Abstract
HDAC3 is a member of the class I histone deacetylase family that regulates gene expression by deacetylation of histones and non-histone proteins. HDAC3 activity has been shown to be modulated by interaction with the co-repressors NCoR and SMRT. Here, we present evidence that the nuclear protein DBC1 is an endogenous inhibitor of HDAC3. DBC1 has been previously identified as a regulator of some nuclear receptors, the methyltransferase SUV39H1, and the NAD-dependent deacetylase SIRT1. Furthermore, DBC1 has been shown to influence transcription regulation and apoptosis, and it may also act as a tumor suppressor. We found that DBC1 interacts and specifically inhibits the deacetylase HDAC3. This interaction depends on the N terminus of DBC1 and the C terminus of HDAC3. Expression of DBC1 not only inhibited HDAC3 activity but also altered its subcellular distribution. In addition, knockdown of endogenous DBC1 in cells and knock-out in mouse tissues increased HDAC3 deacetylase activity. Together, these results identify DBC1 as a new regulator of HDAC3 and demonstrate that DBC1 is a negative regulator of two key distinct deacetylases, SIRT1 and HDAC3. These findings may lead to a better understanding of the biological roles of DBC1 and HDAC3 in metabolic diseases and cancer.
Keywords: Cell Metabolism, Gene Knockout, Histone Deacetylase, Resveratrol, Sirtuins
Introduction
Levels of acetylation in cells are maintained by the complementary activities of two families of enzymes, the histone acetyltransferases and the histone deacetylases (HDACs).2 HDACs are involved in deacetylation of histones and non-histone proteins, including transcription factors, structural proteins, and enzymes, and have an important role in chromatin structure, gene regulation, and control of cell metabolism and cancer (1–3). Because of the vast list of the cellular roles of HDACs, it is imperative that the mechanisms that regulate these enzymes are completely understood. The HDAC superfamily is vast, and these proteins can be classified into four classes, class I, II, III (sirtuins), and IV, based on their homology. Class I includes HDAC1–HDAC3, and HDAC8; class II includes HDAC4–HDAC7, HDAC9, and HDAC10; class III consists of members of the sirtuin family of HDACs; and class IV is represented by HDAC11. In the sirtuin family of deacetylases, special attention has been given to SIRT1 due to its important role in stress responses, cell metabolism, and possibly aging and longevity (4, 5). Understanding the regulation of deacetylases has been the focus of intense investigation. However, the precise mechanisms that regulate HDACs have not been defined.
We and others have recently described that SIRT1 deacetylase is regulated by interaction with the nuclear protein (DBC1 (deleted in breast cancer-1) (6–8). DBC1 was initially described as being absent in certain human breast cancers and has been recently shown to bind and regulate SIRT1, nuclear receptors (such as the estrogen and androgen receptors), and the methyltransferase SUV39H1 (9–11). Furthermore, DBC1 has been implicated as a key component of the molecular mechanism of cellular apoptosis and TNF-α signaling (12). DBC1 binds directly to the catalytic domain of SIRT1, preventing substrate binding to SIRT1 and inhibiting its enzymatic activity (7, 8). Recently, we reported that DBC1 regulates SIRT1 activity in animals during different metabolic conditions and that DBC1 has an important role in the regulation of SIRT1 functions such as control of glucose and lipid homeostasis (6). Despite these findings, much remains unknown about the cellular functions of DBC1.
In higher organisms, it appears that HDACs cooperate with each other or have overlapping roles and deacetylate the same target proteins (2, 13). One deacetylase that shares similar substrates and physiological roles with SIRT1 is HDAC3, a member of the class I family of deacetylases. The members of this class are ubiquitously expressed, but unlike HDAC1 and HDAC2, which are nuclear proteins, HDAC3 can be found in both the nuclei and cytoplasm of cells (14). Similar to other class I HDACs, HDAC3 represses transcription when directed to promoter regions by serving as a co-repressor (2, 15). Interestingly, SIRT1 and HDAC3 share several substrates such as p53, MEF2D (myocyte enhancer factor 2D), and NF-κB, and both associate with p300/CBP (cAMP-responsive element-binding protein-binding protein) acetyltransferase (13, 16–20). Furthermore, both SIRT1 and HDAC3 have been implicated as regulators of several common cellular and physiological functions such as apoptosis, circadian cycle, glucose and lipid metabolism, and cancer (2, 4, 5, 21–23). These observations raise the possibility that HDAC3 and SIRT1 may share not only substrates but also regulatory proteins.
