Abstract
Interaction of the pattern recognition receptor, RAGE with key ligands such as advanced glycation end products (AGE), S100 proteins, amyloid β, and HMGB1 has been linked to diabetic complications, inflammatory and neurodegenerative disorders, and cancer. To help answer the question of how a single receptor can recognize and respond to a diverse set of ligands we have investigated the structure and binding properties of the first two extracellular domains of human RAGE, which are implicated in various ligand binding and subsequent signaling events. The 1.5-Å crystal structure reveals an elongated molecule with a large basic patch and a large hydrophobic patch, both highly conserved. Isothermal titration calorimetry (ITC) and deletion experiments indicate S100B recognition by RAGE is an entropically driven process involving hydrophobic interaction that is dependent on Ca2+ and on residues in the C′D loop (residues 54–67) of domain 1. In contrast, competition experiments using gel shift assays suggest that RAGE interaction with AGE is driven by the recognition of negative charges on AGE-proteins. We also demonstrate that RAGE can bind to dsDNA and dsRNA. These findings reveal versatile structural features of RAGE that help explain its ability to recognize of multiple ligands.
Keywords: Diabetes, Innate immunity, Pattern Recognition Receptor, Receptors, X-ray Crystallography, Advanced Glycation Endproducts, RAGE
Introduction
The receptor for advanced glycation end products (RAGE)3 is a multifunctional cell surface protein of the innate immune system thought to play pivotal roles in diabetes, chronic inflammatory conditions, neurodegenerative diseases, and cancer as well as T-lymphocyte proliferation and priming (1–3). In diabetic patients, abnormally high levels of glucose and accompanying reactive oxygen species (ROS) promote the formation of non-enzymatically glucose-derivatized protein (4, 5), also known as advanced glycation end products (AGE). Interaction of AGE with its primary receptor, RAGE, initiates pro-inflammatory responses from a variety of RAGE-expressing cell types such as vascular cells, monocytes/macrophage, B- and T-lymphocytes, retina Müller cells, kidney podocytes, mesangial cells, glial cells and neurons as well as certain cancer cells (2). Unlike macrophage scavenger receptors that can bind AGE and remove it from the cell environment, RAGE does not accelerate the clearance of AGE. Rather, it induces a sustained pro-inflammatory signal (6), which depending on cell type, results in various responses (7) including the generation of ROS and up-regulation of cytokines, vascular cell adhesion molecule 1 (VCAM1), monocyte chemoattractant protein-1 (MCP-1), matrix metalloproteinases, and RAGE itself, each of which potentially exacerbates diabetic pathology. A critical role of RAGE in diabetic complications has been implicated in a NOD/Scid mouse model where animals treated with sRAGE (soluble RAGE ectodomain) showed significant reduction in the development of diabetes after a transfer of splenocytes from a diabetic NOD donor (8). In addition to AGE, RAGE is also known to bind a number of other protein ligands including various S100/calgranulins, high mobility group protein box-1 (HMGB1), β2-integrin Mac-1, and β-amyloid and promote unique inflammatory responses upon ligand recognition (9–11).
S100B is a member of the S100/calgranulin protein family of EF-hand Ca2+-binding proteins. The expression of S100B is highly brain specific and synthesized and secreted by astrocytes and other glial cells. The high level of S100B expression after traumatic brain injury or during neurodegenerative disorders like Alzheimer disease or encephalytis, has made the serum level of S100B an important biomarker of brain damage (12). RAGE-S100B interaction was elucidated first by Hofmann et al. (9), who showed that RAGE-S100B interaction activated NF-kB and that the activation was inhibited by a RAGE-specific antibody. Recent structural and biochemical studies have shown that S100B can exist as a dimer, tetramer, hexamer, or octamer (12).
RAGE is a 45-kDa cell surface receptor comprising three immunoglobulin (Ig) domains followed by a single transmembrane region, and a short C-terminal cytoplasmic tail (13). Although previous studies have provided insight on how RAGE interacts with various ligands, more detailed studies have been hampered by the lack of high resolution structural information of RAGE. Considering the critical role of RAGE in chronic inflammation process such as diabetes, atherosclerosis, and neurodegenerative disorders, atomic level structural detail of RAGE, and its ligand recognition mechanism will be invaluable in development of receptor antagonists. Here we describe the high-resolution crystal structure of domains 1 and 2 of human RAGE. Isothermal titration calorimetry (ITC) experiments were also conducted to understand thermodynamics of RAGE-S100B interaction. Close examinations of RAGE structure and accompanied biochemical experiments led us to propose ligand recognition mechanisms of RAGE, deepening the understanding of this pleiotropic and clinically important receptor.
MATERIALS AND METHODS
Plasmid Construction and Protein Expression
cDNA encoding human RAGE and S100B were each amplified by PCR from pDNR-LIB plasmids (Open Biosystems) to create the constructs RAGE12 (residues 23–220), MBP-RAGE12 (residues 23–231), MBP-RAGE12 isoform 2 (residues 23–231) and S100B. The PCR products were cloned into pET15b expression vectors (Novagen) for RAGE12 and S100B, and a modified pMAL expression vector incorporating a short Ala-Ala-Ala-Ala-Ser fusion linker (kindly provided by T. Kunkel and X. Zhong, NIEHS/NIH) for the MBP-RAGE fusion proteins. A leaderless E. coli chaperone/disulfide-isomerase (DsbC) was co-expressed with all RAGE constructs as described earlier to aid proper disulfide bond formation (14). Expression plasmids for RAGE proteins were transformed into E. coli Rosettagami pLacI (DE3) strain (Novagen). Plasmid pET15b-S100B was transformed into E. coli BL21 (DE3) strain (Novagen). All proteins were expressed using 0.4 mm isopropyl β-d-thiogalactoside in E. coli cells grown in LB medium containing carbenicillin at 30 °C, with 250 rpm agitation, for 3 h.
