Abstract
In contrast to studies on skeletal and smooth muscles, the identity of kinases in the heart that are important physiologically for direct phosphorylation of myosin regulatory light chain (RLC) is not known. A Ca2+/calmodulin-activated myosin light chain kinase is expressed only in cardiac muscle (cMLCK), similar to the tissue-specific expression of skeletal muscle MLCK and in contrast to the ubiquitous expression of smooth muscle MLCK. We have ablated cMLCK expression in male mice to provide insights into its role in RLC phosphorylation in normally contracting myocardium. The extent of RLC phosphorylation was dependent on the extent of cMLCK expression in both ventricular and atrial muscles. Attenuation of RLC phosphorylation led to ventricular myocyte hypertrophy with histological evidence of necrosis and fibrosis. Echocardiography showed increases in left ventricular mass as well as end-diastolic and end-systolic dimensions. Cardiac performance measured as fractional shortening decreased proportionally with decreased cMLCK expression culminating in heart failure in the setting of no RLC phosphorylation. Hearts from female mice showed similar responses with loss of cMLCK associated with diminished RLC phosphorylation and cardiac hypertrophy. Isoproterenol infusion elicited hypertrophic cardiac responses in wild type mice. In mice lacking cMLCK, the hypertrophic hearts showed no additional increases in size with the isoproterenol treatment, suggesting a lack of RLC phosphorylation blunted the stress response. Thus, cMLCK appears to be the predominant protein kinase that maintains basal RLC phosphorylation that is required for normal physiological cardiac performance in vivo.
Keywords: Calcium Calmodulin-dependent Protein Kinase (CaMK), Cardiac Muscle, Cardiac Hypertrophy, Myosin, Protein Phosphorylation
Introduction
Sarcomeric proteins in myocytes account for contraction of the heart that depends on the molecular motor myosin in the thick filaments binding to actin in thin filaments to initiate shortening and force development (1–3). Myosin cross-bridges contain an actin-binding surface and ATP pocket in the motor domain that taper to an α-helical neck connecting to the myosin rod region responsible for the self-assembly into thick filaments. Two small protein subunits, the essential light chain and the phosphorylatable RLC,2 wrap around each α-helical neck region providing mechanical stability (4). RLC is necessary for assembly of thick filaments in cardiac myocytes, and mutations in RLC are linked to inherited hypertrophic cardiomyopathy (5, 6). There are two types of cardiac RLCs, a ventricular myosin light chain, MLC2v, and an atrium-specific form, MLC2a (7).
In heart and skeletal muscle Ca2+ binds to troponin in the actin thin filament, thereby allowing myosin heads to attach to actin for sarcomeric force development and shortening (8). Additionally, phosphorylation of RLC in fast-twitch skeletal muscle fibers by a skeletal muscle-specific Ca2+/calmodulin-dependent MLCK modulates the contractile response by potentiating frequency-dependent force development (9–11). In the heart, phosphorylation of multiple sarcomeric proteins adjusts myofilament protein interactions and thus fine-tunes the troponin-dependent contraction (3, 12, 13). The basal phosphorylation of RLC (40–50%) in beating hearts is maintained by slow rates of phosphorylation and dephosphorylation (14–17). RLC phosphorylation in skinned fibers increases the extent and rate of force development while decreasing sarcomeric interfilament spacing (9, 12, 18). It is also proposed that altered RLC phosphorylation may contribute to compensatory responses and contractile dysfunction in human diseases (19).
Kinases that phosphorylate RLC in the heart have not been clearly identified, although there are two primary candidates, cMLCK and ZIPK. Skeletal muscle MLCK was reported to be present in heart (20), but its abundance is too low to maintain RLC phosphorylation (10). Cardiac RLC is not a good substrate for the ubiquitous smooth muscle MLCK, which is present in cardiac myocytes and probably phosphorylates nonmuscle cytoplasmic myosin II-B (10, 21–23). Recently, a novel cMLCK was identified in human heart failure and found to be regulated by the cardiac homeobox protein Nkx2-5 during development (24, 25). Suppression of cMLCK expression in zebra fish embryos led to ventricular dilation with incomplete sarcomere formation, whereas overexpression in neonatal myocytes promoted sarcomere organization and increased cell contractility. Cardiac RLC is also a good biochemical substrate for ZIPK, and knockdown of the kinase in neonatal myocytes by siRNA inhibited RLC phosphorylation (26). Thus, ZIPK has emerged as another candidate kinase that may phosphorylate RLC.
We have ablated cMLCK expression in mice to determine its role in RLC phosphorylation. Because it appears to be the primary kinase for basal RLC phosphorylation that may modulate cardiac function, we also evaluated cardiac adaptations resulting from RLC dephosphorylation to obtain insights into the physiological role for cMLCK.
EXPERIMENTAL PROCEDURES
Generation of a Hypomorphic Allele of cMLCK
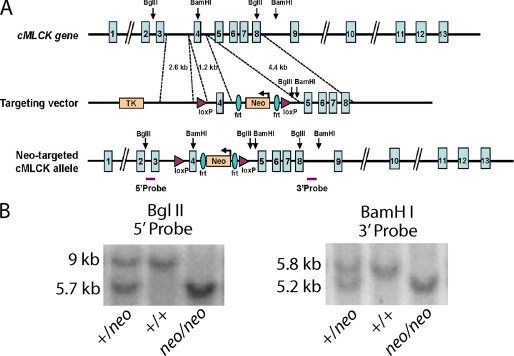
The strategy to study the physiological role of cMLCK in cardiac myocytes was to modify the cMLCK gene to ablate expression at different developmental times from embryonic to adult stages (27). Our approach involved development of a LoxP-targeted allele in mice that will allow different cardiac expressing Cre transgenes to affect cMLCK expression at different developmental times. However, because others suggested that cMLCK was a cardiac myocyte-specific kinase (24, 25), we decided initially to affect kinase expression by retaining the marker cassette containing the neomycin resistance gene (neo) as part of the initial floxed allele to generate a hypomorphic allele (28). Genomic regions of the mouse cMLCK (MYLK3) locus were isolated from 129SvEv genomic DNA by LA TaqTM polymerase (Takara Bio) and cloned into targeting vector OS.DUP/DEL with a TK cassette for negative selection and a neo cassette for positive selection. A 2.6-kb genomic sequence (5′-targeting arm, short arm) upstream of cMLCK exon 4, bounded by an upstream XhoI site and a downstream ClaI site, was cloned upstream of the 5′ loxP sequence in the targeting vector. A 4.4-kb genomic sequence (3′-targeting arm, long arm) downstream of cMLCK exon 4, bounded by an upstream SalI site and a downstream SalI site (including BamHI BglII recognition sequence at the 3′ end for subsequent screening), was cloned downstream of the 3′ loxP sequence. A 1.2-kb region (conditional knock-out region, knock-out arm), including exon 4, bounded by an upstream NdeI site and a downstream AflII site, was cloned between the 5′ loxP sequence and the 5′ FRT sequence. The resulting vector was verified by DNA sequencing and restriction mapping. The vector was linearized at the PvuI site downstream of the 3′-targeting arm and electroporated into 129SvEv-derived embryonic stem cells. Cells were then treated with G418, and negative selection was accomplished by gancyclovir. Southern blot analysis was performed using probes located 5′ of the 5′-targeting arm and 3′ of the 3′-targeting arm (Fig. 1). Accurate recombination was verified by sequencing genomic PCR products derived from primers located 5′ of the 5′-targeting arm and within the neomycin resistance cassette and 3′ of the 3′-targeting arm and within the neomycin resistance cassette. Three cMLCK-targeted embryonic stem clones were identified. Two clones were expanded and injected into C57BL/6 blastocysts that were transferred to the uterus of pseudopregnant females. High percentage chimeric male mice (cMLCK+/neo) were bred into a C57BL/6 background to obtain germ line transmission. We generated mice with a cMLCK hypomorphic allele (cMLCKneo/neo) by intercrossing cMLCK+/neo to each other. All experiments on mice were conducted in a 129SvEv/C57Bl/6 mixed background. Genotyping was performed by Southern blotting with 5′ and 3′ probes. Animals were housed under standard conditions and maintained on commercial mouse chow and water ad libitum. The environment was maintained at 22 °C with a 12-h light/12-h dark cycle. All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center.
