Abstract
Although the mechanisms that regulate folding and maturation of newly synthesized G protein-coupled receptors are crucial for their function, they remain poorly characterized. By yeast two-hybrid screening, we have isolated ANKRD13C, a protein of unknown function, as an interacting partner for the DP receptor for prostaglandin D2. In the present study we report the characterization of this novel protein as a regulator of DP biogenesis and trafficking in the biosynthetic pathway. Co-localization by confocal microscopy with an endoplasmic reticulum (ER) marker, subcellular fractionation experiments, and demonstration of the interaction between ANKRD13C and the cytoplasmic C terminus of DP suggest that ANKRD13C is a protein associated with the cytosolic side of ER membranes. Co-expression of ANKRD13C with DP initially increased receptor protein levels, whereas siRNA-mediated knockdown of endogenous ANKRD13C decreased them. Pulse-chase experiments indicated that ANKRD13C can promote the biogenesis of DP by inhibiting the degradation of newly synthesized receptors. However, a prolonged interaction between ANKRD13C and DP resulted in ER retention of misfolded/unassembled forms of the receptor and to their proteasome-mediated degradation. ANKRD13C also regulated the expression of other GPCRs tested (CRTH2, thromboxane A2 (TPα), and β2-adrenergic receptor), whereas it did not affect the expression of green fluorescent protein, GRK2 (G protein-coupled receptor kinase 2), and VSVG (vesicular stomatitis virus glycoprotein), showing specificity toward G protein-coupled receptors. Altogether, these results suggest that ANKRD13C acts as a molecular chaperone for G protein-coupled receptors, regulating their biogenesis and exit from the ER.
Keywords: Chaperone Chaperonin, G Protein-coupled receptors (GPCR), Prostaglandins, Proteasome, Protein Export, Protein Folding, Protein-Protein Interactions, Receptors
Introduction
As the largest family of membrane receptors, G protein-coupled receptors (GPCRs)4 mediate physiological responses to a vast array of cellular mediators such as hormones, neurotransmitters, lipids, nucleotides, peptides, ions, and photons. To be fully functional, GPCRs need to be delivered to the plasma membrane in a ligand-responsive and signaling-competent form. The synthesis and maturation of these receptors require a complex combination of processes that include protein folding, posttranslational modifications, and transport through distinct cellular compartments of the secretory pathway (1). In recent years, GPCRs were found to interact with many proteins besides G proteins, and such interactions were shown to play important roles in receptor biogenesis and trafficking. For example, the Drosophila cyclophilin protein ninaA (neither inactivation nor afterpotential A) and its mammalian homolog, RanBP2 (Ran-binding protein 2), associate with rhodopsin and opsins, respectively. They have been characterized as chaperones that can facilitate receptor folding and are essential for efficient localization of opsins at the cell surface (2–4). The receptor-activity-modifying proteins represent another example of GPCR-interacting proteins implicated in the trafficking and functionality of receptors. Association of these transmembrane proteins with the calcitonin receptor-like receptor and the calcium-sensing receptor is required for their transport to the plasma membrane (5, 6). The endoplasmic reticulum (ER) membrane-associated dopamine receptor-interacting protein of 78 kDa (DRiP78) is also known to regulate the transport to the plasma membrane of various GPCRs including the D1 receptor for dopamine and the angiotensin II AT1 receptor (7, 8). Expression of DRiP78 causes retention of the D1 receptor in the ER through binding of a conserved FX3FX3F motif found in the C terminus of the receptor.
Our laboratory is interested in the biology of the DP receptor for prostaglandin D2 (PGD2). PGD2 is a lipid mediator produced from the sequential metabolism of arachidonic acid by the cyclooxygenases and prostaglandin D synthases and is mostly known for its role in sleep induction as well as its effects on the immune system and in inflammation (9, 10). PGD2 is critical to the development of allergic diseases such as asthma (11, 12). Recent evidence also suggests new roles for PGD2 in bone physiology (13–15). PGD2 elicits its biological responses by activating two pharmacologically distinct G protein-coupled receptors, the D-prostanoid receptor DP (also known as DP1) and the chemoattractant receptor-homologous molecule expressed on Th2 cells (CRTH2, also known as DP2) (16, 17). The major signaling pathway for DP is through the activation of Gαs, which leads to activation of adenylyl cyclase and results in an increased production of intracellular cAMP (16). Binding of PGD2 to DP results in inhibition of platelet aggregation and causes bronchodilation and vasodilation (11). The activation of DP also inhibits apoptosis of eosinophils and migration and degranulation of basophils (18).
Identification and characterization of accessory proteins confirmed that GPCR-interacting proteins can play crucial roles in the biogenesis and function of GPCRs. Therefore, to characterize putative proteins implicated in the regulation of DP, we used a yeast two-hybrid screen to identify proteins that associate with the C terminus of the receptor. In the present study we demonstrated that DP can interact with a previously uncharacterized protein named ankyrin repeat domain-containing protein 13C (ANKRD13C). We found that this novel protein is localized on ER membranes where it binds to DP to control its biogenesis and trafficking. We also showed that ANKRD13C modulates the expression and maturation of other GPCRs as well, indicating that ANKRD13C is a novel regulator of the biogenesis and trafficking of GPCRs through the biosynthetic pathway.
EXPERIMENTAL PROCEDURES
Reagents
Anti-myc polyclonal antibody, anti-GFP monoclonal antibody, anti-GAPDH polyclonal antibody, anti-GRK2 polyclonal antibody, and protein G-agarose were purchased from Santa Cruz Biotechnology. Anti-GST polyclonal antibody was from Bethyl Laboratories. Monoclonal anti-HA and anti-myc were from Covance. Anti-actin polyclonal antibody and FLAG-specific monoclonal antibodies (M1 and M2) were from Sigma. Rat monoclonal anti-HA -peroxidase High Affinity antibody (clone 3F10) was from Roche Diagnostics. Rabbit anti-calnexin was from Stressgen Bioreagents. Rabbit polyclonal anti-20S proteasome “Core” subunits antibody (PW 8155) was from Enzo Life Sciences. Alexa conjugated secondary antibodies were from Molecular Probes. Easytag Express [35S] protein labeling mix (1175 Ci/mmol) was obtained from PerkinElmer Life Sciences. Goat anti-mouse alkaline phosphatase-conjugated antibody and the alkaline phosphatase substrate kit were purchased from Sigma. The pEGFP-VSVG plasmid was from Addgene (Addgene plasmid 11912) (19).
Yeast Two-hybrid Screen
A pAS2–1 plasmid containing the C terminus of DP (amino acids 332–359) was transformed into the yeast strain pJ69–4α according to the lithium yeast transformation protocol (20). This stably transformed clone was then transformed with a human brain cDNA library or with the empty pGad424 plasmid (Clontech). Screening of 1.6 × 107 transformants yielded five independent clones that were confirmed to interact with the C terminus of DP. Clones showing positive interactions were isolated, and the selected plasmids were sequenced by dideoxy sequencing. Identities of the clones were determined using the NCBI BLAST alignment tool.
Plasmid Construction
The full-length open reading frame (ORF) of ANKRD13C was cloned by RT-PCR using template RNAs extracted from HEK293 cells. Myc- and HA-tagged ANKRD13C constructs were generated by PCR using oligonucleotides containing the myc or HA epitopes in-frame with the N or C terminus of the ANKRD13C ORF. PCR products were inserted in the pcDNA3 vector (Invitrogen) or pSNAP-tag vector (New England Biolabs). cDNA fragments coding for the C terminus of DP were generated by PCR using the Phusion High-Fidelity PCR system (New England Biolabs) and introduced into the pGEX-4T vector (GE Healthcare). To produce the His-myc-ANKRD13C protein, the cDNA coding for full-length ANKRD13C was amplified by PCR and inserted into the pRSETa expression vector (Invitrogen). A SNAP-tag-myc-ANKRD13C construct was also generated by inserting a myc-ANKRD13C fragment in-frame into the pSNAP-tag vector (New England Biolabs). Integrity of the coding sequences of all the constructs was confirmed by dideoxy sequencing.
Cell Culture and Transfection
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum at 37 °C in a humidified atmosphere containing 5% CO2. Transient transfections of HEK293 cells grown to 50–70% confluence were performed using TransIT-LT1 Reagent (Mirus) according to the manufacturer's instructions.
Antibody Generation and Purification
A synthetic peptide corresponding to a sequence in the N terminus of ANKRD13C (residues 7–20) was synthesized and conjugated to a KLH carrier protein by Genscript. This peptide was then used to generate polyclonal antibodies in rabbits following their Express Complete Affinity-purified Peptide Polyclonal Antibody protocol. The specificity of the antibody was established by preincubating it with different concentrations (0–1 mg/ml) of its cognate peptide for 2 h before immunoblotting.