Protein Purification
MBP-RAGE12 and MBP-RAGE12 isoform 2 were purified by passage through an amylose column followed by a cation exchange column and a size exclusion column in an Akta prime FPLC system (GE Healthcare). In brief, cells were harvested by centrifugation and resuspended in sonication buffer (10 mm Tris-HCl, pH 7.5, 200 mm NaCl, 1 mm EDTA), sonicated on an ice-water bath, and centrifuged for 25 min at 22,000 × g. MBP-RAGE12 was isolated from the sonicated supernatant by adsorption to an amylose resin (New England Biolabs) and eluted with 10 mm maltose in sonication buffer. The eluted protein was loaded on a HiPrep S 16/10 column (GE Healthcare) equilibrated with 20 mm HEPES, pH 7.0. Adsorbed protein was eluted with a linear gradient from 100 mm to 2 m NaCl. The protein was purified further by gel filtration through a Superdex 200 16/60 column (GE Healthcare) equilibrated with 10 mm Tris-HCl, pH 7.5, 150 mm NaCl. Fractions containing MBP-RAGE12 were concentrated to 30 mg/ml with a 10 kDa MWCO Amicon ultrafiltration unit (Millipore). For RAGE12 (without a the MBP fusion tag) purification, cells were sonicated in 50 mm NaH2PO4, pH 8.0, 300 mm NaCl, and 15 mm imidazole, and supernatant was applied to a nickel-nitrilotriacetic acid column (Qiagen). The column was then washed with a linear gradient from 20 mm to 250 mm imidazole to elute adsorbed proteins. The subsequent steps for RAGE12 purification were identical to those of MBP-RAGEs. S100B purification was performed as described previously with minimal variations (15).
Crystallization and Structure Determination
Crystals of MBP-RAGE12 were grown by vapor diffusion using 2 μl of 15 mg/ml protein and an equal volume of precipitant containing 200 mm lithium sulfate, 100 mm Tris-HCl, pH 7.5 and 10% (w/v) polyethylene glycol 4,000 and were fully grown within 2 days. The crystals were briefly transferred to reservoir solutions containing an additional 20% glycerol before cryofreezing. A diffraction data set with Bragg spacings to 1.5 Å was collected on a MAR300 CCD detector at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Data were processed with HKL2000 (16). The structure of MBP-RAGE12 was solved by the molecular replacement method using MOLREP (17) with MBP as a search model (PDB entry, 2VGQ). Crystallographic refinement was performed using a combination of REFMAC5.4 (18) and Phenix1.5 (19). Maltotriose and sulfate molecular topologies were obtained from HIC-Up server (20). Manual rebuilding, adjustment of the MBP-RAGE12 structure, and RMSD calculations were carried out using the graphics program Coot (21). Data processing and refinement statistics are shown in Table 1. Molecular figures were created using UCSF Chimera (22). Buried surface area was calculated using CNS (23, 24). The relative domain orientations were determined using the program HINGE (25). Structure validation was carried out with MolProbity (26).
TABLE 1.
Data processing and refinement statistics
| Human RAGE12 C-terminally fused to MBP | |||
|---|---|---|---|
| Space group | P21212 | ||
| Cell dimensions (Å) | a = 81.75, b = 89.31, c = 97.99, α = β = γ = 90 | ||
| Asymmetric unit | 1 molecule | ||
| Resolution (Å) | 42-1.49 | ||
| Unique reflections | 114,554 | ||
| I/σIa | 32.9 (3.47) | ||
| Completeness (%)a | 99.3 (99.3) | ||
| Rsym (I)a,b | 0.057 (0.415) | ||
| Refinement | |||
| Resolution (Å) | 42-1.49 | ||
| Number of reflections | 110,785 | ||
| Rfreea,c | 18.41 (23.44) | ||
| Rcrysta,d | 16.84 (21.09) | ||
| R.m.s deviation | |||
| Bond length (Å) | 0.006 | ||
| Bond angle (°) | 1.047 | ||
| B-factor, average (Å2) | 19.36 | ||
| Number of atoms | |||
| Protein | 4464 | ||
| Water | 985 | ||
| Other (1 maltotriose and 1 sulfate ion) | 39 | ||
| Ramachandran statistics | MolProbity statistics | ||
| Residues in, (%) | MolProbity score | 1.26, 96th % | |
| Most favored regions | 98.1 | Poor rotamers | 0.21% |
| Additionally allowed regions | 1.7 | Bad bonds | 0 |
| Generously allowed regions | 0.2 | Bad angles | 0 |
| Disallowed regions | 0.0 | Cβ deviations | 0 |
| Clashscore | 4.9, 92nd % | ||
a Parentheses refer to statistics for the highest resolution shell.
b Rsym = ΣhklΣi|Ii(hkl)−I(hkl)|/ΣhklΣiIi(hkl).
c Rfree is calculated with removal of 1.8% of the data as the test set at the beginning of refinement.
d Rcryst=Σhkl||Fobs(hkl)|−|Fcalc(hkl)||/Σhkl|Fobs(hkl)|.
Isothermal Titration Calorimetry
The interactions between various RAGE proteins and S100B were carried out on a VP-ITC instrument (MicroCal) at 25 °C. The proteins were dialyzed overnight at 4 °C in 15 mm HEPES, pH 7.5, 150 mm NaCl. S100B protein was added to the calorimetric reaction cell at a concentration of 0.02∼0.05 mm as a dimer with 310 rpm stirring, and an injection syringe was filled with various RAGE protein solutions at a concentration 10–20-fold higher than the reaction cell. Each titration experiment was performed with 27 injections of 10 μl at 300 s. equilibration intervals. The heat of dilution for RAGE proteins were determined by titrating it into the dialysis buffer. Data were fit with the Origin software package (OriginLab Corp).
Homology Modeling of RAGE Isoform 2
A homology model of RAGE isoform 2 was generated with MODELLER (27) using our crystal structure of RAGE12 as a template. A model with the lowest discrete optimized protein energy (DOPE potential) was chosen among 10 models calculated and energy minimized in AMBER force field for 200 steps with UCSF Chimera interface (22).