Until now, it has been known that HDAC3 activity can be regulated by two distinct mechanisms. The principal regulatory mechanism appears to be association with a unique large multisubunit protein complex that contains the NCoR and SMRT co-repressors. Association with this complex not only recruits HDAC3 but also directly stimulates HDAC3 enzymatic activity (24, 25). Another mechanism of regulation is phosphorylation/dephosphorylation. HDAC3 is phosphorylated by protein kinase CK2 and dephosphorylated by protein phosphatase 4 (26). Although HDAC3 activity is up-regulated by phosphorylation by protein kinase CK2, it is negatively regulated by protein phosphatase 4.
To identify new mechanisms of regulation of HDAC3, we investigated whether DBC1 is also a regulator of HDAC3. Here, we report that DBC1 not only specifically interacts with HDAC3 but also modulates its localization and activity. These findings present a novel pathway by which HDAC3 activity can be controlled and demonstrate that DBC1 regulates two key deacetylases, namely SIRT1 and HDAC3. Our study opens a new avenue for understanding the mechanisms of regulation of HDACs and also provides a new role for DBC1 as a key regulator of HDAC3.
MATERIALS AND METHODS
Cell Culture
HEK293T and NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium, and A549 cells were maintained in RPMI 1640 medium supplemented with 10% FBS and penicillin/streptomycin (Invitrogen).
Animal Handling and Experiments
All mice used in this study were maintained in the Mayo Clinic Animal Breeding Facility. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Mayo Clinic (Protocol A33209), and all studies were performed according to the methods approved in the protocol. DBC1 knock-out mice were prepare as described previously (6).
Reagents and Antibodies
Except when specified, all reagents and chemicals were purchased from Sigma. Anti-HDAC3 and anti-HA antibodies were from Abcam (Cambridge, MA), and anti-DBC1 antibody was from Bethyl Laboratories (Montgomery, TX).
Plasmids and Expression and Purification of Recombinant Proteins
Mammalian expression plasmids for full-length DBC1, DBC1 deletion mutants, SIRT1, and p300 and DBC1 baculovirus were kindly provided by Zhenkun Lou. In accordance with his previous study (7) using the DBC1 deletion constructs, we also observed that, although the C3 DBC1 protein is smaller than C4 DBC1, it runs slower than C4 DBC1 in SDS-PAGE. It is also obvious that C3 and C4 both appear as doublets, likely indicating that they are processed in the cell or during cell lysis. We have no direct evidence for why C3 runs slower than C4, but it could be that they are modified and processed in different ways in cells. FLAG-HDAC plasmids were obtained from Addgene (Cambridge, MA). HDAC3 deletion mutants and the H134Q mutant were obtained by site-directed mutagenesis using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). HDAC3 mutant H134Q contains the mutations H134Q and H135A (18). Mutations were verified by DNA sequencing. A full-length cDNA clone of MEF2D was purchased from Open Biosystems, and the coding sequence was subcloned into the pIRES2-EGFP-S-FLAG-SBP and pDNA3-HA/Myc vectors. HDAC3 was subcloned in the pcDNA3-HA/Myc vector. We also generated HDAC3 in the baculovirus/insect cell expression system. FLAG-HDAC3 was cloned into pDEST8. Baculovirus was generated using the Bac-to-Bac system (Invitrogen). Sf9 insect cells (American Type Culture Collection) were infected with the appropriate baculovirus and harvested 48 h after infection.
siRNA and shRNA
siRNA against DBC1 was synthesized by Dharmacon (Lafayette, CO). The siRNA duplexes were 21 bp as follows: DBC1 siRNA sense strand, 5′-AAACGGAGCCUACUGAACAUU. Non-targeting siRNA (D001210-03-20, Dharmacon) was used as a control. HDAC3 siRNA was also from Dharmacon (L-003496-00-0020). Transfections were performed twice, 24 h apart, with 150 nm siRNA using DharmaFECT 1 reagent according to the manufacturer's instructions. Cells were harvested 72 h after the first transfection. DBC1 shRNA was from Open Biosystems (clone V2MM_22238).
Immunoprecipitation and Western Blotting
Cells were transfected using Lipofectamine 2000 (Invitrogen) with the appropriate plasmids for 40 h before immunoprecipitation and immunoblotting. All cultured cells, transfected cells, and mouse tissues were lysed in NETN buffer (100 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl (pH 8.0), and 0.5% Nonidet P-40) supplemented with 5 mm NaF, 50 mm 2-glycerophosphate, and a protease inhibitor mixture (Roche Applied Bioscience). Homogenates were incubated at 4 °C for 20 min under constant agitation. Homogenates were then centrifuged at 11,000 × g for 10 min at 4 °C. 1–2 mg of protein was used for each immunoprecipitation. Samples were incubated with 20 μl of Protein A/G (Santa Cruz Biotechnology, Santa Cruz, CA) and 1–2 μg of antibody for 1–2 h at 4 °C under constant rotation. Nonspecific IgG (Santa Cruz Biotechnology) was used as control. Finally, immunoprecipitates were washed two to three times with cold NETN buffer before the addition of 2× Laemmli buffer. Isolation of liver nuclei and immunoprecipitation from isolated nuclei were performed as described previously (6). 50–100 μg of protein was used for each immunoprecipitation and HDAC3 activity measurement. Cell and tissue lysates and immunoprecipitates were analyzed by Western blotting with the indicated antibodies. Western blots were developed using SuperSignalTM West Pico chemiluminescent substrate (Pierce). Films were scanned, and bands were quantified by densitometry using NIH ImageJ.
