Abstract
Förster resonance energy transfer within a protein-protein complex has previously been invoked to explain emission spectral modulation observed in several bioluminescence systems. Here we present a spatial structure of a complex of the Ca2+-regulated photoprotein clytin with its green-fluorescent protein (cgGFP) from the jellyfish Clytia gregaria, and show that it accounts for the bioluminescence properties of this system in vitro. We adopted an indirect approach of combining x-ray crystallography determined structures of the separate proteins, NMR spectroscopy, computational docking, and mutagenesis. Heteronuclear NMR spectroscopy using variously 15N,13C,2H-enriched proteins enabled assignment of backbone resonances of more than 94% of the residues of both proteins. In a mixture of the two proteins at millimolar concentrations, complexation was inferred from perturbations of certain 1H-15N HSQC-resonances, which could be mapped to those residues involved at the interaction site. A docking computation using HADDOCK was employed constrained by the sites of interaction, to deduce an overall spatial structure of the complex. Contacts within the clytin-cgGFP complex and electrostatic complementarity of interaction surfaces argued for a weak protein-protein complex. A weak affinity was also observed by isothermal titration calorimetry (KD = 0.9 mm). Mutation of clytin residues located at the interaction site reduced the degree of protein-protein association concomitant with a loss of effectiveness of cgGFP in color-shifting the bioluminescence. It is suggested that this clytin-cgGFP structure corresponds to the transient complex previously postulated to account for the energy transfer effect of GFP in the bioluminescence of aequorin or Renilla luciferase.
Keywords: Computer Modeling, NMR, Protein-Protein Interactions, Spectroscopy, X-ray Crystallography, Bioluminescence, Clytia gregaria, GFP, Photoprotein
Introduction
The bioluminescence of many marine coelenterates, well-studied examples being the jellyfish Aequorea and the sea-pansy Renilla, involves the interaction of two proteins, a Ca2+-regulated photoprotein in the jellyfish case, aequorin, and its cognate green-fluorescent protein, Aequorea GFP (1). Addition of Ca2+ to the purified aequorin produces a blue bioluminescence. It was early recognized that, in the jellyfish itself, the in vivo bioluminescence was a green color and after further study, the origin of this green emission was identified as the GFP. A Förster-type resonance energy transfer (FRET)3 mechanism was invoked to explain how this bioluminescence spectrum is shifted (2). However, the well-known Förster theory requires concentrations of the donor-acceptor partners in the millimolar range, whereas in some bioluminescence systems, e.g. from the sea pansy Renilla and also the jellyfish Clytia subject herein, the GFP effect on the in vitro bioluminescence is observed at micromolar concentrations (3). Clearly the bioluminescence interaction has to involve formation of a complex and, in the case of Renilla, the formation of a luciferase-GFP complex has been shown (3).
In this work, we have determined by x-ray crystallography the spatial structures of the recombinant Ca2+-regulated photoprotein clytin and Clytia GFP (cgGFP), which were cloned from a single specimen of Clytia gregaria (syn. Phialidium gregarium). Based on the structures, NMR titration experiments were employed to identify the interaction surfaces in a complex of both proteins. For a mixture of clytin and cgGFP at millimolar concentration, 1H-15N HSQC experiments revealed perturbation of chemical shifts of the separate proteins, which could be mapped to particular residues being affected by complexation. The NMR experiments also indicated that the association was weak but from knowledge of the interaction surface, computational docking was employed to propose an overall three-dimensional structure of the clytin-cgGFP complex.
EXPERIMENTAL PROCEDURES
Molecular Biology
Cloning of the clytin and Clytia GFP genes from a single specimen of the jellyfish C. gregaria, expression, purification, and characterization of recombinant clytin and cgGFP have been published (4). Site-directed and truncation mutagenesis of clytin were done on the template p22-Cl3 E. coli expression plasmid carrying the apo-clytin gene of wild-type C. gregaria. Mutations resulting in the amino acid change: K11A, K13A, N15A, N109A, or N188A were carried out using the QuickChange site-directed mutagenesis kit (Stratagene). N-terminal-truncated clytin mutants 5A (sequence starts from Ala-5) and 10V were amplified by PCR. The plasmids harboring mutations were verified by DNA sequencing.
Crystallography
Crystals of clytin grew at 16 °C within 1 week to a size of 50 × 50 × 300 μm. The crystallization droplet was set up using Mosquito crystallization robot (TTP Labtech) and contained equal volumes of protein (15 mg/ml) and reservoir solution (20% PEG-3350, 0.2 m NaH2PO4, pH 8.8) derived from the Peg/Ion crystallization screen (Hampton Research). The crystal was flash-frozen in liquid nitrogen. Native diffraction data were indexed and scaled to 1.9 Å resolution using HKL2000. The space group of clytin was C2221 with unit cell dimensions (Å), a = 43.39, b = 68.93, c = 115.35. Phases were determined by molecular replacement with PHASER (5), using the structure of obelin (PDB code 1JFO) as a search model. The final models were refined with PHENIX (6). Manual adjustments to the model were done using COOT (7). The quality of the final model was validated with MOLPROBITY (8). The detailed data processing and refinement statistics are shown in Table 1.
TABLE 1.
X-ray structure statistics
| clytin | cgGFP | |
|---|---|---|
| Data collection | ||
| Resolution range (Å) | 50–1.9 (2.0–1.9)a | 27.7–1.47 (1.52–1.47) |
| Wavelength (Å) | 1.54 | 0.979 |
| Space group | C2221 | I212121 |
| Cell dimensions | ||
| a, b, c (Å) | 43.39, 68.93, 115.35 | 53.09, 91.45, 110.61 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Unique reflections | 14018 (1326) | 40289 (3811) |
| Completeness (%) | 99.5 (95.9) | 86.7 (20.3) |
| I/σ(I) | 26.43 (2.82) | 38.77 (2.25) |
| Rsym (%) | 11.2 (60.5) | 4.9 (14.5) |
| Redundancy | 13.0 (6.4) | 4.4 (1.5) |
| Refinement | ||
| Resolution range (Å) | 50–1.9 | 10–1.55 |
| Reflections used | 13990 (728) | 36117 (1877) |
| Rwork, Rfree | 17.07%, 21.94% | 18.2%, 20.2% |
| Mean B factor (Å2) | 19.12 | 14.27 |
| Protein atoms | 1678 | 1827 |
| Solvent atoms | 128 | 235 |
| RMSD bond lengths (Å) | 0.011 | 0.015 |
| RMSD angles (°) | 1.185 | 1.407 |
a Values for the highest resolution shell are given in parentheses.