Native Gel-shift Assays
Native PAGE was performed according to the manufacturer's instruction (Bio-Rad) to examine the interaction of RAGE and modified BSA. These gels were not run under blue native conditions. Therefore the positions of the proteins in the gel are dictated by both protein charge and size and not by size only. The gels were then stained with Coomassie R-250. To investigate oligonucleotide interaction with RAGE, a 1% agarose gel was used with a 10 mm imidazole-HEPES pH 7.5 buffer system and run at a constant 100 volts. Each well contained 30 μl of sample in the following buffer: 138 mm NaCl, 10 mm imidazole-HEPES pH 7.5, 0.02% Tween 20. The 19-bp DNA duplex consisted of the complementary strands 5′-GCATCACCCCTCCAGAATC-3′ and 5′-GATTCTGGAGGGGTGATGC-3′ and the 10-bp RNA duplex consisted of the complementary strands 5′-GGGACACAGG-3′ and 5′-CCUGUCCC-3′. The gel was stained with Sybr-Gold (Invitrogen) according to the manufacturer's instructions.
Derivatization of BSA, Poly-l-Arg, Poly-l-Lys, and Nα-Acetyl Amino Acids
The derivatization of Nα-Acetyl amino acids (Sigma), l-Arg, l-Lys, l-His, and l-Cys with MG (Sigma M0252) was performed following the procedures by Lo et al. (28). BSA (Sigma, A7906) was modified by GA (Sigma, G6805) or ribose (Sigma, R7500) by incubating BSA at 2 mg/ml in 10 mm GA or 500 mm ribose in 200 mm sodium phosphate, pH 7.4 at 37 °C for 24 h. BSA was acetylated by incubating 4 mg/ml BSA in 25 mm acetic anhydride (Sigma, 45830) in 200 mm sodium phosphate, pH 7.4 at 37 °C for 24 h. All BSA samples were then extensively dialyzed into 10 mm sodium phosphate pH 7.4 at 4 °C. Poly-l-Arg (Sigma, P4663) was derivatized at 1 mg/ml by incubating with 50 mm MG in 200 mm HEPES pH 7.5 at 37 °C for 24 h. The sample was then dialyzed overnight against 10,000 volumes of 50 mm Tris pH 8.0 at 4 °C using a 3.5 kDa molecular weight cutoff. Poly-l-Lys (Sigma, P6516) was derivatized in the same way to generate MG-pLys. After derivatization, both samples had a distinct yellow color indicating that derivatization had occurred.
RESULTS AND DISCUSSION
Overall Structure of RAGE Domains 1and 2
A recombinant human RAGE fragment spanning residues 23–231 was successfully crystallized as a maltose-binding protein (MBP) fusion protein (MBP-RAGE12) and its structure was determined by x-ray crystallography using molecular replacement with MBP as a search model. The crystal structure was refined to 1.5 Å resolution and all the residues of both RAGE domains (RAGE12) were modeled unambiguously in the electron density map. The final model consists of 550 amino acid residues, a maltotriose, 985 water molecules, and a sulfate ion. The crystallographic data is summarized in Table 1.
The overall structure of the fusion protein reveals the ligand binding side of MBP facing the concave side of the RAGE12 and the long axes of the two molecules are approximately perpendicular each other (supplemental Fig. S1). Apart from the covalent linkage, there is only one intramolecular van der Waals contact between MBP and RAGE12, which involves side chains of Gln49 of MBP and Pro215 of RAGE12. However, there are numerous intermolecular crystallographic contacts between MBP and RAGE12 that made the crystallization possible.
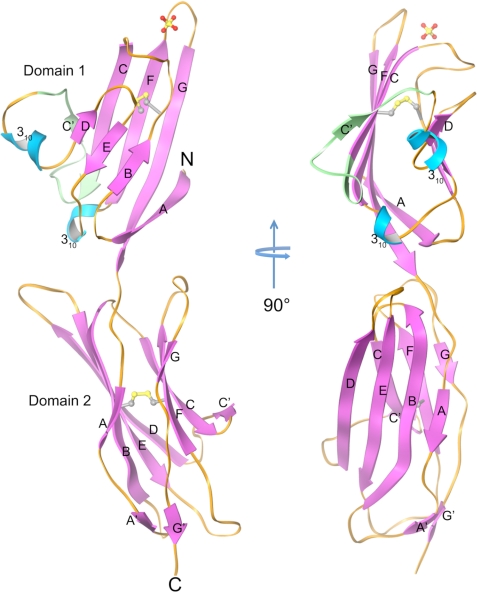
The crystallized fragment of RAGE comprises two Ig domains, domain 1 (residues 23–118) and domain 2 (121–231) connected by a short linker region. Domain 1 has a typical V-type Ig fold with front sheet consisting of β-strands A, G, F, C, and C′ and the back sheet comprising β-strands B, E, and D (Fig. 1). β-strands B and F are connected by a conserved disulfide linkage. However, instead of a C″ strand there is a short four-residue 310 helix and a triple-glycine loop between β-strands C′ and D (C′D loop) (Figs. 1 and 2A). A small one turn 310 helix exists between β-strand E and F as is commonly observed in Ig variable domains. β-strands B, E, and D on the back sheet of the domain 1 are relatively short resulting in longer inter-strand loops. The loops are however well structured through extensive intramolecular hydrogen bonds. A Dali search (29) for domain 1 indicates it is most closely related structurally to λ-light chain variable domains (RMSD of 2.7–2.9 Å over 90 residues) despite having sequence identities of only 14∼20%. Domain 2 adopts a typical C1-type Ig fold with β-strands A, B, E, and D forming the front sheet and β-strands G, F, C, and C′ forming the back sheet. Additionally there are two short β-strands, A′ and G′ appearing after strands A and G, respectively, forming a miniature parallel β-sheet at the end of the domain 2 boundary. A Dali search (29) reveals that the closest structural relative to domain 2 is the second domain of Lutheran blood glycoprotein with which it has a Z-score of 15.0, an RMSD of 2.3 Å over 108 residues and 25% sequence identity (PDB entry 2PET).
FIGURE 1.
Ribbon diagram showing orthogonal views of the crystal structure of the MBP-RAGE 12. MBP is removed from the model for clarity. Helices are cyan, β-strands are magenta, and loops are orange. The part of the C′D loop region lacking in isoform 2 of human RAGE (residues from 54 to 67) is colored green. Disulfide bonds and a sulfate ion are represented by ball and stick models.
