Abstract
α-Synuclein (a-Syn) is a major component of fibrillar aggregates in Lewy bodies (LBs), a characteristic hallmark of Parkinson disease. Almost 90% of a-Syn deposited in LBs is phosphorylated at Ser-129. However, the role of Ser-129-phosphorylated a-Syn in the biogenesis of LBs remains unclear. Here, we investigated the metabolism of Ser-129-phosphorylated a-Syn. In SH-SY5Y cells, inhibition of protein phosphatase 2A/1 by okadaic acid, and inhibition of the proteasome pathway by MG132 or lactacystin accumulated Ser-129-phosphorylated a-Syn. However, these inhibitions did not alter the amounts of total a-Syn within the observation time. Inhibition of the autophagy-lysosome pathway by 3-methyladenine or chloroquine accumulated Ser-129-phosphorylated a-Syn in parallel to total a-Syn during longer incubations. Experiments using cycloheximide showed that Ser-129-phosphorylated a-Syn diminished rapidly (t½ = 54.9 ± 6.4 min), in contrast to the stably expressed total a-Syn. The short half-life of Ser-129-phosphorylated a-Syn was blocked by MG132 to a greater extent than okadaic acid. In rat primary cortical neurons, either MG132, lactacystin, or okadaic acid accumulated Ser-129-phosphorylated a-Syn. Additionally, we did not find that phosphorylated a-Syn was ubiquitinated in the presence of proteasome inhibitors. These data show that Ser-129-phosphorylated a-Syn is targeted to the proteasome pathway in a ubiquitin-independent manner, in addition to undergoing dephosphorylation. The proteasome pathway may play a role in the biogenesis of Ser-129-phosphorylated a-Syn-rich LBs.
Keywords: Parkinson Disease, Proteasome, Protein Degradation, Protein Phosphorylation, Synuclein, Ubiquitination
Introduction
Sporadic Parkinson disease (sPD)3 is characterized pathologically by a loss of dopaminergic neurons in the substantia nigra pars compacta and the presence of intracytoplasmic inclusions called Lewy bodies (LBs) and Lewy neurites (LNs) in surviving neurons. α-Synuclein (a-Syn) is a major component of fibrillar aggregates in LBs and LNs. Accumulating lines of evidence have shown that prefibrillar intermediates of a-Syn, such as soluble oligomers or protofibrils, play a toxic role in degeneration of dopaminergic neurons, and mature fibrils of a-Syn contribute toward this toxicity to a lesser extent (1–4). Therefore, the process of a-Syn aggregation eventually forming LBs is proposed to play a causative role in neuronal degeneration of PD (5, 6). Immunohistochemical and biochemical studies have revealed that ∼90% of a-Syn deposited in LBs is phosphorylated at serine 129 (Ser-129) (7, 8). In contrast, the portion of phosphorylated a-Syn in normal brains is known to be only about 4% (7) or less than the limits of quantification of the assays used (8). This discrepancy implicates a pathogenic role of Ser-129-phosphorylated a-Syn in the biogenesis of LBs (7, 9). One possibility is that the Ser-129-phosphorylation promotes the aggregation-prone property of a-Syn. To elucidate this issue, several in vitro studies have been performed. However, the accelerating effect of phosphorylation on fibril formation of a-Syn is controversial at present (7, 10). Another possibility is that the impairment of the system to degrade Ser-129-phosphorylated a-Syn causes its accumulation. However, the process by which Ser-129-phosphorylated a-Syn is degraded or recycled remains unknown. This study focused on the metabolic fate of Ser-129-phosphorylated a-Syn in cells. We report here that Ser-129-phosphorylated a-Syn undergoes dephosphorylation and degradation by the proteasome pathway. In addition, Ser-129-phosphorylated a-Syn is targeted to the proteasome pathway in a ubiquitin-independent manner.
EXPERIMENTAL PROCEDURES
Plasmid cDNA Construction and Reagents
Wild-type human a-Syn cDNA was described previously (11). S129A, S129E, S129D mutant a-Syn cDNAs were made by the two step PCR mutagenesis method. S9A/S42A/S87A mutant (it abolished Ser residues other than Ser-129) and K12R/K21R/K23R mutant a-Syn cDNAs (it abolished Lys residues for ubiquitination) (8) were generated by applying the two-step PCR mutagenesis method. Human ubiquitin cDNAs with or without a FLAG tag at the N terminus were generated by PCR (clone ID 3879581; Open Biosystems, Huntsville, AL), and they were subcloned into the pcDNA3.1 vector (Invitrogen, Carlsbad, CA). Nucleotide sequences of all constructs were confirmed by direct sequencing. All reagents were purchased from Sigma unless otherwise stated.
Cell Culture and Transfection
Human dopaminergic neuroblastoma SH-SY5Y cells (ECACC 94030304) were maintained in a mixture of F-12 and Eagle's minimum essential medium supplemented with 15% fetal bovine serum (Invitrogen), 1× non-essential amino acids, and 2 mm l-glutamine (Invitrogen) at 37 °C in 5% CO2. The SH-SY5Y cell line stably expressing wild-type a-Syn (wt-aS/SH) was selected against with 1 mg/ml G418 (Invitrogen). For transient transfection, 5 × 106 cells were transfected with 6 μg of cDNA using Nucleofector (Amaxa Cell Line Nucleofector kit V; Lonza Cologne AG, Koln, Germany). The cells were harvested at 48 h post-transfection.
Primary Neuronal Cultures
Primary cortical neuron cultures were prepared from Crl:CD (SD) rats as previously described (11). Briefly, neurons were isolated from the neocortex of embryonic day 18 rats and dissociated cells were plated at a density of 1 × 106 cells on poly-d-lysine-coated 6-well plates (Becton Dickinson, Bedford, MA). Neurons were maintained in serum-free medium, which was composed of neurobasal medium supplemented with B27 and GlutaMAX (Invitrogen). At intervals of 2 days, half of the plating medium was renewed. At 21 days of culture, neurons were harvested for experiments (12).
Chemical Treatments
For inhibition of the proteasome, at 16 h after plating wt-aS/SH cells onto 6-well plates, we checked the cells to be ∼80% confluent, and then the cells were further incubated in fresh medium containing either 10 nm okadaic acid (OA), 10 μm MG132, or 10 μm lactacystin for 4 h. As a vehicle control, cells were treated with the same concentration of DMSO, which was used for dissolving OA, MG132, and lactacystin, or phosphate buffered saline (PBS: 1.06 mm KH2PO4, 2.97 mm Na2HPO4·7H2O, 150 mm NaCl). In rat primary neuronal cultures, neurons were cultured for 21 days and then incubated in fresh medium containing either 10 nm OA, 10 μm MG132, or 10 μm lactacystin for 4 h. For inhibition of the autophagy-lysosome, at 16 h post-plating wt-aS/SH cells onto 6-well plates, we confirmed the cells to be around 50% confluent. The cells were incubated in fresh medium containing 10 mm 3-metyladenine (3-MA) or 100 μm chloroquine for up to 32 h.
To assess protein half-lives in the cells, we performed experiments using the de novo protein synthesis inhibitor, cycloheximide (CHX). At 16 h post-plating wt-aS/SH cells onto 6-well plates, we confirmed the cells to be ∼80% confluent. The cells were incubated in fresh medium containing 100 μm CHX for the indicated times. To test the effect of inhibition of the proteasome pathway or dephosphorylation on the half-lives of target proteins, we treated the cells with CHX plus either MG132 or OA. The 80% confluent wt-aS/SH cells were pre-incubated in fresh medium containing either DMSO, 10 μm MG132, or 10 nm OA for 6 h. After preincubation, CHX was added to a final concentration of 100 μm into medium. The cells were further incubated for the indicated times.
Protein Extract Preparation
For preparation of cell lysates, SH-SY5Y cells were suspended in buffer A (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 10% glycerol, 1 × protease inhibitor mixture (Roche Diagnostic, Mannheim, Germany), 1 mm EDTA, 1 × phosSTOP (Roche Diagnostic)) and kept on ice for 30 min. After centrifugation at 12,000 × g for 30 min, the resultant supernatant was collected and stored at −80 °C until required. In primary neuronal cultures, the cells were suspended in buffer A containing 1 μm OA, and then they were disrupted by passing through a 27-gauge needle 10 times.