HDAC Activity Measurements
HDAC1 and HDAC3 activities were measured from immunoprecipitated samples using a HDAC fluorometric assay (BML-AK500–0001, Enzo Life Sciences). After the last wash of the immunoprecipitation, samples were resuspended in 100 μl of assay buffer (50 mm Tris-HCl (pH 8), 137 mm NaCl, 2.7 mm KCl, and 1 mm MgCl2). The reaction was started by the addition of 100 μl of assay buffer containing 500 μm HDAC substrate. Samples were then incubated at 30 °C with constant agitation, and 50-μl aliquots were taken at different times. Each aliquot was divided into three samples, and activity measurements were done in triplicate. The reaction was stopped by the addition of the assay developer. For the controls, samples were incubated in the presence of 1 μm trichostatin A or 2 mm nicotinamide. Values were determined by reading the fluorescence on a fluorometric plate reader (SpectraMax Gemini XPS, Molecular Devices) with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. In all cases, we confirmed the linearity of the reaction over time.
Protein Acetylation and Deacetylation in Vivo
Expression plasmids for FLAG-MEF2D, HA-p300, HA/Myc-HDAC3, and Myc-DBC1 were transfected in 293T cells. About 40 h post-transfection, cells were lysed in NETN buffer supplemented with phosphatase, protease, and deacetylase inhibitors (2 mm nicotinamide and 3 μm trichostatin A). MEF2D was immunoprecipitated with anti-FLAG antibody, and acetylation levels were determined by immunoblotting with anti-acetyllysine antibody (Cell Signaling Technology, Danvers, MA).
RESULTS
DBC1 Interacts with HDAC3 and Regulates HDAC3 Cellular Distribution
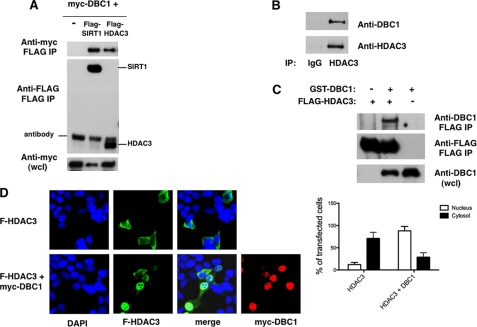
To determine whether HDAC3 interacts with DBC1, we transfected 293T cells with FLAG-HDAC3 and Myc-DBC1. As a control, we also transfected cells with a combination of FLAG-SIRT1 and Myc-DBC1, which are known to interact with each other. As shown in Fig. 1A, Myc-DBC1 specifically interacted with both FLAG-HDAC3 and FLAG-SIRT1. To confirm the interaction between HDAC3 and DBC1, we immunoprecipitated HDAC3 from 293T cells and observed that endogenous DBC1 also interacted with HDAC3 (Fig. 1B). Furthermore, we found that DBC1 interacted specifically with HDAC3 in insect cells infected with baculovirus expressing DBC1 and HDAC3 recombinant proteins (Fig. 1C), suggesting a direct interaction between DBC1 and HDAC3.
FIGURE 1.
Interaction of DBC1 with HDAC3. A, 293T cells were transfected with expression plasmids for Myc-DBC1 and vector control, FLAG-SIRT1, or FLAG-HDAC3. Proteins were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with anti-FLAG and anti-Myc antibodies. wcl, whole cell lysate. B, 293T cell lysates were immunoprecipitated with anti-HDAC3 antibody and immunoblotted with anti-DBC1 and anti-HDAC3 antibodies. C, Sf9 cells were infected with baculovirus expressing GST-DBC1 and FLAG-HDAC3. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-FLAG and anti-DBC1 antibodies. D, 293T cells were transfected with FLAG (F)-HDAC3 in the presence and absence of Myc-DBC1. Cells were fixed and stained with anti-FLAG and anti-Myc antibodies and DAPI. The graph shows the percentage of cells that displayed predominantly cytosolic or nuclear HDAC3 staining in the presence and absence of DBC1. About 30 transfected cells were counted for each condition in each experiment. Each experiment was repeated three times. The differences between HDAC3 distribution in the cytosol and nuclei and between HDAC3 alone and in the presence of DBC1 were statistically significant, with p < 0.05 (t test).
