Abstract
Mitochondrial supercomplexes containing complexes I, III, and IV of the electron transport chain are now regarded as an established entity. Supercomplex I·III·IV has been theorized to improve respiratory chain function by allowing quinone channeling between complexes I and III. Here, we show that the role of the supercomplexes extends beyond channeling. Mutant analysis in Caenorhabditis elegans reveals that complex III affects supercomplex I·III·IV formation by acting as an assembly or stabilizing factor. Also, a complex III mtDNA mutation, ctb-1, inhibits complex I function by weakening the interaction of complex IV in supercomplex I·III·IV. Other complex III mutations inhibit complex I function either by decreasing the amount of complex I (isp-1), or decreasing the amount of complex I in its most active form, the I·III·IV supercomplex (isp-1;ctb-1). ctb-1 suppresses a nuclear encoded complex III defect, isp-1, without improving complex III function. Allosteric interactions involve all three complexes within the supercomplex and are necessary for maximal enzymatic activities.
Keywords: C. Elegans, Electron Transport, Genetics, Mitochondria, Mitochondrial Metabolism, Supercomplex
Introduction
A solid-state model (1) of the mitochondrial respiratory chain within the mitochondrial membrane was proposed a half-century ago. In this model, the respiratory complexes are assembled into multicomplex structures, supercomplexes. Supercomplexes are capable of substrate channeling and thus facilitate transfer of electrons from one complex to the next (2). This is in contrast to the random collision model (3), which proposes that the complexes of the mitochondrial respiratory chain are embedded in the inner mitochondrial membrane as separate entities. Individual complexes are functionally connected to each other by the small, mobile electron carriers, coenzyme Q, and cytochrome c. The random collision model became more generally accepted as kinetic studies demonstrated homogenous pool behavior of coenzyme Q (4), rates of electron transfer that did not require substrate channeling, and the successful isolation of individual respiratory complexes that were enzymatically active (3, 5). However, supercomplex structures containing complexes I, III, and IV, have been investigated by blue native polyacrylamide gel electrophoresis (BN-PAGE),3 sucrose gradient centrifugation, and single particle analysis (6, 7). A recent study by Acin-Perez et al. (8) convincingly showed that supercomplexes are functional units capable of consuming oxygen when provided appropriate electron donors. They concluded that supercomplexes are in fact the functional respiratory unit of the mitochondrion in vivo. Schagger and Pfeiffer (9) demonstrated that supercomplex I·III·IV appears on blue native gels (BNGs) with increasing stoichiometries of complex IV. They termed these entities S0–S4, where the numeral denotes the number of complex IVs within the supercomplex.
Supercomplexes provide a logical explanation for combined deficiencies of the electron transport chain (ETC) observed in patients. It is estimated that ∼30% of all ETC disorders involve multiple complexes (10). Combined I·IV deficiencies (11–14), as well as multiple examples of combined I·III deficiencies have been reported (15–17). In most eukaryotes, complex III is composed of 11 subunits (18, 19), of which only cytochrome b is encoded by the mitochondrial genome. The catalytic core of complex III is comprised of cytochrome b, ISP (iron sulfur protein; Rieske protein), and cytochrome c1. Most of the mutations in cytochrome b cause isolated complex III deficiencies. However, there is a subset of cytochrome b mutations that cause combined I·III deficiencies, both in patients (15–17, 20) and in mammalian cell lines (21). In addition, loss of cytochrome c has been reported to lead to loss of both complexes I and IV in cell lines (22). From these findings, it has been suggested that fully assembled complex III, functional or not, is required for the assembly and stability of complex I. Notably, no structural mutations in ISP or cytochrome c1 have been reported in animals. However, complex III deficiencies in human patients are caused by mutations in BCS1L (23–25), a chaperone of ISP in the nascent complex III (26, 27). There are no reports that defects in complex III can affect the function of fully assembled complex I without decreasing the amount of complex I.
In Caenorhabditis elegans, two different complex III mutants confer intriguing phenotypes. isp-1 changes a highly conserved amino acid (P225S) residue in the head domain of ISP. Hekimi and co-workers (28) reported that isp-1 exhibits low oxygen consumption, decreased sensitivity to reactive oxygen species, extended lifespan, and delayed embryonic development. A mutation (ctb-1) in a complex III subunit cytochrome b, A170V, was discovered as a suppressor of a delayed development phenotype of isp-1. isp-1;ctb-1 shows an improvement in the rate of development compared with isp-1 (28). However, the molecular cause of this improved phenotype is unknown.
We show here that a structural defect of complex III, caused by a mutation in ISP, can reduce the amount of fully assembled complex I without a reduction in the amount of fully assembled complex III. Furthermore, we demonstrate that ctb-1 can decrease complex I function without decreasing either the amount of fully assembled complex I or the distribution of complex I in supercomplexes. In addition, we show that the improved whole animal phenotypes of isp-1;ctb-1 compared with isp-1 alone do not stem from improved complex III function. Rather salutary allosteric effects on complex I result from the introduction of a second mutated subunit into complex III. In addition, this double mutant heightens the weakened association of supercomplex I·II·IV such that the composition of supercomplexes is altered on BNGs. The detailed mechanisms of complex I and III interdependence seen in a model system indicate that supercomplexes play a major role in modulating intrinsic enzymatic activities of complexes I and III. The complexity of the interdependence between components of the ETC has important implications for the diagnosis of patients with defects in multiple complexes.
EXPERIMENTAL PROCEDURES
Nematode, Culture, and Mitochondrial Isolation
All strains were obtained from the Caenorhabditis Genetics Center. To isolate ctb-1, isp-1(qm150);ctb-1(qm189) hermaphrodites were crossed to N2 males. F1s were allowed to self-fertilize, and F2 animals were individually cultured. DNA sequencing of F3 animals from clones identified an isolated ctb-1 mutation with 100% homoplasmy. Worm preparation and mitochondrial isolation were performed using established protocols (29, 30).