Immunofluorescence Staining and Confocal Microscopy
Immunofluorescence staining was done as previously described (21). Briefly, transfected HEK293 cells were transferred onto coverslips coated with 0.1 mg/ml poly-l-lysine (Sigma) and further grown overnight. Cells were then fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), washed with PBS, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 0.1% Triton X-100 in PBS containing 5% nonfat dry milk. Cells were then incubated with primary antibodies diluted in blocking solution, washed twice with PBS, blocked again with 0.1% Triton X-100 in PBS containing 5% non fat dry milk, and incubated with appropriate secondary antibodies diluted in blocking solution. The cells were washed twice with permeabilization buffer and twice with PBS, and coverslips were mounted using Vectashield mounting medium (Vector Laboratories). Confocal microscopy was performed using a scanning confocal microscope (FV1000, Olympus, Tokyo, Japan) coupled to an inverted microscope with a 63× oil immersion objective, and images were processed with FluoView software (Olympus).
Subcellular Fractionation
HEK293 cells were grown overnight before being transfected with a pcDNA3-HA-ANKRD13C construct as described above. Forty-eight hours post-transfection, cells were suspended in hypotonic buffer (PBS 0.1×) supplemented with protease inhibitors (9 mm pepstatin, 9 mm antipain, 10 mm leupeptin, and 10 mm chymostatin) and incubated on ice for 10 min before being broken with 20 strokes of a Dounce homogenizer. After centrifugation at 3000 rpm and 10000 rpm to remove nuclei and large cell debris, the supernatants were incubated in the absence or presence of 0.1 m Na2CO3, pH 11, for 20 min on ice. Lysates were then centrifuged at 100,000 × g for 1 h at 4 °C to give supernatant and pellet fractions. The fractions were subjected to Western blot analysis using specific antibodies.
Recombinant Protein Production and Binding Assays
The pRSETa-Myc-ANKRD13C and pGEX-4-T1 DP-ICLs and -CT constructs were used to produce His- and GST-tagged fusion proteins in OverExpressTM C41(DE3) Escherichia coli strain (Avidis) by following the manufacturer's instructions. The recombinant His-Myc-ANKRD13C protein was purified using nickel-nitrilotriacetic acid-agarose resin (Qiagen), and the GST fusion proteins were purified using glutathione-SepharoseTM 4B (GE Healthcare) as indicated by the manufacturer. Purified recombinant proteins were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue R-250 staining. 5 μg of glutathione-Sepharose bound GST fusion proteins were incubated with 5 μg of purified His-Myc-ANKRD13C in binding buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Igepal) supplemented with protease inhibitors (9 mm pepstatin, 9 mm antipain, 10 mm leupeptin, and 10 mm chymostatin) and 2 mm dithiothreitol. Samples were incubated for 3 h at 4 °C, and the binding reactions were washed three times with binding buffer. SDS sample buffer was added to binding reactions, and samples were boiled for 5 min. Binding reactions were analyzed by SDS-PAGE, and immunoblotting was performed with the indicated specific antibodies.
SNAP Pulldown
SNAP pulldown experiments were performed basically as described by the manufacturer (New England Biolabs). HEK293 cells were transfected with the SNAP-myc-ANKRD13C construct (see Fig. 7B for schematic representation) and FLAG-DP and sonicated 48 h post-transfection in a non-denaturating buffer (50 mm Tris, pH 7.4, 10% glycerol, 150 mm NaCl, and 5 mm MgCl2) supplemented with protease inhibitors (9 nm pepstatin, 9 nm antipain, 10 nm leupeptin, and 10 nm chymostatin) (Sigma). Lysates were cleared by centrifugation for 15 min at 20,000 × g, and the protein concentration was quantified using the Bio-Rad DC Protein Assay kit. 75 μl of washed SNAP-Capture Pulldown Resin (New England Biolabs #S9144S) were added to 1 mg of lysate proteins and incubated overnight at 4 °C. The capture resin was then washed three times in the sonication buffer. Elution of SNAP-tag-myc-ANKRD13C from the SNAP-Capture Pulldown Resin was not possible because the SNAP tag covalently reacts with the capture resin. However, SDS sample buffer was added 60 min at room temperature to denature the interacting proteins and to elute them from ANKRD13C. The eluted proteins along with the original cell extracts were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
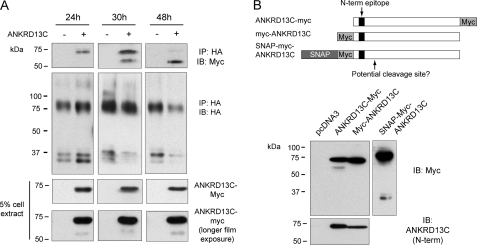
FIGURE 7.
DP receptors targeted for degradation associate with a cleaved form of ANKRD13C. A, HEK293 cells were transiently transfected for 24, 30, or 48 h with HA-DP in combination with pcDNA3 or pcDNA3-ANKRD13C-myc. Immunoprecipitation (IP) of the receptor was performed using an HA specific monoclonal antibody, and immunoblotting (IB) was performed with HA peroxidase or myc antibodies. B, HEK293 cells transiently transfected for 48 h with the indicated myc-tagged ANKRD13C constructs were analyzed by immunoblotting with myc or ANKRD13C antibodies. A schematic representation illustrating the position of the myc and SNAP tags in the different constructs as well as the position of the N-terminal epitope recognized by the ANKRD13C antibody is provided.
Immunoprecipitations
Immunoprecipitation experiments were performed as described (22). Briefly, HEK293 cells were grown overnight in 60-mm plates and transfected on the following day with the indicated constructs. Where indicated, cells were incubated for 30 min at 37 °C in the presence of 1 μm PGD2 before harvesting. Cells were washed with ice-cold PBS and harvested in lysis buffer (150 mm NaCl, 50 mm Tris, pH 8, 1% Igepal, 0.5% deoxycholate, 0.1% SDS, 10 mm Na4P2O7, 5 mm EDTA) supplemented with protease inhibitors (9 nm pepstatin, 9 nm antipain, 10 nm leupeptin, and 10 nm chymostatin) (Sigma). Lysates were clarified by centrifugation, and proteins were immunoprecipitated using 1 μg of specific antibodies and protein A- or protein G-agarose. After being extensively washed with lysis buffer, immunoprecipitated proteins were eluted by the addition of SDS sample buffer followed by incubation at room temperature. Cell extracts and immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotting using specific antibodies.
RNA Interference
The synthetic oligonucleotides targeting the human ANKRD13C gene (siRNA ID s37660 (siRNA#1) and s37661 (siRNA#2)) and the negative control siRNA (catalogue number 4390843) were purchased from Ambion. HEK293 cells stably expressing HA-DP or HT-29 cells were transfected with 15 nm oligonucleotide using Lipofectamine 2000 transfection reagent (Invitrogen).
Metabolic Labeling and Pulse-Chase Experiments
HEK293 cells plated in 60-mm dishes were grown overnight before being transfected with a pcDNA3-FLAG-DP construct in combination with pcDNA3 or pcDNA3-HA-ANKRD13C. On the next day, cells were washed with PBS and incubated in DMEM high glucose without methionine and cysteine for 30 min at 37 °C. One hundred μCi of [35S]methionine/cysteine was added for 90 min at 37 °C. The pulse was terminated by washing the cells with chase medium (complete DMEM supplemented with 5 mm methionine) and chased for the indicated time periods. After the chase, cells were washed with and harvested in PBS, quick-frozen in liquid nitrogen, and stored at −80 °C. After thawing, cells were resuspended in lysis buffer (150 mm NaCl, 50 mm Tris, pH 8, 1% Igepal, 0.1% deoxycholate, 1 mm CaCl2) supplemented with protease inhibitors (9 nm pepstatin, 9 nm antipain, 10 nm leupeptin, and 10 nm chymostatin) and incubated in lysis buffer for 60 min. The lysates were clarified by centrifugation, and protein concentration was determined using the Bio-Rad Protein Assay kit (Bio-Rad). 450 μg of proteins were used for immunoprecipitations. 1 μg of specific antibodies were added, and lysates were incubated for 2 h at 4 °C. Protein G-agarose was then added followed by 60 min of incubation at 4 °C. After being extensively washed with lysis buffer, immunoprecipitated proteins were eluted by the addition of SDS sample buffer and incubation for 60 min at room temperature. Proteins were resolved by SDS-PAGE, and dried gels were exposed to Hyperfilm MP (GE Healthcare). The intensities of radioactive bands were analyzed with the ImageJ program using cells transfected with pcDNA3 alone to determine the background.
Measurement of Cell Surface Receptor Expression
For quantification of cell surface receptor expression, HEK293 cells were plated in 24-well plates precoated with 0.1 mg/ml poly-l-lysine (Sigma). Cells were transfected with the indicated constructs and maintained for an additional 48 h. They were then fixed in 3.7% formaldehyde, TBS (20 mm Tris, pH 7.5, 150 mm NaCl) before being washed twice with TBS. Nonspecific binding was blocked with TBS containing 1% BSA. HA-specific monoclonal antibody diluted in 1% BSA, TBS was added for 60 min. After incubation with the primary antibody, cells were washed with TBS and blocked again with 1% BSA, TBS. Incubation with goat anti-mouse-conjugated alkaline phosphatase diluted in 1% BSA, TBS was carried out for 60 min. After additional washes with TBS, a colorimetric alkaline phosphatase substrate was added. Plates were incubated at 37 °C until a yellow color appeared. Reactions were stopped by the addition of 0.4 n NaOH and read at 405 nm using a Titertek Multiskan MCC/340 spectrophotometer. Cells transfected with pcDNA3 alone were studied concurrently to determine background.
