogens. These receptors bind mannosylated structures and are strong inducers of IL-10 (40, 41). Our results demonstrate that IL-10 induction by P. boydii conidia is dependent of TLR4 signaling, thus it is possible that TLR4 and lectin receptors could cooperate in the recognition of P. boydii conidia and IL-10 induction by this developmental stage, whereas recognition of hyphae would involve different pattern recognition receptors that would not induce IL-10 release by macrophages. IL-10 is an anti-inflammatory cytokine and is involved on inhibition of macrophage activation, so differences in IL-10 induction by conidia and hyphae could contribute for pathogenesis of infection promoting an initial anti-inflammatory response that could allow conidia to establish infection and germinate in tissues.
Previous studies described that A. fumigatus germinating conidia, but not resting conidia, were able to induce a pro-inflammatory macrophage response, being this property is a consequence of the exposure of β-glucans and possibly unknown TLR ligands during the swelling of conidia (29–31). Our results indicated that P. boydii conidia induce cytokine release by macrophages, but we cannot exclude the possibility that the P. boydii conidial preparations used in our experiments contain germinating conidia.
We observed that P. boydii conidia induced the activation of intracellular signaling pathways typical of TLRs, including degradation of IκBα and phosphorylation of MAPKs. Conidia also induced TNF and IP-10/CXCL10. These cytokines are induced by LPS through MyD88 and TRIF pathways upon TLR4 activation (7). The secretion of TNF by macrophages after challenge with P. boydii conidia required MyD88, thus indicating a role for TLRs. Recognition of P. boydii conidia was dependent on TLR4 signaling, whereas TLR2 was dispensable for induction of cytokine release. In contrast, our data demonstrated that macrophage activation by P. boydii hyphae is independent of TLR2 and TLR4 signaling. The Saccharomyces cerevisiae cell wall particle, zymosan, requires TLR2 for induction of cytokines by macrophages (42–44). Similarly C. albicans and A. fumigatus induce activation of immune cells by TLR2 triggering (11, 12, 15, 16). We have previously demonstrated that a highly purified α-glucan obtained from P. boydii is a TLR2 activator (22). Our present results do not discard the possibility that TLR2 participates in P. boydii recognition, but indicate that TLR4 is the major receptor involved on P. boydii conidia recognition. These results also imply that molecules distinct from α-glucans are the major activators of innate immunity induced by P. boydii conidia, and suggest that α-glucans of the cell wall might be inaccessible for recognition by TLR2 or are minor components of the cell wall in P. boydii conidia. Using Cd14−/− macrophages, we observed that these cells also had impaired TNF secretion when compared with WT macrophages, thus indicating an important role for CD14 in macrophage activation by P. boydii conidia. Although CD14 lacks an intracellular signaling tail, it participates on binding of pathogen molecules, increasing responses triggered by TLR2 and TLR4 (33). Possibly, CD14 promotes binding and transference of molecules expressed in the P. boydii conidial surface to TLR4.
Rhamnomannan-enriched preparations isolated from P. boydii induced cytokine release by macrophages, as well as degradation of IκBα and phosphorylation of MAPKs. Our results showed that P. boydii-derived rhamnomannans also required TLR4 signaling for cytokine induction by macrophages. The requirement for TLR4 on macrophage activation by P. boydii-derived rhamnomannans seems to mirror the role of TLR4 in recognition of P. boydii conidia by macrophages. This possibility is supported by our results demonstrating that P. boydii conidia present a strong expression of rhamnomannans on the cell surface, as analyzed by immunofluorescence using labeling with monoclonal antibodies of anti-rhamnomannans from P. boydii (46).
A putative contamination of endotoxin in the preparations of conidia and rhamnomannans as responsible for the observed cytokine secretion is unlikely. We used polymyxin B in a dose that abrogated LPS-induced cytokine production and this treatment had no effect on conidia or rhamnomannans-induced cytokine secretion. Conversely, removal of peptidopolysaccharides in conidia and hydrolysis of rhamnomannans abrogated the production of cytokine, although had no effect on LPS-induced TNF secretion.
Although our preparations of rhamnomannans presented a significant amount of α-glucans (about 28% of total polysaccharides of fraction II), activation of TLR4 seems to be a consequence of the recognition of rhamnomannans, as shown by the following evidence: 1) we have previously demonstrated that highly purified α-glucans are activators of TLR2 but not TLR4; 2) removing terminal rhamnoses from rhamnomannans, by partial acid hydrolysis, abolished cytokine induction by these polysaccharides, whereas α-glucans do not present caps with rhamnoses; and 3) concentrations of contaminant α-glucans are unable to induce the same level of cytokine release observed by a concentration of 50 μg/ml of polysaccharides enriched with rhamnomannans (less than 15 μg/ml of α-glucans).
