Abstract
We have determined the human male specific lethal 3 (hMSL3) chromo-barrel domain structure by x-ray crystallography to a resolution of 2.5 Å (r = 0.226, Rfree = 0.270). hMSL3 contains a canonical methyllysine binding pocket made up of residues Tyr-31, Phe-56, Trp-59, and Trp-63. A six-residue insertion between strands β1 and β2 of the hMSL3 chromo-barrel domain directs the side chain of Glu-21 into the methyllysine binding pocket where it hydrogen bonds to the NH group of a bound cyclohexylamino ethanesulfonate buffer molecule, likely mimicking interactions with a histone tail dimethyllysine residue. In vitro binding studies revealed that both the human and Drosophila MSL3 chromo-barrel domains bind preferentially to peptides representing the mono or dimethyl isoform of lysine 20 on the histone H4 N-terminal tail (H4K20Me1 or H4K20Me2). Mutation of Tyr-31 to Ala in the hMSL3 methyllysine-binding cage resulted in weaker in vitro binding to H4K20Me1. The same mutation in the msl3 gene compromised male survival in Drosophila. Combined mutation of Glu-21 and Pro-22 to Ala in hMSL3 resulted in slightly weaker in vitro binding to H4K20Me1, but the corresponding msl3 mutation had no effect on male survival in Drosophila. We propose MSL3 plays an important role in targeting the male specific lethal complex to chromatin in both humans and flies by binding to H4K20Me1. Binding studies on the related dMRG15 chromo-barrel domain revealed that MRG15 prefers binding to H4K20Me3.
Keywords: Chromatin Regulation, Crystal Structure, Epigenetics, Gene Regulation, Histone Methylation, Chromo-barrel Domain, Drosophila Dosage Compensation, H4K20, MRG15, MSL3
Introduction
Nuclear histone acetyltransferase (HAT)3 enzymes are found in multiprotein complexes that acetylate specific lysine residues on the N-terminal tails of histone proteins, thereby regulating nucleosome structure, chromatin packaging, and gene expression (1–15). MOF, a conserved member of the MYST (Moz, Ysb2, Sas2, Tip60) family of HAT enzymes, functions as the catalytic subunit in a number of distinct HAT complexes that target gene promoters (8), large contiguous domains of chromatin (3, 4, 14, 15), or non-histone proteins such as p53 (5–7). The precise targeting and substrate specificity of MOF relies on the presence of components distinct from the catalytic subunit (3, 4, 7, 8). Specifically, in the MOF-containing Drosophila male specific lethal (MSL) complex, the MSL3 protein is required for chromatin targeting, nucleosome binding, histone tail substrate recognition, and maximal MOF HAT activity (16–20).
The most well studied MOF-containing complex is the Drosophila melanogaster male specific lethal or MSL complex that binds selectively to large regions of the X-chromosome in male flies (14, 15, 21–26) where it is enriched at the 3′ ends of actively transcribed genes (27–30) and acetylates lysine 16 on histone H4 (H4K16Ac) (22, 31), thereby balancing male X-chromosomal gene expression. The Drosophila MSL complex contains the dMSL1, dMSL2, and dMSL3 proteins, the RNA/DNA helicase MLE, the HAT enzyme MOF, and one of two apparently functionally redundant non-coding RNAs (roX1 and roX2) (reviewed in Refs. 14, 15, and 21). The absence of any of the MSL components results in male lethality (14, 15, 23–26). The precise specificity of MOF for H4K16 and the targeting to specific domains of the male X-chromosome is determined by other components of the complex, in particular MSL1 and MSL3 (16–20, 23, 26, 28–30, 32).
A similar MOF-containing human MSL complex has also been identified, and although it contains homologous hMSL1, hMSL2, and hMSL3 subunits, it does not contain the MLE or RNA components, and has nothing to do with dosage compensation (3, 4). However, the human MSL complex does function as a global histone H4K16 acetyltransferase (3–5). Human MOF (hMOF) and the hMSL proteins also play a role in cell-cycle progression and DNA repair (3–5).
The Drosophila and human MSL3 proteins contain a highly conserved N-terminal chromo-barrel domain (CBD) (residues 2–91; 50% identical) plus a conserved C-terminal MRG domain (residues 196–512 in dMSL3; 25% identical to hMSL3) (3, 4, 33–35). The CBD of Drosophila MSL3 is known to contribute to nucleic acid and nucleosome binding in the Drosophila MSL complex (18–20), transcriptional up-regulation on the Drosophila male X-chromosome (18), and proper targeting and spreading of the dosage compensation complex in vivo (20). The CBD of dMSL3 likely contributes to targeting of the MSL complex to chromatin regions containing specific histone tail lysine methylation modifications (19, 20). The C-terminal MRG domain of dMSL3 is responsible for interactions with dMSL1, but does not interact directly with dMOF (16, 17). Through interactions bridged by dMSL1, dMSL3 stimulates the HAT activity of dMOF and controls its substrate specificity (16, 17).
The human MSL3 protein has been less well studied, but two versions of hMSL3 have been identified associated with hMOF, one contains the full-length protein, and the other lacks the N-terminal chromo-barrel domain (3, 4), the particular isoform of hMSL3 found associated with the hMSL complex is tissue-dependent (3). In contrast to the Drosophila MSL complex where dMSL3 interacts directly with dMSL1 (16, 17), hMSL3 has been shown to interact directly with hMOF via its conserved C-terminal MRG domain (4).
To better understand the role played by MSL3 in the MSL HAT complex and histone tail recognition, we have undertaken structural and biochemical studies of the highly conserved MSL3 CBD and compared its in vitro histone tail binding to that of the related MRG15 CBD (9, 35). After submission of this manuscript, an independent structural and biochemical study on the Drosophila and human MSL3 CBDs was published (36). The results and conclusions of that study differ in part from those presented in this report and will be discussed in comparison with our findings.
EXPERIMENTAL PROCEDURES
Cloning and Protein Expression
The D. melanogaster MSL3 CBD (amino acid residues 2–91) was cloned using PCR from a msl3 cDNA. The Homo sapiens MSL3 CBD was cloned using a hMSL3 cDNA kindly provided by Dr. Edwin Smith (Stowers Research Institute). The dMRG15 CBD was cloned from a drmg15 cDNA acquired from the Drosophila Genomics Resource Center, Bloomington, IN. The D. melanogaster msl3 gene, a msl3-TAP tag fusion (20, 28) was kindly provided by Dr. Mitzi Kuroda, Harvard Medical School. The PCR primers used for cloning the dMSL3 (residues 2–91), hMSL3 (residues 2–93), and dMRG15 (residues 2–90) CBDs from their respective cDNAs are provided in the supplemental data (supplemental Table S1). The forward primers all contain a BamHI site and the reverse primers an EcoRI site and stop codon, and were designed for use with the P-GEX-6P3 GST fusion vector (GE Healthcare). PCR amplification was carried out using Pfu high fidelity DNA polymerase using standard reaction conditions. PCR product purification, and ligation with a gel-purified BamHI/EcoRI-digested vector were carried out using standard protocols. The cloning of dMSL3-CBD-GEX6P3, dMRG15-CBD-GEX6P3, and hMSL3-CBD-GEX6P3 was verified by restriction endonuclease digestion of the purified plasmid and DNA sequencing. Competent Escherichia coli BL21(DE3) cells were transformed with the purified plasmid, and an overnight culture was diluted 1/200 into fresh LB containing ampicillin (concentrations = 100 μg/ml). Cells were grown at 37 °C to an optical density (OD) of 0.6 and induced with 0.1 mm isopropyl β-d-thiogalactopyranoside. The temperature was then shifted to 25 °C and cells were grown for approximately 16 h. Cells were pelleted by centrifugation at 8000 × g, suspended in lysis buffer, frozen overnight, and subjected to two passes through a French Press.
