Abstract
The superfamily of prokaryotic inwardly rectifying (KirBac) potassium channels is homologous to mammalian Kir channels. However, relatively little is known about their regulation or about their physiological role in vivo. In this study, we have used random mutagenesis and genetic complementation in K+-auxotrophic Escherichia coli and Saccharomyces cerevisiae to identify activatory mutations in a range of different KirBac channels. We also show that the KirBac6.1 gene (slr5078) is necessary for normal growth of the cyanobacterium Synechocystis PCC6803. Functional analysis and molecular dynamics simulations of selected activatory mutations identified regions within the slide helix, transmembrane helices, and C terminus that function as important regulators of KirBac channel activity, as well as a region close to the selectivity filter of KirBac3.1 that may have an effect on gating. In particular, the mutations identified in TM2 favor a model of KirBac channel gating in which opening of the pore at the helix-bundle crossing plays a far more important role than has recently been proposed.
Keywords: Ion Channels, Membrane Biophysics, Membrane Proteins, Potassium Channels, Potassium Transport, Kir Channel, KirBac
Introduction
Inwardly rectifying (Kir) potassium channels are regulated by many different cellular factors such as G-proteins, phosphatidylinositol 4,5-bisphosphate, ATP, and intracellular pH, and are therefore able to couple channel activity to a wide range of metabolic and physiological stimuli (1, 2). Their importance is illustrated by the fact that inherited mutations in Kir channels underlie a number of diseases including Type II Bartter syndrome (Kir1.1), Andersen syndrome (Kir2.1), SeSAME/EAST syndrome (Kir4.1), and vitreoretinal degeneration (Kir7.1), as well as certain forms of neonatal diabetes and hyperinsulinemia (Kir6.2) (2).
The discovery of a large number of prokaryotic homologs of eukaryotic Kir channels (3, 4) has provided an enormous experimental resource for this family of K+ channels and enables a complementary approach to structural studies of Kir channels (5–7). In particular, the ability to rapidly integrate data from functional, biochemical, and structural studies of KirBac channels represents a major advantage (8–11). However, in comparison with mammalian Kir channels, one of the current limitations is that we still know relatively little about the regulation of KirBac channel activity or their functional role in vivo.
One approach to the study of prokaryotic K+ channel function has been to use K+-auxotrophic strains of Escherichia coli and yeast (Saccharomyces cerevisiae) as an unbiased, high throughput screen for gating mutations (12, 13). Several previous studies have successfully used this “forward genetics” approach to identify activatory mutations in both prokaryotic and eukaryotic potassium channels (12, 14–18). In E. coli, the assay is based upon the fact that K+ uptake is normally mediated by three principal transport pathways: kdp, trk, and kup, and several strains have been generated that have mutations in these genes. These strains are K+-auxotrophic and exhibit no growth in media containing low concentrations of K+ (<10 mm). However, this K+ uptake deficiency can be complemented by recombinant expression of a functional K+ channel that provides an alternative pathway for K+ entry (12). Similar K+-auxotrophic strains of S. cerevisiae have also been generated that have mutations in the trk1 and trk2 genes and can be used in exactly the same type of functional assays (13).
In this study, we have used these K+ uptake-deficient strains to identify activatory mutations in KirBac1.1, KirBac3.1, KirBac4.1, and KirBac6.1. These mutations highlight the importance of several structurally conserved regions in KirBac channel gating, including residues close to the selectivity filter of KirBac3.1. However, the results also indicate a more important role for TM2 in KirBac channel gating than has recently been proposed (5). Finally we addressed the role of KirBac channels in vivo and demonstrate that deletion of the KirBac6.1 gene in its host organism (Synechocystis PCC6803) generates a K+-dependent growth deficiency consistent with a role for this channel in K+ uptake.
EXPERIMENTAL PROCEDURES
Molecular Biology
Wild-type KirBac genes were cloned into the E. coli expression vector pQE60-lac as described previously (4). For expression in yeast, the methionine-regulated pYES2m and p416 vectors were used with a modified Kozak sequence of 6 adenine nucleotides for optimal expression (AAAAAAATG) (19). Randomly mutated libraries were constructed for KirBacs using the GeneMorph II random mutagenesis kit (Stratagene), which uses non-biased, error-prone PCR. The PCR reactions were quantified according to the manufacturer's protocols to produce an error rate of ∼1–3 mutations per open reading frame. PCR products were then subcloned back into their expression vectors. The primers used for amplification overlapped the 5′ and 3′ sequences of the open reading frames to prevent unwanted mutations in these regions. To maximize the transformation efficiency, ligations were purified by phenol:chloroform extraction and ethanol precipitation prior to transformation into Library Efficiency DH5α E. coli (Invitrogen) and growth in culture overnight. Analysis of the transformation efficiency prior to overnight growth indicated that the libraries contained at least 105 independent clones. Randomly selected independent clones were isolated on non-selective media, and upon sequencing, were found to contain an average of 2–5 mutations per open reading frame. All site-directed mutagenesis was performed using the QuikChange II system (Stratagene).