We also demonstrated that O-linked oligosaccharides from the peptidorhamnomanan are not involved on macrophage activation by P. boydii-derived rhamnomannans, because preparations of peptidorhamnomannans that were submitted to β-elimination, a process that removes O-linked but not N-linked carbohydrates, showed a similar induction of cytokines by macrophages and this effect was still dependent on TLR4 activation. In contrast, removal of terminal rhamnopyranosyl units impaired cytokine release by macrophages in response to P. boydii-derived rhamnomannans, implying that structures with terminal rhamnose and/or mannose are structural motifs involved on TLR4 recognition. It has recently been described that a soluble form of TLR4-Fc is able to bind complex mixtures of fungal polysaccharides and the binding is blocked by soluble mannans or fucose (45), a deoxymonosaccharide-like rhamnose, suggesting that TLR4 could recognize structural patterns like fungal polysaccharides through direct interaction with terminal deoxycarbohydrates expressed in complex polysaccharides like rhamnomannans. TLR4 has been described as a receptor involved in C. albicans and A. fumigatus recognition (15–17). Mannans obtained from yeasts like S. cerevisiase and Candida sp. activate macrophages through TLR4 (35). Moreover, O-linked mannans from C. albicans and glucuroxylomannan obtained from Cryptococcus neoformans are also able to induce innate immune activation through TLR4 (14, 36). In these works, TLR4 recognizes C. albicans-derived O-linked mannans, but not N-linked, a result different from ours, that demonstrate that removing O-linked oligosaccharides from P. boydii-derived rhamnomannans does not affect TLR4-mediated recognition. The differences observed could be due to the different experimental settings. The experimental approach employed by Netea et al. (14) was based on genetic deficiency of pathways involved in the N-linked and O-linked formation of oligosaccharide chains in surface proteins. It is possible that the approach used by Netea et al. (14) resulted in an anomalous pattern of glycosylation or in differences on the expression of TLR4 ligands, instead of reflecting a simple absence of recognition of N-linked mannans by TLR4. Another possibility is that the patterns of glycosylation in P. boydii- and C. albicans-derived mannans are distinct in relation to TLR4 activation, with different requirements for N-linked or O-linked mannans/rhamnomannans in these two different fungal pathogens.
Our results indicate that recognition of conidial forms of P. boydii by the innate immune system requires functional TLR4 and CD14 and that P. boydii-derived rhamnomannans are molecular patterns recognized by TLR4. These results add new information on the role of mannan-containing polymers in innate recognition of fungal pathogens. It is possible that these polymers expressed in other filamentous fungi (like A. fumigatus) could be involved in innate immune recognition, possibly by triggering TLR4. Thus, modulation of TLR4 signaling could be an important therapy for inducing resistance in individuals with invasive infections caused by P. boydii. Otherwise, antagonism of TLR4 signaling in association with effective antifungal drugs could control the infection and reduce tissue damage associated with the immune response directed to selected fungal infections including P. boydii.
Supplementary Material
Acknowledgments
We are grateful to Shizuo Akira and Douglas Golenbock for providing genetically deficient mice strains (Myd88, Tlr2, Tlr4, and Cd14), and Patricia Bozza, Maria Bellio, and Ricardo Gazzinelli for providing mice and reagents. We thank Claudia Paiva for the critical reading the manuscript.
The work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior (CAPES), Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ), Programa de Núcleos de Excelência (Pronex), and Universidade Federal do Rio de Janeiro.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TLR
- Toll-like receptors
- MyD88
- myeloid differentiation protein-88
- Pam3Cys
- (S)-(2,3-bis(palmitoyloxy)-(2RS)-propyl)-N-palmitoyl-(R)-Cys-(S)-Ser(S)-Lys(4)-OH, trihydrochloride.