Site-specific Mutagenesis
The Stratagene QuikChange mutagenesis kit was used for generating all point mutants. Mutations were confirmed by DNA sequencing. Primers for mutation of hMSL3 and dMSL3 are provided under supplemental Table S1.
Generation of Transgenic Drosophila
Transgenic Drosophila were generated as described (19). Briefly a mixture of 300 ng/ml of plasmid DNA containing attB and msl3-TAP and 0.5 mg/ml of φC31 capped integrase mRNA were injected into embryos from the y w; attP1; msl31/TM3,Sb line. The injection stock carries an attP1 integration site on chromosome 2R. G0 individuals were crossed with y w; +/CyO; msl31/TM3, Sb, and the transgenic offspring identified by green fluorescent eyes. For complementation, we crossed y w; attP1 y+{p[gfp+ msl3-TAP*-pGreeni]}; msl31/TM3,Sb males with y w; msl31 homozygous females. The rescue rates were calculated by dividing the number of rescued males by the number of their stubble (TM3) brothers. Statistical tests of significance were performed used χ-squared analysis. MSL3TAP* was MSL3 WT, MSL3 Glu21-Pro22-Ala, or MSL3 Tyr31-Ala.
Protein Purification
E. coli cell lysates were pretreated with DNase I and then clarified by centrifugation at 15,000 × g for 1 h and then loaded onto a 10-ml glutathione-Sepharose column pre-equilibrated in PBS buffer and washed extensively. Each fusion protein was eluted with 0.1 mm reduced glutathione, and then dialyzed into Precission protease cleavage buffer. Precission protease was then incubated with the GST fusion overnight at 4 °C, followed by dialysis to remove any remaining glutathione, and then again subjected to glutathione-Sepharose affinity chromatography to remove the GST tag and remaining protease. Partially purified proteins were then subjected to ion-exchange chromatography (dMSL3: Source S, pH 5.0, 50 mm sodium malonate; hMSL3: Source S, pH 6.5, 50 mm bis-tris propane; dMRG15: source Q, pH 8.0, 20 mm Tris) and a 0.1 to 1.0 m NaCl gradient followed by analytical gel-filtration chromatography (Superdex 70), which resulted in highly pure recombinant proteins.
Estimation of Nucleic Acid Contaminants
We used an ethidium bromide fluorescence assay (37) to assess the presence of contaminating double-stranded nucleic acid. 10 μl of a 100 μg/ml of solution of calf thymus DNA was diluted into 2 ml of a 0.5 μg/ml of solution of ethidium bromide in 5 mm Tris-HCl, 0.5 mm EDTA, pH 8.1, yielding a fluorescence of 5.6 units at 600 nm (excitation 430 nm). The assay buffer blank gave a fluorescence of 0.858. 10 μl of each purified protein sample at 2.8 mg/ml concentration was diluted in 2 ml of assay buffer to give fluorescence readings of 0.855 for hMSL3, 0.818 for dMSL3, and 0.881 for dMRG15. Hence, there was less than 0.002 μg of double-stranded nucleic acid per μg of protein in our samples. The far UV absorption spectrum of each protein at 2.8 mg/ml exhibited only a small shoulder absorbance at 260 nm, the observed A280:A260 ratios for the three protein samples (undiluted) was measured to be 1.67 for hMSL3 (1.67 calculated), 1.82 for dMSL3 (1.88 calculated), and 1.86 for dMRG15 (1.84 calculated), indicating less than 2% contamination from nucleic acids. Details of protein absorbance ratio calculations are provided under supplemental data. Extinction values for Phe, Tyr, and Trp at 260 and 280 nm were from the literature (38, 39).
Protein Crystallization
Purified protein was concentrated to 12 mg/ml and crystallization conditions for the purified hMSL3 CBD were found using the Wizard II commercial screen (Emerald Biosciences) from a 2.0 m ammonium sulfate solution buffered at pH 10.5 with 0.1 m CAPS. The initial crystals were clusters of small plates. Single crystals of the hMSL3 CBD were grown by repeated seeding in solutions containing 1.6–2.0 m ammonium sulfate, pH 8.0–9.0, and 0.1 m CHES. Seeded crystals took from 1–2 weeks to grow to a maximal size of 0.02 × 0.1 × 0.1 mm as thin plates. Crystals for flash-freezing were prepared with a mother liquor containing 18% (w/v) glycerol. X-ray diffraction data to 2.5-Å resolution were collected on a crystal flash frozen in a stream of liquid nitrogen vapor at the Canadian Light Source Small-gap undulator beamline (Rmerge = 0.078 to 2.5-Å resolution (see Table 1). Data processing indicated the Laue group was 2/m, space group C2, but with a β angle = 89.6°. The data would not merge in higher symmetry Laue classes or in a primitive monoclinic setting.
TABLE 1.
Structure refinement statistics for H. sapiens MSL3 chromo-barrel domain
| Data collection and refinement statistics | 40.0-2.50 Å (2.568-2.500)a |
|---|---|
| Unit cell parameters (C2) (a, b, c) (Å) β (°) | 179.18, 36.697, 85.556, 90.39 |
| Wavelength (Å) | 0.97934 |
| Resolution (Å) | 40-2.50 (2.59-2.50)a |
| No. unique reflectionsb | 19,514 (1789)a |
| Redundancyb | 3.6 (2.9)a |
| Completeness (%)b | 99.0 (91.8)a |
| Average I/σb | 16.6 (2.1)a |
| Rmergeb,c | 0.078 (0.374)a |
| Refinement resolution limits | 40.0-2.50 Å (2.57-2.50)a |
| No. of reflections in working set | 18,515 (1219)a |
| No. of reflections in test set | 997 (60)a |
| Rworkd | 0.2256 (0.271)a |
| Rfreee | 0.2702 (0.344)a |
| No. of amino acid residues | 361 (3733 atoms) |
| No. of water molecules and sulfates | 57 and 17 |
| No. of ligands (CHES) | 3 |
| Average B-factor (Å2)d,f | 27.8 |
| R.m.s. deviations B bonded MC atoms (Å2)f | 0.39 |
| R.m.s. deviations B bonded SC atoms (Å2)f | 0.76 |
| R.m.s. deviations B angle MC atoms (Å2)f | 1.20 |
| R.m.s. deviations B angle SC atoms (Å2)f | 1.97 |
| R.m.s. deviations bond lengths (Å)f | 0.0090 |
| R.m.s. deviations angles (°)f | 1.09 |
| Residues in preferred Ramachandran (%)g | 93.8 (5.7) |
a Values in parentheses correspond to the highest resolution shell.
b Data processing statistics calculated using Denzo/HKL2000 (37).
c Rmerge = Σhkl Σi | 〈I(hkl)obs〉 − I(hkl)obs, i|/Σhkl,i I(hkl)obs,I, where I(hkl)obs,i is the individual measurement of an hkl intensity and 〈I(hkl)obs〉 = Σi I(hkl)obs,i/n; where i = 1 to n individual reflections are measured.
d Rwork = Σhkl‖Fobs(hkl)‖−|Fcalc(hkl)‖/Σhkl|Fobs(hkl)|, where |Fobs(hkl)| and |Fcalc(hkl)| are the observed and calculated amplitudes, respectively, for the structure factor F(hkl).
e Rfree is the equivalent of Rwork for 5% of the reflections (randomly selected), which were not used in structure refinement.