In the experiments for ubiquitinated proteins, the pellet from above (post-12,000 × g centrifugation step) was resuspended in the same aliquots of buffer A containing 8 m urea and was disrupted by brief sonication. After centrifugation at 12,000 × g for 30 min, the resultant supernatant was collected as the pellet fraction. The protein concentration was measured by the BCA assay (Thermo Scientific, Rockford, IL).
Immunoprecipitation
The cells were suspended in ice-cold lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholic acid, 10% glycerol, 1× protease inhibitor mixture, 1 mm EDTA, 20 mm NaF, 1 mm Na3VO4, 1 μm OA), and kept on ice for 30 min. After centrifugation at 12,000 × g for 30 min, the resultant supernatant was collected. The supernatants were incubated with primary antibodies overnight at 4 °C, and then incubated with Protein G-agarose beads for 2 h. Beads were washed three times with ice-cold lysis buffer, and immunoprecipitates were dissolved from the beads by heating in Laemmli's sample buffer. Equivalent amounts of samples were analyzed by immunoblotting.
Immunoblotting
For SDS-PAGE, protein samples were denatured at 95 °C for 5 min in Laemmli's sample buffer containing 2.5% 2-mercaptoethanol. Samples were applied to a 12.5% SDS-polyacrylamide gel, electrophoresed and then transferred to a PVDF membrane (Millipore, Billerica, MA). After blocking with 5% skim milk in Tris-buffered saline (pH 7.4) containing 0.05% Tween-20 (TBS-T) for 1 h at room temperature, the membrane was incubated with primary antibodies overnight at 4 °C followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Lab., Inc., West Grove, PA) for 1 h at room temperature or overnight at 4 °C. The membrane was reacted with ECL (GE Healthcare, Uppsala, Sweden) for the detection of β-actin, G-protein-coupled receptor kinase (GRK) 2, and total a-Syn, including phosphorylated and non-phosphorylated forms, or ECL plus (GE Healthcare) for the detection of other proteins. The membrane was then visualized using a CCD camera, VersaDog 5000 (Bio-Rad). Relative intensities of detected signals were quantified with Quantity one software (Bio-Rad). For detection of phosphorylated a-Syn, we added 50 mm NaF into TBS-T containing 5% milk or antibodies.
The following primary antibodies were used (11, 13): monoclonal anti-a-Syn antibody (Syn-1, it recognizes total a-Syn including phosphorylated and non-phosphorylated forms, 1:4,000; BD Transduction Laboratories, Franklin Lakes, NJ), monoclonal anti-Ser-129-phosphorylated a-Syn antibody (psyn 64, 1:5,000; Wako, Osaka, Japan), monoclonal anti-β-actin antibody (AC-15, 1:10,000; Sigma), polyclonal anti-GRK2 antibody (sc-562, 1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA), polyclonal anti-GRK3 antibody (sc-563, 1:1,000; Santa Cruz Biotechnology), monoclonal anti-GRK5 antibody (139, 1:5,000) (13), polyclonal anti-GRK6 antibody (sc-566, 1:1,000; Santa Cruz Biotechnology), polyclonal anti-casein kinase (CK) 2α′ antibody (sc-648, 1:1,000; Santa Cruz Biotechnology), and polyclonal anti-ubiquitin antibody (1:1,000, Dako, Glostrup, Denmark).
Quantification of Band Intensities
In experiments for estimating the protein half-lives, we quantified the relative band intensities of phosphorylated or total a-Syn by using protein standards. Recombinant non-phosphorylated a-Syn proteins were purified from Escherichia coli as described previously (14). Phosphorylated a-Syn proteins were made by incubation of 100 μg of non-phosphorylated proteins in reaction buffer (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 200 μm ATP) containing 1,000 units of recombinant CK2 protein (New England Biolabs, Beverly, MA) for 16 h at 37 °C. A set of diluted non-phosphorylated or phosphorylated a-Syn proteins was subjected to SDS-PAGE along with samples. After quantifying band intensities of samples with Quantity one software, they were plotted on a standard curve and corrected relative intensities. Statistical comparisons were made by unpaired Student's t test.
Pulse-chase Metabolic Labeling Experiments
Wt-aS/SH cells were rinsed with PBS and incubated with methionine/cystine-free medium for 1 h. The cells were pulsed with methionine/cystine-free medium containing 100 μCi/ml of [35S]methionine/cysteine (PerkinElmer Life Sciences) for 2 h, and subsequently chased with normal medium containing 1 mm methionine and cysteine for indicated times. After harvesting cells, immunoprecipitation using Syn-1 antibody or anti-Ser-129-phosphorylated a-Syn antibody was carried out. Immunoprecipitates were subjected to 12.5% gels and transferred to the PVDF membrane. Signals were detected by BAS-2000 image analyzer (Fuji Photo Film Co.).
Immunocytochemistry
Wt-aS/SH cells transfected with ubiquitin cDNA were plated on 4-chamber slides (Nunc, Rochester, NY) at 1 × 105 cells per well. After 2 days, the cells were incubated with medium containing either DMSO, 10 μm MG132, or 10 nm OA for 16 h, fixed with 4% paraformaldehyde for 15 min at room temperature and incubated with PBS containing 0.2% Triton X-100 for 5 min. After blocking with 5% skim milk, they were labeled with anti-a-Syn monoclonal antibody (LB509, 1:250; Covance, Emeryville, CA) and anti-ubiquitin polyclonal antibody (1:50) overnight at 4 °C. Then, the cells were incubated with Alexa Fluor 488 anti-mouse IgG and Alexa Fluor 568 anti-rabbit IgG (Invitrogen) for 2 h at 37C°. The slides were analyzed with a LSM510 meta laser confocal microscope (Zeiss, Jena, Germany).
RESULTS
The Effect of Protein Phosphatase 2A/1 Inhibitor on the Levels of Ser-129-phosphorylated a-Syn in SH-SY5Y Cells
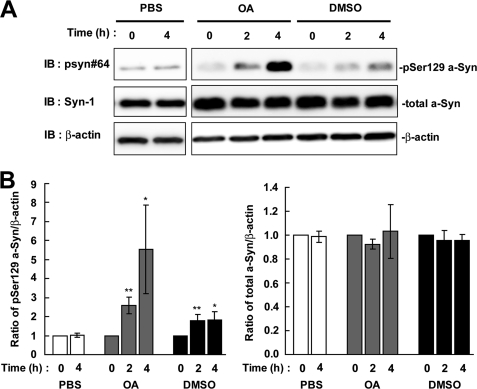
The metabolic fate of a phosphorylated protein is affected by dephosphorylation and/or degradation pathways, such as the proteasome or autophagy-lysosome pathway. Fujiwara et al. (7) has reported that Ser-129-phosphorylated a-Syn protein underwent dephosphorylation. This idea is supported by the findings showing that the protein phosphatase 2A/1 inhibitor, OA, increases the amount of phosphorylated a-Syn in PC12 cells (15) and HEK293 cells (11). To compare the contribution of dephosphorylation to the metabolism of Ser-129-phosphorylated a-Syn with that of degradation pathways, we first assessed the effect of OA on the metabolism of Ser-129-phosphorylated a-Syn. In this study, we used a cell line (wt-aS/SH) stably expressing wild-type a-Syn, because it is difficult to detect Ser-129-phosphorylated a-Syn at endogenous levels in SH-SY5Y cells. As compared with the starting levels, the expression levels of Ser-129-phosphorylated a-Syn were increased 2.60 ± 0.43-fold at 2 h (mean ± S.D., p < 0.01, n = 4) and 5.53 ± 2.32-fold at 4 h (p = 0.03, n = 4) in the presence of 10 nm OA (Fig. 1). In contrast, the expression levels of total a-Syn, including non-phosphorylated and phosphorylated forms, were constant at 0.92 ± 0.04-fold at 2 h (p = 0.125, n = 3) and 1.03 ± 0.23-fold at 4 h (p = 0.861, n = 3), when compared with starting levels (Fig. 1). Unexpectedly, the expression levels of Ser-129-phosphorylated a-Syn were moderately but significantly increased 1.82 ± 0.44-fold at 4 h in the presence of 0.1% DMSO (p = 0.014, n = 5) (Fig. 1). When the cells were incubated in medium containing 0.1% PBS for 4 h, the levels of Ser-129-phosphorylated a-Syn were constant (Fig. 1). The increased effect of OA on the levels of phosphorylated a-Syn was larger than that of DMSO at 4 h of post-treatment (5.53 ± 2.32-fold in OA versus 1.82 ± 0.44-fold in DMSO) (Fig. 1).