Screening Mutant Libraries in E. coli
All media, growth, and propagation of these strains are as described previously (4, 17). To screen the mutant libraries, 200 ng of mutant plasmid DNA was transformed into 100 μl of chemically competent TK2420 cells (1 × 106 cfu/μg) and plated out onto 90-mm Petri dishes containing K0 solid media supplemented to contain 0.5 mm isopropyl-1-thio-β-d-galactopyranoside and (unless otherwise stated) 5 mm KCl. Plates were incubated at 37 °C (30 °C for KirBac3.1), individual colonies were picked and propagated in K115, and plasmid DNA was isolated. The quality of plasmid DNA obtained from the TK2420 strain is relatively low and was therefore retransformed back into DH5α and repurified prior to sequencing. To eliminate false positives, potential activatory mutants were retransformed into TK2420 and confirmed by drop tests on low [K+] before sequencing the entire gene.
E. coli Drop Tests and Growth Curves
Drop tests and growth curves were done as described previously (4, 18). Briefly, overnight cultures were spun down, washed, and resuspended in an equal volume of K0, and then 4 μl of undiluted, 1:10, and 1:1000 dilutions was spotted onto the plates and allowed to dry before being grown overnight at 37 °C. For growth curves, washed overnight cultures were diluted 20-fold into K0 at the indicated KCl concentration, and growth was monitored by following A600 after induction with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside.
Yeast Transformation and Drop Tests
All media, growth, and propagation of these strains are as described previously (16, 18). Wild-type and mutant KirBac channels were transformed into competent SGY1528 cells using a standard lithium acetate yeast transformation protocol. Transformants were grown on APKO-ura plates for ∼48 h before overnight growth in 4 ml of APKO-ura (100 mm KCl). Cultures were washed in APKO-met-ura media (0.5 mm KCl), and 3.5-μl drops of undiluted, 1:10, or 1:1000 dilutions were made onto APKO-met-ura plates (with [KCl] as specified). Drop test plates were incubated at 30 °C for ∼72 h.
Expression and Purification of KirBac Channels for Liposomal Flux Assays
Wild-type and mutant KirBac1.1 proteins were expressed and purified as described previously (20). For expression of KirBac3.1, we utilized a synthetic gene codon optimized for expression in E. coli (GenScript) and cloned into the pET30a vector (Novagen), with a C-terminal His6 tag. BL21 CodonPlus RP cells (Stratagene) were used for expression. The purification protocol was as described previously (21), with the following exceptions. Cells were grown at 19 °C overnight following induction, and the lysis buffer contained 50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 50 mm KCl. Liposomal flux assays were performed as described previously (20).
Homology Modeling and Molecular Dynamics Simulations
Homology models and molecular dynamics (MD)2 simulations were done as has been described before (22). Briefly, a homology model of wild-type KirBac6.1 (residues 21–303) was built using Modeler 9v2 using KirBac3.1 crystal structure (Protein Data Bank (PDB): 1XL6) (5) as a template. Sequence alignments were made using ClustalW2. The selectivity filter was populated with K+ ions at S0, S2, and S4 and interspersed with waters using crystal structure coordinates. The cavity was flooded using packages Voidoo and Flood. The model was inserted into an explicit, pre-equilibrated phosphatidylglycerol:palmitoyl-oleoyl-phosphatidylglycerol bilayer in water, and additional waters were added to accommodate the KirBac6.1 C terminus (box size = 10 × 10 × 12 nm). The protein was optimally positioned in the bilayer, clashing (<2 Å) lipids and waters were removed, and KCl concentration was adjusted to 100 mm. MD simulations were carried out using GROMACS v3.3.3 and GROMOS forcefield. Prior to simulation, energy minimization was carried out followed by incremental (5 ps, 10–20-K steps) warming from 80 to 310 K (37 °C) and a 50-ps equilibration step, both with lipid headgroups restrained. The MD run was 15.2 ns.
Mutant models were created by introducing the mutation (either W48R or G137D) into all four subunits using PyMOL version 0.99 after energy minimization. Further energy minimization, warming, equilibration, and MD simulation used identical parameters to the WT model. Root mean squared deviation analysis showed all three models to be stable between 5 and 15.2 ns of simulation; only data from this period were used in other analyses.
Genetic Deletion of KirBac6.1
The open reading frame encoding the slr5078 (KirBac6.1) gene in Synechocystis PCC6803 was deleted by homologous recombination and replaced with a kanamycin resistance marker (23). Briefly, 1-kb genomic sequences on either side of the slr5078 gene were amplified by PCR and used to flank a kanamycin resistance gene. This targeting vector was then transformed into Synechocystis PCC6803 and selected using 50 μg/ml kanamycin. Successfully recombined colonies were identified by PCR and checked for integration of the kanamycin resistance cassette into every copy of the genome before growth in liquid medium.
Growth of Synechocystis PCC6803
Logarithmically growing cultures were subcultured on alternate days (to 1 × 106 cells/ml) into liquid BG11 medium for a minimum of 7 days to standardize growth rates as described before (24). For growth analysis, cells were inoculated to a density of 1 × 106 cells/ml into acid-washed glass boiling tubes and monitored at 540 nm for 7 days. Growth was examined in standard BG11 growth media or in media supplemented with twice the media concentration of K2HPO4 (175 μm), NaNO3 (17.65 mm), MgSO4 (304 μm), or CaCl2 (245 μm). Growth was also monitored in the presence of twice the normal media concentration of manganese, zinc, copper, and molybdenum or chromium, nickel, and cobalt and in the presence of 175 μm Na2HPO4. Only the addition of K2HPO4 improved the growth of the cells. The assay was carried out in triplicate with independently selected deletion mutants and repeated on two further occasions. Mutants were also not subjected to culture conditions liable to select for second site mutations prior to growth.