Crystals of cgGFP grew at 4 °C within 5 days to a size of 200 × 200 × 250 μm. The crystallization droplet contained equal volumes of protein (9 mg/ml) and reservoir solution (2 m ammonium sulfate, 0.1 m sodium citrate, pH 5.5) derived from the Wizard I crystallization screen (Emerald Biosystems). The crystal was flash-frozen in liquid nitrogen. Native diffraction data were indexed and scaled to 1.55 Å resolution using HKL2000. The space group of cgGFP was I212121 with unit cell dimensions (Å), a = 53.09, b = 91.45, c = 110.61. Phases were determined by molecular replacement with MOLREP (9) using GFP from Aequorea victoria as a search model (PDB code 1EMA). Iterative model validation, rebuilding and refinement, were carried out with MOLPROBITY (8), XFIT (10), and REFMAC5 (11), respectively. The detailed data processing and refinement statistics on cgGFP are shown in Table 1.
Protein concentrations were determined by the dye-binding method of Bradford (12) using an assay kit (Bio-Rad) and bovine serum albumin as a standard. On this basis extinction coefficients for clytin (ϵ280 = 65,200 m−1 cm−1) and cgGFP (ϵ485 = 64,000 m−1 cm−1) were calculated and subsequently, protein concentrations were determined by absorbance.
NMR Sample Preparation
Uniformly 15N- or 15N-, 13C-labeled clytin and cgGFP were obtained from the cells grown in M9 minimal medium containing 15NH4Cl or additional [13C]glucose. To acquire 15N-, 13C-, and 60% 2H-labeled cgGFP, 99.8% 2H2O-based M9 was used. To exchange amide group deuterium to hydrogen, triple-labeled cgGFP was subjected to reversible denaturation in 6 m guanidine-HCl for 10 min, followed by 20-fold droplet dilution in PBS at 25 °C and overnight incubation at 4 °C. Activity of cgGFP was restored upon refolding as indicated by absorbance and fluorescence spectra and bioluminescence color shift assay with clytin.
NMR Spectroscopy
NMR experiments were performed on Bruker DMX 600 MHz and Avance 800 MHz spectrometers equipped with z-gradient triple-resonance cryo-probes. Data were processed in FELIX (Accelrys Inc.) and visualized with NMRVIEW (13). The backbone assignments were obtained by MARS (14). All NMR samples were dissolved in buffer containing 20 mm Tris-HCl, pH 7.0, 10 mm NaCl, 2 mm EDTA, 0.01% (w/v) sodium 2,2-dimethylsilapentane-5-sulfonate (DSS) and 10% (v/v) 2H2O. The experiments for the backbone assignments of clytin include two-dimensional 1H-15N HSQC, and 3D 1H-15N-13C HNCA, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HBHA(CBCA)NH, HBHA(CBCA)(CO)NH (15), all performed at 293 K. The experiments for the backbone assignments of cgGFP include deuterium-decoupled 3D HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO using 15N-, 13C-, and 60% 2H-labeled cgGFP sample and 4D 13C,15N-edited NOESY (16) using 15N-, 13C-labeled cgGFP sample, all performed at 310 K. The backbone assignments for GFP at 298 K were obtained from those at 310 K by following the shift of resonance signals in a series of two-dimensional 1H-15N HSQC spectra recorded at decreasing temperatures.
Chemical shift perturbation analyses were performed at 293 K for clytin and 298 K for cgGFP by monitoring the two-dimensional 1H-15N HSQC spectra of titrated proteins. Unlabeled clytin WT or mutants K11A, K13A, N109A, N188A, and 5A (0.5 mm, 1 mm), were added to 15N- or 15N-, 2H-labeled cgGFP (0.4 mm). Alternatively, unlabeled cgGFP (0.3 mm, 0.6 mm) was added to 15N-labeled clytin (0.2 mm). The amide hydrogen and nitrogen chemical shift changes were calculated according to Equation 1,
where ΔδN and ΔδH represent the changes in the amide nitrogen and proton chemical shifts (in parts per million), respectively.
Calculations of the Clytin-cgGFP Complex Structure
The computational structures of the clytin-cgGFP complex were generated with HADDOCK2.0 (17, 18) in combination with CNS (19). Ambiguous interaction restraints (AIRs) were generated for both clytin and cgGFP based on chemical shift perturbation studies (Table 2) as described (17). The starting structures were the monomer cgGFP (PDB code 2HPW) and the clytin (PDB code 3KPX) with the manually added 2–8 N-terminal segment, which is absent in the deposited structure and was defined as fully flexible during docking. The standard HADDOCK protocol was used. For the rigid-body energy minimization, 1,000 structures were generated, with the 200 lowest energy solutions used for subsequent semi-flexible simulated annealing and water refinement. Resulting structures were sorted according to intermolecular energy and clustered using a 7.5 Å cut-off criterion. Subsequent cluster analysis was performed within a 4.0 Å cut-off criterion. The 10 lowest energy solutions were taken to represent the structure of the complex (supplemental PDB files).
TABLE 2.