Deglycosylation of Immunoprecipitated Receptors
Receptors transiently expressed in HEK293 cells in 60-mm plates were immunoprecipitated as above. Samples were washed three times with lysis buffer. For peptide N-glycosidase F experiments, immunoprecipitated proteins were eluted by denaturation with 0.1% SDS, 0,75% Triton X-100, and 50 mm β-mercaptoethanol in PBS 1×. The peptide N-glycosidase F enzyme (Sigma) was added at a final concentration of 50 units/ml. For Endo H experiments, immunoprecipitated receptors were resuspended in denaturing buffer 1× (New England Biolabs) followed by the addition of reaction buffer (New England Biolabs) and 10,000 units/ml of Endo Hf (New England Biolabs). Samples were incubated with the enzymes for 5 h at 37 °C.
Statistical Analysis
Statistical analyses were performed using Prism v4.0 (GraphPad Software, San Diego, CA) using the Student's t test. Data were considered significant when p values were <0.05 (*) or <0.01 (**).
RESULTS
Identification of ANKRD13C
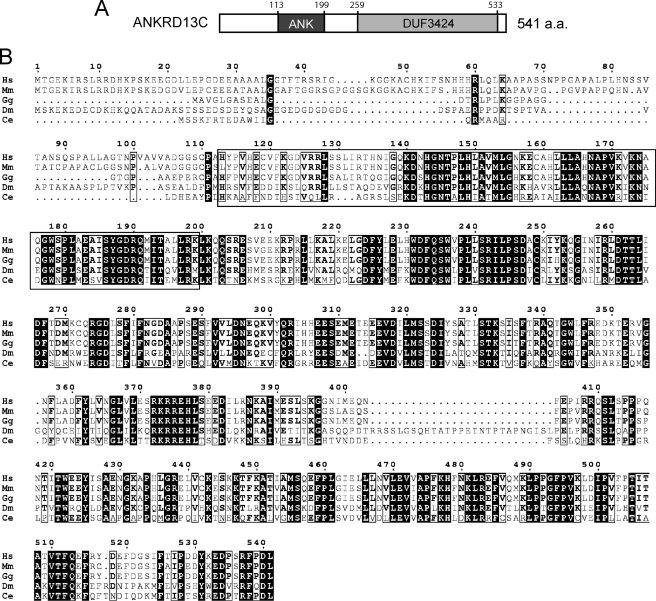
To identify potential DP-interacting proteins, we used a yeast two-hybrid system to screen a human brain cDNA library with the C terminus of DP as bait. We screened a total of 1.6 × 107 independent clones, and of the five galactosidase-positive clones obtained, one encoded a gene called ANKRD13C. The product of the ANKRD13C gene (Entrez Gene ID 81573, accession NM_030816) is a previously uncharacterized molecule. The full-length open reading frame of ANKRD13C was cloned by RT-PCR using template RNAs extracted from HEK293 cells. The main ORF for this gene encodes a 541-amino acid protein (accession NP_110443) containing an ankyrin repeat region (amino acids 113–199) and a C-terminal domain of unknown function termed DUF3424 (amino acids 259–533) (Fig. 1A). Examination of the Aceview data base revealed that ANKRD13C is ubiquitously expressed in body tissues with the most prominent expression levels found in the brain and uterus (23). Through Blast searches of the GenBankTM data base we found ANKRD13C homologs in most eukaryotes, some of which are shown in Fig. 1B. Among them, the amino acid sequences of human ANKRD13C share ∼90% identity with mouse and chicken homologs, indicating that the protein is highly conserved across species.
FIGURE 1.
Homology of ANKRD13C proteins found in different species. A, schematic representation of the ANKRD13C protein is shown. B, sequence alignment of ANKRD13C proteins in Homo sapiens (Hs), Mus musculus (Mm), Gallus gallus (Gg), Drosophila melanogaster (Dm), and Caenorhabditis elegans (Ce) is shown. The region corresponding to the ankyrin repeats is shown in the box. The positions of the ankyrin repeats and DUF3424 domains are indicated.
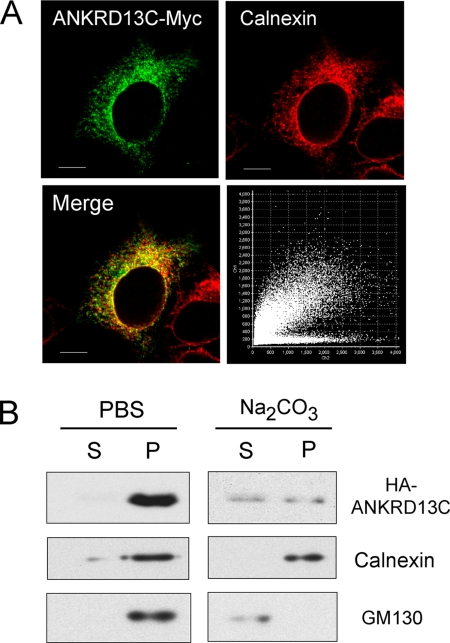
ANKRD13C Is Associated with Endoplasmic Reticulum Membranes
To get insight into ANKRD13C function, we first investigated its subcellular localization by confocal immunofluorescence microscopy and subcellular fractionation studies using HEK293 cells transfected with an ANKRD13C-Myc construct. As can be seen in Fig. 2A, ANKRD13C was found mainly in cytoplasmic punctates and in the perinuclear region. Double-immunofluorescence microscopy revealed significant colocalization of the ANKRD13C protein with calnexin, a marker of the endoplasmic reticulum (ER). To confirm that ANKRD13C is associated with membranes and also to determine whether it is an integral or a peripheral membrane protein, we separated HEK293 lysates into membrane and soluble cytosolic fractions that were used for immunoblot analysis. When cells were lysed in PBS only, ANKRD13C was present in the membrane fraction (Fig. 2B). However, ANKRD13C was partly solubilized by a Na2CO3 treatment, pH 11, known to permeabilize microsome membranes to extract peripheral and luminal proteins but not solubilize integral membrane proteins, suggesting that ANKRD13C is a peripheral protein rather than an integral membrane protein. Calnexin (an integral membrane protein) and GM130 (a peripheral membrane protein) were used as controls. These results suggest that ANKRD13C is a peripheral membrane protein associated with the ER.
FIGURE 2.
ANKRD13C is associated with ER membranes. A, the localization of ANKRD13C in HEK293 cells was determined by immunofluorescence confocal microscopy. HEK293 cells transfected with ANKRD13C-myc were labeled with mouse anti-myc and rabbit anti-calnexin antibodies. Secondary antibodies were Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 546-conjugated anti-rabbit IgG. A merged image of the green-labeled ANKRD13C and red-labeled calnexin signals is shown. The corresponding pixel fluorogram is also shown (abscissa, red color; ordinate, green color). A high degree of colocalization is revealed by a diagonal distribution (at 45°) of the dots on the fluorogram, whereas a lack of colocalization is characterized by two distinct populations with a minimal overlap of dots distributed toward the red and green axes, respectively. Bars, 10 μm. B, lysates of HEK293 cell transfected with HA-ANKRD13C were separated into supernatant (S) and pellet (P) fractions in 1× PBS or in 0.1 m Na2CO3, pH 11, as described under “Experimental Procedures.” These fractions were analyzed by immunoblotting using anti-HA, anti-calnexin, and anti-GM130 antibodies.
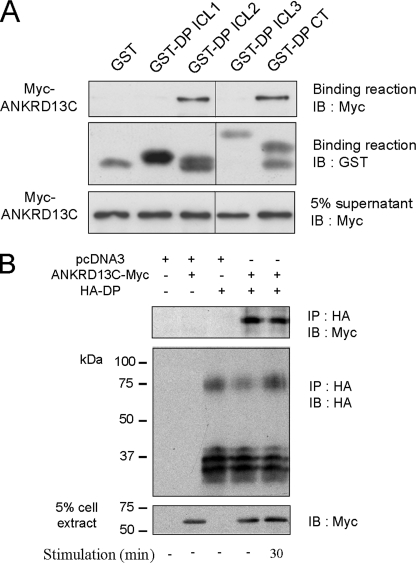
ANKRD13C Interacts with the DP Receptor
To confirm the interaction between ANKRD13C and DP and to determine whether this interaction is direct, we performed in vitro binding assays using the purified recombinant DP intracellular domains fused to glutathione S-transferase along with the purified recombinant myc-ANKRD13C fused to His (His-myc-ANKRD13C). The binding reactions were analyzed by immunoblotting with an Myc-specific monoclonal antibody to detect the presence of the His-myc-ANKRD13C protein. ANKRD13C bound to glutathione-Sepharose-bound GST-DP-CT and GST-ICL2 but not to GST, GST-ICL1, or GST-ICL3 (Fig. 3A). To investigate the interaction between DP and ANKRD13C in a cellular context, we performed immunoprecipitation experiments in HEK293 cells transfected with HA-DP and ANKRD13C-myc. Cell lysates were incubated with a HA-specific monoclonal antibody to immunoprecipitate the receptor, and co-immunoprecipitation of ANKRD13C with DP was detected by Western blot analysis with an myc antibody (Fig. 3B). Agonist stimulation of DP did not modulate the quantity of ANKRD13C that co-immunoprecipitated with DP (Fig. 3B). Taken together, these results demonstrate that the ANKRD13C interaction with DP can be direct, occurs in basal conditions, and is not modulated by DP stimulation in a cellular context.