We therefore considered the possibility that DBC1 may be a regulator of HDAC3 localization and activity. We first analyzed the cellular distribution of HDAC3 in the presence and absence of DBC1. The cellular distribution of HDAC3 is one of the unusual characteristics of this HDAC. Whereas other class I HDACs are predominantly nuclear proteins, HDAC3 appears to be located in both the cytosol and nuclei (14). When we overexpressed HDAC3 alone in 293T, we found that, in the majority of transfected cells (88%), HDAC3 was more concentrated in the cytosol than in the nuclei. In contrast, when HDAC3 was coexpressed with DBC1, we found that HDAC3 was localized primarily in the nuclei (71% of the transfected cells) (Fig. 1D and supplemental Fig. S1). A similar HDAC3 cellular distribution in the presence and absence of DBC1 was also observed in transfected HeLa cells (supplemental Fig. S1). These data indicate not only that the two proteins interact in vivo but also that DBC1 regulates HDAC3 subcellular localization.
The N Terminus of DBC1 and the C Terminus of HDAC3 Are Important for the DBC1-HDAC3 Interaction
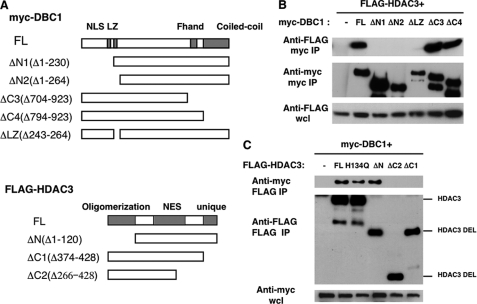
To identify the regions of DBC1 that are responsible for the DBC1-HDAC3 interaction, we transfected cells with FLAG-HDAC3 and deletion mutants of Myc-DBC1 (Fig. 2A). We found that deletion of the N-terminal region of DBC1 (ΔN1 and ΔN2) and also of the leucine zipper motif (ΔLZ) abolished the binding between DBC1 and HDAC3 (Fig. 2B). The leucine zipper motif and the N terminus of DBC1 are also required for the interaction between DBC1 and SIRT1 (7, 8, 10). It thus appears that, akin to the SIRT1-DBC1 interaction (7, 8), both the N terminus and the leucine zipper region of DBC1 contribute to its binding to HDAC3.
FIGURE 2.
The N terminus of DBC1 and the C terminus of HDAC3 are required for the DBC1-HDAC3 interaction. A, shown is a schematic diagram of full-length DBC1 (FL) and deletion mutants of DBC1 and HDAC3. NLS, nuclear localization signal; NES, nuclear export sequence. B, plasmids encoding Myc-tagged full-length DBC1 (FL), deletion mutants, or vector control were transfected in 293T cells along with FLAG-HDAC3. Cell lysates were immunoprecipitated (IP) with anti-Myc antibody and immunoblotted with anti-FLAG and anti-Myc antibodies. wcl, whole cell lysate. C, plasmids encoding FLAG-tagged full-length HDAC3 (FL), mutants, or vector control along with Myc-DBC1 were transfected in 293T cells. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-FLAG and anti-Myc antibodies. Representative experiments of at least three independent experiments are shown. DEL, deletion.
To identify the domains of HDAC3 that interact with DBC1, we transfected 293T cells with full-length Myc-DBC1 and deletion mutants of FLAG-HDAC3 (Fig. 2A). HDAC3 consists of an N-terminal oligomerization region, a central nuclear export region, and a non-conserved C-terminal region that is required for deacetylase and transcriptional repression activity (14, 27). Our results indicate that it is the non-conserved C-terminal region of HDAC3 that is required for the interaction with DBC1. Although a deletion of the N terminus of HDAC3 did not affect its binding to DBC1, a deletion of the last 55 amino acid residues of HDAC3 abolished its binding to DBC1 (Fig. 2C). We also tested whether the HDAC3 mutant H134Q binds to DBC1. This mutant has been described to be deacetylase-deficient (18, 27), and we first investigated whether it had indeed lower HDAC3 activity. Consistent with previous studies (18), we observed reduced deacetylase activity in this mutant (45 ± 21% of wild-type HDAC3 activity, n = 3). Because we observed that this mutant still bound to DBC1 as well as wild-type HDAC3 (Fig. 2C), this indicates that full deacetylase activity of HDAC3 is not necessary for its binding to DBC1 and that DBC1 is probably not a substrate of HDAC3. On the other hand, the fact that DBC1 binds to the C terminus of HDAC3, which is required for the full deacetylase and transcriptional repression activity of this enzyme, suggests that DBC1 may regulate HDAC3 activity.