Structure Determination and Refinement
Diffraction data were processed with HKL2000 (40) (Table 1). The structure was solved using molecular replacement, the search model was the refined x-ray structure of the human MRG15 CBD (PDB 2F5K) (41) and PHASER (42) located five molecules in the asymmetric unit. Subsequent model building and refinement using Coot (43), CNS (44), and finally Refmac 5.5 in CCP4 (45–47) resulted in a refined model with excellent stereochemistry (Table 1) as illustrated by a Ramachandran plot (48) (supplemental Fig. S1). During refinement, 5% of reflections were randomly put aside as a test set for calculation of the free R factor. Figures depicting the contents of the asymmetric unit and electron density for one of the hMSL3 methyllysine binding pockets are provided under supplemental Fig. S2. For Fig. 1, sequence alignments were calculated with T-Coffee (49) and rendered using Espript (50). Molecular superpositions were carried out using LSQKAB in CCP4 (45). All molecular figures were drawn using either Molscript/Raster3D (51, 52) or PyMOL. Qualitative electrostatic surfaces were drawn using the adaptive Poisson-Boltzmann solver as implemented in PyMOL and are contoured at ±75 mV (units of kT/e). The refined atomic coordinates and structure factors for the hMSL3 CBD have been deposited with the RCSB protein data bank (PDB code 3OB9).
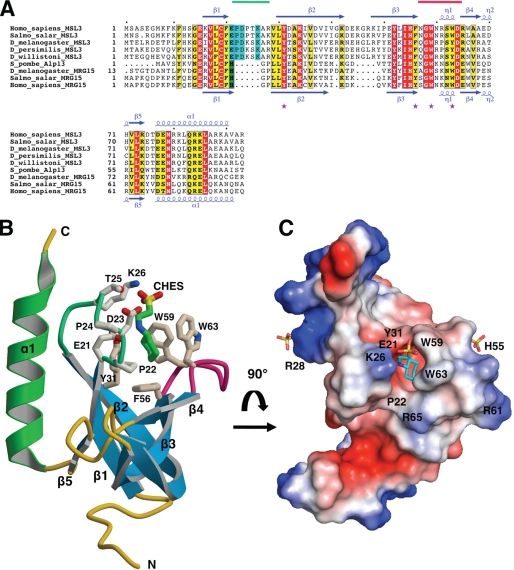
FIGURE 1.
Tertiary structure of human MSL3 chromo-barrel domain. A, multiple sequence alignment of MSL3 and MRG15 CBD sequences from higher eukaryotes. Strictly conserved residues are shaded red, highly conserved residues yellow. A six-residue insertion specific to MSL3 is shaded blue, whereas a histidine residue found in the methyllysine binding cage of MRG15 is shaded green. Secondary structure assignments were derived from x-ray structures of hMSL3 (this work) and hMRG15 CBDs (PDB 2F5K) (41). The locations of the β1-β2 and β3-β4 loops making up the methyllysine binding pocket are marked with light green and magenta lines, respectively, above the sequences. Aromatic cage residues of the methyllysine binding pocket are marked with purple stars. The N-terminal 12 residues (MGEVKPAKVENY) of dMRG15 are not shown for clarity. B, ribbon diagram of hMSL3 residues 5–93 (subunit A in the crystal). The MSL3-specific loop between strands β1 and β2 is colored light green. Amino acid side chains associated with the presumed methyllysine binding pocket and a bound CHES buffer molecule are shown as stick models. C, qualitative electrostatic surface rendering of the hMSL3CBD (red negatively charged; blue positively charged, see “Experimental Procedures”) and bound CHES and sulfate anions are shown as stick representations. The hMSL3 CBD is rotated ∼90 degrees toward the viewer relative to B, looking directly into the methyllysine binding pocket.
Synthetic Peptides
Synthetic peptides with the indicated sequences and post-translational modifications were purchased from Sigma-Genosys or New England Peptide at greater than 95% purity. The peptides used in this study all contained an unmodified N terminus and an amidated C terminus unless indicated otherwise: H4K20Me1–3, GKGGAKRHR(KMe1–3)VLRDY; H3K4Me2, ART(KMe2)QTARKSTGGKAY; H3K36Me1–3, APSTGGV(KMe1–3)KPHRYR; H3K9Me1, KQTAR(KMe1)STGGKAY; H3K18Me1, GGKAPR(KMe1)QLATY; H3K27Me1, TKAAR(KMe1)SAPSTGY; H3K79Me2, AQDY(KMe2)TDLR.
Surface Plasmon Resonance
The dMSL3, hMSL3, and dMRG15 chromo-barrel domains were covalently linked to carboxyl methylcellulose-based sensor chips (CM5, GE-Healthcare) using N-ethyl-N-(3-dimethylaminopropyl)carbodiimide and N-hydroxysuccinimide (NHS), and quenched with ethanolamine according to the manufacturer's instructions. Typically 4000–5000 response units (2000 for dMRG15) of protein were immobilized per experiment. All measurements were carried out on a Biacore X instrument. CBD histone tail peptide dissociation constants were calculated using a Langmuir 1:1 steady state equilibrium binding model (A + B ⇆ AB) with Scrubber. Steady state affinity binding of peptides to the immobilized CBDs was measured for peptide concentrations ranging from either 1 to 800 μm or 1 μm to 3.8 mm. Typically, complete binding data for at least two peptides were measured per CM5 chip, reproducibility from chip to chip was excellent. Peptide concentrations were determined by tyrosine UV absorbance at 276 nm using a Nanodrop device, as each synthetic histone tail peptide was designed to contain a C-terminal tyrosine residue. Peptides were dissolved in a running buffer of 50 mm HEPES pH 7.4, 3 mm EDTA, and 150 or 250 mm NaCl as indicated.
RESULTS AND DISCUSSION
Tertiary Structure of the hMSL3 Chromo-barrel Domain
The structure of the human MSL3 CBD has been fully refined to 2.5-Å resolution (R-factor = 0.226; R-free = 0.270) (Table 1 and supplemental Figs. S1 and S2). There are five independent copies of the hMSL3 CBD in the asymmetric unit of the crystal lattice and the tertiary structures of the five subunits are very similar, (backbone atom pairwise r.m.s. differences of 0.68 to 0.74 Å) (supplemental Fig. S2). The hMSL3 CBD structure is similar to that of the MRG15 CBD (41), folding as a 5-stranded antiparallel β-barrel domain with a C-terminal α-helix (backbone atom rms difference of 0.99 Å for 68 equivalent residues) (Fig. 1 and supplemental Fig. S3). Similar to the CBD of MRG15 and other tudor and chromo-barrel domains (41, 53–57), a canonical methyllysine binding pocket is evident at one end of the hMSL3 β-barrel domain, with important residues coming from surface loops between strands β1-β2 and β3-β4 (Figs. 1 and 2). In hMSL3, there is a highly conserved six-residue insertion relative to MRG15, at the loop between strands β1 and β2, the proximity of this loop to the presumed methyllysine binding pocket suggests this loop may be important for methyllysine binding specificity (Fig. 1). The five hMSL3 CBD molecules observed in the crystal exhibit minimal disorder or heterogeneity at the methyllysine binding pocket and four of the five monomers pack as two almost identically arranged dimers (a modest 670 Å2 is buried at the dimer interface) (supplemental Fig. S2). The fact that the hMSL3 CBD preferentially packs as a dimer in the crystals is unusual as most chromo-barrel domains, tudor domains, and chromodomains, including those of MRG15, and 53BP1, tend to pack as monomers (41, 53). However, we see no evidence of either hMSL3 or dMSL3 CBD dimerization in solution using analytical gel filtration chromatography (not shown). Hence the biological significance of hMSL3 CBD dimerization in the crystal lattice is unclear.