FIGURE 1.
Generation of mice with a hypomorphic cMLCK allele. A, schematic representation of the mouse cMLCK gene and targeting strategy. The targeting vector flanks exon 4 with loxP sites and includes insertion of an frt-neomycin (Neo) cassette as well as BglII and BamHI restriction sites. Positions of 3′ and 5′ probes used for Southern blots are shown below the Neo-targeted cMLCK allele. B, Southern blot analysis of cMLCK mutant alleles. Mouse genomic DNA was digested with BglII and hybridized with 5′ probe (left panel) or digested with BamHI and hybridized with 3′ probe (right panel). Mutant alleles yield shorter fragments, as indicated, due to insertion of restriction sites. Mouse genotypes indicated by +/+, WT; +/neo, heterozygous; neo/neo, homozygous for targeted allele.
Southern Blot Analysis
Southern blot probes were generated by PCR using the following primer sets: 5′ probe forward, 5′-CTGGGACTGGGATTATAGACAATTGTG-3′, reverse, 5′-GGTCTAATTAACAGCATGGCCAATGG-3′; and 3′ probe forward, 5′-GGGTCATAGCCATCATTGCACAG-3′, reverse, 5′-GTTAAAGACCATACTTGAGACTCGAGCC-3′. In brief, tail genomic DNA was digested with BglII (5′ screening) or BamHI (3′ screening) and analyzed using a standard Southern blot protocol.
RNA Analysis
Total RNA was purified from isolated heart ventricles with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Two micrograms of RNA were used as template to synthesize cDNA using random hexamers. Quantitative PCR was performed using the following TaqMan® probes purchased from Applied Biosystems: ANP, Mm01255748_g1; BNP, Mm00435304_g1; Col1a2, Mm00483888_m1; Myh6, Mm00440354_m1; Myh7, Mm00600555_m1; and rodent GAPDH, 4308313. Analyses were performed by the comparative CT method. Initial data were normalized to GAPDH; relative values were obtained by normalizing to the mean for cMLCK+/+ ventricles.
Immunoblotting of Proteins
For Western blot analysis, hearts were isolated for dissection in less than 2 min, frozen in liquid nitrogen, and stored at −80 °C until homogenization. Changes in RLC phosphorylation occur on the order of 30–45 min in heart, so immediate fixation in situ is not essential to measure the extent of phosphorylation that reflects in vivo values (15, 16). Tissues were homogenized in 10% trichloroacetic acid and 10 mm dithiothreitol at 0 °C, and total proteins were collected by centrifugation at 2,000 rpm for 1 min in a tabletop centrifuge. Protein pellets were solubilized into 8 m urea as described previously (10, 29, 30). The protein pellets were readily solubilized following the low centrifugation force for a short time. Muscle samples were subjected to urea/glycerol-PAGE to separate phosphorylated and nonphosphorylated RLC as described previously (31). Because the urea/glycerol-PAGE system separates nonphosphorylated from the phosphorylated RLC, we have a direct quantitative measure of RLC phosphorylation in terms of percent phosphorylation. Because the separation results from a single phosphate, data may also be calculated as mol of phosphate/mol of RLC. Diphosphorylation results in additional migration of RLC in the urea-PAGE system, but cardiac muscle has very little diphosphorylated RLC (26, 31). Quantitative measurements were processed on a Storm PhosphorImager and analyzed by ImageQuant software. Additional Western blotting was performed by SDS-PAGE on other proteins solubilized in the 8 m urea buffer. For preparation of soluble proteins, tissues were homogenized in buffer at pH 7.6 containing (in mm) Tris-HCl 50, EGTA 2, EDTA 2, NaCl 150, dithiothreitol, 1% Nonidet P-40, and 10 μl/ml protease inhibitor mixture (Sigma). Contractile proteins were pelleted by centrifugation at 7,000 × g for 10 min. Equal volumes of total and supernatant fractions were subjected to SDS-PAGE. Antibodies to cMLCK, MYPT2, and MLC2a were raised to bacterially expressed mouse protein (Proteintech Group, Inc.). Antibodies to MLC2v from bovine heart were previously described (31). Antibody to smooth muscle MLCK (K36) was obtained from Sigma. Antibody for GAPDH was obtained from Santa Cruz Biotechnology. Measurements of total cTnI and phosphorylation at Ser-23 and Ser-24 were performed by Western blots with antibodies from Research Diagnostics, Inc. (17, 32).
Histological Analyses
Before histological evaluation, hearts were dissected from anesthetized mice, fixed, and then processed into paraffin according to routine procedures (31). Four-chamber longitudinal views were sectioned at the level of the aortic and pulmonary valves (31). The size of cardiac myocytes was measured following wheat germ agglutinin staining as described previously (23), except an optical fractionator probe of Stereo Investigator software (MBF Bioscience) was used to obtain an unbiased estimate of myocyte areas.
Echocardiography
Echocardiograms were performed on conscious, gently restrained mice using either a Sonos 5500 system with a 15-MHz linear probe or Vevo 2100 system with a MS400C scanhead. Left ventricular internal diameter at end-diastole (LVEDD) and end-systole (LVESD) were measured from M-mode recordings. Fractional shortening was calculated as (LVEDD − LVESD)/LVEDD (%). Measurements of interventricular septum thickness, left ventricular internal diameter, and left ventricular posterior wall thickness were made from two-dimensional parasternal short axis views in diastole. Left ventricular mass was calculated by the cubed method as 1.05 × ((IVS + LVID + LVPW)3 − LVID3) (mg), where IVS is interventricular septum thickness; LVID is left ventricular internal diameter; LVPW is left ventricular posterior wall thickness (33). All measurements were made at the level of papillary muscles.
Animal Protocols
Mice were treated with isoproterenol for 7 days to induce cardiac hypertrophy (31). Isoproterenol at 40 mg/ml/g of mouse in saline or saline itself was injected into an Alzet® mini-osmotic pump (model 2001, Durect Corp.), which releases at 1.0 μl/h. Pumps were surgically implanted on the back during anesthesia. Echocardiographic measurements were performed before and after the isoproterenol infusion.
At the end of the treatment, mice were anesthetized (250 mg/kg Avertin, intraperitoneal) and weighed. Whole hearts were removed, weighed, and quick-frozen in liquid nitrogen. Tibial length was also measured.
Statistical Analyses
Data are expressed as mean ± S.E. Statistical evaluation was carried out by using an unpaired Student's t test for two comparisons or analysis of variance (plus the Newman-Keuls method) for multiple comparisons of data with variance homoscedasticity assessed by the Bartlett method. Kruskal-Wallis rank-sum and Nemenyi tests were used in multiple comparisons for data not meeting the homoscedastic variance test. Significance was accepted at a value of p < 0.05.