Growth Rate
Adult nematodes laid eggs for 2–4 h on lawns of E. coli OP50 and were then removed. The day of hatching was defined as day 0 of life. The first day of adulthood was recorded by the appearance of eggs on the plate. All studies were done at 20 °C.
Egg Laying
Immediately after reaching adulthood, and before any eggs were laid, 25–30 adult nematodes were transferred to new plates and allowed to lay eggs. Worms were transferred to new plates every 24 h, and eggs were counted after adults were removed.
Polarography and ETC Assays
Oxygen consumption in intact mitochondria and mitochondrial enzyme complex activities were measured as described previously (31, 32).
BN-PAGE
BN-PAGE and in-gel activity (IGA) staining of respiratory complexes in BN-PAGE were performed as described previously (32). Supercomplexes from BN-PAGE were eletroeluted using tubes equipped with dialysis membranes with a molecular weight cut-off of 3.5 kDa (Gerard Biotech, Oxford, OH) and an electrode buffer as described (33). The bands from BNGs containing I·III and I·III·IV supercomplexes were excised from the gels and transferred to electroelution tubes that were then submerged in the electrophoresis apparatus containing electrode buffer. Electroelution was performed at 300 V at 4 °C. Eluent was collected from the tubes after 3 h and kept on ice for immediate assay of enzymatic activity.
Two-dimensional BN/hrCNE
Gel strips from BN-PAGE were subjected to two-dimensional BN-PAGE/high resolution clear native electrophoresis (BN/hrCNE) following a previously established protocol (34) with minor modifications. A vertical gel strip excised from a one-dimensional BNG was placed horizontally on the 3.5–11% native gradient polyacrylamide gel. The cathode buffer to run the second dimension contained 0.05% deoxycholate and 0.02% dodecyl maltoside. Electrophoresis was performed at 125 V at 4 °C for 15 h. Complex I IGA and complex IV IGA, Coomassie blue staining, and silver staining (Color Silver Stain Kit, Thermo Fisher Scientific) were performed as described previously (32).
Electroblotting of Native Proteins from BNGs (Native Western Blots)
Supercomplexes resolved by BNGs were transferred to PVDF membranes following as described previously (33). Membranes were blocked with 5% nonfat milk in phosphate-buffered saline/Tween 20 (0.05%) and incubated with monoclonal antibodies as described below.
Proteomic Analysis
ISP-A, -B, and -C from BNGs were analyzed by LC/MS using an LTQ Velos MS coupled to an Eksigent 1D-plus nano-LC. 20 μl of each in-gel tryptic digest of the BNG slices were injected onto a reverse phase column (Magic C18 200 A, Michrom Bioresources, Auburn, CA) and washed for 10 min in stationary phase at a flow rate of 3 μl/min (5% acetonitrile) prior to separation on a 10-cm reverse phase analytical column (HALO C18, Michrom Bioresources) at 300 nl/min over a 30-min gradient of either 10–50% or 10–70% acetonitrile. MS spectra were collected in data-dependent mode, with each full scan followed by MS scans of the 10 most intense ions. MS Peak lists were generated using Xcalibur software (Thermo Fisher Scientific) and ReAdW to produce peak lists in MzXML format. Peak lists were searched against a C. elegans proteome database with SPIRE (Systematic Protein Investigative Research Environment) using the search algorithm X!Tandem, fully tryptic enzyme specificity, and 2.5 Da mass accuracy. A 1% global false discovery rate was used as the basis for protein identification. Protein identifications were made using the methods and tools outlined in Ref. 35. Labels of other bands on BNGs (S0, I, III2, V, etc.) were applied from proteomic analysis of BNGs of wild type mitochondria that have been published previously (32). Different stoichiometries of the supercomplex I·III·IV (labeled S1–S4) were inferred from size on BNGs and corroborated with results shown in Fig. 5.
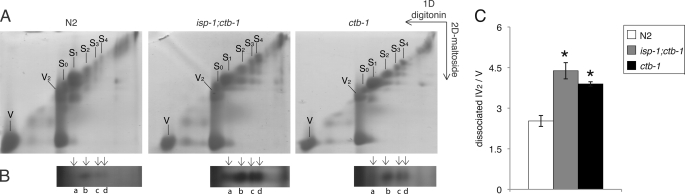
FIGURE 5.
The ability of two-dimensional BN/hrCNE to dissociate wild type and complex III mutant supercomplexes. A, Coomassie stain of the two-dimensional gel over the area of supercomplex dissociation. B, silver stain of the duplicates of the gels in A, which are cut from the 420-kDa region of the second dimension. This size is identical to that of IV2. C, the amount of complex IV2, measured by densitometry scanning of bands a–d in B, relative to the amount of Coomassie staining of monomeric complex V. Error bars represent mean ± S.E. from three independent experiments. An asterisk indicates statistical significance as p < 0.05 in comparison with wild type.
Western Blot Analysis
25 μg of mitochondrial protein were separated by 12% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with 5% nonfat milk in phosphate-buffered saline/Tween 20 (0.05%) and then incubated with the following monoclonal antibodies (Mitosciences, Eugene, OR): anti-complex I subunit NDUFS3 (NUO-2 in C. elegans) (MS112), anti-complex IV subunit I (MS 404), anti-cytochrome c (MSA06), and anti-adenosine nucleotide transporter (MSA02), anti-complex III subunits Rieske (MS305), and anti-Core 2 (both kind gifts from Mitosciences). Secondary antibodies were from Santa Cruz Biotechnology. Chemiluminescence substrate (SuperSignal West Pico, Thermo Fisher Scientific) was used to develop the reactions.