FIGURE 3.
ANKRD13C interacts with the DP receptor. A, binding assays were carried out using purified GST-DP-CT incubated with purified His-myc-ANKRD13C. The binding of ANKRD13C to the receptor was detected by immunoblotting (IB) using an anti-myc antibody and the GST fusion proteins present in the binding reaction were detected using an anti-GST antibody. B, HEK293 cells transiently transfected with HA-DP and ANKRD13C-myc were stimulated or not with 1 μm PGD2 for 30 min. Immunoprecipitation (IP)of the receptor was performed using an HA specific monoclonal antibody, and immunoblotting was performed with a peroxidase-conjugated HA antibody or a myc-specific polyclonal antibody. Blots shown are representative of three independent experiments.
ANKRD13C Regulates the Biogenesis of DP
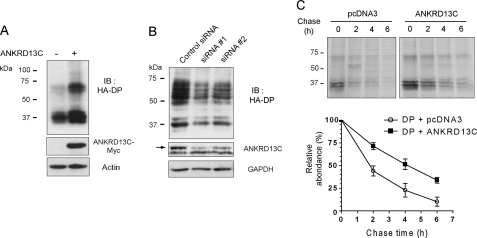
We were next interested in determining the role of ANKRD13C in the function of DP. Because immunofluorescence analysis revealed that ANKRD13C is localized to the ER, we investigated whether ANKRD13C participates in the biogenesis of DP. To do so, we assessed how the modification of ANKRD13C expression would affect the levels of receptor protein produced in cells. Total expression of the receptor in co-transfection with ANKRD13C was determined by Western blot analysis of whole cell lysates at 24 h post-transfection. Co-expression with ANKRD13C led to increased levels of receptor protein produced (Fig. 4A), indicating that ANKRD13C can promote the biogenesis of DP. Conversely, receptor expression in a stable cell line was significantly reduced when cells were transfected with two different siRNAs against endogenous ANKRD13C (siRNA #1 and #2; 63 and 52% reduction in endogenous ANKRD13C expression, respectively) (Fig. 4B), supporting the idea that ANKRD13C is involved in the regulation of DP biogenesis. To investigate whether ANKRD13C affects the kinetics of DP turnover, pulse-chase experiments were performed. HEK293 cells transfected with FLAG-DP in combination with pcDNA3 or pcDNA3-HA-ANKRD13C were metabolically labeled for 1.5 h and chased for the indicated times. Immunoprecipitation of the receptor was carried out, and its turnover rate was calculated as described under “Experimental Procedures.” As shown in Fig. 4C, overexpression of ANKRD13C slowed down the turnover of DP, increasing its half-life from ∼2 to ∼4 h. Because the metabolic labeling of proteins allows us to analyze the degradation of receptors that have been synthesized during the pulse period specifically, this experiment indicates that ANKRD13C increases the half-life of newly synthesized DP receptors. Taken together, these results demonstrate that ANKRD13C promotes the biogenesis of DP by inhibiting the degradation of newly synthesized receptors.
FIGURE 4.
ANKRD13C regulates the biogenesis of DP. A, HEK293 cells transiently transfected for 24 h with HA-DP in combination with pcDNA3 or ANKRD13C-myc were assessed for receptor expression by immunoblotting (IB) using anti-HA peroxidase, anti-myc, and anti-actin antibodies. B, HEK293 cells stably expressing HA-DP were transfected with a negative control siRNA or with ANKRD13C siRNAs. Cell lysates were analyzed by Western blotting using HA peroxidase-, ANKRD13C-, and GAPDH-specific antibodies. The band corresponding to endogenous ANKRD13C is indicated by an arrow. C, the kinetics of DP turnover in the presence of ANKRD13C was determined by pulse-chase experiments as described under “Experimental Procedures” in HEK293 cells transiently transfected with FLAG-DP in combination with pcDNA3 or pcDNA3-HA-ANKRD13C. A representative autoradiogram is shown, and quantification of data from four independent experiments is provided.
Immature Forms of DP Are Retained in the ER by ANKRD13C
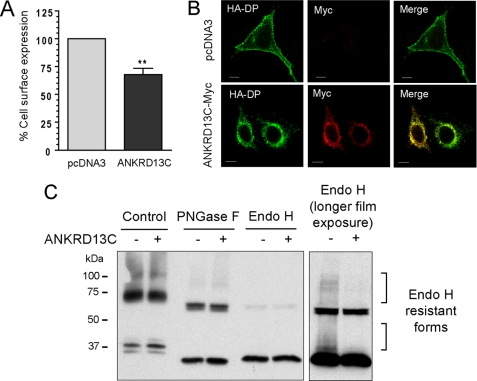
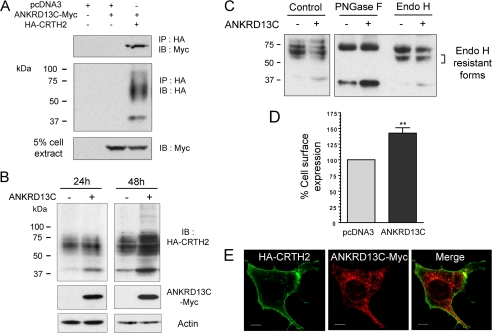
To verify if the increase in total receptor expression in the presence of ANKRD13C led to a greater number of receptors at the plasma membrane, cell surface expression assays were performed by ELISA. HEK293 cells were transfected with HA-DP in combination with pcDNA3 or pcDNA3-ANKRD13C-myc. As shown in Fig. 5A, overexpression of ANKRD13C inhibited DP cell surface expression by 35%, suggesting that DP is retained intracellularly in the presence of ANKRD13C. To visually confirm this result, HEK293 cells transfected with the same combinations of constructs were analyzed by immunofluorescence confocal microscopy. Co-expression of ANKRD13C with DP resulted in a marked shift in the distribution of the receptor from the plasma membrane to intracellular compartments where it co-localized with ANKRD13C (Fig. 5B), consistent with the cell surface expression results obtained by ELISA. The intracellular distribution of endogenous DP and ANKRD13C proteins was also assessed in a cell line that endogenously expresses both proteins (HT-29). The localization of endogenous DP and ANKRD13C in HT-29 cells was similar to that observed in HEK293 cells (supplemental Fig. 1).
FIGURE 5.
DP is retained in the ER in ANKRD13C-overexpressing cells. A, HEK293 cells were transiently transfected with HA-DP in combination with pcDNA3 or a pcDNA3-ANKRD13C-myc construct for 30 h. Cell surface expression of the receptor was measured by ELISA. Results are shown as the percentage of cell surface expression of DP when the cells were transfected with ANKRD13C compared with the cell surface expression of the receptor when they were transfected with pcDNA3 (100%). Results are the mean ± S.E. of five independent experiments. B, the subcellular localization of DP in ANKRD13C-overexpressing cells was determined by immunofluorescence confocal microscopy. HEK293 cells transfected for 30 h with HA-DP in combination with pcDNA3 or a pcDNA3-ANKRD13C-myc construct were labeled with mouse anti-HA and rabbit anti-myc antibodies. Secondary antibodies were Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 546-conjugated anti-rabbit IgG. A merged image of the green-labeled DP and red-labeled ANKRD13C is shown. Bars, 10 μm. C, for deglycosylation assays, HEK293 cells transfected for 30 h with HA-DP in combination with pcDNA3 or pcDNA3-ANKRD13C-myc and processed as described under “Experimental Procedures” were analyzed by Western blotting with an anti-HA antibody. PNGase F, peptide N-glycosidase F.
Because immunofluorescence experiments revealed co-localization of DP with ANKRD13C in ANKRD13C-overexpressing cells (Fig. 5B, lower panel), we hypothesized that DP may be retained in the ER where ANKRD13C is localized. To verify that hypothesis, we analyzed the glycosylation profile of DP. Receptors expressed alone or in combination with ANKRD13C were immunoprecipitated and treated with peptide N-glycosidase F or Endo H. Peptide N-glycosidase F removes all types of N-linked oligosaccharides from glycoproteins, whereas Endo H selectively removes unprocessed high mannose-type N-linked oligosaccharides present on ER-resident glycoproteins. Peptide N-glycosidase F treatment led to the formation of a band with an apparent molecular mass of ∼33 kDa corresponding to the deglycosylated core polypeptide and also produced a ∼65 kDa band that probably represents a dimeric form of the deglycosylated receptor (Fig. 5C), similarly to what we reported for the deglycosylated TP receptors (24). As for Endo H treatment, we obtained the same cleavage products as with peptide N-glycosidase F (Fig. 5C), indicating that most of the N-linked sugars on DP had not undergone the trimming in the Golgi that makes them resistant to Endo H. This high amount of Endo H sensitive forms (which represents ER localized immature forms of the receptor) suggests that the receptor is not efficiently matured in HEK293 cells. However, a fraction of the receptor expressed in the absence of ANKRD13C seems to have reached the Golgi because a longer film exposure revealed the presence of Endo H resistant forms (Fig. 5C, right panel). Co-expression of DP with ANKRD13C led to the disappearance of these Endo H resistant forms, suggesting that ANKRD13C inhibits DP exit from the ER. Altogether, these results revealed that DP is not efficiently transported from the ER to the Golgi and on to the plasma membrane in our system and that this ER retention is amplified by overexpressing ANKRD13C.