FIGURE 1.
The effect of protein phosphatase 2A/1 inhibitor on the levels of Ser-129-phosphorylated a-Syn in SH-SY5Y cells. SH-SY5Y cells stably expressing wild-type a-Syn (wt-aS/SH) were incubated in medium containing either 0.1% PBS, 0.1% DMSO or 10 nm OA for 4 h. A, cell lysates (20 μg/lane) were loaded on SDS-PAGE and analyzed by immunoblotting (IB) with anti-Ser-129-phosphorylated a-Syn antibody (psyn 64) and anti-total a-Syn antibody (Syn-1). For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Representative blots are shown. B, quantitative analysis of the alteration in the expression levels of Ser-129-phosphorylated a-Syn in cells in the absence or presence of OA. Relative ratios of the band intensity of Ser-129-phosphorylated a-Syn to β-actin and total a-Syn to β-actin are shown. Relative ratio was normalized to the starting material just before adding reagents. Data represent means ± S.D. and the p values (*, p < 0.05; **, p < 0.01) were estimated by unpaired Student's t test.
The Effect of Proteasome Inhibitors on the Levels of Ser-129-phosphorylated a-Syn in SH-SY5Y Cells
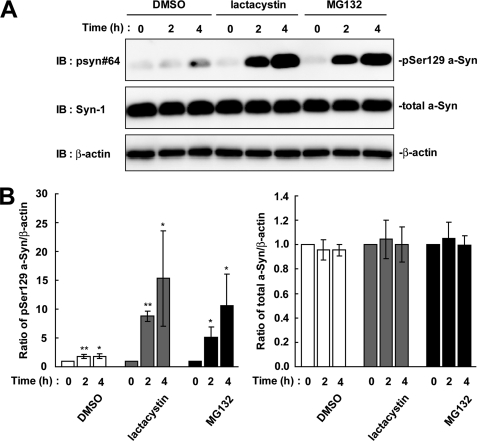
We tested whether Ser-129-phosphorylated a-Syn was degraded by the proteasome pathway, because phosphorylation sometimes acts as a signal to target the proteins, such as IκBα (16) and β-catenin (17), to the ubiquitin-proteasome pathway. We treated wt-aS/SH cells with the proteasome inhibitor, MG132, for 4 h. The expression levels of Ser-129-phosphorylated a-Syn were increased 5.07 ± 1.86-fold at 2 h (p = 0.022, n = 4) and 10.6 ± 5.46-fold at 4 h (p = 0.039, n = 4), as compared with the starting levels (Fig. 2). Treatment with another proteasome inhibitor, lactacystin, also showed a 15.3 ± 8.24-fold increase in the levels of Ser-129-phosphorylated a-Syn at 4 h (p = 0.04, n = 4) (Fig. 2). In contrast, the expression levels of total a-Syn were constant at 1.00 ± 0.08-fold in MG132 (p = 0.929, n = 4) and 1.00 ± 0.14-fold in lactacystin (p = 0.986, n = 4) at 4 h (Fig. 2). The increased effect of proteasome inhibitors on the levels of phosphorylated a-Syn was much larger than that of DMSO at 4 h post-treatment (15.3 ± 8.24-fold in lactacystin versus 1.82 ± 0.44-fold in DMSO) (Fig. 2).
FIGURE 2.
The effect of proteasome inhibitors on the levels of Ser-129-phosphorylated a-Syn in wt-aS/SH cells. The cells were incubated in medium containing either 0.1% DMSO, 10 μm lactacystin, or 10 μm MG132 for 4 h. A. The cell lysates (20 μg/lane) were analyzed by immunoblotting (IB) with psyn 64 and Syn-1 antibodies. For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Representative blots are shown. B, quantitative analysis of the alteration in the expression levels of Ser-129-phosphorylated a-Syn in cells in the absence or presence of proteasome inhibitors. Relative ratios of the band intensity of Ser-129-phosphorylated a-Syn to β-actin and total a-Syn to β-actin are shown. Relative ratio was normalized to the starting material. Data represent means ± S.D. and the p values (*, p < 0.05 and **, p < 0.01) are shown.
The Effect of Autophagy-Lysosome Inhibitors on the Levels of Ser-129-phosphorylated a-Syn in SH-SY5Y Cells
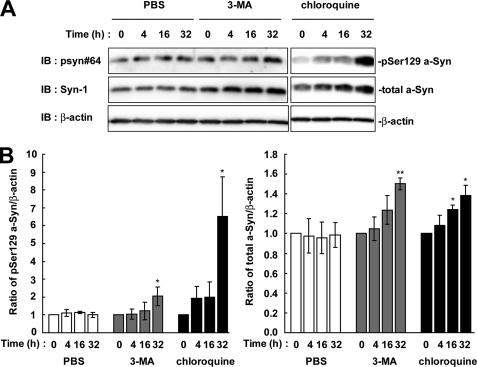
We next tested whether Ser-129-phosphorylated a-Syn was degraded by the autophagy-lysosome pathway. As compared with the starting levels, the expression levels of Ser-129-phosphorylated a-Syn were increased 2.05 ± 0.52-fold at 32 h in the presence of 10 mm 3-MA, which inhibits the formation of the autophagosome (p = 0.027, n = 3) (Fig. 3). The expression levels of total a-Syn were also increased 1.5 ± 0.06-fold at 32 h (p = 0.005, n = 3) (Fig. 3). Additionally, treatment with 100 μm of general lysosomal inhibitor, chloroquine, showed that the expression levels of Ser-129-phosphorylated a-Syn were 6.52 ± 2.20-fold higher than the starting levels at 32 h (p = 0.049, n = 3) (Fig. 3). The expression levels of total a-Syn were found to be increased 1.39 ± 0.10-fold at 32 h (p = 0.022, n = 3) (Fig. 3). When the cells were treated with 0.1% PBS as a vehicle control, the levels of Ser-129-phosphorylated and total a-Syn were constant. Inhibition of the autophagy-lysosome pathway showed that the expression levels of Ser-129-phosphorylated a-Syn were elevated almost in parallel to those of total a-Syn by longer incubations than inhibition of the proteasome pathway.
FIGURE 3.
The effect of autophagy-lysosome inhibitors on the levels of Ser-129-phosphorylated a-Syn in wt-aS/SH cells. The cells were incubated in medium containing either 0.1% PBS, 10 mm 3-MA or 100 μm chloroquine for 32 h. A, cell lysates (20 μg/lane) were analyzed by immunoblotting (IB) with psyn 64 and Syn-1 antibodies. For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Experiments were performed three times. Representative blots are shown. B, quantitative analysis of the alteration in the expression levels of Ser-129-phosphorylated a-Syn in cells in the absence or presence of autophagy-lysosome inhibitors. Relative ratios of the band intensity of Ser-129-phosphorylated a-Syn to β-actin and total a-Syn to β-actin are shown. Relative ratio was normalized to the starting material. Data represent means ± S.D. and the p values (*, p < 0.05; **, p < 0.01) are shown.