RESULTS
Identification of KirBac Activatory Mutations in K+-auxotrophic E. coli
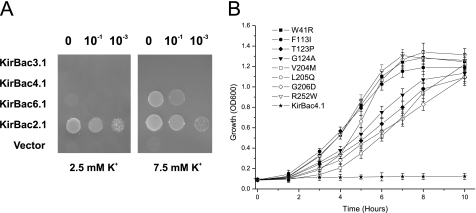
In a previous study, we demonstrated that wild-type KirBac4.1 fails to complement the growth of TK2420 E. coli and that this is not due to a toxic effect on cell growth (4). Fig. 1A shows that KirBac4.1 does not complement growth on low [K+] and is therefore suitable as a candidate gene to screen for activatory mutations. Wild-type KirBac6.1 does complement growth at 7.5 mm [K+], but not at lower concentrations, also making it a suitable candidate if screened on low [K+] media. KirBac3.1 also shows no complementation (Fig. 1A). By contrast, other KirBac channels, such as KirBac2.1, which exhibits robust complementation, or KirBac1.1, which exhibits toxic effects on the long term growth of E. coli, are unsuitable for screening in this assay (4).
FIGURE 1.
KirBac4.1 activatory mutations in TK2420 E. coli. A, drop tests demonstrate that expression of KirBac2.1 complements the growth defect of TK2420 E. coli. However, neither KirBac3.1 nor KirBac4.1 complement growth at low [K+]. KirBac6.1 only complements at higher [K+]. B, screening of a randomly mutated library of KirBac4.1 identifies mutations that activate KirBac4.1. Growth in culture shows that wild-type KirBac4.1 exhibits no complementation of the growth defect in low [K+] media, but eight mutations were identified that exhibit strong complementation. Growth in 2.5 mm [K+] was measured over 10 h. Expression of wild-type and mutant KirBac4.1 was induced at time 0. OD, optical density. Error bars indicate S.D.
We used an error-prone PCR method to create randomly mutated libraries for KirBac3.1, KirBac4.1, and KirBac6.1 and transformed these into TK2420 E. coli. For the KirBac4.1 library grown on 2.5 mm K+, ∼100 colonies were recovered, sequenced, and retransformed to ensure that they were not false positives (see under “Experimental Procedures”). In total, eight separate mutations were identified (Fig. 1B) where they rescue the growth of TK2420 in liquid media. They are also listed in Table 1. For KirBac6.1, we recovered 23 novel mutations, and for KirBac3.1, a total of 13 mutations were recovered (Table 1).
TABLE 1.
Activatory mutations identified in the different KirBac channels
| KirBac1.1 | KirBac3.1 | KirBac4.1 | KirBac6.1 |
|---|---|---|---|
| W60C | I75S | W41R | P21L |
| A109P/G134A | A78P | F113I | R26G |
| S136P | F88L | T123P | W48R |
| G143E | T93I | G124A | L50F |
| L144P | G98D | V204M | S122C |
| V145L | L118Q | L205Q | I125M |
| M121K | G206D | A133P | |
| G123D | R252W | L135I | |
| A125E | G137D | ||
| S129R | L138S | ||
| I150F | M139L | ||
| V181I | F140Y | ||
| S205L | V150L | ||
| A172T | |||
| N173K | |||
| Q174L | |||
| R178H | |||
| S187T | |||
| Q190K | |||
| L209I | |||
| R210C | |||
| H212R | |||
| T213I | |||
| L218S | |||
| R265L | |||
| Q276R |
Identification of KirBac Activatory Mutations in K+-auxotrophic Yeast
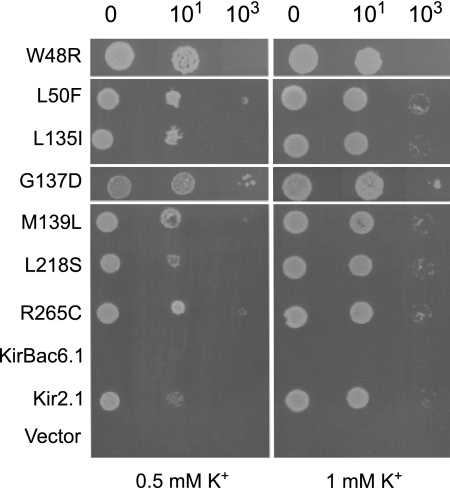
We next investigated the suitability of the SGY1528 K+-auxotrophic strain of S. cerevisiae for the identification of KirBac channel activatory mutations. KirBac1.1, KirBac3.1, KirBac4.1, and KirBac6.1 were subcloned into the pYES2m yeast expression vector and transformed into the SGY1528 strain. No complementation was observed for any of these wild-type channels on 2 mm [K+] (not shown), indicating that random mutagenesis could potentially identify activatory mutations. Mutant libraries for KirBac1.1, KirBac3.1, KirBac4.1, and KirBac6.1 were therefore created, transformed into SGY1528, and grown on 2 mm [K+]. No positive colonies were recovered from the KirBac3.1 and KirBac4.1 libraries. However, 52 clones were recovered and analyzed from the KirBac1.1 library, and ∼100 were recovered and analyzed from the KirBac6.1 library. After retransformation and separation of multiple mutations, a total of nine novel mutations were found in KirBac6.1, and a selection of these is shown in Fig. 2. A total of five novel mutations were identified in KirBac1.1 (Table 1) including one mutant (A109P/G134A) where both mutations are required.