RESULTS
cMLCK Expression in Heart
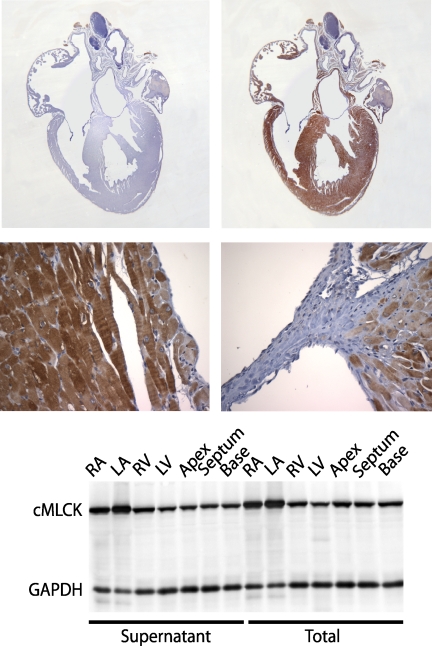
The mRNA for cMLCK was previously shown to be expressed in ventricular and atrial muscle of the heart with no significant expression in other tissues, including skeletal or smooth muscles as well as nonmuscle tissues (24, 25). We have obtained similar results with Northern and Western blotting of cMLCK in diverse tissues, which emphasizes the tissue-specific expression of cMLCK (data not shown). We also immunostained for cMLCK in adult mouse hearts showing specific expression in both ventricular and atrial cardiac myocytes (Fig. 2). The kinase appeared localized in the cytoplasm so we determined biochemically if it was associated with myofilaments (Fig. 2). Comparison of cMLCK in total tissue homogenates with that in supernatant fractions after removal of myofilaments by centrifugation showed that cMLCK was soluble in both ventricular and atrial myocytes, similar to the solubility of skeletal muscle MLCK (34), and in contrast to myofilament binding of smooth muscle MLCK (35). Additionally, the amount of cMLCK expression appeared greater in atria than in ventricles. cMLCK was distributed evenly throughout all portions of the ventricles.
FIGURE 2.
cMLCK expression and distribution in the heart. Upper panels show four-chamber, longitudinal views for sections stained with preimmune serum (left) and serum to cMLCK (right). The lower panels show magnified views of the endocardium (left) and ventricular-aortic valve junction (right) with cMLCK staining in myocytes. The lowest panel shows a Western blot for total and supernatant fractions obtained from various heart locations with cMLCK migrating as a single species at ∼90 kDa. Heart samples were obtained from 18- to 22-week-old male mice. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle.
Disruption of cMLCK Gene Expression Eliminates Basal RLC Phosphorylation in Ventricles and Atria
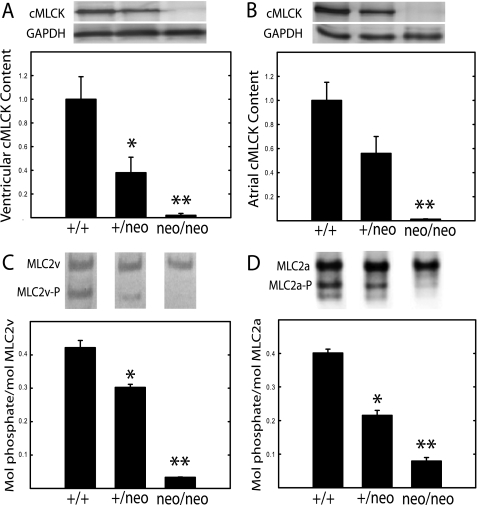
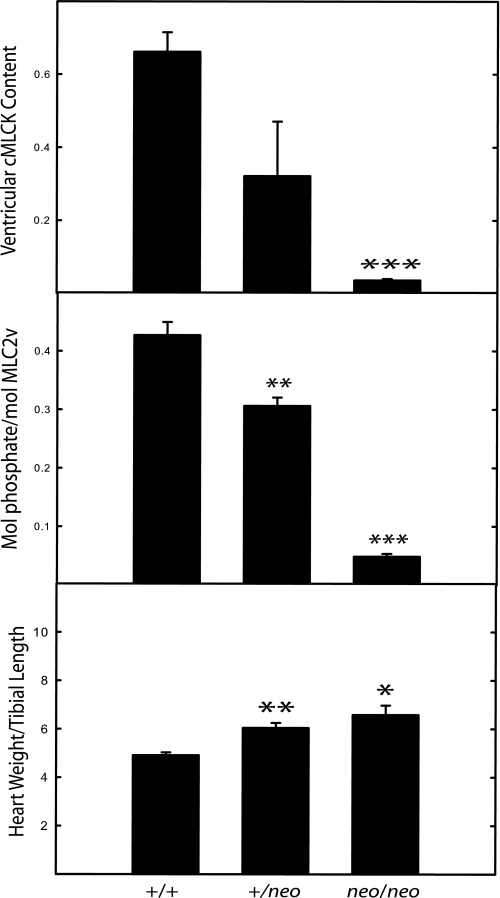
Because cMLCK is a cardiac myocyte-specific kinase, we decided initially to perturb kinase expression by retaining the marker cassette containing the neomycin resistance gene (neo) as part of the initial floxed allele to generate a hypomorphic allele (28). The long range strategy was to have animals in which the neo cassette could be removed and then ablate the gene in adult mice with conditional Cre expression. A targeting vector containing a neo cassette included frt sites as well as loxP sites flanking exon 4 (Fig. 1). Southern analysis of both the 5′ and 3′ arms of the targeted cMLCK gene demonstrated successful recombination and germ line transmission (Fig. 1). Additionally, insertion of the neo cassette disrupted cMLCK expression in both atrial and ventricular myocytes (Fig. 3). Expression of cMLCK protein in ventricular and atrial tissues from cMLCKneo/neo mice was undetectable, whereas the amount in cMLCK+/neo mice was about 50% that found in wild type mice. Interestingly, the partial reduction of cMLCK protein in cMLCK+/neo mice led to a partial reduction of RLC phosphorylation in both ventricular (MLC2v) and atrial (MLC2a) muscles (Fig. 3). The extent of RLC phosphorylation in ventricular and atrial tissues from cMLCKneo/neo mice was less than 5% that obtained for hearts from wild type animals. The extent of basal RLC phosphorylation appears to be dependent on the amount of cMLCK expressed, thus indicating the kinase activity is a limiting factor for RLC phosphorylation.
FIGURE 3.
Relative contents of cMLCK and phosphorylated RLC in ventricular and atrial tissues. Hearts from cMLCK+/+, cMLCK+/neo, and cMLCKneo/neo mice were divided into ventricular and atrial portions and processed for immunoblotting as described under “Experimental Procedures.” A and B, ratio of cMLCK to GAPDH was calculated from densitometry of Western blots with 30 μg of protein loaded per lane (upper panels). Individual ratios were normalized to the mean value for tissue from cMLCK+/+ mice to calculate relative contents in ventricular (A) and atrial (B) tissues (n = 7–13). C and D, molar ratio of phosphorylated RLC was calculated from densitometry of urea-glycerol gel blots with 3 μg of protein loaded per lane (upper panels, phosphorylated forms indicated by P). Average molar ratios are shown for ventricular MLC2v (C) and atrial MLC2a (D) with n = 4–7. Heart samples were obtained from 18- to 22-week-old, male mice. *, p < 0.05; **, p < 0.01 compared with WT.
We removed the neo cassette by crossing Flp-deleter mouse strain to mice containing the neo cassette flanked by frt sites and confirmed crossing results by Southern analysis. The resulting mice containing single (cMLCK+/f) or double (cMLCKf/f) floxed alleles without the neo cassette had similar amounts of cMLCK protein as wild type mice. The relative amounts were 100 ± 1, 104 ± 6, and 99 ± 10% (mean ± S.E., n = 5) for wild type, cMLCK+/f, and cMLCKf/f mice, respectively. The extent of MLC2v phosphorylation was also not different with 0.42 ± 0.2, 0.42 ± 0.01, and 0.41 ± 0.02 for wild type, cMLCK+/f, and cMLCKf/f mice, respectively. The morphological properties of hearts in the three different groups appeared normal. Thus, the insertion of the neo cassette appears to be selective for disrupting cMLCK expression.
We also measured the protein contents of other related proteins, including MLC2v, MLC2a, MYPT2, cTnI, and smooth muscle MLCK (Table 1). There were no differences in the amounts of these proteins in cMLCK+/+, cMLCK+/neo, and cMLCKneo/neo mice. Thus, the loss of cMLCK protein or insertion of the neo cassette did not affect expression of these related proteins.