List of active and passive residues of clytin and cgGFP derived from chemical shift perturbation plots, which comprised AIRs for HADDOCK docking
| Active | Passive | |
|---|---|---|
| clytin | Ala5, Ala9, Val10, Leu12, Lys13, Thr14, Asn15, Glu17, Lys100, Lys104, Ser107, Asn109, Asn188, Gly191 | Thr2, Glu3, Thr4, Ser6, Lys7, Tyr8, Lys11, Pro19, Glu97, Asn108, Lys110, His160, Pro187 |
| cgGFP | Glu55, Lys132, Leu138, Met140, Leu143, His145, Gly175, Gly209, Lys210 | Gly54, Asn103, Asp104, Gly130, Phe131, Ser133, Asn134, Met173, Gly174, Gly176, Phe208, Pro212 |
Bioluminescence Assay
Bioluminescence spectra of clytin and clytin with cgGFP, were measured with a Varioskan Flash Spectrofluorimeter (Thermo Scientific). All measurements were carried out at 25 °C. Luminescence was triggered by injection of 7 μl of 40 mm CaCl2 into the wells containing 150 μl of isolated clytin (final concentration is in the 0.4–1.5 μm range) or clytin mixed with cgGFP (final concentrations from 0 to 9.7 μm) in 2 mm EDTA, 10 mm NaCl, 20 mm Tris-HCl, pH 7.0 buffer. Emission spectra were fully corrected for instrumental spectral sensitivity with the computer program supplied with the instrument, and also for bioluminescence intensity decay over the time for the spectral scan. All spectra were the average of three measurements. The energy transfer efficiency coefficient (KET) for clytin and clytin mutants was determined by plotting the I500/I470 ratio versus total concentration of cgGFP, where I500 and I470 are bioluminescence intensities at 500 nm and 470 nm, respectively. The slope of the linear regression fitted data was taken as the KET value.
Isothermal Titration Calorimetry
Isothermal titration calorimetry measurements were performed on an ITC200 calorimeter (Microcal Inc., Northampton, MA). All experiments were carried out at 25 °C in 20 mm Tris-HCl, 10 mm NaCl, 2 mm EDTA, pH 7.0. The reactant (0.1 mm clytin) was placed in the 200-μl sample chamber and cgGFP (4.68 mm for monomer) in the syringe was added with 20 successive additions of 2 μl for 4 s (with an initial injection of 0.5 μl). The interval between each injection lasted 150 s. The peaks generated were corrected for cgGFP heat of dilution and integrated using the ORIGIN software (Microcal Inc) by plotting the values in microcalories against the ratio of total moles of injectant, monomer cgGFP, to reactant clytin, within the cell. Data were fit using a 1:1 clytin:cgGFP monomer binding model.
RESULTS
Crystal Structures of Clytin and cgGFP
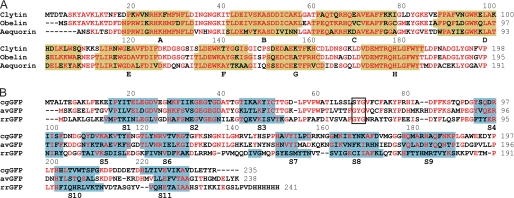
Both proteins were separately crystallized and their structures determined by x-ray crystallography (Table 1). Clytin has molecular mass of 22.4 kDa and shares high structural and sequence similarity with the other Ca2+-regulated photoproteins, obelin (20) (RMSD 0.66 Å, sequence identity 74%) and aequorin (21) (RMSD 1.39 Å, sequence identity 57%) (Fig. 1A). All have four helix-loop-helix motives, three EF-hand Ca2+-binding loops, and the substrate 2-hydroperoxycoelenterazine bound in a hydrophobic cavity (22) (Fig. 2A). Additionally, a contaminant metal ion is found within EF-hand I loop, which approaches the non-standard conformation similar to EF-hand I of obelin, whose crystal was briefly soaked with Ca2+ (23). There is no electron density for the Thr2-Ala9 region implying significant structural flexibility of the N terminus of clytin. cgGFP forms the well-known barrel structure built of 11 β strands (S1–S11) with the chromophore buried inside (24, 25) (Fig. 2B). Despite a low sequence identity (Fig. 1B), the structure of cgGFP highly resembles GFPs from Aequorea (26) (RMSD 1.04 Å, sequence identity 41%), and Renilla (27) (RMSD 1.84 Å, sequence identity 19%). The cgGFP homodimer can be generated from the cgGFP monomer in the crystallographic asymmetric unit by applying a 2-fold symmetry axis. The dimerized form of cgGFP is evident from a mass of 52 kDa determined by analytical ultracentrifugation, close to that of the natural cgGFP (57 kDa) (28, 29), consistent with the 27 kDa monomer mass determined by SDS-PAGE. The cgGFP monomer buries 1,370 Å2 (13% of the total surface area) in the dimer interface.
FIGURE 1.
Protein sequence alignments of (A) photoproteins: clytin, obelin (PDB code 1QVO) and aequorin (PDB code 1EJ3); and (B) GFPs: cgGFP, Aequorea GFP (avGFP) (PDB code 1EMA) and Renilla GFP (rrGFP) (PDB code 2HR7). Identical residues are red. Secondary structure elements are highlighted in yellow (α helices A–H of photoproteins) and light blue (β strands S1–S11 of GFPs). Residues comprising the chromophore of GFPs are enclosed in black box.
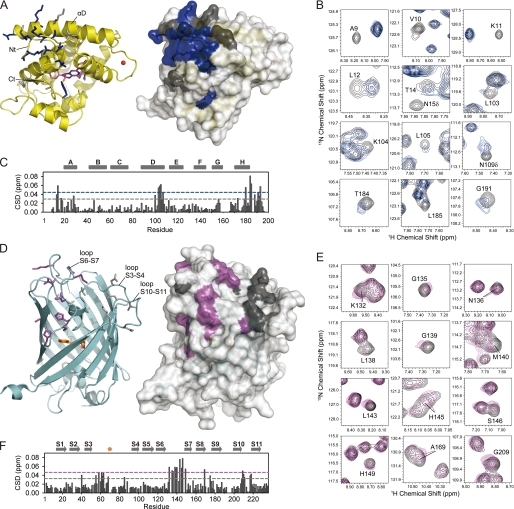
FIGURE 2.
Chemical shift mapping identifies the interaction surfaces of clytin and cgGFP. On crystal structures of clytin (A) and cgGFP (D) the interfacial residues mapped according to cross-peak/intensity shift are shown as sticks and highlighted in color on the surface. B & E, 1H-15N HSQC spectra areas derived from superposition of 15N-labeled clytin (B, black) and 15N, 2H-labeled cgGFP (E, black) with unlabeled cgGFP (blue) and clytin (magenta), respectively. C & F, weighted-average chemical shift differences (CSD) between 15N-clytin (C) or 15N,2H-cgGFP (F) and 1:3 15N-clytin/cgGFP and 1:2 15N,2H-cgGFP/clytin mixtures. The dashed lines represent the one standard deviation (gray) and two standard deviations (blue, magenta) cut-offs. Residues, whose cross-peak shifted more than one or two standard deviations above the average, are mapped on the spatial structures in gray and blue for clytin, and in gray and magenta for cgGFP, respectively. Residues of clytin (mostly N-terminal) with significant peak intensity perturbations, are also mapped in blue.