FIGURE 2.
KirBac6.1 activatory mutations in SGY1528 yeast. Drop tests on low [K+] media are shown. Mammalian Kir2.1 channel complements the growth of SGY1528 K+ uptake-deficient yeast, but wild-type KirBac6.1 does not complement growth.
Clustering of Activatory Mutations
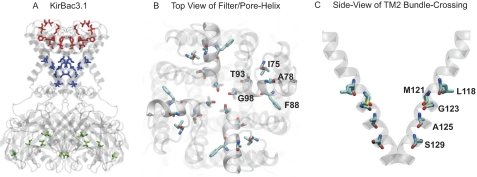
The activatory mutations shown in Table 1 are highlighted in an alignment of KirBac channels (supplemental Fig. S1). Interestingly, a large number of activatory mutations are clustered in TM2 and were found in all of the KirBacs examined. In addition, mutations were found to cluster within the slide helix and at the base of TM1 near the helix-bundle crossing. Intriguingly, a number of activatory mutations were also found near the pore helix and selectivity filter, but only in KirBac3.1. The activatory mutations found in KirBac3.1 were mapped onto the available crystal structure and are illustrated in Fig. 3.
FIGURE 3.
Location of activatory mutations in KirBac3.1. A, mutations cluster into two distinct groups in the filter/pore helix region (red) and in TM2 (blue). Only a small number of mutants are found in the C terminus (green), and the consequence of these is less obvious. B, top-down view of activatory mutations in the filter/pore helix region of KirBac3.1. Mutants are shown as Corey-Pauling-Koltun coloring. Phe-88 has been shown to move during KirBac3.1 channel gating (11), and the F88L mutation directly increases channel activity (Fig. 4B). C, side view of activatory mutations in TM2. For clarity, only two helices are shown. Mutation S129R at the helix-bundle crossing also directly activates KirBac3.1 (Fig. 4B).
The clustering of mutations within the C terminus is less apparent from this linear alignment but becomes clearer when mapped onto a structural model of a KirBac channel. For this purpose, we created a homology model of KirBac6.1 because the largest number of mutations was identified in this channel. Using this model we highlighted all of the mutations shown in Table 1, as well as some of the residues equivalent to those mutated in KirBac3.1 and KirBac4.1. Although these mutations are far apart in the linear sequence, mutations in the CD loop are close to several other clusters of mutations within the C terminus, in particular residues in adjacent loops (βF-βG loop) (see the supplemental movie).
Functional Analysis of TM2/Pore Mutations
The clustering of mutations in TM2 is of particular interest as this helix has been shown to be critically important for K+ channel gating (1, 25). To determine whether the TM2 mutations we identified directly affect channel activity, we used KirBac1.1 and KirBac3.1 because both wild-type channels can be expressed and purified and are therefore suitable for liposomal flux assays (20, 21).
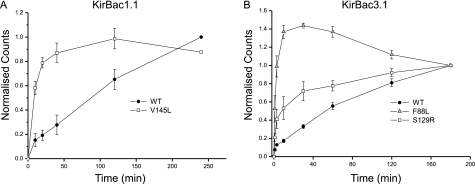
The KirBac1.1 V145L mutation is located within the helix-bundle crossing of KirBac1.1 and is adjacent to the hydrophobic “blocking residue” Phe-146, which has been proposed to form the activation gate in KirBac1.1 (26). This mutant exhibited a dramatically faster rate of 86Rb+ uptake when compared with wild-type KirBac1.1 (Fig. 4A), thus indicating a direct effect on channel activity. Likewise the S129R mutation in TM2 of KirBac3.1 is located in a similar position (Fig. 3) and also showed a faster rate of 86Rb+ uptake when compared with wild-type KirBac3.1 (Fig. 4B).
FIGURE 4.
Mutations increase functional activity of KirBac channels. A, time course of 96Rb+ uptake into liposomes reconstituted with either wild-type or mutant KirBac1.1. The V145L mutation identified in TM2 of KirBac1.1 causes an increase in the rate of uptake when compared with WT KirBac1.1, demonstrating that the activatory effect is due to direct changes in channel activity. B, similar increases in the rate of uptake are seen with the F88L and S219R mutations in KirBac3.1. Error bars indicate S.D.
An additional gating mechanism has also been proposed to exist within the selectivity filter of KirBac3.1 (5). It was therefore intriguing that several activatory mutations were uncovered close to this region in KirBac3.1. The F88L mutation was of particular interest because this residue has previously been reported to undergo conformational changes during KirBac3.1 channel gating (11). This side chain is located within the pore helix that precedes the selectivity filter (Fig. 3) and forms part of an intersubunit interface. The F88L mutation also exhibited a dramatically faster rate of 86Rb+ uptake when compared with wild-type KirBac3.1 (Fig. 4B).