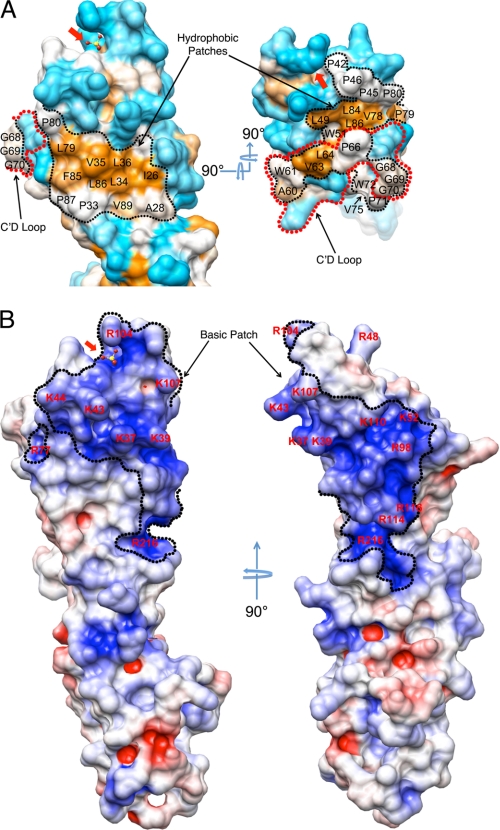
FIGURE 2.
Surface features of RAGE. A, molecular surface representation of RAGE domain 1 highlighting the hydrophobic patches. Side (left) and top view (right) of the hydrophobicity surface were colored using the Kyte and Doolittle scale (49) where hydrophobic residues are colored in orange, hydrophilic residues are colored cyan, and intermediate residues are colored white. The conserved hydrophobic patch is outlined by black dotted lines. The C′D loop is outlined by red dotted lines. B, orthogonal views of the electropotential surface of RAGE domains 1 and 2. The left panel shows the same orientation as the left panel in A. The surface potential was calculated with APBS (30) at an ionic strength of 150 mm NaCl, and the electropotential surface is colored at ±5 kBT. The conserved basic patch is outlined by a black dotted line. The red arrow on all panels points to the bound sulfate.
The domains 1 and 2 of RAGE form an ∼140 degree angle with each other, and are twisted relative to each other along their respective axes almost 70 degrees. The interdomain interface buries 733 Å2 and is predominantly hydrophobic in nature. These interactions suggest that domains 1 and 2 form a rigid unit rather than being domains tethered by a flexible linker. In contrast, a model of the full ectodomain using our crystal structure and the solution NMR structure of RAGE domain 3 (PDB entry 2ENS) suggests that the connection between domains 2 and 3 is a flexible linker composed of a minimum of seven residues (residue from 231 to 237).
During the preparation of this report, three other RAGE-associated structures were released by the PDB. PDB entry 3CJJ is a crystal structure of RAGE domains 1 and 2 similar to our fragment but it has a slightly longer C terminus. PDB entries 2E5E and 2ENS are NMR structures of domain 1 and domain 3, respectively. Superposition of our RAGE12 structure with the 3CJJ structure shows the relative orientation of domains 1 and 2 is well preserved in both structures with a Cα RMSD of 1.93 Å for domain 1, and 0.97 Å for domain 2, respectively. The most noticeable difference is in the C′D loop region where our structure is more open and has a 310 helix while the 3CJJ structure has no 310 helix in this region and the loop has tighter interaction with β-strand D (supplemental Fig. S2). When the C′D loop is omitted from the superposition, the Cα RMSD for domain 1 between the two structures is only 0.86 Å. When our structure is compared with the 2E5E structure, the 310 helix in the C′D loop is preserved. However, the orientation of the preceding C′ β-strand and the triple-glycine portion of the C′D loop differ in 2E5E structure from both our structure and the 3CJJ structure, which indicates flexible nature of C′D loop region (supplemental Fig. S2).
Surface Properties of RAGE Domains 1 and 2
An examination of surface properties of the domain 1 reveals two striking features. First, a large highly conserved slightly recessed hydrophobic patch covers most of the DEB face and extends around the edge of the β-sandwich to include parts of the DE, BC, and C′D loops as well as the edges of strands A, C, and C′ (Fig. 2A). This extensive hydrophobic region includes, but is not limited to residues Ile26, Ala28, Pro33, Leu34, Val35, Leu36, Leu49, Trp61, Val63, Leu64, Trp72, Val75, Val78, Leu79, Pro80, Phe85, Leu86, Pro87, and Val89 as well as the hydrophobic parts of the Lys37 and Tyr113 side chains. Secondly, an electropotential surface map of RAGE generated by APBS (30) reveals a large convex, positively charged region on domain 1 that runs across four strands of the front face (A, G, F, and C) and wraps diagonally around the β-sandwich to the N-terminal end of the opposite DEB face to include the BC loop (Fig. 2B). The residues involved in constructing the positively charged surface include Arg29, Lys37, Lys39, Lys43, Lys44, Arg48, Lys52, Arg98, Arg104, Lys107, Lys110, Arg114, and Arg116 from domain 1 as well as Arg216 from domain 2. Most of the residues arise from BC loop, F and G strands and they are all highly conserved in the RAGE ortholog sequences except for Lys37 which has a Ser or Asn in most other sequences, Arg114 that is replaced by Gln in canine and equine RAGE and Arg116 that is replaced by Gln in pig (supplemental Fig. S3)
Domain 2 shows no distinct surface properties except for a small positively charged patch comprised of four residues Lys123, Arg216, Arg218, and Arg221, on the edge of the β-sandwich. This charged patch is juxtaposed to Arg29 and Arg114 of domain 1, forming a continuation of the positively charged surface of domain 1.