DBC1 Inhibits HDAC3 Activity
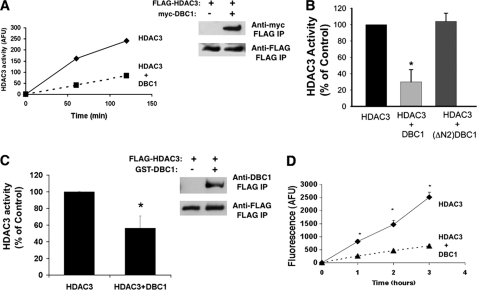
Because DBC1 regulates HDAC3 cellular distribution and interacts with the region of HDAC3 required for its full deacetylase activity, we investigated whether DBC1 can modulate HDAC3 enzymatic activity. We measured HDAC3 deacetylase activity using an in vitro assay after immunoprecipitation of HDAC3 from cells or tissues. As shown in Fig. 3A, full-length HDAC3 overexpressed in 293T cells possessed histone deacetylase enzymatic activity. To confirm that we were indeed measuring HDAC3 activity, we measured this activity in the presence of trichostatin A (a HDAC3 inhibitor) and found that it inhibited HDAC3 activity in our immunoprecipitates (data not shown). When HDAC3 and DBC1 were coexpressed, HDAC3 activity was significantly reduced compared with HDAC3 alone, despite the fact that the same amount of HDAC3 was immunoprecipitated under each condition (Fig. 3, A and B). To further characterize the mechanism of inhibition of HDAC3 activity by DBC1, we determined whether a deletion mutant of the N-terminal region of DBC1 (ΔN2) that does not have the N terminus and the leucine zipper domain of DBC1 and does not interact with HDAC3 (Fig. 2B) had any effect on the HDAC3 activity. Consistent with the interaction experiments (Fig. 2B), deletion of the N-terminal portion from DBC1 completely prevented its inhibitory effect on HDAC3 activity (Fig. 3B).
FIGURE 3.
DBC1 inhibits the HDAC3 deacetylase activity of transfected cells and recombinant proteins. A, 293T cells were transfected with FLAG-HDAC3 in the presence and absence of Myc-DBC1. HDAC3 activity was measured after immunoprecipitation (IP) of FLAG-HDAC3 with anti-FLAG antibody. One representative experiment of a total of five independent experiments is shown. AFU, arbitrary fluorescence units. B, 293T cells were transfected with FLAG-HDAC3 in the presence and absence of Myc-DBC1 or Myc-DBC1-ΔN2. HDAC3 activity was measured after immunoprecipitation with anti-FLAG antibody. C, Sf9 cells were infected with baculovirus expressing FLAG-HDAC3 in the presence and absence of GST-DBC1. HDAC3 activity was measured after immunoprecipitation of FLAG-HDAC3 with anti-FLAG antibody. The immunoblot shows the levels of HDAC3 and DBC1 in immunoprecipitates in a representative experiment. D, FLAG-HDAC3 and GST-DBC1 were purified from baculovirus. FLAG-HDAC3 activity was measured in the presence and absence of purified DBC1 at three time points. Data in B--D are means ± S.D. (n = 3). *, p < 0.05 (t test).
We also measured the effect of DBC1 on HDAC3 activity using recombinant proteins. When both recombinant proteins were expressed in the baculovirus system, we observed an interaction between DBC1 and HDAC3 and an inhibition of HDAC3 activity compared with expression of HDAC3 alone (Fig. 3C). To confirm the effect of recombinant DBC1 on HDAC3 activity, we also purified DBC1 and HDAC3 separately and incubated both proteins together in vitro. Under these conditions, we also observed inhibition of HDAC3 by DBC1 (Fig. 3D), indicating that these proteins directly interact.
DBC1 Inhibits Endogenous HDAC3 Activity
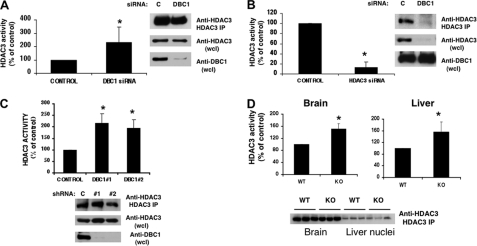
To explore the regulation of HDAC3 by DBC1 in cells and in vivo, we first measured endogenous HDAC3 activity in cells in which DBC1 has been knocked down by siRNA. Consistent with the overexpression results, when DBC1 levels were reduced by siRNA treatment in A549 cells, endogenous HDAC3 activity was higher than in control siRNA-treated cells (Fig. 4A). As a control, we also treated cells with HDAC3 siRNA and found that HDAC3 activity was almost completely inhibited (Fig. 4B). Similar results were obtained when HDAC3 activity was measured in two different clones of NIH3T3 cells that had been stably transfected with DBC1 shRNA. In both DBC1 shRNA clones, HDAC3 activity was higher than in the control shRNA cells (Fig. 4C and supplemental Fig. S2). Western blotting demonstrated that, although DBC1 levels were reduced by both the siRNA and shRNA, the HDAC3 levels remained similar.