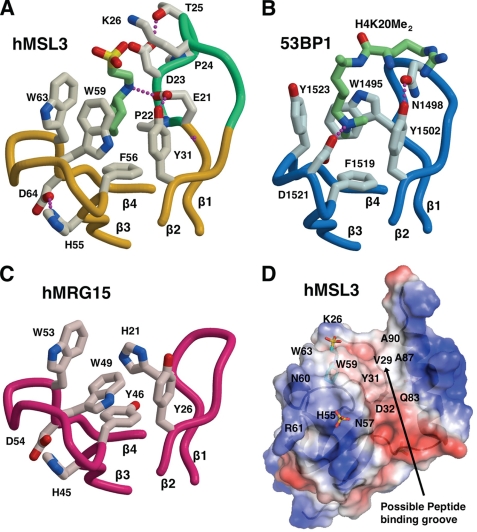
FIGURE 2.
Comparison of methyllysine binding pockets in hMSL3, hMRG15, and h53BP1. A, methyllysine binding pocket in the hMSL3 CBD depicting the bound CHES buffer molecule (green carbons), colored as described in the legend to Fig. 1B. B, methyllysine binding pocket in the 53BP1 tandem tudor domain in blue (53) showing the bound H4K20Me2 peptide with carbons colored light green. Only the first tudor domain is shown. C, methyllysine binding pocket in the hMRG15 CBD drawn in magenta with side chain carbons in light pink (41). Each pocket is drawn in an identical orientation, showing similar structural elements. Hydrogen bonds are drawn as purple dotted lines. Residues are labeled according to the text and the published structures. D, a potential peptide interaction surface on the hMSL3 CBD, the view is rotated ∼180° about the vertical axis relative to Fig. 1B. The hMSL3 CBD is rendered as a semi-transparent electrostatic surface (red negatively charged, blue positively charged) overlaid onto a ribbon diagram of the molecule. The bound CHES molecule (blue carbons) and the sulfate anion near His55 are shown as stick representations. Residues mentioned in the text are labeled accordingly.
An Aromatic Cage Methyllysine Binding Pocket in hMSL3
Residues making up the presumed methyllysine binding pocket of hMSL3 are Glu21, Pro22, and Tyr31 from the loop between strands β1 and β2, and Phe56, Trp59, and Trp63 from the loop between strands β3 and β4 of the β-barrel (Figs. 1 and 2). In the crystal structure, the deep binding pocket is occupied by the piperazine moiety of a CHES buffer molecule for two of the five independent copies of the CBD (Fig. 1 and supplemental Fig. S2). The binding of the piperazine moiety of CHES in a methyllysine pocket is reminiscent of the morpholino ring of MES binding in the methyllysine binding pocket of the h-l(3)mbt repeat protein and is thought to mimic the presence of a mono- or dimethyllysine residue (58). Other conserved residues on the hMSL3 β1-β2 loop near the methyllysine binding pocket are Asp23, Thr25, and Lys26. These residues are conserved in MSL3 sequences and are positioned at the opening of the methyllysine pocket such that their side chains could form hydrogen bonds with the backbone peptide groups flanking the methyllysine residue in a histone tail substrate (Figs. 1 and 2). Importantly, the analogous β1-β2 loop is much shorter in the structure of the hMRG15 CBD and in place of Glu21 and Pro22 in hMSL3, there is a conserved histidine residue that forms one side of the hMRG15 methyllysine binding pocket (41) (Fig. 2). Otherwise, the overall shape and local environment of the hMSL3 methyllysine binding pocket are very similar to that of hMRG15, suggesting that these proteins may recognize a similar ligand (Fig. 2) (41).
The methyllysine binding pocket in hMSL3 also closely resembles the dimethyllysine binding pocket in the tandem tudor domain of human 53BP1 and monomethyllysine binding pockets in mbt repeat proteins that recognize the respective methylated form of lysine 20 on histone H4 (53–55) (Fig. 2). The Glu21 carboxylate side chain that makes up the side of the methyllysine binding pocket on the β1-β2 loop of hMSL3 resembles the positioning of the side chain of Asp1521 in 53BP1, required for dimethyllysine binding, but on the opposite β3-β4 loop (53) (Fig. 2). Based on simple steric and hydrogen bonding arguments, the presence of Glu21 in the hMSL3 methyllysine pocket would be expected to modulate the binding specificity of hMSL3 in the direction of binding mono- or dimethyllsyine residues relative to trimethyllysine. A trimethyllysine residue binding in the pocket would most likely result in unfavorable van der Waals interactions between the carboxylate group of Glu21 and the trimethylammonium group of the trimethyllysine. The side chain of Pro22 on the β1-β2 loop would likely constrict the space inside the methyllysine binding pocket enough so that only a mono- or dimethyllysine residue can easily be accommodated. Neither human MRG15 nor yeast Eaf3 CBDs have a carboxylate residue or a proline residue facing into their respective methyllysine binding pockets (41, 56, 57) (Fig. 2).
The hMSL3 CBD structure exhibits a prominent surface groove between the methyllysine binding pocket and helix α1 that could easily accommodate a short extended peptide (Fig. 2). Residues on a similar surface groove in the structure of the related Eaf3 CBD undergo chemical shift changes upon binding of the H3K36Me3 peptide (57). In hMSL3, this surface depression is also partially negatively charged and contains several polar side chains (Tyr31, Asp32, Asn57, Trp59, and Gln83) that could potentially hydrogen bond to the backbone peptide groups from the amino-terminal residues of a bound histone tail (Fig. 2D). In support of this idea, the side chain indole nitrogen of Trp59 of hMSL3 makes a H-bond to the backbone oxygen from the β2-β3 hairpin loop on a nearby hMSL3 molecule in the crystal lattice, hence possibly mimicking the contact of a histone tail peptide backbone carbonyl group to the CBD (not shown). The hMSL3 surface depression also contains a small hydrophobic patch consisting of residues Val29, Ala87, and Ala90 that may participate in peptide binding (Fig. 2D).