Mapping the Clytin-cgGFP Interface
The backbone NMR resonance assignments of both proteins, which are the basis for our chemical shift perturbation mapping, were obtained by heteronuclear NMR spectroscopy using 15N,13C-labeled clytin and 15N,13C,2H-labeled cgGFP. The backbone resonances of more than 94% residues were assigned for both proteins (supplemental Fig. S1). Lack of assignment for residues Lys159–Asn165 located at the end of α-helix G and the EF-hand IV loop of clytin might be due to the line broadening caused by the structural flexibility in this region.
A similar effect was shown in the NMR study of aequorin (30). Line broadening for the Thr2–Lys7 residues of clytin is in agreement with x-ray crystallography verified structural flexibility of the N terminus. Residues of cgGFP lacking assignments mostly belong to the loop regions comprising the top and bottom of the GFP barrel.
We were unable to crystallize any clytin-cgGFP complex either from a mixture of proteins or for a covalently cross-linked complex under around 300 crystallization conditions available from commercial kits (Hampton Research, Emerald Biosystems). Instead evidence for protein-protein association was inferred from two 1H-15N HSQC titration experiments, first with 15N-labeled clytin and unlabeled cgGFP, then vice versa. NMR titration could not be saturated because of the limited solubility of both proteins and the weak interaction between them, and therefore the Keq could not be derived from the NMR titration data (supplemental Fig. S5). Protein concentrations were 0.3–1 mm, which implies an equilibrium constant Keq in the mm range. Nevertheless, a number of peaks in the HSQC spectra showed chemical shift and/or peak intensity perturbations from which the binding surfaces of clytin and cgGFP were mapped (31–34) (Fig. 2). The perturbed peaks show concentration-dependent chemical shift changes, which indicates a fast exchange on the NMR chemical shift time scale (supplemental Fig. S5).
When mapping the perturbed residues on the structures of clytin and cgGFP, these residues are well clustered on the surfaces of both proteins, which implies specific interactions between the two proteins (Fig. 2). The remaining chemical shift changes upon the titration are relatively small (Fig. 2, C & F), with maximum chemical shift perturbation values for both proteins not exceeding 0.08 ppm, which indicates no large structural rearrangement during the complex formation. For clytin (Fig. 2, B & C), the perturbations are assigned to residues within three segments: 9–17 at the N terminus, 100–109 in the α-helix D, and at the C terminus, 180–193, as well as some adjacent residues. For cgGFP (Fig. 2, E & F), the perturbations are identified as belonging to residues 55–65, mainly in segments of loop S3–S4 and the central α-helix, 132–149 of the longest loop S6–S7 covering the GFP barrel from the top, and 209–218 of the remarkably acidic loop S10–S11, as well as some adjacent residues. For cgGFP, however, chemical shifts are also detected for residues buried inside the protein molecule. These are segment 60–66, “connecting” the interaction surface with the chromophore, and Ser146 and His149, which form hydrogen bonds with Tyr69 of the chromophore through the water molecule. The cgGFP contact surface is less uniform and narrower than that of clytin, which may be explained by overlapping or lack of assignment for some of the resonances in the main interacting regions: excluding prolines these are Ser133, Asn134, Ile137, Arg141, Tyr144 of the loop S6–S7 and Asp215 of the loop S10–S11. On the clytin-cgGFP complex structure these residues are found buried in the protein-protein contact region. cgGFP forms a homodimer in solution, and there is no overlap between the cgGFP dimer interface and the clytin-binding patches.
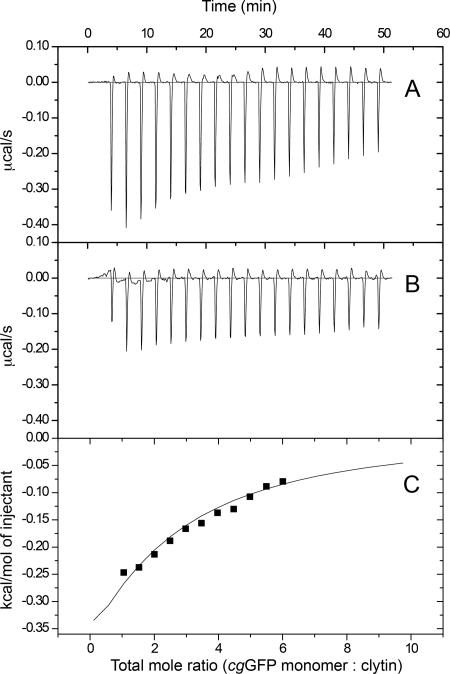
We employed isothermal titration calorimetry (ITC) to obtain an independent assessment of the interaction of clytin with cgGFP. ITC has been shown to be capable of recovering weak binding constants although the accuracy of thermodynamic parameters is problematic compared with protein-protein affinities in the micromolar regime (57). The net heats of interaction of cgGFP as titrant added to clytin are shown in Fig. 7. The experiment and data analysis take into account the precautions suggested by Turnbull et al. (57). Above molar ratio about 3.0 there is large uncertainty due to the mixing signal being hardly different from the control dilution heat of cgGFP alone. The full line is an unweighted fit to a binding model with fixed stoichiometry (n = 1.0) using a 1 clytin:1 cgGFP monomer binding model, and the derived affinity constant is KD = 0.90 ± 0.07 mm. A model with 1 clytin:1 cgGFP dimer yields almost the same result. This affinity is consistent with the millimolar range estimate for Keq from the NMR titration experiment. Interestingly, clytin and cgGFP could be separately concentrated up to 50 mg/ml and 160 mg/ml, respectively, while the mixture of proteins always showed precipitation at concentrations higher than 30 mg/ml under the same conditions. Because the clytin-cgGFP complex buries predominantly hydrophilic residues, (discussed below), the decreased solubility of the protein mixture is more evidence of complexation at these concentrations.
FIGURE 7.