Quantification of Proteins/Protein Complexes
The amount of complex I in each genotype was quantified by densitometry scanning of complex I IGA staining of both digitonin-based and Triton X-100-based BNGs. These measurements were normalized to densitometry scans of Coomassie staining of total complex V. Quantification of complex IV was done by densitometry scanning of complex IV IGA in digitonin-based BNGs, also normalized to Coomassie staining of total complex V. Complex III was quantified from native Western blots of digitonin-based BNGs probed with anti-Rieske mAb then normalized to duplicate native Western blots probed for complex V. The amount of fully assembled complex III was the sum of optical density scanning of complex III in both supercomplexes and in dimeric (III2) forms. In all cases, at least three gels were scanned.
RESULTS
Phenotypes
All complex III mutants that hatched did so within 24 h of being laid. The subsequent rate of development, however, was severely affected. Wild type (N2) and ctb-1 reached adulthood at ∼3.5 days. isp-1 and isp-1;ctb-1 required 7.5 and 5 days to develop from eggs to adults, respectively. Specifically, L3 to L4 development lasted ∼12 h for N2 and ctb-1 and 24 h for isp-1 and isp-1;ctb-1. The development from L4 to adult in N2, isp-1;ctb-1, and ctb-1 was complete within 12 h, whereas isp-1 required another 24 h for L4 animals to develop into adults (supplemental Table S1).
Complex III mutations also affected the number of eggs laid, with isp-1 < isp-1;ctb-1 < ctb-1, approximately = N2 (supplemental Table S1). The maximum rate of egg laying was on the second day of adulthood in wild type, isp-1, and isp-1;ctb-1. For ctb-1, the maximum rate of egg laying was the third day of adulthood (supplemental Fig. S1).
Integrated Mitochondrial Respiration
The state 3 rates of both complex I- and II-dependent respiration of all complex III mutants were significantly decreased compared with that of wild type. Complex IV-dependent respiration rates of isp-1 and isp-1;ctb-1 were greater than wild type (supplemental Fig. S2). Overall, mitochondrial respiration in complex III mutants was consistent with a complex III deficiency.
Respiratory Enzyme Complex Activity
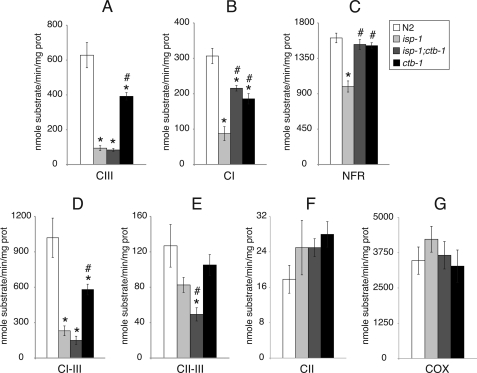
A large decrease in complex III (antimycin A-sensitive decylubiquinol·cytochrome c reductase) activity was observed in isp-1 (Fig. 1A), the mutant that presented the slowest rate of development. Surprisingly, this same decrease in complex III activity was also observed in isp-1;ctb-1, although this mutant had a much improved phenotype compared with isp-1. Complex III activity in ctb-1 was greater than isp-1 or isp-1;ctb-1 but was still significantly less than wild type. Compared with N2, all complex III mutants exhibited significantly decreased rotenone-sensitive complex I (NADH-decylubiquinone reductase) activities (Fig. 1B). However, complex I activities of both isp-1;ctb-1 and ctb-1 were significantly improved from that of isp-1. NADH-ferricyanide reductase activities were decreased only in isp-1, whereas isp-1;ctb-1 and ctb-1 had normal NADH-ferricyanide reductase activities (Fig. 1C). Complex I–III activities (Fig. 1D) were significantly lower in all complex III mutants than in wild type, consistent with a complex III defect. Complexes II–III activity of isp-1;ctb-1 was significantly lower than that of wild type (Fig. 1E). Complex II activities in complex III mutants were normal (Fig. 1F). Complex IV activities, although higher in absolute value, were not significantly different than those for N2 in any complex III mutants (Fig. 1G). In summary, the only improvement we measured in mitochondrial function in isp-1;ctb-1 compared with isp-1 was improved complex I function.
FIGURE 1.
Respiratory chain enzymatic activities of complex III mutants and wild type. A, enzymatic activity of complex III (CIII). B, enzymatic activity of complex I. C, enzymatic activity of NADH-ferricyanide reductase. D, enzymatic activity of complexes I–III. E, enzymatic activity of complexes II–III. F, enzymatic activity of complex II. G, enzymatic activities of complex IV (COX). Data are represented as mean ± S.E. from four to eight independent worm cultures. * and #, statistical significance as p < 0.05 in comparison with wild type and isp-1, respectively. CII, complex II; CI, complex I; prot, protein; NFR, NADH-ferricyanide reductase.
Amounts of Fully Assembled Respiratory Complexes
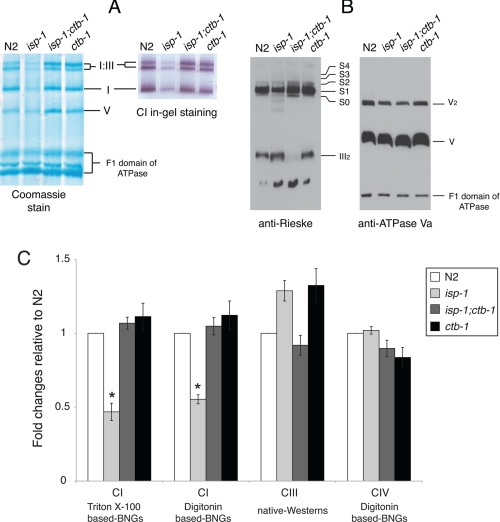
The amount of fully assembled complex I was determined from both Triton X-100-based and digitonin-based BNGs as shown in Figs. 2A and 3B, respectively (see “Experimental Procedures”). The amounts of complex I in supercomplexes were normalized to the total amount of complex V as a loading control and presented as fold changes relative to wild type. Among the complex III mutants, only isp-1 had a decreased amount of fully assembled complex I, which was 50% that of N2 (Fig. 2C).