DP Receptors Retained in the ER by ANKRD13C Are Eventually Degraded by the Proteasome
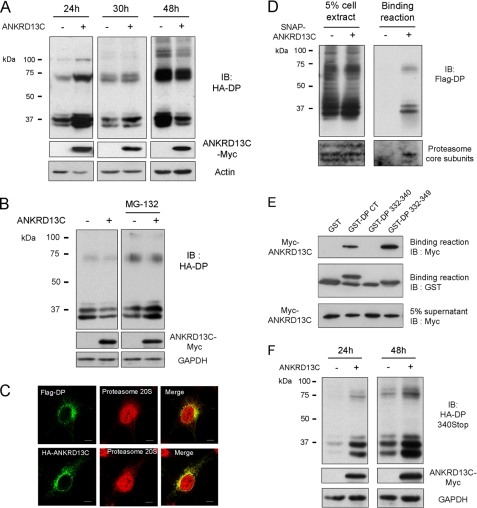
Newly synthesized polypeptides that are not folded or assembled correctly are retained in the ER where a persistent interaction with the quality control machinery generally results in their degradation (25). Because DP is retained in the ER in the presence of ANKRD13C, we wanted to determine whether the receptor was eventually degraded. Therefore, we measured total expression of the receptor at different times after co-transfection with ANKRD13C. As shown in Fig. 6A, co-expressing DP with ANKRD13C for 24 h led to an increase in the expression of the receptor. However, this increase was lost when the two proteins were co-expressed for 30 h and a decrease in the total expression of DP in the presence of ANKRD13C was observed when cells were lysed 48 h post-transfection. This result suggests that, even though ANKRD13C initially promoted the expression of DP, the receptors that were unable to exit the ER were eventually degraded. HT-29 cells were transfected with control or ANKRD13C siRNAs to study the regulation of endogenous DP receptors by ANKRD13C. Inhibition of ANKRD13C expression in these cells resulted in a significant increase in endogenous DP expression after 72 h (supplemental Fig. 2), confirming a role for ANKRD13C in the regulation of DP biogenesis.
FIGURE 6.
DP receptors retained in the ER by ANKRD13C are degraded by the proteasome. A, lysates of HEK293 cells transiently transfected for 24, 30, or 48 h with HA-DP in combination with pcDNA3 or pcDNA3-ANKRD13C-myc were assessed for receptor expression by immunoblotting (IB) with HA peroxidase, myc, and actin antibodies. B, HEK293 cells were transiently transfected for 30 h with HA-DP in combination with pcDNA3 or pcDNA3-ANKRD13C-myc. Cells were then either mock- or MG132 (10 μm)-treated for 5 h before protein extraction followed by SDS-PAGE and immunoblotting using HA peroxidase, myc, or GAPDH antibodies. C, the colocalization of the DP receptor (upper panel) and ANKRD13C (bottom panel) with the proteasome in ANKRD13C-overexpressing cells was determined by immunofluorescence confocal microscopy. HEK293 cells transfected with FLAG-DP and an myc-tagged ANKRD13C construct were labeled with rabbit anti-proteasome 20 S core subunit and mouse anti-FLAG or anti-myc antibodies. Secondary antibodies were Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 633-conjugated anti-rabbit IgG. A merged image of the green-labeled DP or ANKRD13C and red-labeled proteasome is shown. Bars, 10 μm. D, HEK293 cells were transiently transfected with a SNAP-myc-ANKRD13C construct, and pulldown of the protein was performed using a SNAP pulldown resin as described under “Experimental Procedures.” Immunoblotting was performed with FLAG M2 monoclonal and rabbit polyclonal anti-20 S proteasome Core subunits antibodies. E, binding assays were carried out using deletion constructs of DP-CT and purified His-myc-ANKRD13C. The binding of ANKRD13C was detected by immunoblotting (IB) using an anti-myc antibody, and the GST fusion proteins present in the binding reaction were detected using an anti-GST antibody. F, lysates of HEK293 cells transiently transfected for 24 or 48 h with HA-DP340Stop in combination with pcDNA3 or pcDNA3-ANKRD13C-myc were assessed for receptor expression by immunoblotting with HA, myc, and GAPDH antibodies.
Because ER-retained proteins can be retrotranslocated to the cytosol and degraded by the proteasome (26), we wanted to verify if this pathway is involved in the degradation of DP. We co-expressed ANKRD13C with DP for 30 h before treating the cells for 5 h with the proteasomal inhibitor MG-132. Total expression of the receptor was then measured by Western blot analysis of whole cell lysates. In contrast to what was observed in control cells, co-expression of ANKRD13C with DP did not reduce the expression of the receptor in cells treated with MG-132 (Fig. 6B), suggesting a role for the proteasome in the degradation of DP receptors retained in the ER. A possible co-localization between DP and ANKRD13C with the proteasome was assessed by confocal immunofluorescence microscopy. Cells transfected with FLAG-DP and HA-tagged ANKRD13C constructs were labeled to visualize each protein with the 20 S proteasome. As shown in Fig. 6C, co-localization of both DP and ANKRD13C with the 20 S proteasome was observed in conditions where the receptor is retained intracellularly. Pulldown experiments using SNAP-tagged ANKRD13C and the SNAP protein alone as the control expressed in HEK293 cells showed that both DP and at least one of the endogenous proteasome subunits recognized by a 20 S core proteasome antibody were precipitated with SNAP-ANKRD13C but not with SNAP alone (Fig. 6D). These results further support the idea that DP receptors retained in the ER by ANKRD13C are eventually degraded by the proteasome.
To verify if the regulation of DP biogenesis by ANKRD13C requires a direct interaction between the two proteins, we sought to identify ANKRD13C binding sites on DP. However, creating a receptor mutant that would not bind to ANKRD13C revealed to be difficult because ANKRD13C binds to two separate receptor domains, the ICL2 and the C terminus. Nonetheless, we undertook to determine the ANKRD13C binding site on the C terminus, the larger of the two binding domains and the least likely to cause drastic effects on the structure-function of the receptor. Two GST-DP-CT deletion constructs were generated (GST-DP 332–340 and GST-DP 332–349), and the ability of these mutants to bind purified ANKRD13C was tested by GST pulldown assays. Our results show that amino acids 341–349 of DP are essential for ANKRD13C binding to its C terminus (Fig. 6E). An HA-tagged DP deletion mutant lacking the corresponding CT region was then generated (HA-DP340Stop). As shown in Fig. 6F, co-expression of ANKRD13C promoted DP340Stop expression at 24 h of post-transfection, as for the wild-type receptor. However, ANKRD13C did not induce the degradation of DP340Stop at 48 h post-transfection but, rather, still promoted its expression, in contrast to the wild-type receptor (Fig. 6A). These results suggest that a direct interaction between ANKRD13C and the C terminus of DP is important for targeting the receptor for degradation.
A Cleaved Form of ANKRD13C Associates with DP in Conditions Where the Receptor Is Targeted for Degradation
Because of the results described above, we thought it was plausible that ANKRD13C could facilitate the targeting of misfolded/unassembled receptors to the degradation pathway. To investigate that possibility, we first verified if ANKRD13C dissociates from DP before the receptor is degraded or if ANKRD13C remains associated with it throughout the process. To do so, we studied the co-immunoprecipitation of ANKRD13C-myc with HA-DP at 24, 30, and 48 h post-transfection. As shown in Fig. 7A, co-immunoprecipitation of ANKRD13C (∼71 kDa) with DP was observed when the two proteins were co-expressed for 24 and 30 h. However, this co-immunoprecipitation was virtually lost when the two proteins were co-expressed for 48 h. Intriguingly, we observed that a lower molecular mass ANKRD13C band (∼58 kDa) co-immunoprecipitated with the receptor at 30 and 48 h post-transfection. The receptor associated preferentially with the lower molecular mass form as the time progressed even though this form constitutes a small percentage of the ANKRD13C protein found in cells (Fig. 7A, lower panel). To determine whether the reduction in the relative molecular mass of ANKRD13C is caused by cleavage of the protein, we transfected HEK293 cells with constructs of ANKRD13C containing an myc tag at the N or at the C terminus of the protein. If the decrease in electrophoretic mobility is caused by cleavage of the protein, we expected that the ∼58-kDa band would be detected for only one of the two constructs. Western blot analysis of these cell extracts with a myc antibody showed that the ∼58-kDa band is detected only when the myc tag is fused to the C terminus of ANKRD13C, suggesting that the protein is cleaved in its N terminus (Fig. 7B). To confirm the presence of the small cleavage product corresponding to the N-terminal region of the ANKRD13C protein, we used a construct consisting of a SNAP-tag protein fused to the N terminus of myc-ANKRD13C. Fusion with the SNAP tag, which is a 20-kDa mutant of the DNA repair protein O6-alkylguanine-DNA alkyltransferase, allowed us to detect a low molecular mass species (∼33 kDa) that most likely corresponds to the N-terminal cleavage product of the SNAP-myc-ANKRD13C protein (Fig. 7B). Cell extracts were also analyzed using an anti-ANKRD13C raised against an epitope in the N terminus of the protein. The ∼58-kDa band was not detected by this antibody (Fig. 7B), presumably because the epitope recognized by the antibody has been removed from the protein. This is in accordance with the ∼13-kDa cleavage product deduced from the SNAP-mycANKRD13C construct (subtract the SNAP ∼20 kDa from the apparent Mr ∼33 KDa of the cleavage product) and the position of the anti-ANKRD13C antibody epitope on ANKRD13C (residues 7–20). Taken together, these results show that DP associates with an N-terminal-cleaved form of ANKRD13C in conditions where the receptor is targeted for degradation.