ITC titration curves of clytin with cgGFP. A, raw data of heat changes upon addition of cgGFP (4.68 mm monomer) into the cell containing 0.1 mm of clytin. B, corresponding heat of cgGFP dilution. C, processed data corresponding to the heat of each injection plotted against the molar ratio of total cgGFP to total clytin after subtraction of the heat of cgGFP dilution. Buffer contained 20 mm Tris-HCl, 10 mm NaCl, 2 mm EDTA, pH 7.0. The affinity constant (KD = 0.90 ± 0.07 mm) was derived at 1:1 fixed stoichiometry.
Computational Docking of the Clytin-cgGFP Complex
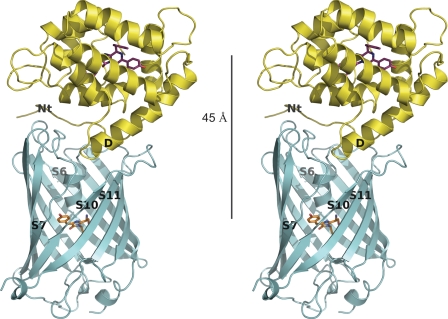
Because of weak interaction between clytin and cgGFP under NMR conditions accompanied by the large molecular size of the proteins, it was not possible to derive accurate spatial restraints from measuring intermolecular NOEs for the complex. Therefore we use a docking approach, named HADDOCK2.0 (17, 18), which relies on ambiguous restraints originating from initial NMR chemical shift perturbation data (Table 2) to derive an accurate model of a protein-protein complex (31, 35–37). A feature of HADDOCK to introduce backbone flexibility was applied to the Thr2-Ala9 N-terminal region of clytin. Fig. 3 is the computational result for the clytin-cgGFP complex dimer, initially based on the identified interaction surfaces which were highly suitable for structure calculation using HADDOCK. The family of final structures had the lowest intermolecular energy (−382.81 kcal/mol) and the highest buried surface area (1,913 Å2) (supplemental Table S1). The average pairwise RMSD in this cluster is 1.29 ± 0.48 Å for backbone atoms. cgGFP forms a homodimer in solution, and the clytin-binding patch on each cgGFP monomer would be distant from each other, thus one cgGFP monomer could accommodate one clytin. As the HSQC chemical shift perturbations and the ITC data both indicate an interaction constant in the millimolar range, this is considered as very weak. The contact surface indeed reveals a relatively low number of hydrophobic contacts and hydrogen bonds, although in total the complex buries 1,913 ± 87 Å2 of surface area, which is average for a protein-protein complex (38).
FIGURE 3.
Stereoview representation of the spatial structure of the clytin-cgGFP complex derived from x-ray structures of clytin and cgGFP, NMR-mapping of the interaction surfaces and computational docking in HADDOCK. 45 Å is the distance between the two chromophores. Structural elements of clytin and cgGFP comprising the interaction surface are labeled.
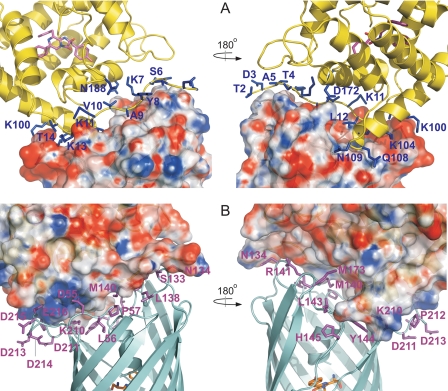
Calculation of the electrostatic potential of clytin and cgGFP reveals that the interfaces have remarkable charge complementarity, which might assist the complex stabilization (Fig. 4). The clytin α-helix D and the proximal N terminus carry the positive charge and occupy, also with good shape complementarity, the negatively charged gutter on the top of the cgGFP barrel formed by the S3–S4, the distal part of the S6–S7, and the S10–S11, loops. The S10–S11 loop is strongly acidic and appears as the least structured region of the cgGFP molecule. This may enable it to adjust for best fit to the clytin interface. In the 10 lowest free energy structures (supplemental Fig. S2 and PDB files), contacts in this region (Fig. 4) are prevalently formed by lysine residues of clytin (Lys11, Leu12, Lys13, Thr14, Lys100, Lys104) and aspartic and glutamic residues of cgGFP (Asp55, Lys210, Asp211, Pro212, Asp213, Asp214, Asp215, Glu216). Also Gln108 of clytin approaches Phe210 of cgGFP, and Asn109 of clytin lies adjacent to His145 and Tyr144 of cgGFP. As expected for electrostatic interactions, the clytin-cgGFP complex formation should be considerably sensitive to ionic strength (Fig. 6). These features of charge complementarity of interfaces together with a low binding affinity, are largely found among transient complexes of various proteins, the well studied examples being redox proteins and Ras or Rap with their signaling effectors (39–41).
FIGURE 4.
The clytin-cgGFP interface. Two views of the molecules are rotated by 180° to allow for viewing of the interaction surfaces. The electrostatic surface (−10kT/e–+10kT/e) of cgGFP (A) and clytin (B) are shown. Poisson-Boltzmann electrostatics calculations were done within PDB2PQR (55) and evaluated in APBS (56). The positively charged, negatively charged, and neutral amino acids are represented in blue, red, and white, respectively. Residues of clytin (A) and of cgGFP (B) buried in the contact surface are shown as blue and magenta sticks, respectively.
FIGURE 6.
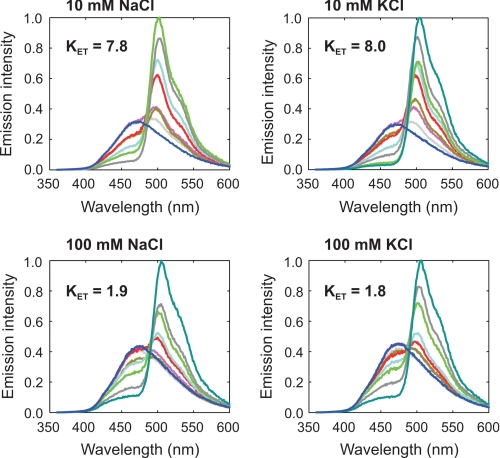
Bioluminescence color-shift assay to show the ionic strength dependence of the clytin-cgGFP energy transfer measured as KET. Bioluminescence spectra of clytin were obtained upon titration with cgGFP (0–3.62 μm; dark blue line, 0 μm; gray line, 0.03 μm; purple line, 0.06 μm; dark yellow line, 0.12 μm; red line, 0.24 μm; light blue line, 0.45 μm; light green line, 0.90 μm; dark gray line, 1.81 μm; dark green line, 3.62 μm). Concentration of clytin was 0.47 μm. Spectra were recorded in the buffer containing 20 mm PIPES, pH 7.2, 1 mm MgCl2, 0.5 mm EDTA, and different concentrations of NaCl or KCl, upon injection of CaCl2. Clytin and cgGFP were from a later batch, indicating some uncertainty in the absolute values of KET values.