The MSL3 CBD Preferentially Binds H4K20Me1 or H4K20Me2 in Vitro
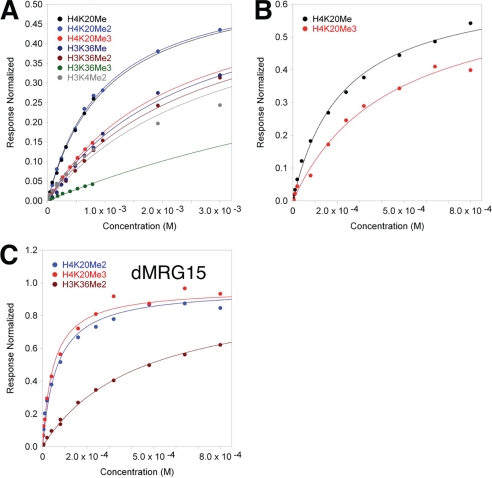
The binding of CHES to the hMSL3 CBD and structural similarities with the 53BP1 methyllysine binding pocket suggest that hMSL3 will have a binding preference for peptides containing mono- or dimethyllysine. Furthermore, the presence of a highly conserved hMSL3 β1-β2 insert at the hMSL3 methyllysine binding pocket relative to MRG15 suggests that there may be different methyllysine binding specificities for these two otherwise closely related CBDs. Hence, we used histone tail peptide sequences containing methylation modifications corresponding to common histone H3 and H4 N-terminal epigenetic modifications in Drosophila and humans (59), and screened peptide binding to the hMSL3, dMSL3, and dMRG15 CBDs using surface plasmon resonance (Figs. 3 and 4 and supplemental Fig. S4). The majority of binding experiments with the MSL3 CBDs were conducted under fairly stringent conditions (250 mm NaCl) to avoid possible nonspecific ionic interactions of the predominantly positively charged histone tail peptides with either the CBD or the CM5 sensor chip surface (see “Experimental Procedures”). For the H4K20Me1 and H4K20Me3 peptides, we also measured the binding affinity for dMSL3 and hMSL3 CBDs at a more physiological salt concentration of 150 mm (Table 2). We carried out a series of single injection experiments for a larger number of peptides for all three CBDs at 50 and 100 μm peptide concentrations in 150 mm NaCl running buffer (100 μm shown) to rule out significant binding by other sequences and ensure that the buffer salt concentration did not affect the relative affinities of peptide binding (supplemental Fig. S4). The results demonstrate that the relative orders of binding affinity of the tested peptides for both the hMSL3 and dMSL3 chromo-barrel domains are highly reproducible and independent of the running buffer salt concentrations tested. The preferential order of peptide binding we observe for both the hMSL3 and dMSL3 chromo-barrel domains is H4K20Me1 ≈ H4K20Me2 > H4K20Me3 > H3K36Me1 ≈ H3K36Me2 > H3K4Me2 > H3K36Me3 (Table 2, Figs. 3 and 4, and supplemental Fig. S4). Hence, the MSL3 CBD in both humans and Drosophila preferentially binds monomethyl or dimethyllysine histone tail peptides over trimethyl-modified lysine peptides and H4 tail sequences over H3, suggesting that the binding specificity is conserved across species, and that the biological function is to recognize and bind H4K20Me1 or H4K20Me2 (Figs. 3 and 4). Based on our fitted Kd values for the relevant peptides in buffer containing 250 mm NaCl, the preference for H4K20Me1 or H4K20Me2 over H3K36Me3 is approximately a factor of 50 (Table 2). The Kd for hMSL3 and H4K20Me1 in 150 mm NaCl buffer is 31 μm, the value for the dMSL3 CBD under identical conditions is 224 μm, a factor of seven weaker (Fig. 4 and Table 2). We do not have an explanation for why the dMSL3 CBD binds to H4K20Me1 more weakly than its human counterpart.
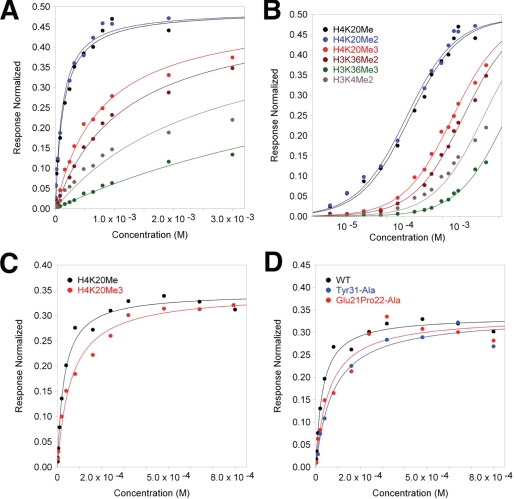
FIGURE 3.
Binding affinity of the human MSL3 chromo-barrel domain for the indicated methyllysine containing histone tail peptides. A, surface plasmon resonance steady state equilibrium binding for methylated histone tail peptides over the hMSL3 chromo-barrel domain (in response units normalized to maximum theoretical occupancy of ligand) in 250 mm NaCl, 3 mm EDTA, 100 mm HEPES pH 7.5. Individual data points and fitted curves based on the calculated Kd values are shown for each peptide series. B, semilog plot of data presented in A. The legend applies to both A and B. C, binding data for hMSL3 CBD with H4K20Me1 and H4K20Me3 in 150 mm NaCl buffer. D, binding data for WT hMSL3, the Glu21-Pro22-Ala and Y31A hMSL3 CBD mutants with H4K20Me1 in 150 mm NaCl running buffer.
FIGURE 4.
Binding affinity of the D. melanogaster MSL3 and MRG15 chromo-barrel domains for methyllysine containing histone tail peptides. A, surface plasmon resonance steady state equilibrium binding (in response units normalized to maximum theoretical occupancy of ligand) for the D. melanogaster CBD and histone tail peptides in 250 mm NaCl, 3 mm EDTA, 100 mm HEPES pH 7.5. Individual data points and fitted curves based on the calculated Kd values are shown for each peptide series. B, as in part A, but 150 mm NaCl and only H4K20Me1 and H4K20Me3. C, SPR binding of the dMRG15 CBD to H4K20Me3, H4K20Me2, and H3K36Me2 in 150 mm NaCl running buffer.
TABLE 2.
Calculated dissociation constants for peptide chromo-barrel domain surface plasmon resonance steady state affinity measurements
| Peptide | [NaCl] | Kd hMSL3a,b | Kd dMSL3a,b |
|---|---|---|---|
| mm | μm | ||
| H4K20Me1 | 250 | 116(4) | 1003(3) |
| H4K20Me2 | 250 | 98(2) | 980(3) |
| H4K20Me3 | 250 | 700(20) | 2230(7) |
| H3K36Me1 | 250 | NDc | 2420(7) |
| H3K36Me2 | 250 | 1070(3) | 2730(8) |
| H3K36Me3 | 250 | 6500(3) | 9400(7) |
| H3K4Me2 | 250 | 2460(7) | 3140(9) |
| H4K20Me1 | 150 | 31(9) | 224(6) |
| H4K20Me3 | 150 | 100(5) | 441(9) |
a Based on a 1:1 Langmuir binding steady state equilibrium, calculated using Scrubber (see “Experimental Procedures”).
b Kd residuals from fitting are provided in parentheses.
c ND, not determined.
Comparison with a hMSL3 CBD:DNA H4K20 Structure
Recently, a structure of the hMSL3 CBD bound to duplex DNA and H4K20Me1 was published (36). In contrast to our results, that study concluded that methylated histone tail binding to hMSL3 or dMSL3 CBDs occurred only in the presence of double-stranded DNA and that histone tail binding was significant only for H4K20Me1 and not other histone tail sequences or higher degrees of methylation. In contrast, our results suggest that hMSL3 is capable of binding H4K20Me1 with reasonable affinity in the absence of nucleic acids as our purified CBDs were deemed essentially nucleic acid free (see “Experimental Procedures”). We do not have an explanation for the reported binding discrepancy between the two studies. Our study used surface plasmon resonance with a fixed amount of immobilized protein and differing concentrations of added peptides to determine dissociation constants. The other study used fluorescence polarization of a fixed amount of fluoresceinated peptide in combination with varying concentrations of the recombinant protein to quantify interactions (36). Although we do not doubt that double-stranded DNA binding to the MSL3 CBD could modulate subsequent in vitro or in vivo methylated histone tail binding, our results suggest that the MSL3 CBD alone is sufficient for in vitro binding to H4K20Me1 and discrimination over other histone tail sequences. We also note that the other study used dMSL3 and hMSL3 constructs each containing an extra nine N-terminal residues (MKKHHHHHH) to facilitate protein purification and an extra eight residues at the C terminus for hMSL3 (94LRSTGRKK101) (36). The hMSL3 and dMSL3 constructs used in our study contain five extra N-terminal residues (GPLGS) from the cloning vector. Least squares superposition of our atomic coordinates with that of the hMSL3 CBD in the hMSL3·DNA·peptide complex (PDB code 3M9P) revealed only modest (0.6 to 1.10 Å) pairwise r.m.s. differences in the positions of the backbone atoms, and very few changes in amino acid side chain conformation, suggesting the tertiary structure of the hMSL3 CBD is essentially the same in the absence or presence of nucleic acids. However, our hMSL3 atomic model does form dimers, and does contain an extra 3–4 residues visible at the N terminus for four of the five subunits, namely Gly6 to Phe9. Differences at the C terminus of the two independent hMSL3 structure determinations are negligible, as residues 92–101 of the hMSL3·DNA complex are not included in the refined model of that structure and hence are likely disordered in solution (36).