RAGE-S100B Interaction
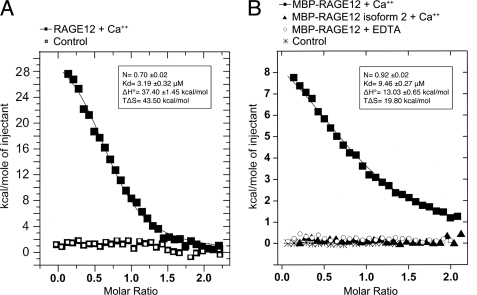
We examined the binding of dimeric S100B to RAGE using ITC. Because of solubility reasons, we used the MBP fusion form of RAGE for most of our ITC experiments. However, addition of MBP to RAGE12 resulted in only a 3-fold reduction in affinity between RAGE12 and S100B (Fig. 3, A and B, and supplemental Fig. S4). Our ITC results on RAGE12-S100B and MBP-RAGE12-S100B showed the interactions occurred with Kd values of 3.2 and 9.4 μm, respectively, in the presence of 1 mm Ca2+, and no interaction occurred in the presence of 1 mm EDTA (Fig. 3B). Thus, the known Ca2+-dependent conformational change of S100B is critical for RAGE binding. Previous surface plasmon resonance (SPR) measurements of RAGE interaction with dimeric S100B have reported values ranging from 0.5 to 8.3 μm for domain 1 alone and 11 nm for a construct containing RAGE domains 1 and 2 (12). The discrepancies with the ITC-derived affinities may potentially be due to immobilization effects in the SPR experiments. The ITC results also demonstrate that the stoichiometry of RAGE to S100B is a 1 to 2 ratio, indicating one molecule of RAGE interacts with a single homodimer of S100B. This stoichiometry and Ca2+ dependence is consistent with SPR and analytical ultracentrifugation measurements performed by Ostendorp et al. (12). The thermodynamics of RAGE-S100B interaction demonstrated in these experiments clearly shows the association between RAGE and S100B involves both positive enthalpy (unfavorable) and entropy (favorable) changes, thus, the association is an entropy driven process (Fig. 3, A and B, supplemental Fig. S4). In general, entropy changes upon protein-protein interaction involve a combination of three factors: hydrophobic solvation effects, conformational changes in protein structure and changes in rotation and translation. Because the last two factors are usually entropically unfavorable in binding reactions, the favorable entropy in this case is most likely due to the burial of hydrophobic surface. This is consistent with the dominant role of hydrophobic interaction observed in several other S100B complexes (31). The presence of Ca2+ is known to induce a large conformational change in S100B that reorients α-helix 3, exposing hydrophobic residues from α-helices 2, 3, and 4 (32). The exposed hydrophobic residues generate a hydrophobic ligand-binding cleft on the surface of the protein (33). On the S100B dimer, the hydrophobic cleft induced by Ca2+ is observed on opposite sides of the molecule. However, for unknown reasons, only one RAGE molecule binds to a single S100B dimer. Apparently RAGE induces asymmetry into the dimer structure, perhaps by sterically occluding part of the other binding site. On the reported octameric S100B structure pairs of adjacent hydrophobic clefts are formed on four sides of the molecule, presenting a total of eight clefts (12). By analogy, tetrameric S100B would be expected to have four clefts. Potentially, the number and orientation of binding clefts presented by various S100B oligomers may allow for very different signaling responses to RAGE recognition of S100B.
FIGURE 3.
ITC titration of S100B to RAGE. The total heat exchanged during each injection of S100B to (A) RAGE12 and (B) MBP-RAGE12 are fit to a single-site binding model with KD and ΔH° as independent parameters, where each fitted value is shown in the inserted panels. No binding was observed for MBP-RAGE12 isoform 2 in the presence of 1 mm Ca2+ or MBP-RAGE12 in the presence of 1 mm EDTA (panel B). The values represent averages from two to three independent experiments.
Importance of the C′D Loop in RAGE-S100B Interaction
RAGE isoform 2 (also known as hRAGEsec) is a human splice variant expressed in both neurons and glial cells, that lacks residues Asn54 to Gln67 in domain 1. This region is highly conserved in all known RAGE sequences. In domain 1 of RAGE, this corresponds to the C-terminal end of β-strand C, all of β-strand C′ and part of the following loop (Figs. 1 and 2A and supplemental Fig. S3). The function of this variant is still under investigation (34). We tested whether this region is critical in S100B recognition by examining S100B binding to RAGE isoform 2 through ITC. RAGE isoform 2 residues spanning from 23 to 231 was expressed as a MBP fusion in the same way as the wild-type form of RAGE. The behavior of MBP-RAGE12 isoform 2 on an analytical size exclusion chromatography was almost identical that of wild-type MBP-RAGE12, which suggested the protein is properly folded (supplemental Fig. S5). An ITC experiment using MBP-RAGE12 isoform 2 under conditions identical to the MBP-RAGE12-S100B binding experiment demonstrated no interaction between RAGE isoform 2 and S100B (Fig. 3B, supplemental Fig. S4). Modeling RAGE isoform 2 with MODELLER (27) using our crystal structure of RAGE12 as a template suggests that the hydrophobic surface generated on the DEB face of the β-sandwich is not significantly affected by this deletion (supplemental Fig. S6). Therefore, we reasoned that the slightly recessed hydrophobic DEB face can be excluded as a S100B binding site and that the interaction between RAGE and S100B requires at least a portion of the Asn54–Gln67 region.
S100B is known to bind many peptides derived from biologically important S100B ligands such as CapZα and p53. Most of these peptides bind in a hydrophobic cleft between α-helices 3 and 4 and require Ca2+ ion coordination to open up the cleft to accommodate incoming peptides. The Ca2+ dependence of S100B interaction with RAGE suggests that a similar binding mechanism is used. The known S100B binding sequences do not appear to follow any particular sequence pattern other than simply containing several hydrophobic and basic residues (31). However, Ivanenkov et al. (35) derived a hydrophobic/basic consensus sequence of (K/R)(L/I)XWXXIL for S100B-binding from a bacteriophage random peptide display library. Interestingly, a portion of the C′D loop of RAGE does bear a resemblance to this consensus sequence in that it contains a conserved Trp61 at the center with a basic residue (Arg57) toward the N terminus and two hydrophobic residues (Val63 and Leu64) at the C-terminal end (Fig. 2A and supplemental Fig. S3). The binding cleft of S100B has been observed to accommodate significantly different peptide ligand conformations and orientations (e.g. p53, NDR-kinase, and TRTK peptides (31, 36, 37)). Although the structure of the C′D loop does not resemble other S100B ligands, the observed flexible nature of the C'D loop suggests it may unfurl and assume a different conformation for S100B binding (supplemental Fig. S2).