FIGURE 2.
Respiratory enzyme complexes. A, Triton X-100-based BNGs demonstrate the fully assembled complex I in an isolated form (I) and in the supercomplex form (I:III). Complex I IGA is performed in a duplicate gel. B, native Western blot (native-Westerns) probed with anti-Rieske mAb (left) or with anti-ATPase subunit Va mAb (right, loading control in duplicate gel). S0, supercomplex I·III; S1, I·III·IV1; S2, I·III·IV2; S3, I·III·IV3; S4, I·III·IV4. C, quantitative analysis of the amount of fully assembled complexes I, III, and IV. Complexes I and IV were measured as densitometry scans of their IGAs on BNGs, complex III as densitometry scans of native Western blots. All values were normalized to complex V (see “Experimental Procedures”). Data are represented as mean ± S.E. from three to four independent experiments. An asterisk indicates statistical significance as p < 0.05 in comparison with wild type.
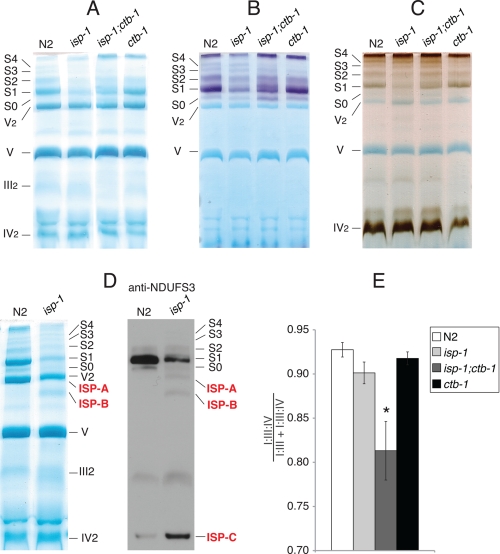
FIGURE 3.
Digitonin-based BNG-PAGE in complex III mutants. A, Coomassie stain. B, complex I IGA. C, complex IV IGA. D (left), the Coomassie stain of gel slices of N2 and isp-1 are compared side by side to reveal ISP-A and ISP-B, which appear only in isp-1. D (right), native Western blots performed using anti-NDUFS3 mAb to better visualize ISP-A, ISP-B, and ISP-C. E, the amounts of complex I in supercomplexes I·III (S0) and I·III·IV (S1–S4) were measured from four independent digitonin-based complex I IGAs and presented as the ratio between them. Error bars represent mean ± S.E. from three independent experiments. An asterisk indicates statistical significance as p < 0.05 in comparison with wild type.
Native Western blots of digitonin-based BNGs (n = 3) probed with anti-Rieske mAb were used to measure fully assembled complex III. (Representative blots are shown in Fig. 2B.) The amount of fully assembled complex III was the same in the mutants as in N2 (Fig. 2C). We observed another signal from the position of dimeric complex IV. This signal was not included in the quantification because its molecular weight indicates that it is a partially formed III2 co-migrating with dimeric complex IV. Identical results were obtained using an antibody to the complex III Core 2 protein (data not shown). In addition, we quantified the amount of fully assembled complex IV by densitometry scanning of in-gel activity of digitonin BNGs. Complex IV was not significantly increased in any of the mutants (Figs. 2C, and 3, A–C). Western blots from SDS-PAGE corroborated our results from native Western blots (supplemental Fig. S3 and Table S3).
Effects of isp-1 on Supercomplexes
isp-1 decreased amounts of both I·III and I·III·IV supercomplexes as seen on digitionin-based BNGs (Fig. 3, A–C). III2 appeared more diffuse in isp-1 than in N2 (Fig. 3, A and D), consistent with native Western blot analysis (Fig. 2B).
Coomassie staining and Western blots of digitonin-based BNGs revealed three unique bands in isp-1 that were not readily discernable in wild type mitochondria (Fig. 3D). These bands were labeled as ISP-A, ISP-B, and ISP-C. The components of ISP-A and ISP-B were identified by mass spectrometry (supplemental Table S2) and consisted of subunits of complexes I, III, and IV. In keeping with our proteomic data, ISP-A had no complex I IGA (Fig. 3B), suggesting that it lacks the N module of complex I (NADH-oxidase module, see (36) for a review). Similarly, ISP-B is an intermediate that consisted of most of the P module (proton pumping module, subcomplex Iβ, (see Ref. 36), and the Q module (coenzyme Q reduction module, (see Ref. 36) as determined by the presence of the NDUFS3 subunit (Fig. 3D, right panel). ISP-C was not characterized by mass spectrometry due to its close proximity to IV2 but was most likely the Q and P modules incorporated into a subcomplex, as determined by its size and the presence of NDUFS3 subunit (Fig. 3D, right panel).
Supercomplex Profiles in Complex III Mutants
Because decreased complex I ETC activities in isp-1;ctb-1 and ctb-1 are not due to decreased amounts of complex I, we asked whether changes in their supercomplex profiles might underlie a complex I deficiency. In mammalian mitochondria, complex I is much less active in the I·III form than in supercomplex I·III·IV (37). Identifications were inferred from our previous proteomic data for wild type and from in-gel activity staining for complexes I and IV (32). isp-1;ctb-1 displayed increased amounts of the I·III supercomplex and decreased amounts of the I·III·IV supercomplexes as determined by optical density scanning of digitonin-based BNGs (representative gels in Fig. 3, A and B, and quantification in E). In contrast, the supercomplex profile of ctb-1 was in every way identical to that of wild type (Fig. 3E).