FIGURE 4.
DBC1 inhibits endogenous HDAC3 deacetylase activity. A and B, HDAC3 activity was measured in HDAC3 immunoprecipitates of A549 cells transfected with control (C), DBC1, and HDAC3 siRNAs. One representative Western blot of A549 cells shows the levels of DBC1 and HDAC3 in cell lysates and immunoprecipitates (IP) used for activity. wcl, whole cell lysate. C, HDAC3 activity was measured in HDAC3 immunoprecipitates of NIH3T3 cells stably expressing a control and two DBC1 shRNA clones. The immunoblot shows one representative experiment blotted with anti-DBC1 and anti-HDAC3 antibodies. Results are means ± S.D. of three independent experiments. *, p < 0.05 (t test). D, HDAC3 activity was measured after immunoprecipitation of HDAC3 from brain homogenates and extracts of liver nuclei from wild-type and DBC1−/− mice. Data show means ± S.D. of three controls and three DBC1−/− mice for liver and six controls and six DBC1−/− mice for brain homogenates. *, p < 0.05 (t test). The immunoblot shows the levels of HDAC3 in three controls and three DBC1−/− mice. KO, knock-out.
To determine whether the regulation of HDAC3 by DBC1 occurs in vivo, we also measured HDAC3 activity in immunoprecipitates of brain lysates and liver nuclei prepared from wild-type and DBC1 knock-out mice (Fig. 4D). HDAC3 activity was higher in tissues from DBC1 knock-out animals compared with wild-type animals, whereas HDAC3 levels remained unchanged. Because the DBC1 knock-out mice also show an increase in SIRT1 activity (6), these results together confirm that DBC1 regulates endogenous SIRT1 and HDAC3 activity in cells and tissues.
DBC1 Specifically Regulates HDAC3
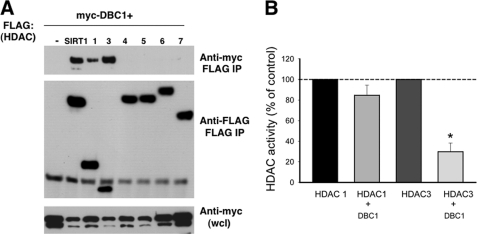
To determine the specificity of the interaction between HDAC3 and DBC1, we investigated whether DBC1 also interacts with other members of the HDAC family. To test this hypothesis, we transfected 293T cells with Myc-DBC1 and FLAG-tagged constructs for other members of the HDAC family of deacetylases. We found that, besides SIRT1, DBC1 interacted only with HDAC3 and HDAC1 and not with HDAC4–HDAC7 (Fig. 5A). This result shows that DBC1 preferentially binds to some types of deacetylases and suggests some specificity for DBC1 as a regulator of deacetylation in cells.
FIGURE 5.
DBC1 specifically regulates HDAC3. A, 293T cells were transfected with Myc-DBC1 and several types of FLAG-tagged HDACs. HDACs were immunoprecipitated (IP) with anti-FLAG antibody, and proteins were immunoblotted with anti-Myc and anti-FLAG antibodies. wcl, whole cell lysate. B, HDAC1 and HDAC3 activities were measured after transfection of 293T cells with FLAG-HDAC1 and FLAG-HDAC3 in the presence and absence of Myc-DBC1. A comparison between the percent inhibition of HDAC1 and HDAC3 activities by DBC1 is shown. Values are means ± S.D. (n = 4). *, p < 0.05 (t test). Inhibition of HDAC1 by DBC1 was not statistically significant.
Because DBC1 also binds to HDAC1, we tested whether DBC1 inhibits HDAC1 activity as well. In contrast to its effect on HDAC3 and SIRT1, the presence of DBC1 promoted a slight, non-statistically significant decrease in HDAC1 activity (Fig. 5B), demonstrating that DBC1 preferentially regulates HDAC3 and SIRT1.