Molecular Dynamics Simulation of Mutations
Using the homology model of KirBac6.1 created above, we examined the possible structural effect of two activatory mutations using MD simulations. Given the relatively short timescale of an atomistic MD simulation, we chose to examine activatory mutations likely to have the most biggest effect on channel structure.
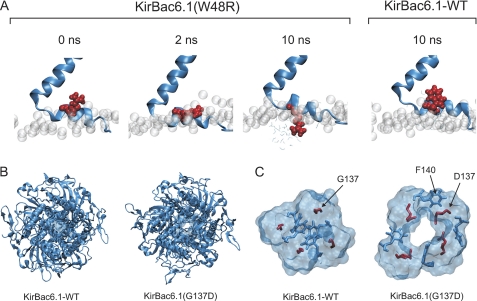
The W48R mutation located within the slide helix of KirBac6.1 was chosen due to the dramatic side-chain substitution involved, as well as the reported role of the slide helix in channel gating (27). The homology model of wild-type KirBac6.1 was found to be stable during a 15-ns simulation (supplemental Fig. S2A). The W48R mutation was next introduced into the model, and a separate 15-ns MD simulation was run. Tryptophan 48 in the slide helix of wild-type KirBac6.1 is firmly embedded within the lipid bilayer. However, the W48R mutation introduces a positively charged residue at this position, and the MD simulation clearly shows that this mutation induces a rotation of the slide helix as the arginine moves away from the lipid environment to a more energetically favorable position (Fig. 5A).
FIGURE 5.
MD simulation of KirBac6.1 activatory mutations. A, snapshots of the slide helix and TM2 during a 10-ns MD simulation KirBac6.1. Highlighted in red is the W48R mutation, whereas the phospholipid head groups of the bilayer are shown as gray spheres. After 2 ns, Arg-48 begins to twist the slide helix. After 10 ns, there is a >90° rotation of the slide helix, and Arg-48 becomes fully exposed to water in the cytoplasm. By contrast, after 10 ns, the wild-type Trp-48 remains firmly embedded within the lipid bilayer. B, top-down view of WT and mutant KirBac6.1 after a 10-ns MD simulation. The wild-type channel remains in a closed conformation, whereas the G137D mutant channel opens at the helix-bundle crossing. C, expanded view of residues at the helix-bundle crossing of KirBac6.1 after a 10-ns MD simulation. Residue 137 is highlighted in red, and Phe-140 is in blue. The Asp-137 mutant residue causes an opening of the channel at the helix-bundle crossing. Further details are show in supplemental Fig. S3.
We next examined the effect of the G137D mutation in the MD simulation. Glycine 137 in KirBac6.1 is equivalent to the TM2 “lower glycine hinge” residue that is highly conserved in the KirBac/Kir channel superfamily (28, 29). It is also equivalent to Ser-129 in KirBac3.1, which is unique in possession of a serine at this position (supplemental Fig. S1) (4, 29). Some previous studies have suggested that this position in TM2 may form a hinge point at the helix-bundle crossing. However, others have indicated that its small size is critical for the TM2 helices to come into close proximity at the helix-bundle crossing (28, 29). Activatory mutations at this position were found in all four KirBacs tested, and in most cases, a charged amino acid was substituted at this position (Table 1). The MD simulation (Fig. 5, B and C) shows that introduction of a negatively charged aspartate residue at this position causes a significant widening of the pore at this constriction point. The bundle crossing opens to 2.3 Å ± 0.2 in KirBac6.1(G137D) when compared with 0.6 Å ± 0.1 in wild-type KirBac6.1 (supplemental Fig. S2B). Although it might be presumed that simple electrostatic repulsion between 4 aspartates in close proximity could be responsible for pushing these helices apart, the simulation clearly shows that the side chains are pointing away from the cavity and not into it. Further examination also shows that the aspartate side chain forms a hydrogen bond with a highly conserved tyrosine residue (Tyr-46) located within the slide helix (supplemental Fig. S4).
Genetic Deletion of KirBac6.1
In general, the functional role of microbial K+ channels is still poorly understood (30, 31). We therefore decided to examine the phenotypic effects of deleting a KirBac channel in its host organism. KirBac6.1 was originally identified in the genome of the freshwater cyanobacterium Synechocystis PCC 6803, a model photosynthetic organism that is readily amenable to genetic manipulation. We created a targeting vector that was used to replace the coding sequence of the KirBac6.1 gene (denoted slr5078 in the Synechocystis genome) with a kanamycin resistance cassette (see under “Experimental Procedures” for details).
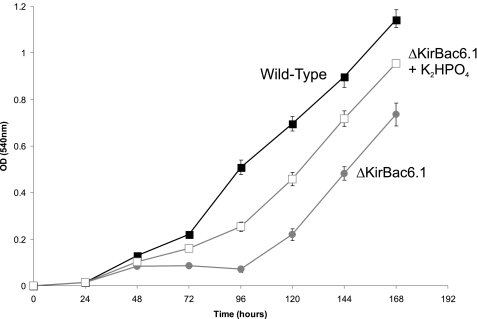
Fig. 5 shows that the ΔKirBac6.1 strain grows at a significantly slower rate than wild-type Synechocystis. Importantly, this growth defect could be partially reversed by growth in medium that contained twice the normal concentration of K2HPO4 (175 μm) in the medium (Fig. 6). As a control, supplementation with 175 μm Na2HPO4 was without effect on the growth rate, as was media supplemented with twice the standard concentration of NaNO3 (17.65 mm), MgSO4 (304 μm), or CaCl2 (245 μm) (not shown). Growth was also monitored in the presence of twice the normal media concentration of manganese, zinc, copper, and molybdenum or chromium, nickel, and cobalt and had no effect on growth of the mutant strain (not shown). Only the addition of K+ ions improved the growth of the ΔKirBac6.1 strain. The phenotype was also identical in three independently selected deletion mutants observable, thus removing the possibility that it the result of a second site mutation.