Another major interacting region is moderately polar and comes from overlapping of the surface accessible region of the S6–S7 loop of cgGFP (residues Ser133, Asn134, Leu138, Gly139, Met140, Arg141) and some adjacent residues (Met173, Met174, Gly174) by the distal part of the N terminus of clytin (Thr2, Asp3, Thr4, Ala5, Ser6, Lys7, Tyr8, Ala9, Val10). The exact contacts in this region are highly variable because of flexibility of the residues 2–9 of clytin, which together with mutagenesis data discussed below implies a less important role for the N terminus of clytin in binding cgGFP compared with the charge complementarity region (Fig. 4). Superimposition of the 10 best structures of the complex demonstrates flexibility of structural elements comprising the interface (RMSD for Thr2–Ala9 region of clytin 4.51 ± 1.57 Å, RMSD for the S10–S11 loop of cgGFP 1.13 ± 0.21 Å) (supplemental Fig. S2).
Clytin Mutants
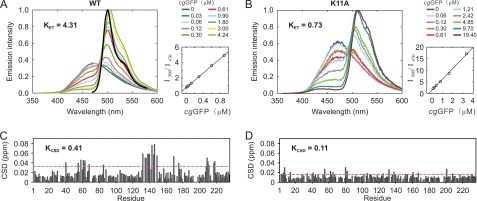
Site-directed and N-terminal truncation mutagenesis of clytin was introduced to assess the contribution of the residues found at the interface to the degree of protein association and to determine if there was a correlation with the effectiveness of cgGFP in producing the well-known bioluminescence color shift in the reaction. These mutants of clytin were K11A, K13A, N15A, N109A, N188A, and two N-terminal truncations (5A, 10V). All substitutions had negligible effect on the bioluminescence properties of clytin, implying no significant rearrangements in spatial structures of clytin mutants. Fig. 5 shows the bioluminescence spectral titration with cgGFP for one of the point mutants compared with native clytin. The cgGFP effectiveness in producing a color shift was measured by an interaction constant, KET, and shown to decrease for all the mutants (supplemental Fig. S4) and most significantly for the substitution K11A (Fig. 5A). The degree of cgGFP HSQC chemical shift perturbations, obtained upon titration with clytin mutants, was used to calculate a quantitative parameter of protein association, named KCSD. Values of chemical shift perturbation after subtracting the average perturbation plus one standard deviation were summed to give the KCSD value in ppm units. Peak tables of cgGFP upon titration with clytin mutants are shown in supplemental Fig. S4 and for K11A mutant in Fig. 4B. The apparent association constant KCSD correlates well with KET, all mutants show a decrease in KET with a lessening of the degree of association (KCSD). Substitutions K11A, K13A, and the 10V truncation, had the strongest effect, reducing KET up to 4-fold from clytin along with a strong reduction in binding affinity. These positions are of interest because of their contribution to the surface charge complementarity of the interacting proteins. On the other hand it indicates the important role of the charge complementarity region compared with the flexible N-terminal segment, because the effects of K11A and K13A substitutions are comparable to that of deleting the first 9 N-terminal residues of clytin. It also implies a minimal role of any small structural rearrangement of clytin mutants in affecting binding to cgGFP. Substitutions N109A, N15A, and N188A had strong, moderate and the smallest effect, respectively.
FIGURE 5.
Mutations of interfacial residues correspondingly decrease the affinity of the complex (KCSD) together with the energy transfer efficiency (KET). A, bioluminescence spectra of wild-type clytin (left) and K11A clytin (right) obtained upon titration with cgGFP (0–19.4 μm). The fluorescence spectrum of cgGFP is shown in black on the wild-type clytin spectrum (left). KET was determined from the corresponding plots as the slope of the I500/I470 ratio versus cgGFP concentration, where I500 and I470 are bioluminescence intensities at 500 nm and 470 nm, respectively. B, weighted-average chemical shift differences (CSD) between 15N,2H-cgGFP and mixtures of 15N,2H-cgGFP with 1:2 molar excess of clytin (left), and K11A clytin (right), respectively. KCSD was determined as a sum of CSD above the average CSD plus one standard deviation cut-off (purple dashed line) in ppm units.
DISCUSSION
Energy transfer or the bioluminescence color shift on the addition of GFP, has previously received detailed study for two bioluminescence systems, that of aequorin and of Renilla luciferase (3, 42, 44–46). The mechanism has been proposed to be by FRET within a transient protein-protein complex. According to the well-known FRET equation, the probability or rate of energy transfer, from the excited donor to the acceptor, depends on several parameters, most critical being the donor-acceptor separation and the spectral overlap of donor fluorescence and acceptor absorption. A convenient measure is the “Förster separation” where the probability of the donor radiative S1 → S0 transition equals the probability of energy transfer populating the acceptor S1 state; in almost all cases this distance is less than 10 nm. This means that for the partners randomly distributed in free solution, they need to be in the millimolar concentration range. The bioluminescence color shifts however, are observed at micromolar protein concentrations so for FRET to be feasible the donor-acceptor separation must be constrained within a protein-protein complex.
For the Renilla luciferase bioluminescence in particular, the addition of Renilla GFP at micromolar concentrations, not only produced the green color shift but enhanced the bioluminescence quantum yield about three times. This is conclusive evidence for FRET indicating that efficient excited state coupling in the transient complex competes with both radiative and non-radiative deactivation pathways of the primary excited S1 state formed by the reaction on the luciferase. A stable complex was not observed by direct methods, chromatography, fluorescence anisotropy, at these micromolar concentrations. However, using the Hummell-Dryer chromatographic method, Ward and Cormier (45) reported the presence of a Renilla luciferase-Renilla GFP complex. Further evidence that such a complex must be involved for the bioluminescence shift was that the energy transfer was specific for the type of GFP, it occurred with GFPs from other species of Renilla but not from GFPs of more distantly related organisms. Also, the shift effect was negated by amino acid modification in the GFP and by higher ionic strength in the buffer (>100 mm) (Fig. 6).