Role of Dephosphorylation and Proteasomal Degradation in the Metabolism of Ser-129-phosphorylated a-Syn in SH-SY5Y Cells
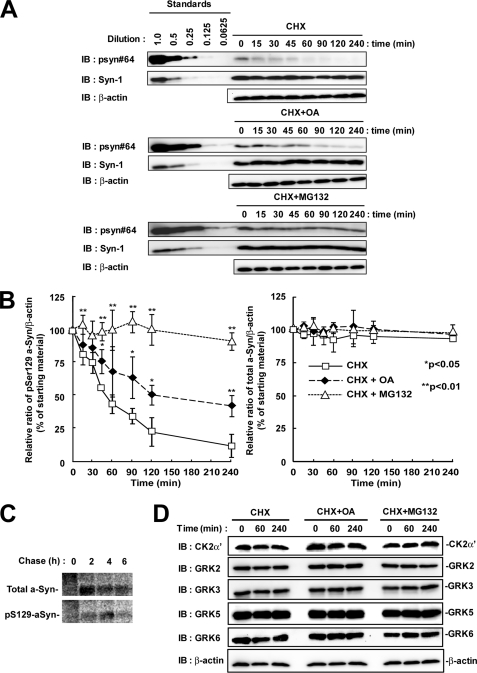
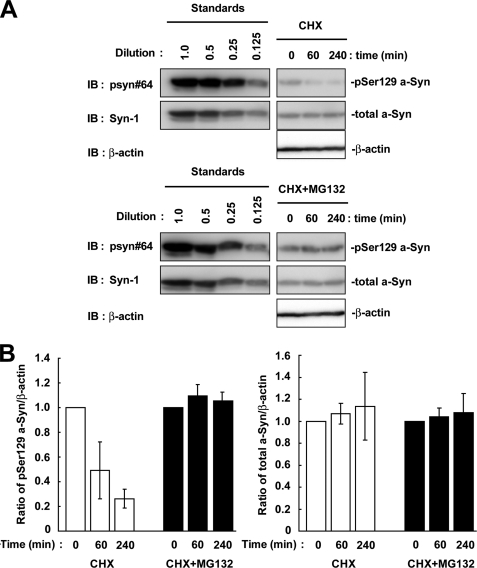
To elucidate the contribution of dephosphorylation and degradation pathways to the metabolism of Ser-129-phosphorylated a-Syn, we determined the half-life of Ser-129-phosphorylated a-Syn by using the de novo protein synthesis inhibitor, CHX, in wt-aS/SH cells. As shown in Fig. 4, A and B, the experiment using CHX showed that the amounts of Ser-129-phosphorylated a-Syn were rapidly decreased. Its estimated half-life (t½) was 54.9 ± 6.4 min. In contrast, the expression levels of total a-Syn were stable within the observation time of up to 240 min (Fig. 4, A and B). Because this finding suggested that Ser-129-phosphorylated a-Syn was rapidly processed by the specific pathway that differed from the non-phosphorylated form, we then focused on the role of dephosphorylation and degradation by the proteasome pathway. We treated wt-aS/SH cells with CHX plus either OA or MG132. To ensure that OA or MG132 exerts its inhibitory effect during the entire course of the experiment, we pre-treated the cells with the reagents until accumulation of Ser-129-phosphorylated a-Syn was easily detectable. Under these conditions, treatment with OA suppressed the decrease in the amounts of Ser-129-phosphorylated a-Syn (Fig. 4, A and B). The amounts of Ser-129-phosphorylated a-Syn were decreased to about 50% at 240 min in the presence of OA (Fig. 4, A and B). Treatment with MG132 remarkably blocked the decrease in the amounts of Ser-129-phosphorylated a-Syn (Fig. 4, A and B). The amounts of Ser-129-phosphorylated a-Syn were almost constant in the presence of MG132 (Fig. 4, A and B). The amounts of total a-Syn did not change in the presence of OA or MG132 (Fig. 4, A and B). These data suggested that Ser-129-phosphorylated a-Syn underwent degradation mainly by the proteasome pathway rather than dephosphorylation in SH-SY5Y cells. We performed pulse-chase experiments after metabolic labeling. Ser-129-phosphorylated a-Syn was decreased between 4 and 6 h of the chase periods, while total a-Syn was almost stable in the chase period (Fig. 4C). This finding was consistent with the data from experiments using CHX.
FIGURE 4.
The effect of protein phosphatase 2A/1 and proteasome inhibitors on the half-life of Ser-129-phosphorylated a-Syn in SH-SY5Y cells. The cells were pre-incubated in medium containing either 0.1% DMSO, 10 nm OA, or 10 μm MG132 for 6 h. Then, the chase experiment was started by adding 100 μm CHX. The cells were collected at the indicated times. A, cell lysates (20 μg/lane) were analyzed by immunoblotting (IB) with psyn 64 and Syn-1. For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Experiments were performed three times. Representative blots are shown. B, quantitative analysis of the alteration in the expression levels of Ser-129-phosphorylated and total a-Syn in the chase experiments using CHX. Relative band intensities of Ser-129-phosphorylated a-Syn and total a-Syn were corrected by plotting them on the standard curves, as described in the Experimental procedures. Relative ratios of the band intensity of Ser-129-phosphorylated a-Syn to β-actin and total a-Syn to β-actin are shown by normalizing to the starting materials as percentage. Data represent means ± S.D. and the p values (*, p < 0.05; **, p < 0.01) are shown. C, cells were pulsed with 100 μCi/ml of [35S]methionine/cysteine for 2 h, and subsequently chased for indicated times. Immunoprecipitation using Syn-1 antibody or anti-Ser-129-phosphorylated a-Syn antibody was carried out. D, expression levels of the members of G-protein-coupled receptor kinase (GRK) family and casein kinase (CK) 2 in experiments using CHX. The cell lysates (10 μg/lane) were analyzed by immunoblotting with CK2 α′ subunit antibody and antibodies against each member of GRK family (GRK2, GRK3, GRK5, and GRK6). Experiments were performed four times.
To exclude a possibility that the decrease in Ser-129-phosphorylated a-Syn was due to degradation of kinases, we examined whether treatment with CHX altered the expression levels of GRKs and CK2, which have been known to contribute to the Ser-129-phosphorylation of a-Syn (11, 13, 15, 18, 19). The expression levels of ubiquitously expressing members of GRKs (GRK2, -3, -5, and -6) and CK2 α′-subunit were stable in the presence of CHX and in the presence of CHX plus either OA or MG132 during the observation times (Fig. 4D).
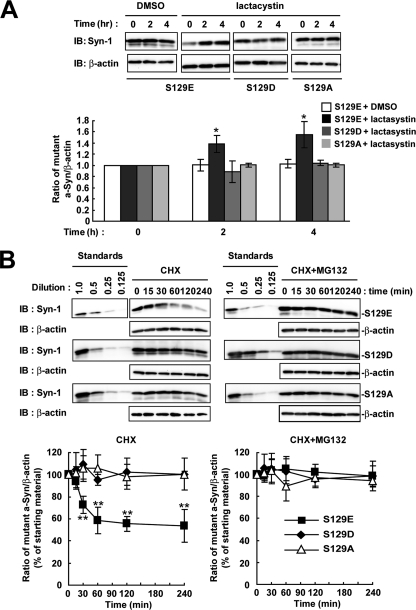
We next assessed the metabolism of phosphorylation-mimic mutants (S129E and S129D) and phosphorylation-abolished mutant (S129A) of a-Syn. Each mutant cDNA was transfected into SH-SY5Y cells, and the cells were treated with lactacystin for 4 h. The expression levels of S129A mutant a-Syn were stable at 4 h, as compared with the starting levels (Fig. 5A). In phosphorylation-mimic mutants, the expression levels of S129E mutant a-Syn were significantly increased 1.39 ± 0.15-fold at 2 h (p = 0.014, n = 4) and 1.55 ± 0.23-fold at 4 h (p = 0.016, n = 4), as compared with the starting levels, whereas S129D mutant a-Syn did not alter the expression levels (Fig. 5A). In the experiments using CHX, S129A and S129D mutants were not decreased within 4 h (Fig. 5B). On the other hand, S129E mutant a-Syn showed a rapid decrease (Fig. 5B). Treatment with MG132 suppressed the rapid decrease in the levels of S129E mutant a-Syn (Fig. 5B). The data of S129A and S129E mutants supported that Ser-129-phosphorylation played a role in the degradation of a-Syn by the proteasome pathway. However, S129D mutant a-Syn did not reproduce the metabolic fate of Ser-129-phosphorylated form.