TABLE 1.
Relative expression of proteins related to cardiac myosin
Values are means ± S.E., n = 3 or more. Heart samples were obtained from 18- to 22-week-old, male mice.
| Measurementsa | cMLCK+/+ | cMLCKneo/neo |
|---|---|---|
| % | % | |
| MLC2v | 100 ± 20 | 99 ± 9 |
| MLC2a | 100 ± 7 | 102 ± 4 |
| MYPT2 | 100 ± 11 | 88 ± 12 |
| cTnI | 100 ± 7 | 96 ± 10 |
| Smooth muscle MLCK | 100 ± 16 | 87 ± 12 |
a p < 0.05 for comparisons to cMLCK+/+ mice.
The thin filament protein cTnI plays an important role in Ca2+ sensitivity of myofilaments, and it was recently reported that overexpression of a nonphosphorylatable MLC2v resulted in a marked compensatory decrease in cTnI phosphorylation (17). We thus determined if cTnI phosphorylation was changed at Ser-23 and -24 in cMLCK hypomorphic hearts. The relative phosphorylation of cTnI was 100 ± 5.7, 94 ± 9.9, and 87 ± 10.5% (mean ± S.E., n = 4) for hearts from wild type, cMLCK+/neo, and cMLCKneo/neo mice, respectively, with no significant differences among groups. Thus, the loss of cMLCK activity with attenuation of MLC2v phosphorylation resulted in no changes in basal cTnI phosphorylation.
Lack of cMLCK and RLC Phosphorylation Leads to Ventricular Hypertrophy
Disruption of cMLCK expression with diminished RLC phosphorylation resulted in ventricular hypertrophy (Table 2 and Fig. 4). The body weights and left tibial lengths were not different, but there were significant differences in heart weights for both cMLCK+/neo and cMLCKneo/neo male mice compared with hearts from wild type animals. These differences are also apparent when heart weights were normalized to tibial lengths showing 16 and 47% increases in the ratios (p < 0.05).
TABLE 2.
Morphometric and echocardiographic parameters
Values are means ± S.E. for at least six samples. Heart samples were obtained from 18- to 22-week-old, male mice.
| Measurements | cMLCK+/+ | cMLCK+/neo | cMLCKneo/neo |
|---|---|---|---|
| Body weight | 28.0 ± 1.5 g | 29.1 ± 0.9 g | 29.7 ± 0.8 g |
| Heart weight | 119 ± 4.6 mg | 145 ± 5.0 mga | 181 ± 3.6 mgb |
| Tibial length | 18.8 ± 0.4 mm | 19.0 ± 0.2 mm | 19.2 ± 0.24 mm |
| Heart weight/tibial length | 6.4 ± 0.16 | 7.4 ± 0.31c | 9.4 ± 0.21a |
| Heart rate | 560 ± 20 beats/min | 540 ± 41 beats/min | 492 ± 20 beats/min |
| Interventricular septum | 0.66 ± 0.02 mm | 0.66 ± 0.02 mm | 0.84 ± 0.02 mmc |
| Posterior wall | 0.92 ± 0.04 mm | 0.80 ± 0.03 mm | 0.90 ± 0.05 mm |
| Left ventricular mass | 70.9 ± 8.92 mg | 79.09 ± 4.25 mg | 148.76 ± 13.28 mgc |
a p < 0.01 for comparisons with cMLCK+/+ mice.
b p < 0.001 for comparisons with cMLCK+/+ mice.
c p < 0.05 for comparisons with cMLCK+/+ mice.
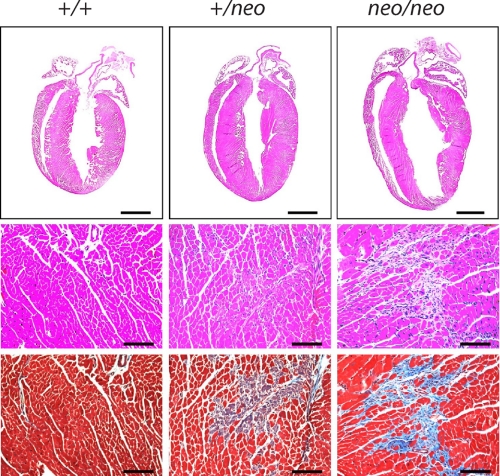
FIGURE 4.
Hypertrophy of hearts from cMLCK hypomorphic mice. Representative histological sections stained with H&E (top two rows) or Masson's trichrome (bottom row). Scale bars, 2 mm (upper row) or 100 μm (lower rows). Hearts were obtained from 18- to 22-week-old male mice with genotypes indicated.
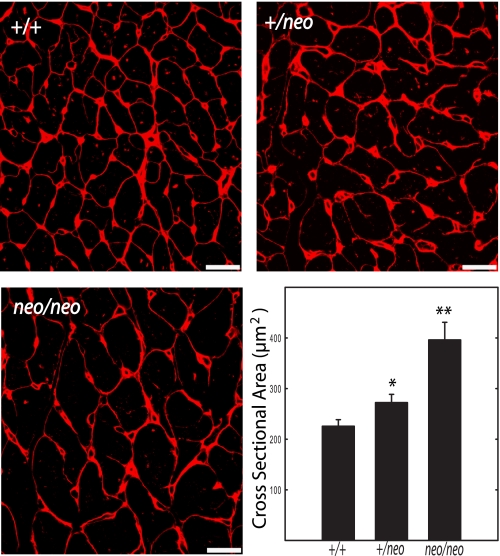
Histological sections confirmed enlargement in both right and left ventricles (Fig. 4). Fig. 5 shows wheat germ agglutinin staining to visualize and quantify cardiac myocyte sizes in hearts from the mice. There was a progressive increase in the cross-sectional area of the myocytes in cMLCK+/+, cMLCK+/neo, and cMLCKneo/neo mice, confirming the hypertrophic response. Hearts from cMLCK+/neo mice manifested ventricular myocyte cell necrosis, although hearts from cMLCKneo/neo mice had interstitial fibrosis by 19–22 weeks of age (Fig. 4). We noted no necrosis or interstitial fibrosis in histological sections of atria from cMLCK+/neo or cMLCKneo/neo mice (data not shown). Thus, evidence was obtained for pathological remodeling with attenuation of cMLCK expression and MLC2v phosphorylation.
FIGURE 5.
Wheat germ agglutinin staining of cardiac myocytes. Staining of transverse sections of hearts from 18- to 22-week-old male cMLCK+/+, cMLCK+/neo, and cMLCKneo/neo mice with wheat germ agglutinin (red) shows a progressive increase in myocyte size. Quantification of myocyte cross-sectional areas indicates significant increases in cMLCK+/neo and cMLCKneo/neo mice. *, p < 0.05; **, p < 0.01 (n = 3 mice). Bar, 20 μm.
Similar to responses obtained in male mice, hearts from female cMLCK+/neo and cMLCKneo/neo mice showed proportional decreases in cMLCK content and MLC2v phosphorylation associated with larger hearts compared with female cMLCK+/+ mice (Fig. 6). Thus, although hearts from female mice are smaller than hearts from male mice, there is no gender difference in the pathological remodeling with the loss of cMLCK.
FIGURE 6.
Characterization of hearts from female hypomorphic mice. Hearts were obtained from 19- to 26-week-old female mice (n = 4–10) for analysis of cMLCK content (upper panel), MLC2v phosphorylation (middle panel), and ratio of heart weight to tibial length (lower panel). *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with values for cMLCK+/+ mice.
Molecular markers of cardiac hypertrophy were analyzed by real time PCR as shown in Table 3. Transcriptional levels of brain natriuretic peptide, but not other hypertrophy markers, were increased in hearts from cMLCKneo/neo mice. The extent of change was modest but consistent with re-activation of a fetal gene program associated with pathological remodeling (36, 37), including changes associated with decreased MLC2v phosphorylation with overexpression of MYPT2 (38).