Complex I Function from Electroeluted Supercomplexes
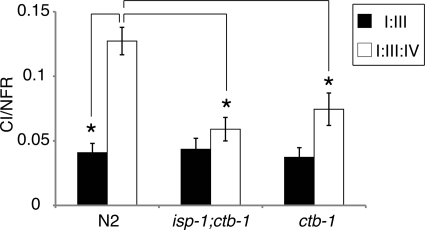
The decrease in complex I activity in ctb-1 was not explained by a decrease in either total complex I or in the ratio of complex I·III·IV to total complex I. However, as in mammalian mitochondria (37), after electroelution from BNGs, complex I in I·III·IV supercomplexes was significantly more active than complex I in the I·III supercomplex from N2. Complex I in the I·III supercomplex of isp-1;ctb-1 and ctb-1 was as active as that of wild type. Surprisingly however, complex I in I·III·IV supercomplexes from isp-1;ctb-1 or ctb-1 were significantly less active than in N2 (Fig. 4).
FIGURE 4.
Complex I activity in supercomplex I·III and I·III·IV. Electroeluted I·III and I·III·IV supercomplexes were assayed for complex I (CI) and NADH-ferricyanide reductase (NFR) activities. Data are represented as mean ± S.E. from four independent experiments. An asterisk indicates statistical significance as p < 0.05.
Stability of Supercomplexes
If components of the supercomplex are displaced because of altered subunits, the supercomplex might dissociate more easily than normal. Dodecyl maltoside dissociated the I·III·IV supercomplexes (S1–S4 differ by the number of complex IVs in the supercomplex) of isp-1;ctb-1 and ctb-1 from the one-dimensional gel (digitonin-treated) into smaller supercomplexes in the second dimension (Fig. 5A, middle and right panel). The original S4 from the one-dimensional gel was partially dissociated into three smaller supercomplexes whose molecular weights were comparable to the original S3, S2, and S1 that migrated from the one-dimensional gel. Similarly, S3, S2, and S1 were partially dissociated and gave the smaller supercomplexes. All dissociated supercomplexes stained positive for complex I by in-gel staining (data not shown). Silver staining visualized a band in the second dimension (Fig. 5B) at the size expected for dimeric complex IV, below the smaller, disassociated supercomplexes. In wild type mitochondria, dissociated supercomplexes and dimeric IV also appeared in the second dimension, but not to the extent of the mutants (Fig. 5, A and B). Quantitative measurement of the amount of complex IV2 (normalized to the amount of complex V) indicated a significant loss of complex IV from the I·III·IV supercomplexes in isp-1;ctb-1 and ctb-1, compared with the wild type (Fig. 5C).
DISCUSSION
We have shown that, in the nematode, primary changes in complex III can significantly affect the function of complex I. This can occur without decreasing the amount of complex III (isp-1) or the amount or the distribution of complex I in supercomplexes (ctb-1).
Originally, we hypothesized that in isp-1;ctb-1, ctb-1 would suppress the slow development of isp-1 (28) by restoring complex III activity. Surprisingly, compared with isp-1, complex III activity in isp-1;ctb-1 was not improved by the suppressor mutation ctb-1. Rather, complex I activity was improved, which was the only improvement in mitochondrial function that we could measure. Our results indicate that an allosteric interaction between complexes I and III is the basis of ctb-1 suppression of the severe developmental phenotype of isp-1.
Development from L3 to L4 and L4 to adult is accompanied by a dramatic increase in mitochondrial DNA content (38, 39), which has been linked to increased cellular energy expenditure (39). isp-1 and isp-1;ctb-1 spent twice the time in L3–L4 as did ctb-1 or wild type, implying that the mitochondrial deficiencies in isp-1 and isp-1;ctb-1 are each more severe than in ctb-1. These results were corroborated by decreased fecundity in isp-1 and isp-1;ctb-1. Interestingly, although ctb-1 developed at the same rate and laid a similar number of eggs as wild type, (supplemental Table 1), its maximum rate of egg laying was 1 day later than wild type. This was the only whole animal phenotype observed in ctb-1, which we interpret as a mild form of mitochondrial deficiency.
The isp-1 allele we studied is a serine substitution of a highly conserved proline, which is located in the head domain of ISP (18). This proline residue has been shown to be crucial for the secondary structure of the [2Fe-2S] cluster, for it creates an inward folding of the backbone of the Rieske head domain immediately preceding strand β5 (40). Such an amino acid change could negatively affect redox properties of the ISP by altering the position of the [2Fe-2S] residues and significantly impede complex III activity. Furthermore, a P146L substitution (a position that is equivalent to the isp-1 allele of the worm in our study) in Saccharomyces cerevisiae was found to alter the midpoint potential of the [2Fe-2S] cluster (41). Therefore, it was not unexpected to observe a large decrease in complex III activity of the isp-1 mutant. isp-1(qm150) is the first example of a defect in the ISP itself that affects complex III function in animals. All of the complex III deficiencies in animals, which are associated with the ISP, are reportedly caused by the mutations of BCS1L (23–25), a chaperone protein involved in ISP synthesis.