FIGURE 5.
The metabolism of phosphorylation-mimic and phosphorylation-abolished mutant a-Syn in SH-SY5Y cells. We used S129E and S129D as phosphorylation-mimic mutants, and S129A as phosphorylation-abolished mutant of a-Syn. A, cells were transiently transfected with one of the a-Syn mutant cDNAs, and treated with or without 10 μm lactacystin for 4 h. The cell lysates (10 μg/lane) were analyzed by immunoblotting (IB) with Syn-1 antibodies or anti-β-actin antibody. The graph shows relative ratios of the band intensity of mutant a-Syn to β-actin by normalizing to the starting materials. B, at 42 h after transfection, the cells were pre-incubated in medium containing either 0.1% DMSO or 10 μm MG132 for 6 h. Then, 100 μm CHX was added into medium. The cells were collected at the indicated times. The cell lysates (10 μg/lane) were analyzed by immunoblotting (IB) with Syn-1 antibody or anti-β-actin antibody. Experiments were performed three times. Graphs show quantitative analysis of the alteration in the expression levels of mutant a-Syn. Relative ratios of the band intensity of mutant a-Syn to β-actin are shown by normalizing to the starting materials. Data represent means ± S.D. and the p values (*, p < 0.05; **, p < 0.01) are shown.
To test whether the phosphorylation of other Ser residues contributed to the metabolism of Ser-129-phosphorylated a-Syn, we made S9A/S42A/S87A mutant a-Syn cDNA, whose product abolished the possible Ser-phosphorylation sites except for Ser-129. In the experiment using CHX, Ser-129-phosphorylated form of this mutant a-Syn was rapidly decreased (Fig. 6). The expression levels of total a-Syn in the mutant were stable (Fig. 6). Treatment with MG132 suppressed the rapid decrease in the levels of Ser-129-phosphorylated form of the mutant a-Syn (Fig. 6).
FIGURE 6.
The effect of Ser-9, Ser-42, and Ser-87 residues on the metabolism of Ser-129-phosphorylated a-Syn in SH-SY5Y cells. The cells were transiently transfected with S9A/S42A/S87A mutant a-Syn cDNA. At 42 h after transfection, the cells were pre-incubated in medium containing either 0.1% DMSO or 10 μm MG132 for 6 h. Then, 100 μm CHX was added into medium. The cells were collected at the indicated times. A, cell lysates (10 μg/lane) were analyzed by immunoblotting (IB) with psyn 64 and Syn-1 antibodies. For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Experiments were performed three times. Although the blots of standards are separated from those of samples, these blots are originally derived from the same blot. B, quantitative analysis of the alteration in the expression levels of Ser-129-phosphorylated and total a-Syn in experiments using CHX. Relative ratios of the band intensity of Ser-129-phosphorylated a-Syn to β-actin and total a-Syn to β-actin are shown by normalizing to the starting materials. Data represent means ± S.D.
Effects of Dephosphorylation and the Proteasome Pathway on the Metabolism of Ser-129-phosphorylated a-Syn in Rat Primary Cortical Neurons
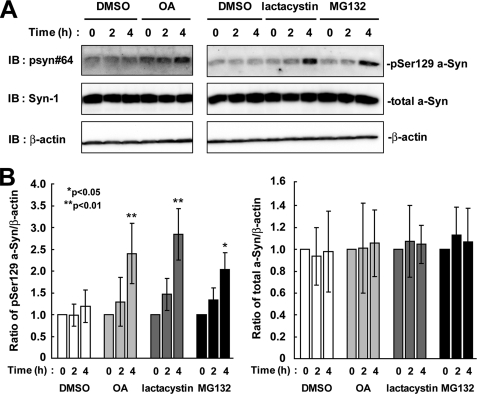
To assess whether or not overexpression of a-Syn artificially targeted Ser-129-phosphorylated a-Syn toward degradation by the proteasome pathway, we investigated the effect of the proteasome inhibitors on the endogenous a-Syn protein in rat primary cortical neurons. When 21 day cultured neurons were incubated in the presence of proteasome inhibitors for 4 h, the expression levels of Ser-129-phosphorylated a-Syn were increased 2.84 ± 0.59-fold in the presence of lactacystin (p = 0.008, n = 4) and 2.03 ± 0.39-fold in the presence of MG132 (p = 0.013, n = 4), as compared with starting levels (Fig. 7). The expression levels of total a-Syn were constant in the presence of lactacystin (1.04 ± 0.17-fold, p = 0.637, n = 4) and MG132 (1.07 ± 0.30-fold, p = 0.730, n = 4) (Fig. 7). Additionally, when the primary neurons were treated with OA, the expression levels of Ser-129-phosphorylated a-Syn were increased 2.40 ± 0.69-fold (p = 0.007, n = 4), as compared with starting levels (Fig. 7). Although treatment with DMSO showed a tendency to elevate the levels of phosphorylated a-Syn, there was no significant difference between 0 and 4 h. The data demonstrated that Ser-129-phosphorylated a-Syn underwent degradation by the proteasome pathway and dephosphorylation at the endogenous level in neurons.
FIGURE 7.
The effects of protein phosphatase 2A/1 and proteasome inhibitors on the levels of Ser-129-phosphorylated a-Syn in rat primary cortical neurons. A, 21-day cultured primary neurons were incubated in fresh medium containing either 0.1% DMSO, 10 μm MG132, 10 μm lactacystin, or 10 nm OA for 4 h. The cell lysates (50 μg/lane) were loaded on SDS-PAGE and analyzed by immunoblotting (IB) with psyn 64 and Syn-1 antibodies. For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Experiments were performed four times. Representative blots are shown. B, quantitative analysis of the alteration in the expression levels of Ser-129-phosphorylated a-Syn in the cells. Relative ratios of the band intensity of Ser-129-phosphorylated a-Syn to β-actin and total a-Syn to β-actin are shown by normalizing to the starting material. Data represent means ± S.D., and the p values (*, p < 0.05; **, p < 0.01) are shown.
Role of Ubiquitination in the Degradation of Ser-129-phosphorylated a-Syn by the Proteasome Pathway
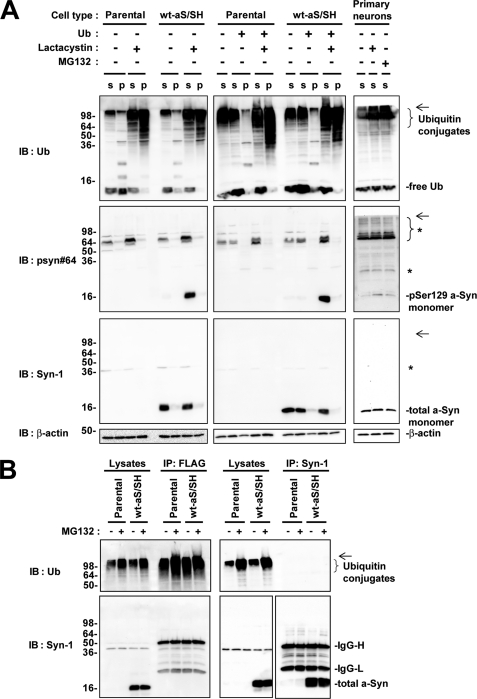
To assess whether polyubiquitination plays a role in targeting Ser-129-phosphorylated a-Syn to the proteasome pathway, we investigated wt-aS/SH cells and parental SH-SY5Y cells in the absence or presence of lactacystin for 16 h. Immunoblotting of post-12,000 × g centrifuged supernatant fractions with anti-ubiquitin antibody showed that polyubiquitin conjugates were increased in the presence of lactacystin in both wt-aS/SH and parental cells (Fig. 8A). In post-12,000 × g centrifuged pellet fractions, polyubiquitin conjugates were also increased in the presence of lactacystin in these cells (Fig. 8A). These findings indicated that lactacystin treatment effectively blocked protein degradation by the proteasome pathway, resulting in accumulation of polyubiquitinated conjugates in the cells. However, there was no difference in the pattern of ubiquitin-positive bands between wt-aS/SH cells and parental cells in both supernatant and pellet fractions (Fig. 8A). In supernatant fractions of lactacystin-treated wt-aS/SH cells, immunoblotting with anti-Ser-129-phosphorylated a-Syn antibody showed that monomeric Ser-129-phosphorylated a-Syn was increased; however, there was no appearance of the specific bands migrating at a position of higher molecular weight than its monomer (Fig. 8A). Although we analyzed the pellet fractions of wt-aS/SH cells, the specific bands corresponding to polyubiquitinated Ser-129-phosphorylated a-Syn were not detectable (Fig. 8A). We also found no specific band in immunoblotting with anti-total a-Syn antibody (Fig. 8A). We next investigated wt-aS/SH cells transiently overexpressing ubiquitin. Although polyubiquitinated conjugates were clearly increased in supernatant and pellet fractions by treatment with lactacystin, there was no appearance of the specific bands corresponding to polyubiquitinated a-Syn in the immunoblots of these fractions with anti-ubiquitin, anti-Ser-129 phosphorylated a-Syn or anti-total a-Syn antibody (Fig. 8A). Additionally, we did not find polyubiquitinated a-Syn in rat primary cortical neurons in the presence of MG132 or lactacystin, as compared with the neurons in the absence of inhibitor (Fig. 8A).