TABLE 3.
Relative expression of cardiac hypertrophy marker genes
Values are means ± S.E., n = 3. Heart samples were obtained from 18- to 22-week-old, male mice.
| Measurements | cMLCK+/+ | cMLCKneo/neo |
|---|---|---|
| ANP | 1.0 ± 1.2 | 1.5 ± 0.57 |
| BNP | 1.0 ± 0.08 | 1.8 ± 0.23a |
| Myh7 (bMHC) | 1.0 ± 1.1 | 1.9 ± 0.50 |
| Myh6 (aMHC) | 1.0 ± 0.09 | 1.2 ± 0.22 |
| Col1a2 | 1.0 ± 0.08 | 1.2 ± 0.14 |
a p < 0.05 for comparisons with cMLCK+/+ mice.
Disruption of cMLCK Expression Leads to Compromised Cardiac Performance
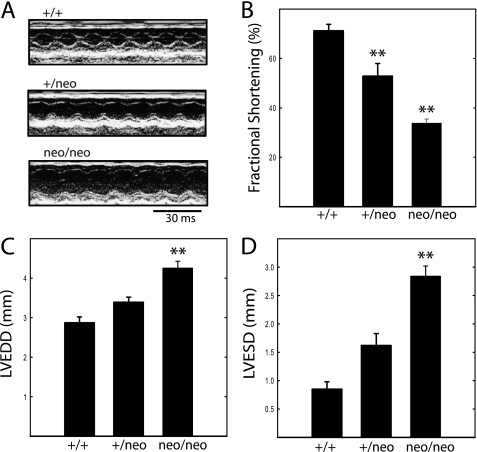
Cardiac function was monitored by echocardiography in nonsedated animals. Attenuation of cMLCK expression with loss of MLC2v phosphorylation led to compromised cardiac function as shown by echocardiography (Fig. 7). There was a proportional decrease in systolic performance assessed as percent fractional shortening from 71% in wild type animals to 53% (p < 0.01) and 34% (p < 0.01) for hearts from cMLCK+/neo and cMLCKneo/neo mice, respectively. However, heart rates among all groups were not different (Table 2). Declines in cardiac function were associated with ventricular dilation with progressive increases in left ventricular end-systolic and end-diastolic dimensions (Fig. 7). Both echocardiographic estimate of LV mass and necropsy evaluation of heart weight/tibia length revealed a significant degree of hypertrophic growth due to disruption of cMLCK expression.
FIGURE 7.
Echocardiographic analysis of hearts from mice of indicated genotype. A, representative M-mode echocardiograms recorded in unanesthetized mice. Details of the analysis are shown in the larger images in Fig. 8. B, left ventricular fractional shortening ((LVEDD − LVESD)/LVEDD) expressed as percent. C, LVEDD. D, LVESD. Data were obtained on 18- to 22-week-old male mice. n = 9, 6, and 15 for cMLCK+/+, MLCK+/neo, and MLCKneo/neo mice, respectively. **, p < 0.01 compared with cMLCK+/+ mice.
Stress-induced Cardiac Hypertrophy Is Altered in cMLCKneo/neo Mice
Isoproterenol infusion for 7 days induced a cardiac hypertrophic response in wild type mice similar to results described previously (31). Isoproterenol treatment increased the heart weight/tibial length ratio in cMLCK+/+ mice from 6.4 ± 0.16 to 11.6 ± 0.33, showing a 1.8-fold increase (Tables 2 and 4). Isoproterenol treatment increased the heart weight/tibial length ratio in cMLCKneo/neo mice from 9.4 ± 0.21 to 11.3 ± 1.27, showing a smaller 1.2-fold increase. The left ventricular masses measured by echocardiography were similar in cMLCK+/+ mice after isoproterenol infusion (136 ± 12 mg) and cMLCKneo/neo mice with or without isoproterenol treatment (147 ± 14 and 132 ± 10 mg; Table 4); all were greater than left ventricular masses for cMLCK+/+ mice without infusion (71.8 ± 8.7; Table 4). Thus, loss of cMLCK itself results in marked hypertrophy, which is not increased with the isoproterenol treatment.
TABLE 4.
Effect of isoproterenol infusion on cardiac morphometric and echocardiographic parameters
Mice were treated with isoproterenol for 7 days to induce cardiac hypertrophy, and echocardiographic measurements were made before and after infusion as described under “Experimental Procedures.” Values are means ± S.E. for at least four samples. Heart samples were obtained from 18- to 22-week-old male mice.
| cMLCK+/+ | cMLCKneo/neo | |
|---|---|---|
| Before isoproterenol infusion | ||
| Heart rate | 664 ± 15 beats/min | 575 ± 31 beats/min |
| Interventricular septum | 0.52 ± 0.04 mm | 0.84 ± 0.01 mma |
| Posterior wall | 0.81 ± 0.04 mm | 0.95 ± 0.1 mm |
| Left ventricular mass | 71.8 ± 8.69 mg | 147.11 ± 13.9 mga |
| After isoproterenol infusion | ||
| Body weight | 30.9 ± 2.1 g | 30.9 ± 2.7 g |
| Heart weight | 212.6 ± 6.6 mg | 208 ± 25.3 mg |
| Tibial length | 18.3 ± 0.17 mm | 18.4 ± 0.17 mm |
| Heart weight/tibial length | 11.6 ± 0.33 | 11.3 ± 1.27 |
| Heart rate | 618 ± 25 beats/min | 687 ± 25 beats/min |
| Interventricular septum | 0.54 ± 0.09 mm | 0.63 ± 0.00 mm |
| Posterior wall | 1.16 ± 0.08 mm | 1.02 ± 0.05 mm |
| Left ventricular mass | 136.1 ± 11.8 mg | 131.9 ± 9.83 mg |
a p < 0.01 for comparisons with cMLCK+/+ mice.
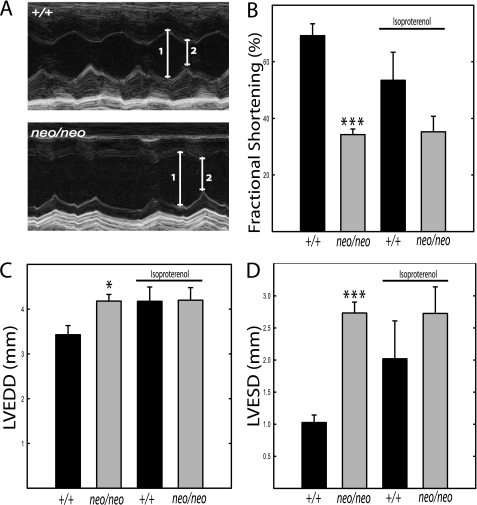
Functional measurements were made by echocardiography in mice infused with isoproterenol (Fig. 8). The diminished fractional shortening observed in cMLCKneo/neo mice was not significantly affected by the isoproterenol infusion. There was also no decrease in animal survival associated with the stress induced by isoproterenol infusion in cMLCKneo/neo mice that already had compromised cardiac performance.
FIGURE 8.
Echocardiographic analysis of hearts from cMLCK+/+ and cMLCKneo/neo mice before and after isoproterenol infusion. A, representative M-mode echocardiograms recorded in unanesthetized mice treated with isoproterenol; 1 and 2 indicate LVEDD and LVESD, respectively. B, left ventricular fractional shortening ((LVEDD − LVESD)/LVEDD) expressed as percent. C, LVEDD. D, LVESD. Data were obtained from 18- to 22-week-old male mice. cMLCK+/+ (black bar, n = 4) and cMLCKneo/neo (gray bar, n = 4), respectively. *, p < 0.05; ***, p < 0.001 compared with cMLCK+/+ mice. Data were obtained from 18- to 22-week-old male mice.