The Ankyrin Repeats and Amino Acids 489–498 of ANKRD13C Are Essential for Its Function
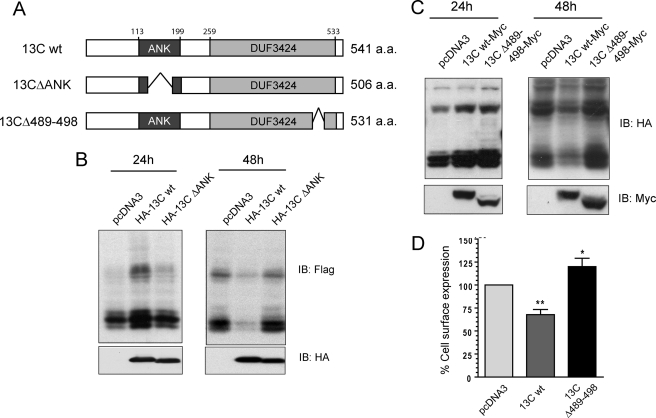
Because ankyrin repeats have been directly implicated in numerous biological processes through their ability to mediate protein-protein interactions (27–30), we wanted to assess if the ankyrin repeats of ANKRD13C are essential for its function. To do so, we deleted part of the ankyrin repeats region by generating a mutant lacking amino acids 158–193 (Fig. 8A, 13CΔANK). This mutant corresponds to a putative isoform of ANKRD13C generated by alternative splicing (UniProtKB/Swiss-Prot Q8N6S4, Isoform 2). To evaluate if this variant is still able to regulate the biogenesis of DP, we analyzed lysates of HEK293 cells transfected with FLAG-DP in combination with wild-type ANKRD13C or 13CΔANK for 24 and 48 h. In contrast to the wild-type ANKRD13C protein, 13CΔANK failed to increase total expression of DP after 24 h of transfection (Fig. 8B, left panel). Moreover, degradation of the receptor was not observed when DP was co-expressed with 13CΔANK for 48 h (Fig. 8B, right panel). These results indicate that the ankyrin repeats of ANKRD13C are essential for its function.
FIGURE 8.
The ankyrin repeats and amino acids 489–498 of ANKRD13C are essential for its function. A, shown is schematic diagram of the 13CΔANK variant and 13CΔ489–498 mutant ANKRD13C constructs. a.a., amino acids. B, lysates from HEK293 cells transiently transfected for 24 or 48 h with FLAG-DP in combination with pcDNA3 or the indicated pcDNA3-HA-ANKRD13C constructs were analyzed by immunoblotting (IB) with FLAG or HA peroxidase antibodies. C, lysates of HEK293 cells transiently transfected for 24 or 48 h with HA-DP in combination with pcDNA3 or the indicated pcDNA3-ANKRD13C-myc constructs were analyzed by immunoblotting using HA peroxidase or myc antibodies. D, HEK293 cells were transiently transfected for 30 h with HA-DP in combination with pcDNA3 or the indicated pcDNA3-ANKRD13C constructs. Cell surface expression of the receptor was measured by ELISA. Results are shown as the percentage of cell surface expression of DP receptors when cells were transfected with ANKRD13C compared with the cell surface expression of the receptors when they were transfected with pcDNA3 (100%). Results are the mean ± S.E. of four independent experiments.
Next, we searched for conserved motifs present in the DUF3424 domain to explore its role. Using the program FingerPRINTScan, we found a region of 10 amino acids (residues 489–498) that is highly similar to one of the nine-element fingerprint of the amphiphysin family signature (PRINTS accession PR01251, InterPro accession IPR003005). This motif is highly conserved in the proteins of the amphiphysin family, but its function is unknown. To determine whether this region is important for the function of ANKRD13C, we generated a mutant of the protein in which amino acids 489–498 were deleted (Fig. 8A, 13CΔ489–498)). As shown in Fig. 8C, this mutant was still able to promote the biogenesis of DP when co-expressed with the receptor for 24 h. However, contrary to the wild-type ANKRD13C protein, co-expression of 13CΔ489–498 with DP for 48 h also increased the expression of the receptor (Fig. 8C, right panel), suggesting that 13CΔ489–498 does not induce ER retention and subsequent degradation of DP. To confirm that DP is not retained intracellularly in the presence of 13CΔ489–498, we measured cell surface expression of the receptor by ELISA. In contrast to what we observed with the wild-type ANKRD13C protein, co-expression of 13CΔ489–498 with DP promoted its cell surface expression (Fig. 8D). Taken together, these results demonstrate that a mutant of ANKRD13C lacking amino acids 489–498 can promote the biogenesis of DP without causing its retention in the ER and subsequent degradation.
ANKRD13C Regulates the Biogenesis and Cell Surface Expression of Other GPCRs
The same set of previously described experiments was conducted on CRTH2 (the other PGD2 receptor) to assess whether or not ANKRD13C would have the same effect as on DP. Co-immunoprecipitation experiments in HEK293 cells showed an interaction between ANKRD13C and CRTH2 (Fig. 9A). Co-expression of ANKRD13C increased total expression of CRTH2 after 24 h and, in contrast to what we observed with DP, after 48 h also (Fig. 9B). To determine whether CRTH2 is exported more efficiently from the ER than DP, we analyzed its maturation using Endo H. As shown in Fig. 9C, the presence of Endo H-resistant forms after treatment with the enzyme indicates that a significant fraction of the receptor is properly matured. Because the presence of ANKRD13C did not reduce the levels of Endo H resistant forms, these results also suggest that, in contrast to what was observed with DP, CRTH2 is not retained in the ER by ANKRD13C. To confirm that ANKRD13C does not inhibit the transport of CRTH2 to the plasma membrane, we measured cell surface expression of the receptor in cells co-expressing the two proteins. Co-expression of ANKRD13C with CRTH2 for 48 h increased cell surface expression of the receptor by 40% (Fig. 9D). Immunofluorescence microscopy analysis confirmed that CRTH2 is not retained in the ER in the presence of ANKRD13C and also showed that ANKRD13C did not co-localize with CRTH2 at the plasma membrane (Fig. 9E). Taken together, these results indicate that ANKRD13C can promote the biogenesis of CRTH2 and its trafficking to the plasma membrane.
FIGURE 9.
ANKRD13C promotes the biogenesis and cell surface expression of CRTH2. A, HEK293 cells were transiently transfected with HA-CRTH2 in combination with pcDNA3 or pcDNA3-ANKRD13C-myc. Immunoprecipitation (IP) of the receptor was performed using an HA specific monoclonal antibody, and immunoblotting (IB) was performed with HA peroxidase or myc antibodies. B, lysates from HEK293 cells transiently transfected for 24 or 48 h with HA-CRTH2 in combination with pcDNA3 or pcDNA3-ANKRD13C-myc were analyzed by immunoblotting using HA peroxidase, myc, or actin antibodies. C, for deglycosylation assays, HEK293 cells were transfected with HA-CRTH2 in combination with a pcDNA3 or pcDNA3-ANKRD13C-myc. Immunoprecipitation of the receptor was performed using an HA specific monoclonal antibody. Receptors were incubated for 5 h at 37 °C with or without peptide N-glycosidase F (PNGase F; 50 units/ml) or Endo H (10000 units/ml) as described under “Experimental Procedures.” Reactions were stopped by adding SDS sample buffer followed by SDS-PAGE and immunoblotting using an HA antibody. D, cell surface expression of CRTH2 was measured by ELISA in cells transiently transfected for 48 h with HA-CRTH2 in combination with pcDNA3 or pcDNA3-ANKRD13C-myc. Results are shown as the percentage of cell surface expression of CRTH2 when cells were transfected with ANKRD13C compared with cell surface expression of the receptors when they were transfected with pcDNA3 (100%). Results are the mean ± S.E. of five independent experiments. E, intracellular distribution of CRTH2 and ANKRD13C was determined by immunofluorescence confocal microscopy. HEK293 cells co-transfected with HA-CRTH2 and ANKRD13C-myc were labeled with mouse HA and rabbit myc antibodies. Secondary antibodies were Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 546-conjugated anti-rabbit IgG. A merged image of the green-labeled CRTH2 and red-labeled ANKRD13C is shown. Bars, 10 μm.