Implications for the Recognition of AGE and other Anionic Ligands
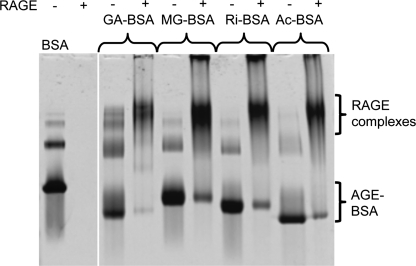
Using native polyacrylamide gel electrophoresis (PAGE) band-shift assays we observed that RAGE12 efficiently recognizes several different AGE-BSA derivatizations including methyl glyoxal (MG)-BSA, glycoaldehyde (GA)-BSA, ribose (Ri)-BSA, and acetylated (Ac)-BSA (Fig. 4). MG-BSA binding to RAGE12 was also demonstrated using size exclusion chromatography (supplemental Fig. S7). Most AGE is generated on chemically reactive surface residues such as Arg, Lys, His, and Cys (28). In an attempt to identify an AGE epitope for RAGE recognition, we chemically modified Nα-acetylated forms of these four amino acids with MG and then tested the displacement of MG-modified BSA (MG-BSA) from RAGE by the MG-derivatized amino acids in native PAGE. However, none of the MG-modified amino acids alone or in combination were able to displace MG-BSA from RAGE even at 29-fold molar excess (supplemental Fig. S8A). Additionally, MG modified poly-l-Lys and poly-l-Arg also failed to displace MG-BSA or Ac-BSA (supplemental Fig. S8B). On the other hand, poly-l-Glu, an overtly polyanionic molecule was able to displace MG-BSA, GA-BSA, and Ac-BSA (supplemental Fig. S8C). Taken together, these lines of evidence suggest that instead of recognizing specific amino acid modifications of AGE-BSA, RAGE recognizes negatively charged patches of the protein surface where AGE modification has resulted in the removal of positive charges from the protein surface and in some cases introduced negative charges (e.g. carboxymethyllysine (CML) and carboxyethyllysine (CEL)) (38, 39). The large positively charged region we note on the surface of domain 1 and part of domain 2 (Fig. 2B) is a likely binding site for this type of interaction. Indeed, recent NMR spectroscopy and mutation experiments imply the involvement of positively charged residues such as Lys37, Lys43, Arg48, Lys44, Arg98, and Arg104 within this region on domain 1 for binding AGE-modified BSA (40, 41). The positively charged surface on RAGE covers an area of ∼20 × 10 Å2. Conservatively, an extended β-strand of only seven residues or a 12–14 residue β-hairpin from an AGE ligand could fit across this region. However, it is also very likely that a collection of neighboring, discontinuous, AGE-derivatized regions such as surface loops on the target protein would form a typical binding site for RAGE. Furthermore, a number of such sites on the protein surface may be required to achieve sufficient avidity for binding RAGE.
FIGURE 4.
Native PAGE of RAGE12/AGE-BSA complexes. The presence of RAGE up-shifted the bands for GA-BSA, MG-BSA, Ri-BSA, and Ac-BSA. Unliganded RAGE is not visible since it is a basic protein and forced out of the top of the gel by the applied voltage. The concentrations of RAGE12 and modified BSA in labeled lanes are 47 and 14 μm, respectively.
Direct RAGE Interaction with DNA and RNA
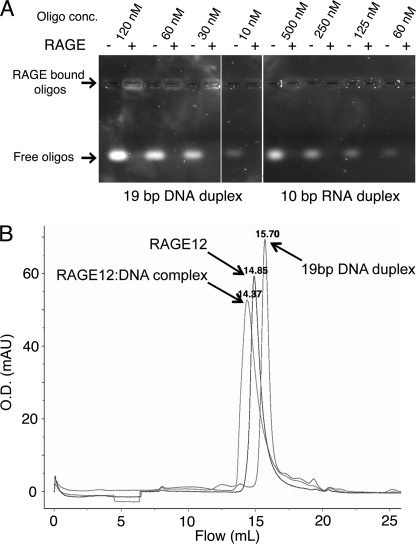
Intrigued by the large positively charged surface of RAGE and noting that other pattern recognition receptors such as TLR3, TLR9, SP-A, and SP-D recognize nucleic acids (42, 43), we tested whether RAGE could also recognize dsDNA and dsRNA. Using agarose gel-shift assays we observed that RAGE12 could bind a 19-bp DNA duplex at a concentration as low as 10 nm and a 10 bp RNA duplex bound RAGE at a concentration as low as 60 nm (Fig. 5A). Addition of EDTA or divalent ions such as Mg2+ did not affect binding (data not shown). Using gel filtration we also observed shifts in the elution profiles of RAGE12 the 19-bp DNA duplex when mixed together indicating complex formation (Fig. 5B). To our knowledge, this is the first time that RAGE has been reported to bind oligonucleotides. During the refinement of the RAGE12 crystal structure, we identified a 9.5σ of tetrahedral electron density sandwiched between BC and FG loops (Figs. 1, 2, A and B, and supplemental Fig. S9). Because 200 mm lithium sulfate was present in the crystallization condition, this electron density was assigned as a sulfate ion. The sulfate ion interacts with two residues that arise from the FG loop, namely Asn103 and Arg104. Further stabilization of the sulfate ion is gained through highly ordered water molecules bonded to the ion. Additionally, we identified four conserved basic residues, Lys37, Lys39, Lys43, and Lys44, of the BC loop running diagonally across the positively charged region (Fig. 2B). Because sulfate-coordinated regions often represent the binding site for a phosphate group on nucleic acids, it is possible that the sulfate binding region along with the Lys residues on BC loop are involved in coordination of the phosphate backbone of oligonucleotides. However, elucidation of the exact nature of RAGE recognition of oligonucleotides will require further investigation.
FIGURE 5.