A complex has also been reported for the aequorin-Aequorea GFP bioluminescence using the Hummel-Dryer method (47). In that case no bioluminescence quantum yield increase accompanying the energy transfer was observed (42) as also the case here for the clytin bioluminescence in Fig. 5A. Morise et al. (42) however, demonstrated that energy transfer was significantly enhanced in a suspension of DEAE particles on which the aequorin and Aequorea GFP had been co-adsorbed, presumably bringing the two partners to proximity, but the color shift was also observed to an unrelated acceptor, FMN, meaning that it was nonspecific.
The observations on the clytin bioluminescence system reported here bear similarity to these earlier reports. The clytin bioluminescence spectrum is shifted to the fluorescence of cgGFP by only micromolar concentrations of cgGFP, the effect is diminished by modification of amino acid residues in the clytin, which otherwise affect no change in the clytin bioluminescence properties, the cgGFP shift is an order of magnitude less effective using the distantly related photoprotein obelin, even though with this pair, the spectral overlap is significantly higher (4), and the KET is reduced but not eliminated at increased ionic strength.
Additional similarity to earlier reports was that no clytin-cgGFP interaction in the micromolar range could be detected by the methods of fluorescence anisotropy, analytical ultracentrifugation, or plasmon resonance (results not shown). However, in contrast to the cases of Renilla and aequorin just mentioned, Markova et al. (4) recently observed no complex by Hummel-Dryer chromatography using a starting concentration ten times higher than Ward and Cormier used for their Renilla experiment. Altogether, we estimate here a 0.9 mm value for the clytin-cgGFP affinity constant, consistent with the weak Keq in the mm range inferred from the NMR perturbations.
Although the computational model in Fig. 3 needs to be interpreted with appropriate reservation, we point out that the spatial arrangement of the donor and acceptor makes it attractive to consider this complex as the functional bioluminescence unit in vitro. There is a very favorable spectral overlap, 1.3 × 10−13 m−1 cm3, between the bioluminescence from clytin, maximum 470 nm, and the absorption of cgGFP, having a monomer extinction coefficient of 64,000 m−1 cm−1 at 485 nm (4). Combined with the 45 Å separation of the donor and acceptor in the structure of the complex (Fig. 3), and the fact that the cgGFP will be dimerized in the complex, the electronic transitions are very strongly coupled. The energy from the bioluminescence reaction of the clytin will be quantitatively deposited into the excited state of the acceptor, the cgGFP. However, as the protein-protein complex is weak with a dissociation constant (Keq) in the millimolar range according to the NMR chemical shift and ITC methods (Fig. 7), the mechanism by which added cgGFP at only micromolar concentrations is able to shift the bioluminescence toward the fluorescence of cgGFP, remains to be established.
The computational structure of the clytin-cgGFP complex resembles features of a weak protein-protein complex predominantly governed by electrostatic forces, with a low number of total intermolecular contacts (39–41). For a weak protein interaction the relatively high value of the clytin-cgGFP buried surface area (1,913 Å) derives from the impact of the distal (flexible) part of the clytin N terminus interacting with the top cgGFP barrel loops which together account for 30% of the total buried surface. However, intermolecular contacts in this region are minimal and the position of the clytin N terminus itself is highly variable among the best batch of structures, which implies a less significant impact of the distal part of the clytin N terminus compared with its proximal part and the α-helix D carrying the positive charge. This conclusion is supported by mutagenesis of clytin where we observe that deletion of the flexible part of the N terminus has the same effect on complex affinity and cgGFP color shift efficiency as the single substitutions K11A and K13A. These substitutions evidently affect electrostatics similarly to the high ionic strength conditions.
The question arises as to the physiological relevance of this clytin-cgGFP computational structure in Fig. 3. The photocytes of the jellyfish Aequorea and Clytia can be assumed to be the same, contain concentrations of the bioluminescence proteins estimated to be in the millimolar range (42, 47), similar to the concentrations required to form the complex detected by the NMR and ITC experiments. The in vivo bioluminescence spectra of several animals or their tissue samples, reveal nearly exact correspondence to the fluorescence of GFP, i.e. no contribution from the blue emission implying near 100% FRET efficiency (50, 52, 53). This demands that the origin of the emission is from a complex where the donor and acceptor have restricted separation and orientation. The inhibition of the GFP shift at increased salt concentration is consistent with electrostatic forces at the protein-protein interface driving the clytin-cgGFP complexation. This would argue against this same spatial structure existing in vivo if within the photocytes, the ionic strength approaches that of sea-water, or is even as low as that characteristic of eukaryotic cells, 100–150 mm because of potassium ions. On the other hand, several bioluminescent organisms are found to contain their bioluminescence systems within membrane enclosed vesicles, “lumisomes” in Renilla (54) and “scintillons” in the dinoflagellates (43). Such vesicles apparently modulate the intracellular environment for the benefit of the bioluminescence function (50). Because cgGFP itself is a tight dimer it is probable that in vivo the clytin-cgGFP complex is a heterotetramer. It should be noted that this supposition was advanced for in vivo aequorin-Aequorea GFP complex (51).
For a heterotetrameric complex of this size, >100 kDa, and weakly interacting, there is little prospect that further NMR experiments will yield unambiguous distance constraints for model refinement. Whether the spatial structure of the in vivo complex relates to that determined here at low ionic strength, hopefully will be proven by crystallography, although for a weak protein-protein complex this methodology presents its own set of impediments.
Supplementary Material
Acknowledgments
We thank Professor W. W. Ward (Rutgers University) for informative discussions of GFP interactions, and Professors Changwen Jin and Bin Xia (Beijing NMR Center, Peking University) for providing the 800 MHz NMR spectrometer facility.
This work was supported by the National Natural Science Foundation of China, Ministry of Science and Technology of China, CAS Research Grant, CAS Fellowship for Young International Scientists Grant, Russian Foundation for Basic Research (08-09-92209 RFBR-China joint grant), SB RAS Grant 2, “Molecular and Cell Biology” program of RAS, Bayer AG (Germany), and by the University of Georgia Research Foundation and the Georgia Research Alliance.