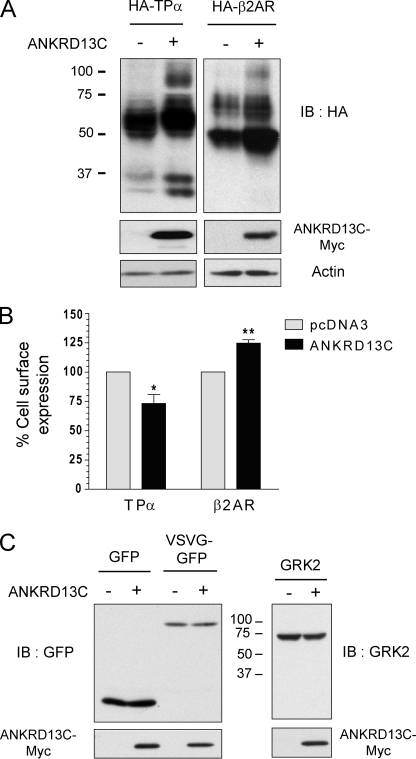
We next wished to establish whether the function of ANKRD13C could be extended to other GPCRs. Like what we observed for DP and CRTH2, co-expression of ANKRD13C increased total protein expression of the thromboxane A2 (TPα) and β2-adrenergic receptors after 24 h of transfection (Fig. 10A). However, co-expression of ANKRD13C enhanced cell surface expression of the β2-adrenergic receptor but inhibited that of TPα (Fig. 10B). These results indicate that the ability of ANKRD13C to regulate the biogenesis and trafficking of GPCRs is not restricted to DP and CRTH2. Finally, we wanted to determine whether ANKRD13C could affect the expression of other classes of proteins. We studied the effect of ANKRD13C co-transfection on the expression of the green fluorescent protein (GFP), the cytosolic and membrane-associated protein GRK2, and the membrane protein vesicular stomatitis virus glycoprotein fused to GFP (VSVG-GFP). As shown in Fig. 10C, total expression of these three proteins was not affected by co-transfection of ANKRD13C.
FIGURE 10.
ANKRD13C regulates the expression of other GPCRs but not of other tested types of proteins. A, lysates from HEK293 cells transiently transfected for 24 h with HA-TPα or HA-β2-adrenergic receptor in combination with pcDNA3 or pcDNA3-ANKRD13C-myc were analyzed by immunoblotting (IB) using HA peroxidase, myc, or actin antibodies. B, cell surface expression of TPα and the β2-adrenergic receptor (β2AR) was measured by ELISA in cells transiently transfected for 30 h with the HA-tagged receptors in combination with pcDNA3 or pcDNA3-ANKRD13C-myc. Results are shown as the percentage of cell surface expression of the receptors when cells were transfected with ANKRD13C compared with cell surface expression of the receptors when they were transfected with pcDNA3 (100%). Results are the mean ± S.E. of three independent experiments. C, lysates from HEK293 cells transiently transfected for 24 h with GFP, VSVG-GFP, or GRK2 in combination with pcDNA3 or pcDNA3-ANKRD13C-myc were analyzed by immunoblotting using GFP, GRK2, or myc antibodies.
DISCUSSION
Although the mechanisms that regulate folding, assembly and maturation of newly synthesized GPCRs are crucial for their efficient cell surface expression, they remain poorly characterized. In this study we identified ANKRD13C as a novel regulator of GPCR biogenesis and trafficking through the biosynthetic pathway. This previously uncharacterized protein displays a high degree of conservation among eukaryotes and is ubiquitously expressed in tissues, suggesting that it plays an important role in cellular biology. Several lines of evidence indicate that ANKRD13C is a protein associated with the cytosolic side of ER membranes. First, our confocal immunofluorescence microscopy data demonstrated that ANKRD13C co-localizes with ER markers. These results are in accordance with a study by Simpson et al. (31) who analyzed the subcellular localization of novel proteins in which they reported ER localization for ANKRD13C. Second, subcellular fractionation experiments and treatment of isolated membranes with Na2CO3 revealed that ANKRD13C is a peripheral membrane protein. Last, our interaction studies showed a direct interaction between ANKRD13C and the cytoplasmic ICL2 and C terminus of DP, indicating that ANKRD13C is located on the cytosolic side of ER membranes.
Modulation of ANKRD13C levels uncovered a role for this novel protein in the regulation of DP biogenesis as well as in ER retention of immature forms of the receptor. Co-expression of ANKRD13C with DP for 24 h increased receptor protein levels, whereas siRNA-mediated knockdown of endogenous ANKRD13C decreased them. Accordingly, pulse-chase experiments showed that ANKRD13C can promote the biogenesis of DP by inhibiting the degradation of newly synthesized receptors. Even though we clearly demonstrated a role for ANKRD13C in promoting the biogenesis of DP, the exact mode of action is still unclear. One possibility would be that ANKRD13C acts as a molecular chaperone for the receptor. Like what is observed for molecular chaperones such as calnexin and Hsp70 proteins, ANKRD13C binding to DP may promote proper folding of the receptor and prevent the accumulation and aggregation of misfolded proteins (32). ANKRD13C would, therefore, increase the half-life of newly synthesized receptors by enhancing their proper folding/assembly, thus, preventing their degradation.
However, in the case of DP, its inefficient folding/assembly led to its retention in the ER by ANKRD13C. Indeed, our cell surface expression and confocal microscopy experiments showed that when the two proteins are co-expressed for a prolonged period, DP receptors are retained intracellularly where they co-localized with ANKRD13C. This co-localization was suggestive of ER retention as ANKRD13C localized at the ER and deglycosylation experiments using Endo H indicated that co-expression of ANKRD13C with DP prevented its transport out of the ER. Retention of misfolded/unassembled receptors in the ER by ANKRD13C may, therefore, represent an important step in the quality control mechanism, ensuring that only properly folded receptors are transported to the Golgi. A similar function has been reported for molecular chaperones involved in quality control at the ER such as calnexin and BiP/GRP98. Like what we observed with ANKRD13C, overexpression of these chaperones can induce retention in the ER of their associated substrates (33, 34). Although we do not know exactly how the binding of ANKRD13C to DP induces its retention in the ER, one possibility would be that ANKRD13C impairs the transport of the receptor by binding to an ER retention motif. On the other hand, ANKRD13C could induce ER retention by masking an ER export motif on DP. In the case of the dopamine D1 receptor, binding of the ER-localized chaperone DRiP78 to an export motif of the receptor inhibits receptor cell surface expression by blocking its transport beyond the ER (7). In both cases, the retention could be relieved by the dissociation of ANKRD13C after appropriate receptor folding/assembly, therefore, allowing transport of the receptor to the plasma membrane.
Interestingly, the effect of ANKRD13C co-expression on DP receptor protein expression gradually shifted from increasing at 24 h to neutral at 30 h to inhibiting at 48 h post-transfection. The fact that co-expression of ANKRD13C did not induce the degradation of a receptor mutant lacking the C-terminal region involved in ANKRD13C binding (DP340Stop) at 48 h post-transfection, contrary to the wild-type receptor, suggests that a direct interaction with the C terminus of DP is required to observe this effect. Treatment with the proteasome inhibitor MG132 revealed that the decrease in DP expression was due to proteasomal degradation of the receptor in the presence of ANKRD13C at 48 h post-transfection. Accordingly, confocal microscopy showed that DP co-localized with ANKRD13C and the 20 S proteasome in these conditions. Moreover, pulldown experiments using SNAP-tagged ANKRD13C expressed in HEK293 cells showed that proteasome subunits can be purified in a complex with ANKRD13C. Altogether, our data indicate that ANKRD13C can promote folding/assembly of newly synthesized receptors but can also retain misfolded/unassembled receptors in the ER and eventually target them for degradation by the proteasome.
The concept of a dual function for ANKRD13C is further supported by the results obtained with the 13CΔ489–498 mutant which was still able to promote the biogenesis of DP but did not induce the retention of the receptor in the ER nor its subsequent degradation. On the other hand, our data with the 13CΔANK variant, lacking amino acids 158–193 and corresponding to a putative isoform of ANKRD13C generated by alternative splicing, showed that the ANK repeats are necessary for the ANKRD13C functions. Given that the localization of the 13CΔANK variant and the 13CΔ489–498 mutant is similar to the wild-type protein and that they still bind to the DP receptor (data not shown), we presume that the two corresponding regions of ANKRD13C are implicated in the binding to other proteins essential for its functions. Studies under way in our laboratory to identify ANKRD13C binding partners will be useful in addressing that question. We were also particularly intrigued by the interaction between DP and the ∼58-kDa cleaved form of ANKRD13C in conditions where the receptor is targeted for degradation. It will be interesting to study how the cleavage of an N-terminal fragment of ANKRD13C affects its function. It is possible that the removal of the ANKRD13C N terminus results in an altered conformation of the protein and/or in differential interactions with potential binding partners that would modify its function. Like what has been described for substrates of the N-end rule degradation pathway (35), this cleavage could also act as a signal to target the receptors associated with the cleaved form of ANKRD13C for degradation.