FIGURE 6.
Genetic deletion of KirBac6.1. Growth curves of the cyanobacterium Synechocystis PCC6803 in which the KirBac6.1 gene has been deleted (ΔKirBac6.1) exhibit a reduced rate of growth (gray circles) when compared with the wild-type organism (black squares). This growth defect can be partially restored by supplementation of the growth media with K2HPO4 (open squares). OD, optical density. Error bars indicate S.D.
DISCUSSION
In this study, we have used a forward genetics approach to identify a large number of activatory mutations in several different KirBac channels, which highlight structural domains likely to be important for the control of KirBac channel activity. Our results demonstrate that KirBac channels represent an attractive experimental system that enables the rapid integration of functional, biochemical, and structural studies in a way that can be difficult to achieve with eukaryotic Kir channels.
A Functional role for KirBac Channels?
Despite the wealth of structural and mechanistic data that now exist for many prokaryotic potassium channels, their biological role remains largely unclear (30, 31). K+ channel genes are found in almost every prokaryotic genome that has been sequenced, and although some studies have used genetic deletion to understand their functional role, in most cases, no specific phenotype has been observed (32). For example, deletion of KcsA in Streptomyces lividans produced no obvious phenotype, and neither did deletion of the 6-TM Kch channel gene in E. coli, suggesting either that significant functional redundancy exists or that these channels are only required to counter very specific environmental stresses (31). However, the hpKch K+ channel in Helicobacter pylori is essential for the ability of this organism to colonize the murine gut and plays an important role in K+ uptake at low [K+] (33). Interestingly the K+-dependent growth defect we observed when KirBac6.1 was deleted from Synechocystis is very similar to the growth defect seen when hpKch was deleted in H. pylori (33).
In most microorganisms, high affinity K+ uptake occurs through ATP-dependent transport pathways, but at higher concentrations of K+, uptake can be mediated through K+-selective channels due to the hyperpolarized membrane potentials. With a typical membrane potential of about −200 mV, most microorganisms are capable of accumulating K+ ∼1000-fold via such a K+-specific diffusion pathway (31). Therefore, although further studies will be required to demonstrate its specific role, we propose that KirBac6.1 may contribute to low affinity K+ uptake in Synechocystis and that similar functional roles may exist for KirBac channel genes. It may be of some significance that KirBac channels have been identified in every cyanobacterial genome that has been sequenced so far (www.jgi.doe.gov) yet are not common in other microorganisms (4).
TM2 and KirBac Channel Gating
The parallel screening of multiple different KirBac channels used in this study has enabled us to visualize the clustering of mutations that may not have been apparent if only one channel had been examined. The identification of similar mutations in several different KirBac channels highlights several functionally sensitive domains. In particular, the clustering of activatory mutations in TM2 close to the helix-bundle crossing is consistent with an important role for this domain in channel gating, and functional studies of eukaryotic Kir channels have also shown that mutations in this region can have a dramatic effect on channel gating. For example, in human Kir6.2, several activatory mutations causing neonatal diabetes are located in this region of TM2 (34). Furthermore, previous complementation studies of GIRK/Kir3.0 channels in K+-auxotrophic yeast also identified multiple mutations in TM2, many of which resulted in an increase in channel open probability (14, 15, 35).
The structural and functional consequences of the mutations we have identified in TM2 therefore support a key role for the bundle crossing in the control of channel activity. However, this contrasts with suggestions (based on recent x-ray crystal structures of KirBac3.1) (5) that movement of the cytoplasmic domains regulates channel conduction via structural changes at the selectivity filter without movement of the helix-bundle crossing. It still remains possible that the selectivity filter plays an important role in channel gating, and our results also lend some support to this hypothesis (see below). However, our findings demonstrate that the helix-bundle crossing is also clearly important for the regulation of channel activity and is consistent with several previous structural studies of KirBac3.1, which indicate opening of the channel at the bundle crossing (11, 21, 36, 37), as well as access studies that indicate ligand gating occurs at this region in eukaryotic Kir channels (38, 39).
Other Mutation-sensitive Domains
In addition to mutations in TM2, activatory mutations were also identified within the slide helix and at the base of TM1. The slide helix is thought to be critical for the regulation of channel activity in both Kir and KirBac channels (27, 40, 41), and the identification of mutations within these regions supports this. In particular, our MD simulation of the W48R mutation in KirBac6.1 suggests that rotational movement of the slide helix can induce channel opening and that this may therefore represent an important step in the activatory mechanism observed in wild-type channels. This is consistent with functional studies showing that movement of the slide helix induced by lipid tethering can dramatically enhance KirBac1.1 activity (27). Furthermore, mutations that disrupt the interaction of the slide helix with the cytoplasmic domains can also give rise to a loss of function in Kir channels (42).