The extent of phosphorylation of MLC2v in hearts from isoproterenol-treated wild type mice (0.43 ± 0.01 mol of phosphate/mol of MLC2v) was similar to that obtained with hearts from noninfused animals (0.42 ± 0.02 mol of phosphate/mol of MLC2v). Notably, the isoproterenol treatment increased MLC2v phosphorylation in hearts from cMLCKneo/neo mice (0.13 ± 0.02 versus 0.03 ± 0.00 mol of phosphate/mol of MLC2v), perhaps by activating ZIPK. However, the increase in MLC2v phosphorylation by this particular treatment did not reach values obtained in cMLCK+/+ mice.
DISCUSSION
The attenuation of cMLCK expression eliminates RLC phosphorylation in both atrial and ventricular myocytes where the basal phosphorylation is thought to fine-tune or modulate normal Ca2+-dependent contraction. These results provide a definitive identification of the primary kinase responsible for cardiac RLC phosphorylation, similar to the identification of other MLCKs responsible for phosphorylation of RLCs in skeletal and smooth muscles (10, 39, 40). Although it was suggested that skeletal muscle MLCK may phosphorylate cardiac RLC in vivo, our results are consistent with the previous observation that knock-out of skeletal muscle MLCK affected RLC phosphorylation only in skeletal muscle and not cardiac muscle (10). ZIPK may also phosphorylate RLC in cardiac muscle (26), but it does not appear to be involved in the basal phosphorylation. ZIPK, a member of the family of death-associated protein kinases, is activated by upstream kinases responsive to different signaling pathways (41–44). The potential signaling pathways that activate ZIPK to phosphorylate cardiac RLC are not known at this time but may involve G protein-coupled receptors and RhoA (42, 45).
Reported biochemical properties of cMLCK show that its maximal specific kinase activity is much lower than skeletal or smooth muscle MLCKs (21, 24, 25). cMLCK contains a high affinity calmodulin-binding sequence, but there were differences in reported Ca2+/calmodulin-dependent kinase activity that will need to be resolved with additional investigations (24, 25). However, the low kinase activity is consistent with the slow turnover of phosphate in RLC (t½ = 250 min) that results in the 40–50% basal phosphorylation in contracting heart muscle (15, 16). With continuous contractions at high frequencies such as those found in rodent hearts, it is predicted that cMLCK would be saturated with bound Ca2+/calmodulin (9, 11, 30). However, spatial gradients resulting in compartmented and local control of Ca2+-signaling pathways in cardiac myocytes could modulate Ca2+/calmodulin signaling to cMLCK (46, 47).
Because there is an attenuation of RLC phosphorylation with a partial decrease in kinase content in cMLCK+/neo animals, cMLCK itself appears limiting for RLC phosphorylation, similar to skeletal but not smooth muscle RLC phosphorylation (11, 48). The phosphorylation of RLC in cardiac muscle occurs with a kinase not tightly bound to myofilaments, which is similar to skeletal muscle (9). In smooth muscle, the availability of Ca2+/calmodulin is limiting so that only a fraction of the kinase can be activated even at high cytosolic Ca2+ concentrations. Smooth muscle MLCK is also bound tightly to actin filaments due to a specific repeat sequence at its N terminus that is not present in either of the striated muscle MLCKs (49–52). The tightly bound kinase extends to myosin thick filaments where the catalytic domain phosphorylates smooth muscle RLC upon Ca2+/calmodulin activation. This spatial organization provides a rapid RLC phosphorylation because the amount of kinase is more abundant relative to the amount of myosin in smooth muscle cells compared with striated myocytes.
Disruption of cMLCK expression results in loss of RLC phosphorylation that leads to inhibition of cardiac performance in vivo. Mice with attenuated cMLCK had increased heart weight/tibial length ratios, increased LV mass, dilation of the LV, impaired LV function, and evidence of fetal gene activation. Animals with cMLCK+/neo genotype exhibited intermediate values between WT and cMLCKneo/neo mice. Together, these observations are consistent with a model where diminished contractile performance provokes a compensatory hypertrophic growth response. As is typical, this response manifests maladaptive features, culminating ultimately in heart failure. Our observations of substantial fibrotic change are consistent with this. That we also observed evidence of necrosis suggests that cell death pathways are activated. The phenotype resembles that described for transgenic mice where cardiac RLC phosphorylation but not cTnI phosphorylation was reduced by overexpression of myosin phosphatase target subunit 2 (38). Together, these studies strongly suggest that the observed ventricular dysfunction results from depressed cardiac RLC phosphorylation.
These results support predictions made from skinned striated muscle fibers where RLC phosphorylation is a positive modulator of Ca2+ sensitivity that shifts the pCa-force relationship to the left and increases the maximal force response (12, 17, 18, 53). Phosphorylation of myosin-binding protein C has similar effects. Phosphorylation of cTnI has the opposite effect where it decreases myofilament sensitivity to calcium by shifting the pCa-force curve to the right. Mice expressing a nonphosphorylatable MLC2v in the heart did not change the Ca2+ sensitivity of force (17). It was observed that phosphorylation of cTnI was also reduced, which would counteract the rightward shift in the pCa-force relation due to reduced MLC2v phosphorylation. Interestingly, these transgenic mice developed atrial but not ventricular hypertrophy (54). The atrial hypertrophy observed with overexpression of nonphosphorylatable MLC2v may have been due to displacement of the endogenous MLC2a subunit of atrial myosin resulting in myofibrillar dysfunction (54).
With the selective attenuation of cMLCK expression, we observed a decrease in RLC but not cTnI phosphorylation, which was associated with development of ventricular hypertrophy. The extent of ventricular enlargement and functional impairment was dependent on the extent of decrease in cMLCK and hence the amount of MLC2v phosphorylation. Overexpression of the nonphosphorylatable MLC2v did not eliminate all MLC2v phosphorylation, but the decrease was sufficient to impair cardiac contractility (17, 54, 55). However, the counterbalancing loss of cTnI phosphorylation may have diminished the impairment of cardiac contractility due to decreased MLC2v phosphorylation. The ventricular hypertrophic response with loss of cMLCK was not associated with decreased cTnI phosphorylation and would thus lead to a greater impairment of cardiac contractility associated with selective cell death, necrosis, and fibrosis with the hypertrophic response.
Pathophysiological stresses induce cardiac hypertrophy through different signaling pathways (56–58). Therefore, it was of interest to determine whether stress would affect hypertrophic hearts with compromised performance from cMLCKneo/neo mice. Isoproterenol infusion did not significantly change the size or performance of hearts from cMLCKneo/neo mice, although hearts from cMLCK+/+ showed hypertrophic responses as reported previously (29). Because the heart has the capacity to increase its mass 2.8–3.0-fold (59), we consider the possibility that the lack of cMLCK and the marked reduction of RLC phosphorylation have dual effects. First, hypertrophy is induced due to negative energetic effects on contractile performance. However, further increases in heart size to a stress such as isoproterenol infusion are attenuated in cMLCKneo/neo mice due to negative effects on myofibril assembly, perhaps because of attenuated RLC phosphorylation (24–25, 60, 61). Additional investigations are needed to establish specific cellular mechanisms involved, but recent results provide new perspectives. Infusion of neuregulin, a peptide that activates ErbB receptor tyrosine kinases in cardiac myocytes, increases expression of cMLCK with increased MLC2v phosphorylation associated with improved cardiac performance after myocardial infarction in rats (62). Administration of neuregulin to patients with stable chronic heart failure improves hemodynamic responses acutely and chronically (63).