The ctb-1 mutation is a conservative alanine to valine substitution at the N-terminal of helix αD of cytochrome b that is adjacent to the docking site (helix αC, helix αcd1, and loop EF) of the Rieske head domain during electron transfer (18). The mutated alanine is not a highly conserved residue and may not significantly alter the conformation of cytochrome b. Thus, it is not surprising that the decrease in complex III activity in ctb-1 was not as dramatic as in isp-1. In addition, the locations of the ctb-1 and isp-1 mutations are not close (18, 19) and not predicted to directly interact. Therefore, it may not be surprising that the two mutations do not combine to repair complex III function. All complex III mutants had normal amounts of fully assembled complex III but decreased complex III activity, including the conservative change in ctb-1, which significantly affected complex III function.
Only isp-1 decreased the amount of fully assembled complex I, which undoubtedly led to its decreased complex I activity compared with N2. Complex I activity in isp-1;ctb-1 is decreased relative to N2 despite having normal amounts of complex I. However, we have shown that complex I is normally more active in the I·III·IV supercomplex than in the I·III form, as in bovine heart (37). Thus, in the double mutant, the decreased amount of supercomplex I·III·IV relative to supercomplex I·III is likely responsible for the decreased complex I activity, as is the case for complex IV mutations in C. elegans (32). It most likely represents a increase of the detrimental effect of ctb-1 on complex IV binding within the supercomplex (see below). Nevertheless, the increased amount of total complex I in the double mutant is sufficient to improve complex I function compared with isp-1 alone.
ctb-1 has decreased complex I activity despite a wild type distribution of supercomplex I·III and I·III·IV and a normal amount of complex I. Therefore, ctb-1 negatively affected complex I in its most active form, the I·III·IV supercomplex. The simplest explanation may be that this mutation changes the conformation of the supercomplex I·III·IV such that complex IV is less tightly bound to the supercomplex, reducing complex I activity toward enzymatic rates of its I·III form. If so, the increased proportion of supercomplex I·III relative to I·III·IV in isp-1;ctb-1 may be a reflection of this effect of ctb-1 in the double mutant and the reason that the activity in the double mutant was approximately the same as in ctb-1.
We hypothesized that if complex III altered interactions between components of the I·III·IV supercomplex in this manner, we could detect an increased vulnerability of the supercomplex to disassociation. Both complex III mutants lost complex IV from supercomplexes (S1–S4) more readily than the wild type. In addition, S4 was partially dissociated into smaller supercomplexes whose molecular weights were comparable to S3, S2, and S1 suggesting, as we originally inferred, that S4 was supercomplex I·III·IV4 containing four copies of complex IV. The wild type supercomplex resisted dissociation relative to either isp-1;ctb-1 and ctb-1, suggesting that these two mutations produce conformational changes that disrupt supercomplex integrity.
How can we link the finding that complex III mutations alter supercomplexes such that complex IV loosens its grip on supercomplex I·III·IV and decreases complex I activity? According to a current three-dimensional map of supercomplex I·III·IV, complex III and IV saddle the membranous arm of complex I (37) where the proton-pumping machinery of complex I is thought to reside (42, 43). NADH·coenzyme Q reductase in the matrix arm provides the driving force for proton pumping via conformational coupling (43–45). Mutations in the homologues of ND subunits in the membranous arm of complex I, which form a core of the proton-pumping machinery, result in decreased complex I activity but normal NADH-ferricyanide reductase activity (46–49). The relaxed interaction between complex IV and other components of the supercomplexes in the mutants may lead to an imperfect conformation and dysfunction of the proton-pumping module, which then impedes NADH·coenzyme Q reductase in the matrix arm. A similar logic may explain how complex I in wild type supercomplex I·III is not as active as complex I in wild type supercomplex I·III·IV.
As expected, complex III defects affected both complex I and complex II-dependent respiration; complex IV respiration was increased in all mutants, most dramatically in isp-1 and isp-1;ctb-1, even though the amount of complex IV appeared unchanged in these mutants. Cytochrome c oxidase controls mitochondrial energy metabolism (50, 51) by acting as a sensor of extramitochondrial ATP/ADP (52). A low ATP/ADP ratio, as is expected in the complex III mutants, may increase complex IV driven respiration. However, complex IV activity in ETC assays was not significantly increased in complex III mutants compared with wild type. Because the mitochondrial membranes are solubilized in ETC assays, ATP/ADP differences between strains are eliminated. A recent study by Yang and Hekimi (53) demonstrated decreased enzymatic activities of complexes I–III and complexes II–III in isp-1, although rates of complexes II–III were depressed more than complexes I–III. This is qualitatively different than our data. They did not measure complex III activity per se or complex I activity, and all rates were normalized to citrate synthase. It is difficult to compare techniques between our studies and those of Yang and Hekimi (53), so the differences between them are not fully interpretable.
We also identified three unique bands in the BNGs of isp-1 that may represent intermediates in complex I assembly or degradation. A current model of supercomplex assembly proposes that the subcomplex Iβ of complex I associates first with the partially assembled complex III and IV, leading to a module upon which the mature supercomplex forms (36). ISP-A represents a nearly complete subcomplex consisting of the P and Q modules already combined with ND4/ND5 intermediates and partially assembled complex III and IV. This suggests that the mutation in the ISP halted supercomplex maturation at the final step prior to incorporation of the NADH-oxidase module (N module). In turn, two smaller intermediates accumulate, ISP-B (an 830-kDa intermediate P/Q complex, with partially assembled complexes III and IV), and ISP-C (likely a 400-kDa intermediate consisting of the Q module and an early form of P, the membranous arm). Even though the bands we see correspond well to proposed assembly modules of complex I, we have not ruled out that they may represent steps in degradation of a supercomplex.
The physical association between complexes I and III is thought to involve ISP of complex III and the NDUFS2 and NDUFS4 subunits of the Q module of complex I. Missense mutations in those complex I subunit decreased levels of complex III (54). Because the mutation in isp-1(qm150) alters an amino acid in the highly mobile peripheral domain of the protein, it is difficult to envision how this might alter a complex I-III interaction. However, such a change could easily affect assembly or stability of the supercomplex resulting in misaligned complexes. The appearance of abnormal subcomplexes in the isp-1 BNGs supports this model.