DBC1 Inhibits the Effect of HDAC3 on MEF2D Acetylation
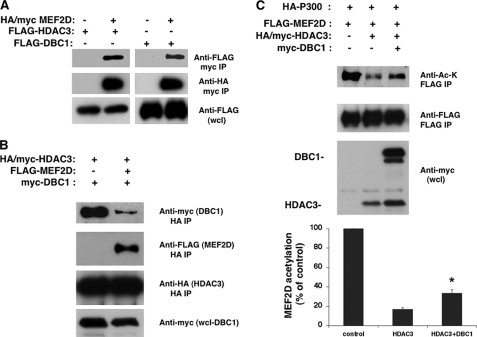
We further explored the role of DBC1 in HDAC3 activity by assessing whether DBC1 affects the ability of HDAC3 to deacetylate MEF2D in cells. The MEF2D transcription factor was initially identified as a nuclear factor important for muscle cell differentiation and has been shown to interact with and be deacetylated by HDAC3 (18). We first tested whether MEF2D also interacts with DBC1. As shown if Fig. 6A, MEF2D interacted with both HDAC3 and DBC1. To test whether both DBC1 and MEF2D compete to bind to HDAC3, we measured the ability of HDAC3 to interact with DBC1 when MEF2D was present. Whereas DBC1 bound to HDAC3 in the absence of MEF2D, the binding between DBC1 and HDAC3 was dramatically reduced when MEF2D was present (Fig. 6B), suggesting that DBC1 competes with HDAC3 substrates to bind to HDAC3. This indicates that DBC1 interaction with HDAC3 is reversible and that the inhibition of HDAC3 by DBC1 may be of a competitive nature. We next addressed whether DBC1 interferes with deacetylation of MEF2D by HDAC3. When cells were transfected with MEF2D and p300, we detected acetylation of MEF2D (Fig. 6C). If cells were also transfected with HDAC3, MEF2D was deacetylated. However, when DBC1 was also present, it inhibited the ability of HDAC3 to deacetylate MEF2D. These results together confirm our hypothesis that DBC1 specifically inhibits HDAC3 deacetylase activity.
FIGURE 6.
DBC1 inhibits the effect of HDAC3 in MEF2D acetylation. A, HA/Myc-MEF2D was transfected in 293T cells in the presence of FLAG-HDAC3 or FLAG-DBC1. MEF2D was immunoprecipitated (IP) with anti-Myc antibody, and proteins were immunoblotted with anti-FLAG and anti-HA antibodies. B, HA/Myc-HDAC3 and Myc-DBC1 were transfected in 293T cells in the presence and absence of FLAG-MEF2D. Protein lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-Myc, anti-HA, and anti-FLAG antibodies. C, FLAG-MEF2D and HA-p300 were transfected in 293T cells in the presence of HA/Myc-HDAC3 and/or Myc-DBC1. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-acetyllysine (Anti-Ac-K) and anti-FLAG antibody. Whole cell lysates (wcl) were immunoblotted for HDAC3 and DBC1 with anti-Myc antibody. The graph shows the percent change in the levels of MEF2D acetylation from three independent experiments as determined by densitometry. Values are the mean ± S.D. *, p < 0.05 (t test).
DISCUSSION
DBC1 was recently identified as a regulator of several proteins, including the deacetylase SIRT1 (6–8). Because of the functional similarities between the deacetylases SIRT1 and HDAC3, we investigated whether DBC1 also regulates HDAC3. The results described in this study identify HDAC3 as a new target of DBC1 and also uncover a new regulatory mechanism of HDAC3 activity.
Until recently, the main mechanism described for regulation of HDAC3 activity was the interaction with the co-repressors NCoR and SMRT. Interaction with these co-repressors is necessary for recruitment and activation of HDAC3 (24, 25). In addition, it was shown that HDAC3 activity is inhibited by interaction with protein phosphatase 4 (26). Beside these two mechanisms, not much is known about HDAC3 regulation in cells. Because we now identified DBC1 as a new negative regulator of HDAC3, it will be important to investigate the exact mechanism by which DBC1 inhibits HDAC3 activity. Because DBC1 binds to the C-terminal region of HDAC3, which is required for both the deacetylase and transcriptional repression functions of HDAC3, it is possible that DBC1 interferes with binding between HDAC3 and substrates or regulators. The possibility that DBC1 prevents binding between HDAC3 and its substrates is supported by the fact that both DBC1 and MEF2D compete for binding to HDAC3. Alternatively, DBC1 could interfere with the interaction between HDAC3 and its activators or inhibitors, such as NCoR/SMRT or kinases and phosphatases. It will also be interesting to investigate how relocation of HDAC3 by DBC1 affects HDAC3 function as a regulator of various cellular processes. In addition, we would like to determine the upstream regulatory events in this DBC1-HDAC3 pathway.