Histone Tail Binding by the dMRG15 CBD
The MRG15 protein contains an N-terminal CBD highly similar to MSL3 (Figs. 1 and 2), and a more distantly related C-terminal MRG domain (9, 35). MRG15 also associates with Tip60, a MYST HAT highly similar to MOF as part of a large multiprotein complex that functions in gene regulation and DNA repair through acetylation of histones phospho-H2Av and H4 (9–13). Hence, the conserved domain organization of MSL3 and MRG15, plus their association with highly similar HAT enzymes, suggest a related biological function for the two proteins (3, 4, 9). However, the CBD of hMRG15 was previously reported to bind H3K36Me3, although binding of methylated H4K20 peptides to MRG15 was not tested in that study (41). Similar binding studies on the related yeast Eaf3 CBD also reported preferential binding to H3K36Me2 or H3K36Me3 (56, 57), but one of the studies also suggested that H4-based peptides bound nearly as well (57). Given the high degree of structural similarity between the hMSL3 and hMRG15 CBDs, we therefore measured the in vitro binding of methylated histone tail sequences to the dMRG15 CBD to see if there was any similarity with the MSL3 CBD binding profile. The results are presented in Fig. 4C and supplemental Fig. S4. We found that the CBD of dMRG15 binds preferentially to H4K20Me3 (Kd = 48 μm) and with similar affinity for H4K20Me2 (Kd = 61 μm) in 150 mm NaCl containing buffers, although the single injection profile suggests consistently stronger binding for H4K20Me3 (supplemental Fig. S4). Binding to H3K36Me3 or H3K36Me2 (Kd = 430 μm) is significantly weaker (Fig. 4C and supplemental Fig. S4), indicating that the dMRG15 CBD prefers binding H4K20Me3 or H4K20Me2 over H3K36Me3 or H3K36Me2. Hence both MRG15 and MSL3 CBDs have similar histone H4 tail binding profiles, preferring H4K20 over H3 sequences, although MSL3 prefers a lower degree of methylation of Lys20, and MRG15 prefers trimethylation. As the structures of the hMSL3 and hMRG15 CBDs are highly similar (Figs. 1 and 2), the different binding affinities for H4K20Me1 versus H4K20Me3 suggest that subtle differences in the methyllysine binding pockets of these proteins confer discrimination for the degree of methylation of H4K20.
Effect of MSL3 Methyllysine Binding Pocket Mutations on Histone Tail Binding in Vitro and on Male Survival in D. melanogaster
To test the possible contribution of Glu21 and Pro22 of MSL3 to binding mono- or dimethyllysine at H4K20, we simultaneously mutated both residues to alanine in hMSL3. As measured by SPR, the binding of the EP to AA mutant to H4K20Me1 was somewhat weaker than for wild type hMSL3 (Kd = 59 μm) (Fig. 3D), suggesting that these residues make a modest contribution to the binding of H4K20Me1. We followed up on this observation by making the same point mutations in a TAP-tagged version of the Drosophila msl3 gene (20, 28) and observed the male versus female survival ratios in homozygous offspring carrying a stably integrated mutated msl3-TAP gene (Table 3). The EPAA double mutant had essentially wild type male survival rates, again suggesting that Glu21 and Pro22 in MSL3 play a minimal role in dosage compensation or targeting of MSL3 to a particular histone modification on the Drosophila male X-chromosome. However, the overall structure of the MSL3 β1-β2 loop or other nearby residues may also contribute to the observed H4K20Me1 in vitro binding preference.
TABLE 3.
Rescue frequencies of transgenic Drosophila carrying msl3-TAP point mutations
| Construct | Non-Sb males (rescued) | Sb males | Rescue frequency | p value |
|---|---|---|---|---|
| % | ||||
| msl3-TAP WT | 456 | 544 | 83.8 | 0.0542 |
| Y31A | 103 | 323 | 31.9 | 1.069e-14 |
| Gly21-Pro22-Ala | 349 | 313 | 111 | 0.350 |
To test the contribution of residue Tyr31 to methyllysine binding in hMSL3, we made a Tyr31 to Ala mutation. The in vitro binding of the Y31A mutant to H4K20Me1 was significantly weaker than the wild-type chromo-barrel domain, suggesting a larger contribution to methyllysine binding (Fig. 3; Kd = 78 μm). Male flies homozygous for the Y31A mutant did not survive as well as females (30.9% male survival; Table 3), suggesting that Tyr31 does play a role in dosage compensation, consistent with its contribution to H4K20Me1 binding in vitro. In a previously published study of msl3 chromo-barrel domain mutants, the Tyr31 residue was mutated to alanine along with two adjacent residues (Leu30 and Thr32) (20). However, the triple amino acid mutant had a 0% rescue frequency and was unstable. The resultant mutant dMSL3-LYTA protein could not be detected by Western blots of heterozygous LYT30A cell extracts (20). The instability of MSL3 produced by this triple mutation deemed it uninformative in terms of specific dosage compensation function analysis. As the mutation of Leu30 is likely to disrupt the hydrophobic core of the dMSL3 CBD, we chose to test a less drastic mutation. In our Y31A transgenic line only 30.9% of msl3 null homozygous males are rescued by the mutant construct. Furthermore, we could easily purify the corresponding hMSL3 Y31A mutant CBD and conduct SPR binding studies, suggesting that the single mutant has a much less severe effect on the integrity and folding of the MSL3 protein.
Possible Nucleic Acid Binding Surfaces on the MSL3 CBD
In addition to binding methylated histone tails, we observed that both hMSL3 and dMSL3 chromo-barrel domains bound strongly to nucleic acids when purified from bacterial cell lysates, and remained bound after glutathione-Sepharose affinity purification, an observation consistent with the recent determination of a hMSL3 CBD·DNA complex (36). We could only effectively remove contaminating nucleic acid using ion-exchange chromatography (results not shown). Previously published studies have more specifically shown that the dMSL3 chromo-barrel domain is necessary for efficient in vitro binding of dMSL3 to nucleosomes (18, 19). Analysis of the hMSL3 CBD electrostatic surface revealed two positively charged surface patches (Fig. 5) and three sites where sulfate anions preferentially bind in the five hMSL3 monomers (Fig. 5 and supplemental Fig. S2). These three sulfate-binding sites involve residues Lys10, Arg28, and His55, respectively (Fig. 5). Residues Lys10, His55, Arg74, Asn79, and Arg84 on the hMSL3 CBD that contact sulfate anions in the crystal structure are highly conserved in both MSL3 and MRG15 sequences (Fig. 1), suggesting that the MRG15 CBDs may also bind nucleic acids or nucleosome core particles in a similar manner to MSL3 (supplemental Fig. S3). We also note that the sulfate ion bound consistently at His55 in the five subunits of our structure overlaps almost perfectly with the position of a phosphate group of bound duplex DNA in the recently published hMSL3 CBD·DNA structure (36). Hence the observed sulfate binding sites in our hMSL3 CBD structure could represent binding sites for the phosphodiester backbone of nucleic acids (Fig. 5 and supplemental Fig. S2). Based on the conservation of amino acid residues in the vicinity of these sites in MSL3 and MRG15 sequences, nucleosome recognition could be important in both MSL3 and MRG15 function in their respective HAT complexes.