RAGE interaction with short duplex DNA and RNA oligomers. A, native agarose gel electrophoresis. Oligomers of various concentrations were applied with or without 1 μm of RAGE12. No free oligomers were visible when RAGE12 was added to the wells. When applied to agarose gels, RAGE12 was consistently observed stay in or near the well, perhaps due to interaction with the agarose (data not shown). Therefore, RAGE12 bound oligomers were visible in the application wells when higher concentrations were used. B, gel filtration. Fifty microliters of 20 μm RAGE12 or 6.7 μm 19 bp dsDNA (same DNA as in A) or both together were injected onto a Superdex 200 10/30 column (GE Healthcare) equilibrated with 150 mm NaCl, 25 mm HEPES, pH 7.5. A repeatable elution shift was observed for both RAGE12 and dsDNA when injected together. Labels indicate the elution volumes (in ml) for each peak.
A recent report that class A CpG oligodeoxynucleotides (CpG-A ODN) directly augment HMGB1-RAGE interaction and induce RAGE-dependent cytokine secretion from plasmacytoid dendritic cells and B cells suggests the potential for direct interaction between CpG-A ODN and RAGE (44). Thus, oligonucleotide interaction with RAGE may play a role in immune regulation. Moreover, many pattern recognition receptors of the innate immune system including macrophage scavenger receptors and several toll-like receptors (TLRs) show broad specificity against polyanionic ligands such as oligonucleotides, anionic polysaccharides and lipoteichoic acid (LTA) (45, 46). These proteins are specialized in detecting pathogen-associated molecular patterns (PAMPs) often associated with various microbes as well as cellular stress and consequently activating signaling pathways in the innate and adaptive immune systems. The recognition characteristics of RAGE we have observed suggest that it may fulfill a similar role in the innate immune system, perhaps identifying polyanionic ligands as a danger signal (45, 47, 48).
CONCLUSIONS
Interaction of the pattern recognition receptor RAGE with diverse sets of ligands initiate various cellular responses unique to each ligand. To help understand the recognition mechanisms for this large ligand repertoire, we have determined the high resolution crystal structure of the first two domains of RAGE. Our crystal structure shows large hydrophobic and positively charged regions on the surface of domain 1 likely having direct implications for ligand binding to RAGE. We propose that RAGE employs at least two completely different mechanisms for ligand binding based on the structure and biochemical experiments.
First, we used ITC to show that S100B recognizes RAGE by an entropically driven process dependent on the presence of Ca2+ ions. This strongly suggests a mechanism involving hydrophobic interaction, which is consistent with other known S100B-protein recognition mechanisms (31). Furthermore, we demonstrate that C′D loop-deficient RAGE isoform 2 has no measureable affinity for S100B indicating that the C′D loop is or is very close to the primary point of recognition for RAGE-S100B interaction.
Second, we present evidence that AGE-RAGE recognition occurs primarily through binding negatively charged regions of AGE-modified proteins rather than interaction with distinct glycation moieties on the amino acid side chains of proteins. Consistent with this is our observation that RAGE recognizes BSA that has been derivatized by different types of small molecules such as MG, GA, ribose, and acetic anhydride as well as anionic ligands such as dsDNA, dsRNA, and polyglutamic acid.
Many RAGE ligands are expected to have multiple binding sites due to their oligomeric or polymeric nature or multiple sites of AGE derivatization. It is conceivable that the level of RAGE clustering induced by bound ligands as well as the spatial arrangement of RAGE within these clusters may control the signaling intensities and the ultimate downstream target of these signaling events. Our structural and biochemical information from this study will serve to further the understanding of biology surrounding RAGE.
Supplementary Material
Acknowledgments
We thank Mack Sobhany (NIEHS) and Janeen L. Vanhooke (University of North Carolina) for assistance with ITC experiments; Floyd G. Adsit Jr. (NIEHS) for experimental assistance; colleagues for helpful comments on the manuscript; and the staff at the SER-CAT beamline at the Advanced Photon Source, Argonne National Laboratories for synchrotron support.
Note Added in Proof
While our article was in press, Koch et al. published an article featuring PDB entry 3CJJ (Koch, M., Chitayat, S., Dattilo, B. M., Schiefner, A., Diez, J., Chazin, W. J., and Fritz, G. (2010) Structure 18, 1342–1352).
This work was supported, in whole or in part, by the Intramural Research Program of the National Institutes of Health, NIEHS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
The atomic coordinates and structure factors (code 3O3U) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- RAGE
- receptor for advanced glycation end products
- AGE
- advanced glycation end products
- ITC
- isothermal titration calorimetry
- MBP
- maltose-binding protein
- RMSD
- root mean standard deviation
- SPR
- surface plasmon resonance
- MG
- methyl glyoxal
- GA
- glycoaldehyde
- Ri
- ribose
- Ac
- acetylated
- CML
- carboxymethyllysine
- CEL
- carboxyethyllysine.