Within the cytoplasmic domains themselves, a clustering of mutations only became apparent when visualized on a structural model of the KirBac channel. In particular, several mutations within the CD loop had a potent activatory effect in several different KirBac channels and are in close proximity to several other activatory mutations, which are far apart in the linear sequence and form a potential network of residues between adjacent subunits (see the supplemental material). The CD loop has already been implicated in the control of Kir channel gating (43, 44), and our results would therefore support the importance of this domain in the regulation of channel activity.
A Gate within the Selectivity Filter of KirBac3.1?
Recent x-ray crystal structures of KirBac3.1 have been used to suggest that, similar to KcsA, KirBac3.1 has a gating mechanism located close to the selectivity filter (5). It is therefore highly significant that our complementation screen also identified a cluster of mutations close to, and within, the selectivity filter of KirBac3.1. One of these residues (Phe-88) is located in the pore helix at an intersubunit interface (Fig. 3B) and was also identified in our previous x-ray footprinting study of KirBac3.1 as a side chain that exhibits dramatic changes in solvent accessibility during channel gating (11). The identification of Phe-88 in two independent assays therefore provides strong evidence that this side chain must undergo structural changes during channel gating, and is supported by the increased functional activity seen in the F88L mutant channel (Fig. 4B). Intriguingly, Phe-88 in KirBac3.1 is also equivalent to Trp-67 in KcsA, which has recently been shown to be involved in coupling movement of TM2 to an inactivation mechanism in the selectivity filter of KcsA (25, 45). A similar gating mechanism may therefore exist in KirBac3.1, and movement of the pore helix at this intersubunit interface may also play an important role in channel gating.
Interestingly, one of the mutations we identified in KirBac3.1 (G98D) is located directly within the GYG selectivity filter. In most K+ channels, such mutations are either non-functional or non-selective. Clearly this channel is functional, but we cannot determine selectivity using our complementation assay. If the mutation allowed both Na+ and K+ to permeate, then this would still permit complementation as any excess Na+ would be removed through a range of active transport systems (46). Unfortunately, this mutation dramatically decreased the stability of purified KirBac3.1 protein, preventing detailed functional studies (not shown).
Our study indicates a high sensitivity of both the bundle crossing and the selectivity filter to mutagenesis. Although it is possible that some of the mutations permit complementation through mechanisms that do not alter channel gating (e.g. an increase in the unitary conductance of the channel or an increase in trafficking to the membrane), the overall clustering in these locations is consistent with proposed models of Kir gating, in which movement of the cytoplasmic domains is coupled through the slide helix to opening of the helix-bundle crossing and thence to the selectivity filter.
Supplementary Material
Acknowledgments
We thank Man-Jiang Xie, Si Sun, Richard Wheeler, Laxmi Parajuli, and Maria Mogni for contributions to the initial stages of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants HL54171 (to C. G. N.), DK069424 (to D. E.) and by grants from the British Heart Foundation, the Wellcome Trust, the Biotechnology and Biological Sciences Research Council (BB/H006052/1).

The on-line version of this article (available at http://www.jbc.org) contains a supplemental movie and Figs. S1–S4.
- MD
- molecular dynamics.
REFERENCES
- 1.Bichet D., Haass F. A., Jan L. Y. (2003) Nat. Rev. Neurosci. 4, 957–967 [DOI] [PubMed] [Google Scholar]
- 2.Hibino H., Inanobe A., Furutani K., Murakami S., Findlay I., Kurachi Y. (2010) Physiol. Rev. 90, 291–366 [DOI] [PubMed] [Google Scholar]
- 3.Durell S. R., Guy H. R. (2001) BMC Evol. Biol. 1, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun S., Gan J. H., Paynter J. J., Tucker S. J. (2006) Physiol. Genomics 26, 1–7 [DOI] [PubMed] [Google Scholar]
- 5.Clarke O. B., Caputo A. T., Hill A. P., Vandenberg J. I., Smith B. J., Gulbis J. M. (2010) Cell 141, 1018–1029 [DOI] [PubMed] [Google Scholar]
- 6.Nishida M., Cadene M., Chait B. T., MacKinnon R. (2007) EMBO J. 26, 4005–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng W. W., Enkvetchakul D., Nichols C. G. (2009) J. Gen. Physiol. 133, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh D. K., Rosenhouse-Dantsker A., Nichols C. G., Enkvetchakul D., Levitan I. (2009) J. Biol. Chem. 284, 30727–30736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Alimi Y., Tong A., Nichols C. G., Enkvetchakul D. (2009) J. Biol. Chem. 284, 2854–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta S., Bavro V. N., D'Mello R., Tucker S. J., Vénien-Bryan C., Chance M. R. (2010) Structure 18, 839–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parfenova L. V., Rothberg B. S. (2006) Methods Mol. Biol. 337, 157–165 [DOI] [PubMed] [Google Scholar]