FIGURE 5.
An electropositive surface on the hMSL3 CBD binds sulfate anions. A, the hMSL3 CBD is rendered as a opaque qualitative electrostatic surface (red negatively charged, blue positively charged, see “Experimental Procedures”). Sulfate anions bound at Lys10-Asn79, His55, and Arg28 are shown as stick models (sulfur, yellow; oxygen, red). Green arrows point to regions that may interact with nucleic acids. B, ball and stick representation of two sulfate anions bound by conserved residues near Arg28 and Lys10 on the surface represented in A. Backbone trace is colored as described in the legend to Fig. 1B.
MSL3 CBD Function in MSL HAT Complex Targeting and Dosage Compensation
Previous studies in yeast have suggested an interaction between the Eaf3 CBD and H3K36Me3 (60, 61). However, the preferential in vitro association of the related MSL3 and MRG15 CBDs with methylated H4K20 sequences suggests that the binding of H3K36Me3 by the individual proteins is not significant in metazoans. The in vitro binding of H4K20Me1 or H4K20Me2 over H3K36Me2 or other histone tail modifications associated with gene activation presented in this study strongly points to the N terminus of histone H4 being the likely in vivo binding target for the MSL3 and MRG15 CBDs. Supporting a role for H4K20Me1 in the recruitment of the MSL complex, H4K20Me1 is an abundant modification on histone tails in Drosophila and humans, and its presence correlates positively with the level of transcriptional activity along human genes, at least as well as for H3K36Me3 (59, 62–67). Furthermore, other studies have shown that H4K20Me1 is found to be preferentially deposited on nucleosomes residing within exons (68, 69), a pattern mirrored in the X-chromosome binding of the Drosophila MSL complex (30). However, there is no published polytene chromosome immunofluorescence evidence for H4K20Me1 being enriched at the sites of Drosophila MSL complex binding (70, 71). Consistent with a role for the dMSL3 CBD in histone tail recognition in vivo, we see dosage compensation effects in a point mutation of the dMSL3 CBD (Y31A) that weakens H4K20Me1 binding in vitro. In addition, the related MRG15 CBD prefers binding methylated H4K20, albeit the trimethyl modification, suggesting that recognition of H4K20 methylation marks is a conserved feature of the MSL3/MRG15 gene family in higher eukaryotes.
As a primary function of the MSL complex is to acetylate lysine 16 on histone H4, it is not entirely surprising that the MSL3 CBD binds preferentially to the two most abundant H4 tail modifications found in vivo in humans and Drosophila (59, 62, 63). It also seems plausible that the binding of histone H4 tails by the MSL3 chromo-barrel domain may play a role in presenting H4 tails for acetylation by MOF within the MSL complex. Given we have shown that the dMSL3 and hMSL3 CBDs bind preferentially to H4K20Me1 in vitro, MSL3 may help to target the MSL complex to specific chromatin regions at least in part by binding to H4K20Me1.
Similarly, the demonstrated in vitro binding of H4K20Me3 to the MRG15 component of the Tip60 complex may assist the targeting of Tip60 to regions of heterochromatin, as H4K20Me3 is highly enriched in human and Drosophila heterochromatin (61, 70, 71) and Tip60 prefers binding to heterochromatic regions enriched in H3K9Me3 (and presumably H4K20Me3) during double-stranded DNA repair (13). Importantly, Tip60 and MRG15 are associated with gene repression functions in Drosophila and are classified as part of the Polycomb group (12). Work presented in this study suggests that the MSL complex may be recruited to active genes in part by MSL3 binding to H4K20Me1, whereas the repressive Tip60 HAT complex may be targeted to heterochromatin by MRG15 being recruited to sites enriched in H4K20Me3. Hence, chromo-barrel domain containing HAT subunits appear capable of exquisitely discriminating the methylation status of lysine 20 on histone H4 and the corresponding biological context.
Supplementary Material
Acknowledgments
We thank Bradley McLellan, Massey University, New Zealand, for cloning the dMSL3 chromo-barrel domain fragment; George Wong for purification conditions for the dMSL3 chromo-barrel domain; and Andrew Welham who carried out the initial characterization of the dMSL3 and hMSL3 chromo-barrel domains. Ewa Kerc contributed to the cloning of the hMSL3 chromo-barrel domain. Robert Shearer cloned the D. melanogaster MRG15 chromo-barrel domain and conducted the initial purification studies. We thank Edwin Smith for generously providing cDNAs for the hMSL proteins, and Prof. Mitzi Kuroda, Harvard Medical School, for providing the msl3-TAP construct for Drosophila transgenic studies. David Sanders (Dept of Chemistry, University of Saskatchewan) provided access to the Nanodrop device. Jeremy Lee graciously conducted the ethidium bromide fluorescence assays. SPR data were collected at the Saskatchewan Structural Sciences Centre, University of Saskatchewan. We thank Pawel Grochulski and James Gorin of the Canadian Light Source CMCF small-gap undulator beamline for timely access and expert assistance with data collection. The Canadian Light Source is funded by the Canadian Institutes for Health Research (CIHR), the National Science and Engineering Council of Canada (NSERC), the Canadian foundation for innovation (CFI) and the University of Saskatchewan.
This work was supported by Canadian Institutes for Health Research Grant MOP 79377 (to S. M.), the Saskatchewan Health Research Foundation regional partnership program, and bridge funding from the University of Saskatchewan College of Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- HAT
- histone acetyltransferase
- MSL
- male specific lethal
- CBD
- chromo-barrel domain
- bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CAPS
- 3-(cyclohexylamino)-1-propanesulfonic acid
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- r.m.s.
- root mean square.