In summary, cMLCK is the cardiac specific kinase responsible for the basal phosphorylation of both MLC2v and MLC2a in vivo. The selective loss of MLC2v phosphorylation leads to contractile dysfunction, ventricular hypertrophy, necrosis, and fibrosis. Considering cMLCK was identified in human heart failure (25), it may play an important role not only in the physiological performance of the heart but also adaptive responses to pathophysiological stresses. Mutations in multiple sarcomeric proteins, including MLC2v, lead to ventricular enlargement and failure (6, 64). There is a direct correlation between decreased MLC2v phosphorylation and expression in mice of some MLC2v mutations linked to familial hypertrophic cardiomyopathy (65, 66). Interestingly, the down-regulation of the phosphorylated form of MLC2v by the D166V mutation was associated with an increase in TnI phosphorylation. The R58Q in MLC2v mutation also reduced RLC phosphorylation, thereby decreasing the kinetics of myosin cross-bridges and increasing myofilament Ca2+ sensitivity leading to changes in intracellular Ca2+ homeostasis. Mutation of Glu-22 to Lys in human MLC2v is associated with hypertrophic cardiomyopathy. This mutation inhibits phosphorylation and Ca2+ binding to the light chain (67). Additionally, it increased Ca2+ sensitivity of myofibrillar ATPase activity and force development (68). It is thus predicted cMLCK mutations that cause a loss of kinase activity and reduced MLC2v phosphorylation will also lead to sarcomeric dysfunction and cardiac failure. The identification of the central importance of cMLCK provides a new clinical target for discovery of its role in human cardiac pathophysiology.
Acknowledgments
We thank Tara Billman for experimental, surgical, and mouse assistance, the Histology Core with John Shelton and James Richardson for histological analyses, and Rhonda Bassel-Duby for assistance with RNA analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants HL080536, HL-075173, HL-080144, and HL-090842. This work was also supported by American Heart Association Grant 0640084N, American Diabetes Association Grant 7-08-MN-21-ADA, the American Heart Association-Jon Holden DeHaan Foundation Grant 0970518N, and the Moss Heart Fund.
- RLC
- regulatory light chain
- MLC2v
- ventricular RLC
- MLC2a
- atrial RLC
- cMLCK
- cardiac myosin light chain kinase
- MLCK
- myosin light chain kinase
- ZIPK
- zipper-interacting protein kinase
- cTnI
- inhibitory subunit of cardiac troponin
- MYPT2
- myosin protein targeting subunit 2 of myosin light chain phosphatase
- LV
- left ventricular
- LVEDD
- left ventricular end-diastole dimension
- LVESD
- left ventricular end-systole dimension
- LVID
- left ventricular internal diameter
- LVPW
- left ventricular posterior wall.
REFERENCES
- 1.Gordon A. M., Homsher E., Regnier M. (2000) Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 2.Moss R. L., Razumova M., Fitzsimons D. P. (2004) Circ. Res. 94, 1290–1300 [DOI] [PubMed] [Google Scholar]
- 3.Solaro R. J. (2008) J. Biol. Chem. 283, 26829–26833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayment I. (1996) J. Biol. Chem. 271, 15850–15853 [DOI] [PubMed] [Google Scholar]
- 5.Rottbauer W., Wessels G., Dahme T., Just S., Trano N., Hassel D., Burns C. G., Katus H. A., Fishman M. C. (2006) Circ. Res. 99, 323–331 [DOI] [PubMed] [Google Scholar]
- 6.Poetter K., Jiang H., Hassanzadeh S., Master S. R., Chang A., Dalakas M. C., Rayment I., Sellers J. R., Fananapazir L., Epstein N. D. (1996) Nat. Genet. 13, 63–69 [DOI] [PubMed] [Google Scholar]
- 7.Collins J. H. (2006) J. Muscle Res. Cell Motil. 27, 69–74 [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T., Solaro R. J. (2005) Annu. Rev. Physiol. 67, 39–67 [DOI] [PubMed] [Google Scholar]
- 9.Sweeney H. L., Bowman B. F., Stull J. T. (1993) Am. J. Physiol. 264, C1085–C1095 [DOI] [PubMed] [Google Scholar]
- 10.Zhi G., Ryder J. W., Huang J., Ding P., Chen Y., Zhao Y., Kamm K. E., Stull J. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17519–17524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryder J. W., Lau K. S., Kamm K. E., Stull J. T. (2007) J. Biol. Chem. 282, 20447–20454 [DOI] [PubMed] [Google Scholar]
- 12.Olsson M. C., Patel J. R., Fitzsimons D. P., Walker J. W., Moss R. L. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H2712–H2718 [DOI] [PubMed] [Google Scholar]
- 13.Sweeney H. L., Stull J. T. (1986) Am. J. Physiol. 250, C657–C660 [DOI] [PubMed] [Google Scholar]
- 14.High C. W., Stull J. T. (1980) Am. J. Physiol. 239, H756–H764 [DOI] [PubMed] [Google Scholar]
- 15.Herring B. P., England P. J. (1986) Biochem. J. 240, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silver P. J., Buja L. M., Stull J. T. (1986) J. Mol. Cell. Cardiol. 18, 31–37 [DOI] [PubMed] [Google Scholar]
- 17.Scruggs S. B., Hinken A. C., Thawornkaiwong A., Robbins J., Walker L. A., de Tombe P. P., Geenen D. L., Buttrick P. M., Solaro R. J. (2009) J. Biol. Chem. 284, 5097–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colson B. A., Locher M. R., Bekyarova T., Patel J. R., Fitzsimons D. P., Irving T. C., Moss R. L. (2010) J. Physiol. 588, 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Velden J., Papp Z., Boontje N. M., Zaremba R., de Jong J. W., Janssen P. M., Hasenfuss G., Stienen G. J. (2003) Cardiovasc. Res. 57, 505–514 [DOI] [PubMed] [Google Scholar]
- 20.Davis J. S., Hassanzadeh S., Winitsky S., Lin H., Satorius C., Vemuri R., Aletras A. H., Wen H., Epstein N. D. (2001) Cell 107, 631–641 [DOI] [PubMed] [Google Scholar]
- 21.Stull J. T., Nunnally M. H., Michnoff C. H. (1986) in The Enzymes (Krebs E. G., Boyer P. D. eds) pp. 113–166, Academic Press, Orlando [Google Scholar]
- 22.Dudnakova T. V., Stepanova O. V., Dergilev K. V., Chadin A. V., Shekhonin B. V., Watterson D. M., Shirinsky V. P. (2006) Cell. Motil. Cytoskeleton 63, 375–383 [DOI] [PubMed] [Google Scholar]
- 23.Ma X., Takeda K., Singh A., Yu Z. X., Zerfas P., Blount A., Liu C., Towbin J. A., Schneider M. D., Adelstein R. S., Wei Q. (2009) Circ. Res. 105, 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seguchi O., Takashima S., Yamazaki S., Asakura M., Asano Y., Shintani Y., Wakeno M., Minamino T., Kondo H., Furukawa H., Nakamaru K., Naito A., Takahashi T., Ohtsuka T., Kawakami K., Isomura T., Kitamura S., Tomoike H., Mochizuki N., Kitakaze M. (2007) J. Clin. Invest. 117, 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan J. Y., Takeda M., Briggs L. E., Graham M. L., Lu J. T., Horikoshi N., Weinberg E. O., Aoki H., Sato N., Chien K. R., Kasahara H. (2008) Circ. Res. 102, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang A. N., Chen G., Gerard R. D., Kamm K. E., Stull J. T. (2010) J. Biol. Chem. 285, 5122–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maillet M., Davis J., Auger-Messier M., York A., Osinska H., Piquereau J., Lorenz J. N., Robbins J., Ventura-Clapier R., Molkentin J. D. (2010) J. Biol. Chem. 285, 6716–6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewandoski M. (2001) Nat. Rev. Genet. 2, 743–755 [DOI] [PubMed] [Google Scholar]