REFERENCES
- 1.Beck-Sagué C., Jarvis W. R. (1993) J. Infect. Dis. 167, 1247–1251 [DOI] [PubMed] [Google Scholar]
- 2.Baddley J. W., Stroud T. P., Salzman D., Pappas P. G. (2001) Clin. Infect. Dis. 32, 1319–1324 [DOI] [PubMed] [Google Scholar]
- 3.Warris A., Bjorneklett A., Gaustad P. (2001) N. Engl. J. Med. 344, 1099–1100 [PubMed] [Google Scholar]
- 4.Peter E., Bakri F., Ball D. M., Cheney R. T., Segal B. H. (2002) Clin. Infect. Dis. 35, e54–56 [DOI] [PubMed] [Google Scholar]
- 5.Zelante T., Montagnoli C., Bozza S., Gaziano R., Bellocchio S., Bonifazi P., Moretti S., Fallarino F., Puccetti P., Romani L. (2007) Adv. Exp. Med. Biol. 590, 209–221 [DOI] [PubMed] [Google Scholar]
- 6.Hohl T. M., Rivera A., Pamer E. G. (2006) Curr. Opin. Immunol. 18, 465–472 [DOI] [PubMed] [Google Scholar]
- 7.Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 8.Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A. (1996) Cell 86, 973–983 [DOI] [PubMed] [Google Scholar]
- 9.Netea M. G., Van der Graaf C., Van der Meer J. W., Kullberg B. J. (2004) Eur. J. Clin. Microbiol. Infect. Dis. 23, 672–676 [DOI] [PubMed] [Google Scholar]
- 10.Levitz S. M. (2010) PLoS Pathog. 6, e1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mambula S. S., Sau K., Henneke P., Golenbock D. T., Levitz S. M. (2002) J. Biol. Chem. 277, 39320–39326 [DOI] [PubMed] [Google Scholar]
- 12.Meier A., Kirschning C. J., Nikolaus T., Wagner H., Heesemann J., Ebel F. (2003) Cell. Microbiol. 5, 561–570 [DOI] [PubMed] [Google Scholar]
- 13.Netea M. G., Warris A., Van der Meer J. W., Fenton M. J., Verver-Janssen T. J., Jacobs L. E., Andresen T., Verweij P. E., Kullberg B. J. (2003) J. Infect. Dis. 188, 320–326 [DOI] [PubMed] [Google Scholar]
- 14.Netea M. G., Gow N. A., Munro C. A., Bates S., Collins C., Ferwerda G., Hobson R. P., Bertram G., Hughes H. B., Jansen T., Jacobs L., Buurman E. T., Gijzen K., Williams D. L., Torensma R., McKinnon A., MacCallum D. M., Odds F. C., Van der Meer J. W., Brown A. J., Kullberg B. J. (2006) J. Clin. Invest. 116, 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villamón E., Gozalbo D., Roig P., O'Connor J. E., Fradelizi D., Gil M. L. (2004) Microbes Infect. 6, 1–7 [DOI] [PubMed] [Google Scholar]
- 16.Netea M. G., Van Der Graaf C. A., Vonk A. G., Verschueren I., Van Der Meer J. W., Kullberg B. J. (2002) J. Infect. Dis. 185, 1483–1489 [DOI] [PubMed] [Google Scholar]
- 17.Balloy V., Si-Tahar M., Takeuchi O., Philippe B., Nahori M. A., Tanguy M., Huerre M., Akira S., Latgé J. P., Chignard M. (2005) Infect. Immun. 73, 5420–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellocchio S., Montagnoli C., Bozza S., Gaziano R., Rossi G., Mambula S. S., Vecchi A., Mantovani A., Levitz S. M., Romani L. (2004) J. Immunol. 172, 3059–3069 [DOI] [PubMed] [Google Scholar]
- 19.O'Bryan T. A. (2005) Expert Rev. Anti-infect. Ther. 3, 765–773 [DOI] [PubMed] [Google Scholar]
- 20.Cortez K. J., Roilides E., Quiroz-Telles F., Meletiadis J., Antachopoulos C., Knudsen T., Buchanan W., Milanovich J., Sutton D. A., Fothergill A., Rinaldi M. G., Shea Y. R., Zaoutis T., Kottilil S., Walsh T. J. (2008) Clin. Microbiol. Rev. 21, 157–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roilides E., Simitsopoulou M., Katragkou A., Walsh T. J. (2009) Med. Mycol. 47, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bittencourt V. C., Figueiredo R. T., da Silva R. B., Mourão-Sá D. S., Fernandez P. L., Sassaki G. L., Mulloy B., Bozza M. T., Barreto-Bergter E. (2006) J. Biol. Chem. 281, 22614–22623 [DOI] [PubMed] [Google Scholar]
- 23.Gilgado F., Cano J., Gené J., Guarro J. (2005) J. Clin. Microbiol. 43, 4930–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubois M., Gilles K. A., Hamilton J. K., Rebers P. A., Swith E. (1956) Anal. Chem. 28, 350–356 [Google Scholar]