REFERENCES
- 1.Carrozza M. J., Utley R. T., Workman J. L., Côté J. (2003) Trends Genet. 19, 321–329 [DOI] [PubMed] [Google Scholar]
- 2.Rea S., Xouri G., Akhtar A. (2007) Oncogene 26, 5385–5394 [DOI] [PubMed] [Google Scholar]
- 3.Smith E. R., Cayrou C., Huang R., Lane W. S., Côté J., Lucchesi J. C. (2005) Mol. Cell. Biol. 25, 9175–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taipale M., Rea S., Richter K., Vilar A., Lichter P., Imhof A., Akhtar A. (2005) Mol. Cell. Biol. 25, 6798–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta A., Sharma G. G., Young C. S., Agarwal M., Smith E. R., Paull T. T., Lucchesi J. C., Khanna K. K., Ludwig T., Pandita T. K. (2005) Mol. Cell. Biol. 25, 5292–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., McMahon S. B. (2006) Mol. Cell 24, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., Wu L., Corsa C. A., Kunkel S., Dou Y. (2009) Mol. Cell 36, 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raja S. J., Charapitsa I., Conrad T., Vaquerizas J. M., Gebhardt P., Holz H., Kadlec J., Fraterman S., Luscombe N. M., Akhtar A. (2010) Mol. Cell 38, 827–841 [DOI] [PubMed] [Google Scholar]
- 9.Doyon Y., Selleck W., Lane W. S., Tan S., Côté J. (2004) Mol. Cell. Biol. 24, 1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Squatrito M., Gorrini C., Amati B. (2006) Trends Cell Biol. 16, 433–442 [DOI] [PubMed] [Google Scholar]
- 11.Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., Yates J. R., 3rd, Abmayr S. M., Washburn M. P., Workman J. L. (2004) Science 306, 2084–2087 [DOI] [PubMed] [Google Scholar]
- 12.Qi D., Jin H., Lilja T., Mannervik M. (2006) Genetics. 174, 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y., Jiang X., Xu Y., Ayrapetov M. K., Moreau L. A., Whetstine J. R., Price B. D. (2009) Nat. Cell Biol. 11, 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelbart M. E., Kuroda M. I. (2009) Development 136, 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucchesi J. C. (2009) Curr. Opin. Genet. Dev. 19, 550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morales V., Straub T., Neumann M. F., Mengus G., Akhtar A., Becker P. B. (2004) EMBO J. 23, 2258–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales V., Regnard C., Izzo A., Vetter I., Becker P. B. (2005) Mol. Cell. Biol. 25, 5947–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buscaino A., Legube G., Akhtar A. (2006) EMBO Rep. 7, 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larschan E., Alekseyenko A. A., Gortchakov A. A., Peng S., Li B., Yang P., Workman J. L., Park P. J., Kuroda M. I. (2007) Mol. Cell 28, 121–133 [DOI] [PubMed] [Google Scholar]
- 20.Sural T. H., Peng S., Li B., Workman J. L., Park P. J., Kuroda M. I. (2008) Nat. Struct. Mol. Biol. 15, 1318–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott M. J., Li F. (2008) RNA Biol. 5, 13–16 [DOI] [PubMed] [Google Scholar]
- 22.Bone J. R., Lavender J., Richman R., Palmer M. J., Turner B. M., Kuroda M. I. (1994) Genes Dev. 8, 96–104 [DOI] [PubMed] [Google Scholar]
- 23.Gu W., Wei X., Pannuti A., Lucchesi J. C. (2000) EMBO J. 19, 5202–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demakova O. V., Kotlikova I. V., Gordadze P. R., Alekseyenko A. A., Kuroda M. I., Zhimulev I. F. (2003) Chromosoma 112, 103–115 [DOI] [PubMed] [Google Scholar]
- 25.Copps K., Richman R., Lyman L. M., Chang K. A., Rampersad-Ammons J., Kuroda M. I. (1998) EMBO J. 17, 5409–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman M., Franke A., Baker B. S. (1995) Development 121, 463–475 [DOI] [PubMed] [Google Scholar]
- 27.Smith E. R., Allis C. D., Lucchesi J. C. (2001) J. Biol. Chem. 276, 31483–31486 [DOI] [PubMed] [Google Scholar]
- 28.Alekseyenko A. A., Larschan E., Lai W. R., Park P. J., Kuroda M. I. (2006) Genes Dev. 20, 848–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legube G., McWeeney S. K., Lercher M. J., Akhtar A. (2006) Genes Dev. 20, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilfillan G. D., Straub T., de Wit E., Greil F., Lamm R., van Steensel B., Becker P. B. (2006) Genes Dev. 20, 858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith E. R., Pannuti A., Gu W., Steurnagel A., Cook R. G., Allis C. D., Lucchesi J. C. (2000) Mol. Cell. Biol. 20, 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott M. J., Pan L. L., Cleland S. B., Knox A. L., Heinrich J. (2000) EMBO J. 19, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonin E. V., Zhou S., Lucchesi J. C. (1995) Nucleic Acids Res. 23, 4229–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prakash S. K., Van den Veyver I. B., Franco B., Volta M., Ballabio A., Zoghbi H. Y. (1999) Genomics 59, 77–84 [DOI] [PubMed] [Google Scholar]
- 35.Bertram M. J., Pereira-Smith O. M. (2001) Gene 266, 111–121 [DOI] [PubMed] [Google Scholar]
- 36.Kim D., Blus B. J., Chandra V., Huang P., Rastinejad F., Khorasanizadeh S. (2010) Nat. Struct. Mol. Biol. 17, 1027–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan A. R., Lee J. S., Pulleyblank D. E., Murray N. L., Evans D. H. (1979) Nucleic Acids Res. 7, 547–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wetlaufer D. B. (1962) Adv. Protein Chem. 17, 303–391 [DOI] [PubMed] [Google Scholar]
- 39.Edelhoch H. (1967) Biochemistry 6, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 41.Zhang P., Du J., Sun B., Dong X., Xu G., Zhou J., Huang Q., Liu Q., Hao Q., Ding J. (2006) Nucleic Acids Res. 34, 6621–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 44.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 45.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 46.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 47.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 48.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 49.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 50.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 51.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 52.Merritt E. A., Bacon D. J. (1997) Methods Enzymol. 277, 505–524 [DOI] [PubMed] [Google Scholar]
- 53.Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Fischle W., Wang W., Duncan E. M., Liang L., Murakami-Ishibe S., Allis C. D., Patel D. J. (2007) Mol. Cell 28, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun B., Hong J., Zhang P., Dong X., Shen X., Lin D., Ding J. (2008) J. Biol. Chem. 283, 36504–36512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu C., Cui G., Botuyan M. V., Mer G. (2008) Structure 16, 1740–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W. K., Tereshko V., Boccuni P., MacGrogan D., Nimer S. D., Patel D. J. (2003) Structure 11, 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 60.Joshi A. A., Struhl K. (2005) Mol. Cell 20, 971–978 [DOI] [PubMed] [Google Scholar]
- 61.Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J. L. (2007) Science 316, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 62.Yang H., Mizzen C. A. (2009) Biochem. Cell Biol. 87, 151–161 [DOI] [PubMed] [Google Scholar]
- 63.Pesavento J. J., Yang H., Kelleher N. L., Mizzen C. A. (2008) Mol. Cell. Biol. 28, 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Talasz H., Lindner H. H., Sarg B., Helliger W. (2005) J. Biol. Chem. 280, 38814–38822 [DOI] [PubMed] [Google Scholar]
- 65.Vakoc C. R., Sachdeva M. M., Wang H., Blobel G. A. (2006) Mol. Cell. Biol. 26, 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang H., Pesavento J. J., Starnes T. W., Cryderman D. E., Wallrath L. L., Kelleher N. L., Mizzen C. A. (2008) J. Biol. Chem. 283, 12085–12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakaguchi A., Karachentsev D., Seth-Pasricha M., Druzhinina M., Steward R. (2008) Genetics 179, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwartz S., Meshorer E., Ast G. (2009) Nat. Struct. Mol. Biol. 16, 990–995 [DOI] [PubMed] [Google Scholar]
- 69.Tilgner H., Nikolaou C., Althammer S., Sammeth M., Beato M., Valcárcel J., Guigó R. (2009) Nat. Struct. Mol. Biol. 16, 996–1001 [DOI] [PubMed] [Google Scholar]
- 70.Nishioka K., Rice J. C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Lis J. T., Allis C. D., Reinberg D. (2002) Mol. Cell 9, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 71.Ebert A., Lein S., Schotta G., Reuter G. (2006) Chromosome Res. 14, 377–392 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.