- 13.Tang W., Ruknudin A., Yang W. P., Shaw S. Y., Knickerbocker A., Kurtz S. (1995) Mol. Biol. Cell 6, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi B. A., Lin Y. F., Jan Y. N., Jan L. Y. (2001) Neuron 29, 657–667 [DOI] [PubMed] [Google Scholar]
- 15.Bichet D., Lin Y. F., Ibarra C. A., Huang C. S., Yi B. A., Jan Y. N., Jan L. Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4441–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minor D. L., Jr., Masseling S. J., Jan Y. N., Jan L. Y. (1999) Cell 96, 879–891 [DOI] [PubMed] [Google Scholar]
- 17.Irizarry S. N., Kutluay E., Drews G., Hart S. J., Heginbotham L. (2002) Biochemistry 41, 13653–13662 [DOI] [PubMed] [Google Scholar]
- 18.Paynter J. J., Sarkies P., Andres-Enguix I., Tucker S. J. (2008) Channels 2, 413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton R., Watanabe C. K., de Boer H. A. (1987) Nucleic Acids Res. 15, 3581–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enkvetchakul D., Bhattacharyya J., Jeliazkova I., Groesbeck D. K., Cukras C. A., Nichols C. G. (2004) J. Biol. Chem. 279, 47076–47080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo A., Domene C., Johnson L. N., Doyle D. A., Vénien-Bryan C. (2005) Structure 13, 1463–1472 [DOI] [PubMed] [Google Scholar]
- 22.Haider S., Khalid S., Tucker S. J., Ashcroft F. M., Sansom M. S. (2007) Biochemistry 46, 3643–3652 [DOI] [PubMed] [Google Scholar]
- 23.Hagemann M., Zuther E. (1992) Arch. Microbiol. 158, 429–434 [DOI] [PubMed] [Google Scholar]
- 24.Borrelly G. P., Rondet S. A., Tottey S., Robinson N. J. (2004) Mol. Microbiol. 53, 217–227 [DOI] [PubMed] [Google Scholar]
- 25.Cuello L. G., Jogini V., Cortes D. M., Pan A. C., Gagnon D. G., Dalmas O., Cordero-Morales J. F., Chakrapani S., Roux B., Perozo E. (2010) Nature 466, 272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo A., Gulbis J. M., Antcliff J. F., Rahman T., Lowe E. D., Zimmer J., Cuthbertson J., Ashcroft F. M., Ezaki T., Doyle D. A. (2003) Science 300, 1922–1926 [DOI] [PubMed] [Google Scholar]
- 27.Enkvetchakul D., Jeliazkova I., Bhattacharyya J., Nichols C. G. (2007) J. Gen. Physiol. 130, 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaoka Y., Shang L., Banerjee A., Bayley H., Tucker S. J. (2008) Chembiochem. 9, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 29.Shang L., Tucker S. J. (2008) Eur. Biophys. J. 37, 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kung C., Blount P. (2004) Mol. Microbiol. 53, 373–380 [DOI] [PubMed] [Google Scholar]
- 31.Kuo M. M., Haynes W. J., Loukin S. H., Kung C., Saimi Y. (2005) FEMS Microbiol. Rev. 29, 961–985 [DOI] [PubMed] [Google Scholar]
- 32.Hegermann J., Overbeck J., Schrempf H. (2006) Microbiology 152, 2831–2841 [DOI] [PubMed] [Google Scholar]
- 33.Stingl K., Brandt S., Uhlemann E. M., Schmid R., Altendorf K., Zeilinger C., Ecobichon C., Labigne A., Bakker E. P., de Reuse H. (2007) EMBO J. 26, 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flanagan S. E., Edghill E. L., Gloyn A. L., Ellard S., Hattersley A. T. (2006) Diabetologia 49, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 35.Sadja R., Smadja K., Alagem N., Reuveny E. (2001) Neuron 29, 669–680 [DOI] [PubMed] [Google Scholar]
- 36.Domene C., Doyle D. A., Vénien-Bryan C. (2005) Biophys. J. 89, L01–L03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grottesi A., Domene C., Hall B., Sansom M. S. (2005) Biochemistry 44, 14586–14594 [DOI] [PubMed] [Google Scholar]
- 38.Phillips L. R., Enkvetchakul D., Nichols C. G. (2003) Neuron 37, 953–962 [DOI] [PubMed] [Google Scholar]
- 39.Phillips L. R., Nichols C. G. (2003) J. Gen. Physiol. 122, 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proks P., Antcliff J. F., Lippiat J., Gloyn A. L., Hattersley A. T., Ashcroft F. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17539–17544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Männikkö R., Jefferies C., Flanagan S. E., Hattersley A., Ellard S., Ashcroft F. M. (2010) Hum. Mol. Genet. 19, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Decher N., Renigunta V., Zuzarte M., Soom M., Heinemann S. H., Timothy K. W., Keating M. T., Daut J., Sanguinetti M. C., Splawski I. (2007) Cardiovasc. Res. 75, 748–757 [DOI] [PubMed] [Google Scholar]
- 43.Epshtein Y., Chopra A. P., Rosenhouse-Dantsker A., Kowalsky G. B., Logothetis D. E., Levitan I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8055–8060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenhouse-Dantsker A., Leal-Pinto E., Logothetis D. E., Levitan I. (2010) Channels 4, 63–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuello L. G., Jogini V., Cortes D. M., Perozo E. (2010) Nature 466, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Epstein W. (2003) Prog. Nucleic Acid Res. Mol. Biol. 75, 293–320 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.