FIGURE 8.
Immunoblotting analysis of ubiquitination and proteasomal degradation of Ser-129-phosphorylated a-Syn in SH-SY5Y cells and rat primary cortical neurons. A, immunoblotting (IB) analysis of ubiquitination of a-Syn in SH-SY5Y cells and primary cortical neurons. Parental SH-SY5Y cells and wt-aS/SH cells were transfected with or without ubiquitin cDNA and they were incubated in medium containing either 0.1% DMSO or 10 μm lactacystin for 16 h. In SH-SY5Y cells, the collected cells were fractionated to supernatant (S) and pellet (P) fractions. Rat primary cortical neurons were incubated in medium either 0.1% DMSO, 10 μm lactacystin, or 10 μm MG132 for 4 h, and collected supernatant (S) fractions. Samples (20 μg/lane) were loaded on SDS-PAGE and analyzed by immunoblotting with anti-ubiquitin, psyn 64 and Syn-1 antibodies. For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Left and middle panels show the immunoblots of SH-SY5Y cells without overexpression of ubiquitin and SH-SY5Y cells transiently overexpressing ubiquitin, respectively. Right panels show the immunoblots of rat primary cortical neurons. * indicates nonspecific bands. The arrows indicate the interface of resolving and stacking gels. B, immunoprecipitation (IP) analysis of ubiquitination of a-Syn in SH-SY5Y cells. Parental SH-SY5Y cells or wt-aS/SH cells were transfected with FLAG-tagged ubiquitin cDNA and incubated in the presence or absence of MG132 for 4 h. Cell lysates were subjected to immunoprecipitation with anti-FLAG (left panels) and Syn-1 antibodies (right panels). Equivalent amounts of immunoprecipitated products were analyzed by immunoblotting with anti-ubiquitin (upper panels) and Syn-1 antibodies (lower panels). The arrows indicate the interface of resolving and stacking gels.
We further examined ubiquitination of a-Syn by immunoprecipitation of wt-aS/SH and parental cells transfected with FLAG-tagged ubiquitin cDNA (Fig. 8B). In products immunoprecipitated with anti-total a-Syn antibody, we detected the a-Syn monomer by immunoblotting with anti-total a-Syn antibody (Fig. 8B). However, there was no signal showing polyubiquitination of a-Syn in the immunoprecipitated products of wt-aS/SH cells in the absence or presence of MG132 (Fig. 8B). Additionally, in products immunoprecipitated with anti-FLAG antibody, we detected polyubiquitinated conjugates by immunoblotting with anti-ubiquitin antibody (Fig. 8B). However, there was no signal showing polyubiquitination of a-Syn in the immunoprecipitated products of wt-aS/SH cells in the absence or presence of MG132 (Fig. 8B).
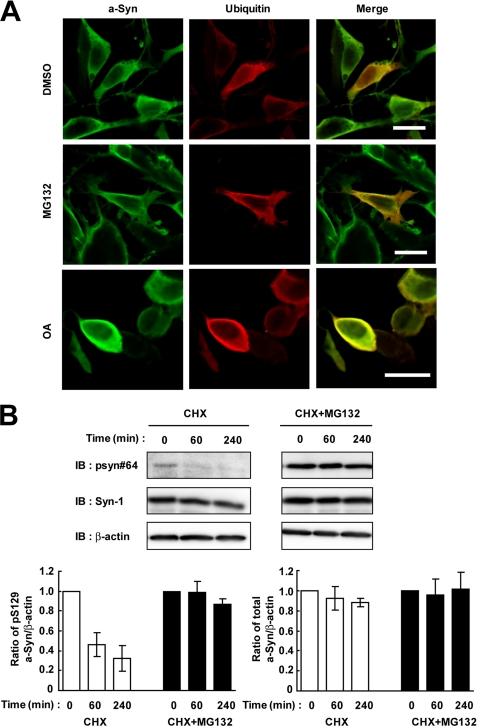
To clarify whether the present biochemical fractionation experiments failed to detect insoluble ubiquitinated a-Syn by the aggregate formation, we performed immunocytochemical analysis using wt-aS/SH cells. The cells were transiently transfected with ubiquitin cDNA and treated with 10 μm MG132 for 16 h. Overexpressed a-Syn and ubiquitin proteins were diffusely distributed in the cytoplasm in the absence or presence of MG132 (Fig. 9A). The formation of a-Syn- or ubiquitin-positive inclusions was not found in the presence of MG132 (Fig. 9A). These findings were consistent with the biochemical fractionation data that there were no ubiquitinated a-Syn proteins in the pellet fractions under the present condition. Also, the formation of a-Syn- or ubiquitin-positive inclusions was not detected in the presence of OA (Fig. 9A).
FIGURE 9.
Immunocytochemical and biochemical analysis of ubiquitination and proteasomal degradation of Ser-129-phosphorylated a-Syn in SH-SY5Y cells. A, immunocytochemical analysis of a-Syn and ubiquitin in proteasomal inhibitor-treated SH-SY5Y cells. Wt-aS/SH cells were transfected with ubiquitin cDNA, and they were incubated with medium containing either 0.1% DMSO, 10 μm MG132, or 10 nm OA for 16 h. Immunostainings of a-Syn (green, left panels) and ubiquitin (red, middle panels), and merged images (left panels) are shown. Scale bars represent 20 μm. B, role of Lys-12, Lys-21, and Lys-23 residues in proteasomal degradation of Ser-129-phosphorylated a-Syn in cells. The cells were transiently transfected with K12R/K21R/K23R mutant a-Syn cDNA. At 42 h after transfection, the cells were pre-incubated in medium containing either 0.1% DMSO or 10 μm MG132 for 6 h. Then, 100 μm CHX was added into medium. The cells were collected at the indicated times. The cell lysates (10 μg/lane) were analyzed by immunoblotting (IB) with psyn 64 and Syn-1 antibodies. For loading control, the same amounts of samples were immunoblotted with anti-β-actin antibody. Experiments were performed three times. The graphs show the alteration in the expression levels of Ser-129-phosphorylated and total a-Syn. Relative ratios of the band intensity of Ser-129-phosphorylated a-Syn to β-actin and total a-Syn to β-actin are shown by normalizing to the starting materials. Data represent means ± S.D.
To elucidate whether ubiquitination of lysine residues of a-Syn contributed to the metabolism of Ser-129-phosphorylated a-Syn, we made a K12R/K21R/K23R mutant a-Syn cDNA, whose product abolished the previously reported ubiquitination sites of a-Syn (8). In the experiment using CHX, Ser-129-phosphorylated form of this mutant a-Syn was rapidly decreased (Fig. 9B). The expression levels of total a-Syn in the mutant were constant (Fig. 9B). Treatment with MG132 suppressed the rapid decrease in the levels of Ser-129-phosphorylated form of the mutant a-Syn (Fig. 9B).