It is intriguing that ctb-1 improves complex I function in an isp-1 background without affecting complex III function. The effects of complex III subunits on complex I represent an allosteric mechanism wherein subunits of the catalytic core can regulate supercomplex assembly (isp-1), ratios of supercomplexes (isp-1;ctb-1) or complex I electron flow in the face of normal amounts and profiles of supercomplexes (ctb-1). This is the first report of a complex III mutation that can affect complex I in this manner.
This study provides a new insight into the role of supercomplexes in ETC defects. Complexes I and III have intricate structural and allosteric interactions revealing a complicated inter-relatedness between different complexes of the mitochondrial respirasome (9, 15). Diagnosis of patients with mitochondrial disease, which is clearly problematic, must take into account the myriad of consequences in the ETC that may result from a single mutation. Clearly, a defect in a single subunit of the ETC can have wide ranging and unanticipated effects on mitochondrial function.
Supplementary Material
Acknowledgments
We thank Drs. Charles Hoppel and Sihoun Hahn for very helpful suggestions and discussions. We also thank Drs. Eugene Kolker and Andrew Bauman for assistance with the proteomic studies.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S3.
- BN-PAGE
- blue native polyacrylamide gel electrophoresis
- ETC
- electron transport chain
- IGA
- in-gel activity
- BNG
- blue native gel
- hrCNE
- high resolution clear native electrophoresis
- mAb
- monoclonal antibody
- ISP
- iron sulfur protein
- L3
- larval stage 3
- L4
- larval stage 4.
REFERENCES
- 1.Chance B., Williams G. R. (1955) Nature 176, 250–254 [DOI] [PubMed] [Google Scholar]
- 2.Bianchi C., Genova M. L., Parenti Castelli G., Lenaz G. (2004) J. Biol. Chem. 279, 36562–36569 [DOI] [PubMed] [Google Scholar]
- 3.Hackenbrock C. R., Chazotte B., Gupte S. S. (1986) J. Bioenerg. Biomembr. 18, 331–368 [DOI] [PubMed] [Google Scholar]
- 4.Kröger A., Klingenberg M. (1973) Eur. J. Biochem. 39, 313–323 [DOI] [PubMed] [Google Scholar]
- 5.Hatefi Y., Haavik A. G., Griffiths D. E. (1962) J. Biol. Chem. 237, 1676–1680 [PubMed] [Google Scholar]
- 6.Dudkina N. V., Eubel H., Keegstra W., Boekema E. J., Braun H. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schäfer E., Dencher N. A., Vonck J., Parcej D. N. (2007) Biochemistry 46, 12579–12585 [DOI] [PubMed] [Google Scholar]
- 8.Acín-Pérez R., Fernández-Silva P., Peleato M. L., Pérez-Martos A., Enriquez J. A. (2008) Mol. Cell 32, 529–539 [DOI] [PubMed] [Google Scholar]
- 9.Schägger H., Pfeiffer K. (2000) EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits P., Mattijssen S., Morava E., van den Brand M., van den Brandt F., Wijburg F., Pruijn G., Smeitink J., Nijtmans L., Rodenburg R., van den Heuvel L. (2010) Eur. J. Hum. Genet. 18, 324–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benecke R., Strümper P., Weiss H. (1993) Brain 116, 1451–1463 [DOI] [PubMed] [Google Scholar]
- 12.Brass E. P., Hiatt W. R., Gardner A. W., Hoppel C. L. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H603–609 [DOI] [PubMed] [Google Scholar]
- 13.Korenke G. C., Bentlage H. A., Ruitenbeek W., Sengers R. C., Sperl W., Trijbels J. M., Gabreels F. J., Wijburg F. A., Wiedermann V., Hanefeld F., et al. (1990) Eur. J. Pediatr. 150, 104–108 [DOI] [PubMed] [Google Scholar]
- 14.van Straaten H. L., van Tintelen J. P., Trijbels J. M., van den Heuvel L. P., Troost D., Rozemuller J. M., Duran M., de Vries L. S., Schuelke M., Barth P. G. (2005) Neuropediatrics 36, 193–199 [DOI] [PubMed] [Google Scholar]
- 15.Schägger H., de Coo R., Bauer M. F., Hofmann S., Godinot C., Brandt U. (2004) J. Biol. Chem. 279, 36349–36353 [DOI] [PubMed] [Google Scholar]
- 16.Budde S. M., van den Heuvel L. P., Janssen A. J., Smeets R. J., Buskens C. A., DeMeirleir L., Van Coster R., Baethmann M., Voit T., Trijbels J. M., Smeitink J. A. (2000) Biochem. Biophys. Res. Commun. 275, 63–68 [DOI] [PubMed] [Google Scholar]
- 17.Lamantea E., Carrara F., Mariotti C., Morandi L., Tiranti V., Zeviani M. (2002) Neuromuscul. Disord. 12, 49–52 [DOI] [PubMed] [Google Scholar]
- 18.Iwata S., Lee J. W., Okada K., Lee J. K., Iwata M., Rasmussen B., Link T. A., Ramaswamy S., Jap B. K. (1998) Science 281, 64–71 [DOI] [PubMed] [Google Scholar]
- 19.Xia D., Yu C. A., Kim H., Xia J. Z., Kachurin A. M., Zhang L., Yu L., Deisenhofer J. (1997) Science 277, 60–66 [DOI] [PubMed] [Google Scholar]