- 25.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 26.Ames B. N. (1966) Methods Enzymol. 8, 115–118 [Google Scholar]
- 27.Belcher R. A., Nutten A. J., Sambrook C. M. (1954) Analyst 79, 201–208 [Google Scholar]
- 28.Leitao E. A., Bittencourt V. C., Haido R. M., Valente A. P., Peter-Katalinic J., Letzel M., de Souza L. M., Barreto-Bergter E. (2003) Glycobiology 13, 681–692 [DOI] [PubMed] [Google Scholar]
- 29.Gersuk G. M., Underhill D. M., Zhu L., Marr K. A. (2006) J. Immunol. 176, 3717–3724 [DOI] [PubMed] [Google Scholar]
- 30.Hohl T. M., Van Epps H. L., Rivera A., Morgan L. A., Chen P. L., Feldmesser M., Pamer E. G. (2005) PLoS Pathog. 1, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steele C., Rapaka R. R., Metz A., Pop S. M., Williams D. L., Gordon S., Kolls J. K., Brown G. D. (2005) PLoS Pathog. 1, e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gantner B. N., Simmons R. M., Underhill D. M. (2005) EMBO J. 24, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pugin J., Heumann I. D., Tomasz A., Kravchenko V. V., Akamatsu Y., Nishijima M., Glauser M. P., Tobias P. S., Ulevitch R. J. (1994) Immunity 1, 509–516 [DOI] [PubMed] [Google Scholar]
- 34.Barbosa F. M., Fonseca F. L., Figueiredo R. T., Bozza M. T., Casadevall A., Nimrichter L., Rodrigues M. L. (2007) Clin. Vaccine Immunol. 14, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tada H., Nemoto E., Shimauchi H., Watanabe T., Mikami T., Matsumoto T., Ohno N., Tamura H., Shibata K., Akashi S., Miyake K., Sugawara S., Takada H. (2002) Microbiol. Immunol. 46, 503–512 [DOI] [PubMed] [Google Scholar]
- 36.Shoham S., Huang C., Chen J. M., Golenbock D. T., Levitz S. M. (2001) J. Immunol. 166, 4620–4626 [DOI] [PubMed] [Google Scholar]
- 37.Saijo S., Ikeda S., Yamabe K., Kakuta S., Ishigame H., Akitsu A., Fujikado N., Kusaka T., Kubo S., Chung S. H., Komatsu R., Miura N., Adachi Y., Ohno N., Shibuya K., Yamamoto N., Kawakami K., Yamasaki S., Saito T., Akira S., Iwakura Y. (2010) Immunity 32, 681–691 [DOI] [PubMed] [Google Scholar]
- 38.Gorin P. A. (1981) Adv. Carbohydr. Chem. Biochem. 38, 13–104 [DOI] [PubMed] [Google Scholar]
- 39.Pinto M. R., Mulloy B., Haido R. M., Travassos L. R., Barreto Bergter E. (2001) Microbiology 147, 1499–1506 [DOI] [PubMed] [Google Scholar]
- 40.Chieppa M., Bianchi G., Doni A., Del Prete A., Sironi M., Laskarin G., Monti P., Piemonti L., Biondi A., Mantovani A., Introna M., Allavena P. (2003) J. Immunol. 171, 4552–4560 [DOI] [PubMed] [Google Scholar]
- 41.Wieland C. W., Koppel E. A., den Dunnen J., Florquin S., McKenzie A. N., van Kooyk Y., van der Poll T., Geijtenbeek T. B. (2007) Microbes Infect. 9, 134–141 [DOI] [PubMed] [Google Scholar]
- 42.Brown G. D., Herre J., Williams D. L., Willment J. A., Marshall A. S., Gordon S. (2003) J. Exp. Med. 197, 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gantner B. N., Simmons R. M., Canavera S. J., Akira S., Underhill D. M. (2003) J. Exp. Med. 197, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Underhill D. M., Ozinsky A., Hajjar A. M., Stevens A., Wilson C. B., Bassetti M., Aderem A. (1999) Nature 401, 811–815 [DOI] [PubMed] [Google Scholar]
- 45.Hsu T. L., Cheng S. C., Yang W. B., Chin S. W., Chen B. H., Huang M. T., Hsieh S. L., Wong C. H. (2009) J. Biol. Chem. 284, 34479–34489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopes L. C., Rollin-Pinheiro R., Guimaraes A. J., Bittencourt V. C., Martinez L. R., Koba W., Farias S. E., Nosanchuk J. D., Barreto-Bergter E. (2010) PLoS Negl. Trop. Dis. 4, e853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.