DISCUSSION
The present data demonstrated that Ser-129-phosphorylated a-Syn underwent dephosphorylation and degradation. In SH-SY5Y cells stably expressing a-Syn, inhibition of the proteasome pathway or the autophagy-lysosome pathway resulted in the accumulation of Ser-129-phosphorylated a-Syn. However, the inhibitory effect of the proteasome pathway was different in two points from that of the autophagy-lysosome pathway. First, inhibition of the proteasome pathway did not accompany the alteration in the levels of total a-Syn. Second, inhibition of the proteasome pathway increased the levels of Ser-129-phosphorylated a-Syn faster than that of the autophagy-lysosome pathway. To elucidate these differences, we assessed the half-life of Ser-129-phosphorylated a-Syn using CHX. The result showed that the half-life of Ser-129-phosphorylated a-Syn was much shorter than that of total a-Syn. Inhibition of the proteasome pathway remarkably prolonged the short half-life of Ser-129-phosphorylated a-Syn with no alteration in that of total a-Syn. In rat primary cortical neurons, inhibition of the proteasome pathway also accumulated phosphorylated a-Syn at endogenous levels. These findings suggest that Ser-129-phosphorylated a-Syn specifically undergoes degradation by the proteasome pathway, and that the portion of Ser-129-phosphorylated a-Syn is too small to affect the levels of total a-Syn. On the other hand, contribution of the autophagy-lysosome pathway to degradation of phosphorylated a-Syn remains to be elucidated. One may speculate that the autophagy-lysosome pathway selectively degrades non-phosphorylated a-Syn. The increase in the levels of phosphorylated a-Syn may be a consequence of accumulation of non-phosphorylated proteins in the cytosol due to inhibition of the autophagy-lysosome pathway. Alternatively, the autophagy-lysosome pathway may degrade both non-phosphorylated and phosphorylated a-Syn. In this case, the portion of phosphorylated a-Syn is estimated to be very small, because this corresponds to phosphorylated a-Syn, which remains undiminished in the present experiments using CHX.
Since Bennett et al. (20) initially reported that inhibition of the proteasome pathway led to the accumulation of overexpressed a-Syn in SH-SY5Y cells, this finding has also been shown in more recent studies (21–24). However, other studies have failed to detect accumulation of endogenous or overexpressed a-Syn by inhibition of the proteasome pathway (25–28). In contrast, a-Syn is reported to accumulate in cells by inhibition of the autophagy-lysosome pathway (29–31). At present, the involvement of the proteasome pathway or the autophagy-lysosome pathway in degradation of a-Syn is still debated. In general, proteins with short half-lives are mostly degraded by the proteasome pathway, whereas most cytosolic proteins with long half-lives (> 10 h) are degraded by the lysosome pathway (30). In contrast to this principle, a previous study utilized long incubation times over 48 h to detect accumulation of a-Syn by inhibition of the proteasome pathway (21). Additionally, treatment with a selective proteasome inhibitor, epoxomicin, only showed a 2.3-h increase in the half-life of a-Syn (16.8 ± 2 h) in rat ventral midbrain cultures (30). The findings suggest that a portion of a-Syn, which is targeted to the proteasome pathway, is small. These studies may have seen accumulation of Ser-129-phosphorylated a-Syn by inhibition of the proteasome pathway. Also, the present data were consistent with the finding that a-Syn was degraded by the autophagy-lysosome pathway. Ser-129-phosphorylation may be a key for resolving whether the metabolism of a-Syn fulfills this general principle.
In the present study, we investigated the role of Ser residues in targeting a-Syn to the proteasome pathway. a-Syn has four Ser residues and is known to be phosphorylated at Ser-87, as well as at Ser-129 (15). To assess the effect of Ser residues other than Ser-129 on proteasomal degradation, we constructed the S9A/S42A/S87A mutant of a-Syn. In experiments using CHX, overexpressed mutant a-Syn demonstrated a rapid decrease in the levels of the Ser-129-phosphorylated form and the stable expression of total protein. The rapid decrease in the levels of the Ser-129-phosphorylated form was inhibited by MG132. These findings were similar to wild-type a-Syn, suggesting that Ser-129 may play a central role in targeting the protein to the proteasome pathway. However, this study could not exclude a possibility that targeting of a-Syn to the proteasome pathway depends on the phosphorylation of other residues, because a-Syn is also known to be phosphorylated at three tyrosine residues.
We also assessed the metabolism of phosphorylation-mimic (S129E and S129D) and phosphorylation-abolished (S129A) mutants of a-Syn. S129A mutant a-Syn did not show the degradation through the proteasome pathway. In contrast, S129E mutant a-Syn was degraded by the proteasome pathway. These findings were consistent with the proteasomal degradation of Ser-129-phosphorylated a-Syn. However, S129D mutant a-Syn was not targeted to the proteasome pathway. This finding indicated that substitution of aspartic acid for Ser-129 did not mimic the phosphorylated residue. The previous work reported that phosphorylation-mimic mutants of a-Syn did not reproduce the effect of the phosphorylation on the structural properties of a-Syn in vitro (10). One may speculate that conformation changes induced by the phosphate group, rather than negative charge, are responsible for the effect of phosphorylation on the metabolism of a-Syn. Further studies are required to elucidate the determinants that lead to the difference in the metabolism between S129E and S129D mutants of a-Syn.
It is well established that conjugation of at least four ubiquitins on a protein is necessary for ubiquitin-dependent degradation to occur (32). However, previous studies have shown that ubiquitination of unmodified a-Syn does not occur after inhibition of the proteasome in transfected cells (20, 27, 28). In vitro experiments using purified recombinant proteins have shown that unmodified a-Syn is degraded by the 20 S (21, 23, 24) and 26 S proteasomes (23). These findings indicate that a-Syn is degraded by the proteasome pathway in a ubiquitin-independent manner (21). Although the present data support the previous findings, we propose an idea that Ser-129-phosphorylated a-Syn is targeted to the proteasome pathway in a ubiquitin-independent manner. What is the role of Ser-129-phosphorylation in the proteasomal degradation of a-Syn? The physiological function of a-Syn is thought to require its association with lipid vesicles where it adopts an α-helical conformation from a natively unstructured one (33). Structural studies have shown that a-Syn dynamically binds to vesicles and is promoted to dissociate from the vesicle by mutation (34), oxidative stress (35) and Ser-129-phosphorylation (18). Liu et al. (24) reported that 20 S proteasome degraded free, unstructured a-Syn, but not vesicle-bound, α-helical a-Syn. The Ser-129-phosphorylation may trigger the dissociation of a-Syn proteins from vesicles and accumulate free, unstructured proteins in the cytosol, resulting in targeting them to the proteasome pathway.
In summary, our data suggest that Ser-129-phosphorylation plays a role in the rapid degradation of a-Syn by targeting the protein to the proteasome pathway. Bedford et al. reported that depletion of the 26 S proteasome in mouse neurons caused extensive neurodegeneration in the nigrostriatal pathway and the formation of LB-like inclusions containing a-Syn (36). This study strongly indicates that 26 S proteasome dysfunction in neurons is involved in aggregation of a-Syn (36). Ser-129-phosphorylation might provide a clue for linking the degradation pathway of a-Syn with the formation of LBs. However, the present study does not resolve the question why Ser-129-phosphorylated a-Syn proteins deposited in LBs are also ubiquitinated (9, 37). Our data suggest that phosphorylated a-Syn proteins undergo ubiquitination in the pathway independent of its physiological degradation. Ubiquitination might represent an unsuccessful “last-ditch stand” of cells in their attempt to unfold and/or degrade misfolded proteins in a disease-specific pathway (37). Alternatively, ubiquitination of a-Syn might occur after polymerization of the molecule (38). Further studies are required to clarify whether the proteasome dysfunction accumulates Ser-129-phosphorylated a-Syn selectively, resulting in generation of LBs, which abundantly contain Ser-129-phosphorylated a-Syn.
This work was supported in part by a Grant-in-Aid from the Global COE Program (F03) of the Japan Society for the Promotion of Science (to T. K.).
- sPD
- sporadic Parkinson disease
- CHX
- cycloheximide
- a-Syn
- α-synuclein
- OA
- okadaic acid
- 3-MA
- 3-methyladenine.