- 29.Huang G., Yao J., Zeng W., Mizuno Y., Kamm K. E., Stull J. T., Harding H. P., Ron D., Muallem S. (2006) J. Cell Sci. 119, 153–161 [DOI] [PubMed] [Google Scholar]
- 30.Ding H. L., Ryder J. W., Stull J. T., Kamm K. E. (2009) J. Biol. Chem. 284, 15541–15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Shelton J. M., Richardson J. A., Kamm K. E., Stull J. T. (2008) J. Biol. Chem. 283, 19748–19756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomes A. V., Harada K., Potter J. D. (2005) J. Mol. Cell. Cardiol. 39, 754–765 [DOI] [PubMed] [Google Scholar]
- 33.Collins K. A., Korcarz C. E., Shroff S. G., Bednarz J. E., Fentzke R. C., Lin H., Leiden J. M., Lang R. M. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H1954–H1962 [DOI] [PubMed] [Google Scholar]
- 34.Nunnally M. H., Stull J. T. (1984) J. Biol. Chem. 259, 1776–1780 [PubMed] [Google Scholar]
- 35.Smith L., Parizi-Robinson M., Zhu M. S., Zhi G., Fukui R., Kamm K. E., Stull J. T. (2002) J. Biol. Chem. 277, 35597–35604 [DOI] [PubMed] [Google Scholar]
- 36.Rosenkranz S., Flesch M., Amann K., Haeuseler C., Kilter H., Seeland U., Schlüter K. D., Böhm M. (2002) Am. J. Physiol. Heart Circ. Physiol. 283, H1253–H1262 [DOI] [PubMed] [Google Scholar]
- 37.Xiang W., Kong J., Chen S., Cao L. P., Qiao G., Zheng W., Liu W., Li X., Gardner D. G., Li Y. C. (2005) Am. J. Physiol. Endocrinol. Metab. 288, E125–E132 [DOI] [PubMed] [Google Scholar]
- 38.Mizutani H., Okamoto R., Moriki N., Konishi K., Taniguchi M., Fujita S., Dohi K., Onishi K., Suzuki N., Satoh S., Makino N., Itoh T., Hartshorne D. J., Ito M. (2010) Circ. J. 74, 120–128 [DOI] [PubMed] [Google Scholar]
- 39.He W. Q., Peng Y. J., Zhang W. C., Lv N., Tang J., Chen C., Zhang C. H., Gao S., Chen H. Q., Zhi G., Feil R., Kamm K. E., Stull J. T., Gao X., Zhu M. S. (2008) Gastroenterology 135, 610–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W. C., Peng Y. J., Zhang G. S., He W. Q., Qiao Y. N., Dong Y. Y., Gao Y. Q., Chen C., Zhang C. H., Li W., Shen H. H., Ning W., Kamm K. E., Stull J. T., Gao X., Zhu M. S. (2010) J. Biol. Chem. 285, 5522–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacDonald J. A., Borman M. A., Murányi A., Somlyo A. V., Hartshorne D. J., Haystead T. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haystead T. A. (2005) Cell. Signal. 17, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 43.Graves P. R., Winkfield K. M., Haystead T. A. (2005) J. Biol. Chem. 280, 9363–9374 [DOI] [PubMed] [Google Scholar]
- 44.Hagerty L., Weitzel D. H., Chambers J., Fortner C. N., Brush M. H., Loiselle D., Hosoya H., Haystead T. A. (2007) J. Biol. Chem. 282, 4884–4893 [DOI] [PubMed] [Google Scholar]
- 45.Ihara E., MacDonald J. A. (2007) Can. J. Physiol. Pharmacol. 85, 79–87 [DOI] [PubMed] [Google Scholar]
- 46.Saucerman J. J., Bers D. M. (2008) Biophys. J. 95, 4597–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song Q., Saucerman J. J., Bossuyt J., Bers D. M. (2008) J. Biol. Chem. 283, 31531–31540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isotani E., Zhi G., Lau K. S., Huang J., Mizuno Y., Persechini A., Geguchadze R., Kamm K. E., Stull J. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6279–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin P., Luby-Phelps K., Stull J. T. (1999) J. Biol. Chem. 274, 5987–5994 [DOI] [PubMed] [Google Scholar]
- 50.Lin P., Luby-Phelps K., Stull J. T. (1997) J. Biol. Chem. 272, 7412–7420 [DOI] [PubMed] [Google Scholar]
- 51.Smith L., Stull J. T. (2000) FEBS Lett. 480, 298–300 [DOI] [PubMed] [Google Scholar]
- 52.Stull J. T., Lin P. J., Krueger J. K., Trewhella J., Zhi G. (1998) Acta Physiol. Scand. 164, 471–482 [DOI] [PubMed] [Google Scholar]
- 53.Szczesna D., Zhao J., Jones M., Zhi G., Stull J., Potter J. D. (2002) J. Appl. Physiol. 92, 1661–1670 [DOI] [PubMed] [Google Scholar]
- 54.Sanbe A., Fewell J. G., Gulick J., Osinska H., Lorenz J., Hall D. G., Murray L. A., Kimball T. R., Witt S. A., Robbins J. (1999) J. Biol. Chem. 274, 21085–21094 [DOI] [PubMed] [Google Scholar]
- 55.Dias F. A., Walker L. A., Arteaga G. M., Walker J. S., Vijayan K., Peña J. R., Ke Y., Fogaca R. T., Sanbe A., Robbins J., Wolska B. M. (2006) J. Mol. Cell. Cardiol. 41, 330–339 [DOI] [PubMed] [Google Scholar]
- 56.Dorn G. W., 2nd, Force T. (2005) J. Clin. Invest. 115, 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinsey T. A., Olson E. N. (2005) J. Clin. Invest. 115, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hill J. A., Olson E. N. (2008) N. Engl. J. Med. 358, 1370–1380 [DOI] [PubMed] [Google Scholar]
- 59.Antos C. L., McKinsey T. A., Frey N., Kutschke W., McAnally J., Shelton J. M., Richardson J. A., Hill J. A., Olson E. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferrari M. B., Podugu S., Eskew J. D. (2006) Cell. Biochem. Biophys. 45, 317–337 [DOI] [PubMed] [Google Scholar]
- 61.Li H., Cook J. D., Terry M., Spitzer N. C., Ferrari M. B. (2004) Dev. Dyn. 229, 231–242 [DOI] [PubMed] [Google Scholar]
- 62.Gu X., Liu X., Xu D., Li X., Yan M., Qi Y., Yan W., Wang W., Pan J., Xu Y., Xi B., Cheng L., Jia J., Wang K., Ge J., Zhou M. (2010) Cardiovasc. Res. 88, 334–343 [DOI] [PubMed] [Google Scholar]
- 63.Jabbour A., Hayward C. S., Keogh A. M., Kotlyar E., McCrohon J. A., England J. F., Amor R., Liu X., Li X. Y., Zhou M. D., Graham R. M., Macdonald P. S. (2010) Eur. J. Heart Fail., in press [DOI] [PubMed] [Google Scholar]
- 64.Wang L., Seidman J. G., Seidman C. E. (2010) Ann. Intern. Med. 152, 513–520, W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kerrick W. G., Kazmierczak K., Xu Y., Wang Y., Szczesna-Cordary D. (2009) FASEB J. 23, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abraham T. P., Jones M., Kazmierczak K., Liang H. Y., Pinheiro A. C., Wagg C. S., Lopaschuk G. D., Szczesna-Cordary D. (2009) Cardiovasc. Res. 82, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szczesna D., Ghosh D., Li Q., Gomes A. V., Guzman G., Arana C., Zhi G., Stull J. T., Potter J. D. (2001) J. Biol. Chem. 276, 7086–7092 [DOI] [PubMed] [Google Scholar]
- 68.Szczesna-Cordary D., Guzman G., Zhao J., Hernandez O., Wei J., Diaz-Perez Z. (2005) J. Cell. Sci. 118, 3675–3683 [DOI] [PubMed] [Google Scholar]