REFERENCES
- 1.Sasahira T., Kirita T., Bhawal U. K., Yamamoto K., Ohmori H., Fujii K., Kuniyasu H. (2007) Histopathology 51, 166–172 [DOI] [PubMed] [Google Scholar]
- 2.Brett J., Schmidt A. M., Yan S. D., Zou Y. S., Weidman E., Pinsky D., Nowygrod R., Neeper M., Przysiecki C., Shaw A. (1993) Am. J. Pathol. 143, 1699–1712 [PMC free article] [PubMed] [Google Scholar]
- 3.Lue L. F., Walker D. G., Brachova L., Beach T. G., Rogers J., Schmidt A. M., Stern D. M., Yan S. D. (2001) Exp. Neurol. 171, 29–45 [DOI] [PubMed] [Google Scholar]
- 4.Schmidt A. M., Vianna M., Gerlach M., Brett J., Ryan J., Kao J., Esposito C., Hegarty H., Hurley W., Clauss M. (1992) J. Biol. Chem. 267, 14987–14997 [PubMed] [Google Scholar]
- 5.Schmidt A. M., Hasu M., Popov D., Zhang J. H., Chen J., Yan S. D., Brett J., Cao R., Kuwabara K., Costache G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8807–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramasamy R., Yan S. F., Schmidt A. M. (2005) Trends Cardiovasc Med 15, 237–243 [DOI] [PubMed] [Google Scholar]
- 7.Bierhaus A., Humpert P. M., Morcos M., Wendt T., Chavakis T., Arnold B., Stern D. M., Nawroth P. P. (2005) J. Mol. Med. 83, 876–886 [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Yan S. S., Colgan J., Zhang H. P., Luban J., Schmidt A. M., Stern D., Herold K. C. (2004) J. Immunol. 173, 1399–1405 [DOI] [PubMed] [Google Scholar]
- 9.Hofmann M. A., Drury S., Fu C., Qu W., Taguchi A., Lu Y., Avila C., Kambham N., Bierhaus A., Nawroth P., Neurath M. F., Slattery T., Beach D., McClary J., Nagashima M., Morser J., Stern D., Schmidt A. M. (1999) Cell 97, 889–901 [DOI] [PubMed] [Google Scholar]
- 10.Yan S. D., Chen X., Fu J., Chen M., Zhu H., Roher A., Slattery T., Zhao L., Nagashima M., Morser J., Migheli A., Nawroth P., Stern D., Schmidt A. M. (1996) Nature 382, 685–691 [DOI] [PubMed] [Google Scholar]
- 11.Hori O., Brett J., Slattery T., Cao R., Zhang J., Chen J. X., Nagashima M., Lundh E. R., Vijay S., Nitecki D. (1995) J. Biol. Chem. 270, 25752–25761 [DOI] [PubMed] [Google Scholar]
- 12.Ostendorp T., Leclerc E., Galichet A., Koch M., Demling N., Weigle B., Heizmann C. W., Kroneck P. M., Fritz G. (2007) EMBO J. 26, 3868–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neeper M., Schmidt A. M., Brett J., Yan S. D., Wang F., Pan Y. C., Elliston K., Stern D., Shaw A. (1992) J. Biol. Chem. 267, 14998–15004 [PubMed] [Google Scholar]
- 14.Park H., Adsit F. G., Boyington J. C. (2005) J. Biol. Chem. 280, 13593–13599 [DOI] [PubMed] [Google Scholar]
- 15.Baudier J., Holtzscherer C., Gerard D. (1982) FEBS Lett. 148, 231–234 [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z., Minor W. (1997) Macromolecular Crystallography, Part A, Academic Press [Google Scholar]
- 17.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 18.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 19.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 20.Kleywegt G. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 23.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 24.Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 25.Snyder G. A., Brooks A. G., Sun P. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3864–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) in Current Protocols in Bioinformatics, Unit 5.6, John Wiley & Sons, Inc., New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo T. W., Westwood M. E., McLellan A. C., Selwood T., Thornalley P. J. (1994) J. Biol. Chem. 269, 32299–32305 [PubMed] [Google Scholar]
- 29.Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 30.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya S., Large E., Heizmann C. W., Hemmings B., Chazin W. J. (2003) Biochemistry 42, 14416–14426 [DOI] [PubMed] [Google Scholar]
- 32.Smith S. P., Shaw G. S. (1998) Structure 6, 211–222 [DOI] [PubMed] [Google Scholar]
- 33.Drohat A. C., Tjandra N., Baldisseri D. M., Weber D. J. (1999) Protein Sci 8, 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malherbe P., Richards J. G., Gaillard H., Thompson A., Diener C., Schuler A., Huber G. (1999) Brain Res. Mol. Brain Res. 71, 159–170 [DOI] [PubMed] [Google Scholar]
- 35.Ivanenkov V. V., Jamieson G. A., Jr., Gruenstein E., Dimlich R. V. (1995) J. Biol. Chem. 270, 14651–14658 [DOI] [PubMed] [Google Scholar]
- 36.Inman K. G., Yang R., Rustandi R. R., Miller K. E., Baldisseri D. M., Weber D. J. (2002) J. Mol. Biol. 324, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 37.Rustandi R. R., Baldisseri D. M., Weber D. J. (2000) Nat. Struct. Biol. 7, 570–574 [DOI] [PubMed] [Google Scholar]
- 38.Kislinger T., Fu C., Huber B., Qu W., Taguchi A., Du Yan S., Hofmann M., Yan S. F., Pischetsrieder M., Stern D., Schmidt A. M. (1999) J. Biol. Chem. 274, 31740–31749 [DOI] [PubMed] [Google Scholar]
- 39.Ahmed M. U., Brinkmann Frye E., Degenhardt T. P., Thorpe S. R., Baynes J. W. (1997) Biochem. J. 324, 565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie J., Reverdatto S., Frolov A., Hoffmann R., Burz D. S., Shekhtman A. (2008) J. Biol. Chem. 283, 27255–27269 [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto S., Yoshida T., Murata H., Harada S., Fujita N., Nakamura S., Yamamoto Y., Watanabe T., Yonekura H., Yamamoto H., Ohkubo T., Kobayashi Y. (2008) Biochemistry 47, 12299–12311 [DOI] [PubMed] [Google Scholar]
- 42.Horner A. A., Redecke V., Raz E. (2004) Curr. Opin. Allergy Clin. Immunol. 4, 555–561 [DOI] [PubMed] [Google Scholar]
- 43.Palaniyar N., Nadesalingam J., Clark H., Shih M. J., Dodds A. W., Reid K. B. (2004) J. Biol. Chem. 279, 32728–32736 [DOI] [PubMed] [Google Scholar]
- 44.Tian J., Avalos A. M., Mao S. Y., Chen B., Senthil K., Wu H., Parroche P., Drabic S., Golenbock D., Sirois C., Hua J., An L. L., Audoly L., La Rosa G., Bierhaus A., Naworth P., Marshak-Rothstein A., Crow M. K., Fitzgerald K. A., Latz E., Kiener P. A., Coyle A. J. (2007) Nat. Immunol. 8, 487–496 [DOI] [PubMed] [Google Scholar]
- 45.Platt N., Gordon S. (1998) Chem. Biol. 5, R193–203 [DOI] [PubMed] [Google Scholar]
- 46.Takeda K., Kaisho T., Akira S. (2003) Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 47.Gordon S. (2002) Cell 111, 927–930 [DOI] [PubMed] [Google Scholar]
- 48.Vorup-Jensen T., Carman C. V., Shimaoka M., Schuck P., Svitel J., Springer T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1614–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.