REFERENCES
- 1.Volles M. J., Lee S. J., Rochet J. C., Shtilerman M. D., Ding T. T., Kessler J. C., Lansbury P. T., Jr. (2001) Biochemistry 40, 7812–7819 [DOI] [PubMed] [Google Scholar]
- 2.Xu J., Kao S. Y., Lee F. J., Song W., Jin L. W., Yankner B. A. (2002) Nat. Med. 8, 600–606 [DOI] [PubMed] [Google Scholar]
- 3.Sharon R., Bar-Joseph I., Frosch M. P., Walsh D. M., Hamilton J. A., Selkoe D. J. (2003) Neuron 37, 583–595 [DOI] [PubMed] [Google Scholar]
- 4.Chen L., Periquet M., Wang X., Negro A., McLean P. J., Hyman B. T., Feany M. B. (2009) J. Clin. Invest. 119, 3257–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eriksen J. L., Dawson T. M., Dickson D. W., Petrucelli L. (2003) Neuron 40, 453–456 [DOI] [PubMed] [Google Scholar]
- 6.Ross C. A., Poirier M. A. (2004) Nat. Med. 10, (suppl.), S10–S17 [DOI] [PubMed] [Google Scholar]
- 7.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M. S., Shen J., Takio K., Iwatsubo T. (2002) Nat. Cell Biol. 4, 160–164 [DOI] [PubMed] [Google Scholar]
- 8.Anderson J. P., Walker D. E., Goldstein J. M., de Laat R., Banducci K., Caccavello R. J., Barbour R., Huang J., Kling K., Lee M., Diep L., Keim P. S., Shen X., Chataway T., Schlossmacher M. G., Seubert P., Schenk D., Sinha S., Gai W. P., Chilcote T. J. (2006) J. Biol. Chem. 281, 29739–29752 [DOI] [PubMed] [Google Scholar]
- 9.Hasegawa M., Fujiwara H., Nonaka T., Wakabayashi K., Takahashi H., Lee V. M., Trojanowski J. Q., Mann D., Iwatsubo T. (2002) J. Biol. Chem. 277, 49071–49076 [DOI] [PubMed] [Google Scholar]
- 10.Paleologou K. E., Schmid A. W., Rospigliosi C. C., Kim H. Y., Lamberto G. R., Fredenburg R. A., Lansbury P. T., Jr., Fernandez C. O., Eliezer D., Zweckstetter M., Lashuel H. A. (2008) J. Biol. Chem. 283, 16895–16905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arawaka S., Wada M., Goto S., Karube H., Sakamoto M., Ren C. H., Koyama S., Nagasawa H., Kimura H., Kawanami T., Kurita K., Tajima K., Daimon M., Baba M., Kido T., Saino S., Goto K., Asao H., Kitanaka C., Takashita E., Hongo S., Nakamura T., Kayama T., Suzuki Y., Kobayashi K., Katagiri T., Kurokawa K., Kurimura M., Toyoshima I., Niizato K., Tsuchiya K., Iwatsubo T., Muramatsu M., Matsumine H., Kato T. (2006) J. Neurosci. 26, 9227–9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai Y., Fujita S. C., Iwatsubo T., Hasegawa M. (2004) FEBS Lett. 572, 227–232 [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto M., Arawaka S., Hara S., Sato H., Cui C., Machiya Y., Koyama S., Wada M., Kawanami T., Kurita K., Kato T. (2009) Biochem. Biophys. Res. Commun. 384, 378–382 [DOI] [PubMed] [Google Scholar]
- 14.Karube H., Sakamoto M., Arawaka S., Hara S., Sato H., Ren C. H., Goto S., Koyama S., Wada M., Kawanami T., Kurita K., Kato T. (2008) FEBS Lett. 582, 3693–3700 [DOI] [PubMed] [Google Scholar]
- 15.Okochi M., Walter J., Koyama A., Nakajo S., Baba M., Iwatsubo T., Meijer L., Kahle P. J., Haass C. (2000) J. Biol. Chem. 275, 390–397 [DOI] [PubMed] [Google Scholar]
- 16.Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., Maniatis T. (1995) Genes Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 17.Orford K., Crockett C., Jensen J. P., Weissman A. M., Byers S. W. (1997) J. Biol. Chem. 272, 24735–24738 [DOI] [PubMed] [Google Scholar]
- 18.Pronin A. N., Morris A. J., Surguchov A., Benovic J. L. (2000) J. Biol. Chem. 275, 26515–26522 [DOI] [PubMed] [Google Scholar]
- 19.Ishii A., Nonaka T., Taniguchi S., Saito T., Arai T., Mann D., Iwatsubo T., Hisanaga S., Goedert M., Hasegawa M. (2007) FEBS Lett. 581, 4711–4717 [DOI] [PubMed] [Google Scholar]
- 20.Bennett M. C., Bishop J. F., Leng Y., Chock P. B., Chase T. N., Mouradian M. M. (1999) J. Biol. Chem. 274, 33855–33858 [DOI] [PubMed] [Google Scholar]
- 21.Tofaris G. K., Layfield R., Spillantini M. G. (2001) FEBS Lett. 509, 22–26 [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Fallon L., Lashuel H. A., Liu Z., Lansbury P. T., Jr. (2002) Cell 111, 209–218 [DOI] [PubMed] [Google Scholar]
- 23.Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. (2003) Science 299, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C. W., Giasson B. I., Lewis K. A., Lee V. M., Demartino G. N., Thomas P. J. (2005) J. Biol. Chem. 280, 22670–22678 [DOI] [PubMed] [Google Scholar]
- 25.Ancolio K., Alves da Costa C., Uéda K., Checler F. (2000) Neurosci. Lett. 285, 79–82 [DOI] [PubMed] [Google Scholar]
- 26.Paxinou E., Chen Q., Weisse M., Giasson B. I., Norris E. H., Rueter S. M., Trojanowski J. Q., Lee V. M., Ischiropoulos H. (2001) J. Neurosci. 21, 8053–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rideout H. J., Larsen K. E., Sulzer D., Stefanis L. (2001) J. Neurochem. 78, 899–908 [DOI] [PubMed] [Google Scholar]
- 28.Rideout H. J., Stefanis L. (2002) Mol. Cell Neurosci. 21, 223–238 [DOI] [PubMed] [Google Scholar]
- 29.Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 30.Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. (2004) Science 305, 1292–1295 [DOI] [PubMed] [Google Scholar]
- 31.Vogiatzi T., Xilouri M., Vekrellis K., Stefanis L. (2008) J. Biol. Chem. 283, 23542–23556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickart C. M. (2000) Trends Biochem. Sci. 25, 544–548 [DOI] [PubMed] [Google Scholar]
- 33.Davidson W. S., Jonas A., Clayton D. F., George J. M. (1998) J. Biol. Chem. 273, 9443–9449 [DOI] [PubMed] [Google Scholar]
- 34.Jensen P. H., Nielsen M. S., Jakes R., Dotti C. G., Goedert M. (1998) J. Biol. Chem. 273, 26292–26294 [DOI] [PubMed] [Google Scholar]
- 35.Hodara R., Norris E. H., Giasson B. I., Mishizen-Eberz A. J., Lynch D. R., Lee V. M., Ischiropoulos H. (2004) J. Biol. Chem. 279, 47746–47753 [DOI] [PubMed] [Google Scholar]
- 36.Bedford L., Hay D., Devoy A., Paine S., Powe D. G., Seth R., Gray T., Topham I., Fone K., Rezvani N., Mee M., Soane T., Layfield R., Sheppard P. W., Ebendal T., Usoskin D., Lowe J., Mayer R. J. (2008) J. Neurosci. 28, 8189–8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tofaris G. K., Razzaq A., Ghetti B., Lilley K. S., Spillantini M. G. (2003) J. Biol. Chem. 278, 44405–44411 [DOI] [PubMed] [Google Scholar]
- 38.Sampathu D. M., Giasson B. I., Pawlyk A. C., Trojanowski J. Q., Lee V. M. (2003) Am. J. Pathol. 163, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]