The discovery that DBC1 regulates HDAC3, but not other HDACs, suggests that DBC1 is not a general regulator of deacetylases but specifically regulates HDAC3 and SIRT1. HDAC3 has been shown to regulate a wide variety of cellular processes, including differentiation, proliferation, and apoptosis. At the organismal level, it plays a critical role in development, metabolism, and inflammation through its ability to regulate a number of different transcription factors (2, 3). Although not much is known about the cellular functions of DBC1, it appears that DBC1 is also involved in some of these processes. For instance, both proteins appear to be involved in cancer regulation. Whereas DBC1 has been implicated as a tumor suppressor and expression of DBC1 appears to be lost in several cancer cell lines and in breast cancer (28), HDAC3 expression appears to be elevated in some breast cancer tumors, and HDAC3 inhibitors are being considered as promising candidates for cancer therapy (29). These observations together suggest that the deletion of DBC1 in tumor cells might increase HDAC3 activity, and this could contribute to tumorigenesis. Further insights into how HDAC3 is regulated by DBC1 may provide novel therapeutic uses for both HDAC3 and DBC1 in cancer and metabolic diseases.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK-084055 from NIDDK. This work was supported by grants from the American Federation for Aging Research and from the Mayo Foundation and by the Strickland Career Development Award.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HDAC
- histone deacetylase.
REFERENCES
- 1.Haberland M., Montgomery R. L., Olson E. N. (2009) Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karagianni P., Wong J. (2007) Oncogene 26, 5439–5449 [DOI] [PubMed] [Google Scholar]
- 3.Villagra A., Sotomayor E. M., Seto E. (2010) Oncogene 29, 157–173 [DOI] [PubMed] [Google Scholar]
- 4.Donmez G., Guarente L. (2010) Aging Cell 9, 285–290 [DOI] [PubMed] [Google Scholar]
- 5.Kim E. J., Um S. J. (2008) BMB Rep. 41, 751–756 [DOI] [PubMed] [Google Scholar]
- 6.Escande C., Chini C. C., Nin V., Dykhouse K. M., Novak C. M., Levine J., van Deursen J., Gores G. J., Chen J., Lou Z., Chini E. N. (2010) J. Clin. Invest. 120, 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J. E., Chen J., Lou Z. (2008) Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 8.Zhao W., Kruse J. P., Tang Y., Jung S. Y., Qin J., Gu W. (2008) Nature 451, 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J., Jiang J., Li J., Wang S., Shi G., Feng Q., White E., Qin J., Wong J. (2009) J. Biol. Chem. 284, 6832–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z., Chen L., Kabra N., Wang C., Fang J., Chen J. (2009) J. Biol. Chem. 284, 10361–10366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trauernicht A. M., Kim S. J., Kim N. H., Boyer T. G. (2007) Mol. Endocrinol. 2, 1526–1536 [DOI] [PubMed] [Google Scholar]
- 12.Sundararajan R., Chen G., Mukherjee C., White E. (2005) Oncogene 24, 4908–4920 [DOI] [PubMed] [Google Scholar]
- 13.Zhao X., Sternsdorf T., Bolger T. A., Evans R. M., Yao T. P. (2005) Mol. Cell. Biol. 25, 8456–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W. M., Tsai S. C., Wen Y. D., Fejer G., Seto E. (2002) J. Biol. Chem. 277, 9447–9454 [DOI] [PubMed] [Google Scholar]
- 15.Ishizuka T., Lazar M. A. (2003) Mol. Cell. Biol. 23, 5122–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouras T., Fu M., Sauve A. A., Wang F., Quong A. A., Perkins N. D., Hay R. T., Gu W., Pestell R. G. (2005) J. Biol. Chem. 280, 10264–10276 [DOI] [PubMed] [Google Scholar]
- 17.Chen L., Fischle W., Verdin E., Greene W. C. (2001) Science 293, 1653–1657 [DOI] [PubMed] [Google Scholar]
- 18.Grégoire S., Xiao L., Nie J., Zhang X., Xu M., Li J., Wong J., Seto E., Yang X. J. (2007) Mol. Cell. Biol. 27, 1280–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng L., Xiao Q., Margariti A., Zhang Z., Zampetaki A., Patel S., Capogrossi M. C., Hu Y., Xu Q. (2006) J. Cell Biol. 174, 1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alenghat T., Meyers K., Mullican S. E., Leitner K., Adeniji-Adele A., Avila J., Bućan M., Ahima R. S., Kaestner K. H., Lazar M. A. (2008) Nature 456, 997–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., Sassone-Corsi P. (2008) Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto H., Schoonjans K., Auwerx J. (2007) Mol. Endocrinol. 21, 1745–1755 [DOI] [PubMed] [Google Scholar]
- 24.Guenther M. G., Barak O., Lazar M. A. (2001) Mol. Cell. Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. (2000) EMBO J. 19, 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Ozawa Y., Lee H., Wen Y. D., Tan T. H., Wadzinski B. E., Seto E. (2005) Genes Dev. 19, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen Y. D., Cress W. D., Roy A. L., Seto E. (2003) J. Biol. Chem. 278, 1841–1847 [DOI] [PubMed] [Google Scholar]
- 28.Kim J. E., Chen J., Lou Z. (2009) Cell Cycle 8, 2933–2936 [Google Scholar]
- 29.Marks P. A., Xu W. S. (2009) J. Cell. Biochem. 107, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.