The ability of ANKRD13C to regulate the biogenesis of proteins was not limited to DP as it also affected total and cell surface expression of CRTH2, TPα, and the β2-adrenergic receptor. Even though ANKRD13C promoted total expression of these three receptors, it differentially affected their transport to the plasma membrane. In the case of CRTH2, according to data obtained from deglycosylation experiments as well as cell surface expression levels that are considerably higher than DP (data not shown), its ER to Golgi transfer appears to be more efficient than DP. The fact that ANKRD13C did not induce ER retention of this efficiently matured receptor supports the idea that ANKRD13C is involved in the quality control process through retention of immature forms of receptors in the ER. This concept is further supported by the results obtained with TPα and the β2-adrenergic receptor. Like what we observed for DP, ANKRD13C inhibited cell surface expression of TPα, a receptor that is not matured efficiently in HEK293 cells (data not shown). On the other hand, ANKRD13C increased membrane expression of the β2-adrenergic receptor, a GPCR that is properly matured in our system. These results indicate that, similarly to what is observed for molecular chaperones like calnexin (33), ANKRD13C can either promote cell surface expression (in the case of efficiently matured receptors like CRTH2 and β2-adrenergic receptor) or inhibit it (in the case of inefficiently matured receptors like DP and TP). The study of receptor sequences failed to provide evidence of a common motif that could explain the effect of ANKRD13C on the receptors included in the present report. Notably, ANKRD13C did not interact in GST pulldown experiments with the C-tail of TPα (data not shown) but still affected its expression. It seems most likely that the chaperone-like effect of ANKRD13C on GPCR expression is indirect, whereas its effect on degradation requires the direct interaction with DP. Work is under way in our laboratory to identify molecular partners of ANKRD13C that could participate in its chaperone-like function. Alternatively, one might also consider the possibility that ANKRD13C binds to or recognizes a particular fold in the intracellular loops of the native structure of receptors.
Even though ANKRD13C regulated the expression of all the GPCRs that we tested, it did not change the expression of co-expressed GFP, cytosolic and membrane-associated GRK2 protein, and VSVG membrane protein. Because of the localization of ANKRD13C on ER membranes, it is not surprising that co-expression of ANKRD13C did not affect the expression levels of GFP and GRK2. The fact that the expression of the single-transmembrane VSVG membrane protein is not affected by ANKRD13C may imply that ANKRD13C is involved in the biology of polytopic membrane proteins such as GPCRs. Whether ANKRD13C strictly affects the biogenesis of GPCRs or whether it can also influence the biogenesis of other types of polytopic membrane proteins remains to be determined.
Supplementary Material
Acknowledgments
We thank Dr. Jennifer Lippincott-Schwartz (National Institutes of Health) for the pEGFP-VSVG plasmid provided through Addgene. We also thank Dr. Leonid Volkov for assistance with the confocal microscope.
This work was supported by a grant from the Canadian Institutes of Health Research (to J.-L. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GPCR
- G protein-coupled receptor
- ANKRD13C
- ankyrin repeat domain-containing protein 13C
- CRTH2
- chemoattractant receptor-homologous molecule expressed on Th2 cells
- CT
- C-terminal tail
- DP
- prostaglandin D2 receptor
- endo H
- endoglycosidase H
- ER
- endoplasmic reticulum
- GRK2
- G protein-coupled receptor kinase 2
- PGD2
- prostaglandin D2
- TPα
- thromboxane A2 receptor α-isoform
- VSVG
- vesicular stomatitis virus glycoprotein
- SNAP
- soluble N-ethylmaleimide-sensitive factor attachment protein.
REFERENCES
- 1.Achour L., Labbé-Jullié C., Scott M. G., Marullo S. (2008) Trends Pharmacol. Sci. 29, 528–535 [DOI] [PubMed] [Google Scholar]
- 2.Colley N. J., Baker E. K., Stamnes M. A., Zuker C. S. (1991) Cell 67, 255–263 [DOI] [PubMed] [Google Scholar]
- 3.Baker E. K., Colley N. J., Zuker C. S. (1994) EMBO J. 13, 4886–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira P. A., Nakayama T. A., Pak W. L., Travis G. H. (1996) Nature 383, 637–640 [DOI] [PubMed] [Google Scholar]
- 5.McLatchie L. M., Fraser N. J., Main M. J., Wise A., Brown J., Thompson N., Solari R., Lee M. G., Foord S. M. (1998) Nature 393, 333–339 [DOI] [PubMed] [Google Scholar]
- 6.Bouschet T., Martin S., Henley J. M. (2005) J. Cell Sci. 118, 4709–4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bermak J. C., Li M., Bullock C., Zhou Q. Y. (2001) Nat. Cell Biol. 3, 492–498 [DOI] [PubMed] [Google Scholar]
- 8.Leclerc P. C., Auger-Messier M., Lanctot P. M., Escher E., Leduc R., Guillemette G. (2002) Endocrinology 143, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 9.Hayaishi O., Urade Y. (2002) Neuroscientist 8, 12–15 [DOI] [PubMed] [Google Scholar]
- 10.Kostenis E., Ulven T. (2006) Trends Mol. Med. 12, 148–158 [DOI] [PubMed] [Google Scholar]
- 11.Pettipher R., Hansel T. T., Armer R. (2007) Nat. Rev. Drug Discov. 6, 313–325 [DOI] [PubMed] [Google Scholar]
- 12.Pettipher R. (2008) Br. J. Pharmacol. 153, S191–S199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallant M. A., Samadfam R., Hackett J. A., Antoniou J., Parent J. L., de Brum-Fernandes A. J. (2005) J. Bone Miner. Res. 20, 672–681 [DOI] [PubMed] [Google Scholar]
- 14.Durand M., Gallant M. A., de Brum-Fernandes A. J. (2008) J. Bone Miner. Res. 23, 1097–1105 [DOI] [PubMed] [Google Scholar]
- 15.Gallant M. A., Chamoux E., Bisson M., Wolsen C., Parent J. L., Roux S., de Brum-Fernandes A. J. (2010) J. Rheumatol. 37, 644–649 [DOI] [PubMed] [Google Scholar]
- 16.Boie Y., Sawyer N., Slipetz D. M., Metters K. M., Abramovitz M. (1995) J. Biol. Chem. 270, 18910–18916 [DOI] [PubMed] [Google Scholar]
- 17.Hirai H., Tanaka K., Yoshie O., Ogawa K., Kenmotsu K., Takamori Y., Ichimasa M., Sugamura K., Nakamura M., Takano S., Nagata K.) (2001) J. Exp. Med. 193, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oguma T., Asano K., Ishizaka A. (2008) Allergol. Int. 57, 307–312 [DOI] [PubMed] [Google Scholar]
- 19.Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. (1997) Nature 389, 81–85 [DOI] [PubMed] [Google Scholar]
- 20.Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 21.Parent A., Hamelin E., Germain P., Parent J. L. (2009) Biochem. J. 418, 163–172 [DOI] [PubMed] [Google Scholar]
- 22.Parent A., Laroche G., Hamelin E., Parent J. L. (2008) Traffic 9, 394–407 [DOI] [PubMed] [Google Scholar]
- 23.Thierry-Mieg D., Thierry-Mieg J. (2006) Genome Biol. 7, S12.1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laroche G., Lépine M. C., Thériault C., Giguère P., Giguère V., Gallant M. A., de Brum-Fernandes A., Parent J. L. (2005) Cell. Signal. 17, 1373–1383 [DOI] [PubMed] [Google Scholar]
- 25.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakatsukasa K., Brodsky J. L. (2008) Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huxford T., Huang D. B., Malek S., Ghosh G. (1998) Cell 95, 759–770 [DOI] [PubMed] [Google Scholar]
- 28.Mosavi L. K., Cammett T. J., Desrosiers D. C., Peng Z. Y. (2004) Protein Sci. 13, 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arniges M., Fernández-Fernández J. M., Albrecht N., Schaefer M., Valverde M. A. (2006) J. Biol. Chem. 281, 1580–1586 [DOI] [PubMed] [Google Scholar]
- 30.Li J., Mahajan A., Tsai M. D. (2006) Biochemistry 45, 15168–15178 [DOI] [PubMed] [Google Scholar]
- 31.Simpson J. C., Wellenreuther R., Poustka A., Pepperkok R., Wiemann S. (2000) EMBO Rep. 1, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buck T. M., Wright C. M., Brodsky J. L. (2007) Semin. Cell Dev. Biol. 18, 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brothers S. P., Janovick J. A., Conn P. M. (2006) J. Mol. Endocrinol. 37, 479–488 [DOI] [PubMed] [Google Scholar]
- 34.Susuki S., Sato T., Miyata M., Momohara M., Suico M. A., Shuto T., Ando Y., Kai H. (2009) J. Biol. Chem. 284, 8312–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tasaki T., Kwon Y. T. (2007) Trends Biochem. Sci. 32, 520–528 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.