- 20.Blakely E. L., Mitchell A. L., Fisher N., Meunier B., Nijtmans L. G., Schaefer A. M., Jackson M. J., Turnbull D. M., Taylor R. W. (2005) FEBS J. 272, 3583–3592 [DOI] [PubMed] [Google Scholar]
- 21.Acín-Pérez R., Bayona-Bafaluy M. P., Fernández-Silva P., Moreno-Loshuertos R., Pérez-Martos A., Bruno C., Moraes C. T., Enríquez J. A. (2004) Mol. Cell 13, 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vempati U. D., Han X., Moraes C. T. (2009) J. Biol. Chem. 284, 4383–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Vizarra E., Bugiani M., Goffrini P., Carrara F., Farina L., Procopio E., Donati A., Uziel G., Ferrero I., Zeviani M. (2007) Hum. Mol. Genet. 16, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 24.de Lonlay P., Valnot I., Barrientos A., Gorbatyuk M., Tzagoloff A., Taanman J. W., Benayoun E., Chrétien D., Kadhom N., Lombès A., de Baulny H. O., Niaudet P., Munnich A., Rustin P., Rötig A. (2001) Nat. Genet. 29, 57–60 [DOI] [PubMed] [Google Scholar]
- 25.De Meirleir L., Seneca S., Damis E., Sepulchre B., Hoorens A., Gerlo E., García Silva M. T., Hernandez E. M., Lissens W., Van Coster R. (2003) Am. J. Med. Genet. A. 121A, 126–131 [DOI] [PubMed] [Google Scholar]
- 26.Cruciat C. M., Hell K., Fölsch H., Neupert W., Stuart R. A. (1999) EMBO J. 18, 5226–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobrega F. G., Nobrega M. P., Tzagoloff A. (1992) EMBO J. 11, 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J., Bussière F., Hekimi S. (2001) Dev. Cell 1, 633–644 [DOI] [PubMed] [Google Scholar]
- 29.Kayser E. B., Sedensky M. M., Morgan P. G. (2004) Mech. Ageing Dev. 125, 455–464 [DOI] [PubMed] [Google Scholar]
- 30.Kayser E. B., Morgan P. G., Hoppel C. L., Sedensky M. M. (2001) J. Biol. Chem. 276, 20551–20558 [DOI] [PubMed] [Google Scholar]
- 31.Hoppel C., DiMarco J. P., Tandler B. (1979) J. Biol. Chem. 254, 4164–4170 [PubMed] [Google Scholar]
- 32.Suthammarak W., Yang Y. Y., Morgan P. G., Sedensky M. M. (2009) J. Biol. Chem. 284, 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 34.Wittig I., Karas M., Schägger H. (2007) Mol. Cell Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 35.Higdon R., Hogan J. M., Van Belle G., Kolker E. (2005) OMICS 9, 364–379 [DOI] [PubMed] [Google Scholar]
- 36.Lazarou M., Thorburn D. R., Ryan M. T., McKenzie M. (2009) Biochim. Biophys. Acta. 1793, 78–88 [DOI] [PubMed] [Google Scholar]
- 37.Schäfer E., Seelert H., Reifschneider N. H., Krause F., Dencher N. A., Vonck J. (2006) J. Biol. Chem. 281, 15370–15375 [DOI] [PubMed] [Google Scholar]
- 38.Bratic I., Hench J., Henriksson J., Antebi A., Bürglin T. R., Trifunovic A. (2009) Nucleic Acids Res. 37, 1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang W. Y., Lemire B. D. (2002) Biochem. Biophys. Res. Commun. 291, 8–16 [DOI] [PubMed] [Google Scholar]
- 40.Iwata S., Saynovits M., Link T. A., Michel H. (1996) Structure 4, 567–579 [DOI] [PubMed] [Google Scholar]
- 41.Gatti D. L., Meinhardt S. W., Ohnishi T., Tzagoloff A. (1989) J. Mol. Biol. 205, 421–435 [DOI] [PubMed] [Google Scholar]
- 42.Brandt U. (1997) Biochim. Biophys. Acta. 1318, 79–91 [DOI] [PubMed] [Google Scholar]
- 43.Efremov R. G., Baradaran R., Sazanov L. A. (2010) Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 44.Friedrich T. (2001) J. Bioenerg. Biomembr. 33, 169–177 [DOI] [PubMed] [Google Scholar]
- 45.Brandt U. (2006) Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 46.Kao M. C., Di Bernardo S., Nakamaru-Ogiso E., Miyoshi H., Matsuno-Yagi A., Yagi T. (2005) Biochemistry 44, 3562–3571 [DOI] [PubMed] [Google Scholar]
- 47.Kao M. C., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2005) Biochemistry 44, 9545–9554 [DOI] [PubMed] [Google Scholar]
- 48.Torres-Bacete J., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2007) J. Biol. Chem. 282, 36914–36922 [DOI] [PubMed] [Google Scholar]
- 49.Kao M. C., Di Bernardo S., Perego M., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2004) J. Biol. Chem. 279, 32360–32366 [DOI] [PubMed] [Google Scholar]
- 50.Kadenbach B., Frank V., Rieger T., Napiwotzki J. (1997) Mol. Cell Biochem. 174, 131–135 [PubMed] [Google Scholar]
- 51.Napiwotzki J., Kadenbach B. (1998) Biol. Chem. 379, 335–339 [DOI] [PubMed] [Google Scholar]
- 52.Arnold S., Goglia F., Kadenbach B. (1998) Eur. J. Biochem. 252, 325–330 [DOI] [PubMed] [Google Scholar]
- 53.Yang W., Hekimi S. (2010) Aging Cell 9, 433–447 [DOI] [PubMed] [Google Scholar]
- 54.Ugalde C., Janssen R. J., van den Heuvel L. P., Smeitink J. A., Nijtmans L. G. (2004) Hum. Mol. Genet. 13, 659–667 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.