The atomic coordinates and structure factors (codes 3KPX and 2HPW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The backbone resonance assignments of clytin and cgGFP are available at the Biological Magnetic Resonance Data Bank with accession codes BMRB 16599 for clytin and BMRB 16600 for cgGFP.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and PDB data.
- FRET
- Förster resonance energy transfer
- cgGFP
- green-fluorescent protein from jellyfish C. gregaria
- HSQC
- heteronuclear single quantum coherence spectroscopy
- ITC
- isothermal titration calorimetry
- RMSD
- root mean square deviation
- AIR
- ambiguous interaction restraint.
REFERENCES
- 1.Shimomura O. (2006) Bioluminescence: Chemical Principles and Methods, World Scientific, Singapore [Google Scholar]
- 2.Morin J. G., Hastings J. W. (1971) J. Cell Physiol. 77, 313–318 [DOI] [PubMed] [Google Scholar]
- 3.Ward W. W., Cormier M. J. (1976) J. Phys. Chem. 80, 2289–2291 [Google Scholar]
- 4.Markova S. V., Burakova L. P., Frank L. A., Golz S., Korostileva K. A., Vysotski E. S. (2010) Photochem. Photobiol. Sci. 9, 757–765 [DOI] [PubMed] [Google Scholar]
- 5.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Iorger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. Sect. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 7.Emsley P., Cowtan K. (2004) Acta Crystallogr. Sect. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 8.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vagin A. A., Isupov M. N. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 10.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 11.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. Sect. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 12.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 13.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 14.Jung Y. S., Zweckstetter M. (2004) J. Biomol. NMR 30, 11–23 [DOI] [PubMed] [Google Scholar]
- 15.Sattler M., Schleucher J., Griesinger C. (1999) Prog. NMR Spectrosc. 34, 93–158 [Google Scholar]
- 16.Xu Y., Zheng Y., Fan J. S., Yang D. (2006) Nat. Methods 3, 931–937 [DOI] [PubMed] [Google Scholar]
- 17.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 18.de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) Proteins: Struct., Funct., Bioinf. 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 19.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 20.Liu Z. J., Vysotski E. S., Chen C. J., Rose J. P., Lee J., Wang B. C. (2000) Protein Sci. 9, 2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Head J. F., Inouye S., Teranishi K., Shimomura O. (2000) Nature 405, 372–376 [DOI] [PubMed] [Google Scholar]
- 22.Vysotski E. S., Lee J. (2004) Acc. Chem. Res. 37, 405–415 [DOI] [PubMed] [Google Scholar]
- 23.Liu Z. J., Vysotski E. S., Deng L., Lee J., Rose J., Wang B. C. (2003) Biochem. Biophys. Res. Commun. 311, 433–439 [DOI] [PubMed] [Google Scholar]
- 24.Remington S. J. (2006) Curr. Opin. Struct. Biol. 16, 714–721 [DOI] [PubMed] [Google Scholar]
- 25.Wachter R. M. (2006) Photochem. Photobiol. 82, 339–344 [DOI] [PubMed] [Google Scholar]
- 26.Ormö M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J. (1996) Science 273, 1392–1395 [DOI] [PubMed] [Google Scholar]
- 27.Loening A. M., Fenn T. D., Gambhir S. S. (2007) J. Mol. Biol. 374, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine L. D., Ward W. W. (1982) Comp. Biochem. Physiol., Part B: Comp. Biochem. 72, 77–85 [Google Scholar]
- 29.Ward W. W. (1998) in Green Fluorescent Protein (Chalfie M., Kain S. eds), pp. 45–75, Wiley-Liss, New York [Google Scholar]
- 30.Ohashi W., Inouye S., Yamazaki T., Doi-Katayama Y., Yokoyama S., Hirota H. (2005) J. Biomol. NMR 31, 375–376 [DOI] [PubMed] [Google Scholar]
- 31.Nicastro G., Menon R. P., Masino L., Knowles P. P., McDonald N. Q., Pastore A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10493–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang N., Liu L., Liu F., Wagner C. R., Hanna P. E., Walters K. J. (2006) J. Mol. Biol. 363, 188–200 [DOI] [PubMed] [Google Scholar]
- 33.Koglin A., Mofid M. R., Löhr F., Schäfer B., Rogov V. V., Blum M. M., Mittag T., Marahiel M. A., Bernhard F., Dötsch V. (2006) Science 312, 273–276 [DOI] [PubMed] [Google Scholar]
- 34.Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., Groll M. (2008) Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan X., Simpson P., McKeown C., Kondo H., Uchiyama K., Wallis R., Dreveny I., Keetch C., Zhang X., Robinson C., Freemont P., Matthews S. (2004) EMBO J. 23, 1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Card P. B., Erbel P. J., Gardner K. H. (2005) J. Mol. Biol. 353, 664–677 [DOI] [PubMed] [Google Scholar]
- 37.Banci L., Bertini I., Ciofi-Baffoni S., Kandias N. G., Robinson N. J., Spyroulias G. A., Su X. C., Tottey S., Vanarotti M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8320–8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 39.Reichmann D., Rahat O., Cohen M., Neuvirth H., Schreiber G. (2007) Curr. Opin. Struct. Biol. 17, 67–76 [DOI] [PubMed] [Google Scholar]
- 40.Prudêncio M., Ubbink M. (2004) J. Mol. Recognit. 17, 524–539 [DOI] [PubMed] [Google Scholar]
- 41.Kiel C., Selzer T., Shaul Y., Schreiber G., Herrmann C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9223–9228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morise H., Shimomura O., Johnson F. H., Winant J. (1974) Biochemistry 13, 2656–2662 [DOI] [PubMed] [Google Scholar]
- 43.Nicolas M. T., Nicolas G., Johnson C. H., Bassot J. M., Hastings J. W. (1987) J. Cell Biol. 105, 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward W. W., Cormier M. J. (1978) Photochem. Photobiol. 27, 389–396 [Google Scholar]
- 45.Ward W. W., Cormier M. J. (1978) Methods Enzymol. 57, 257–267 [Google Scholar]
- 46.Ward W. W., Cormier M. J. (1979) J. Biol. Chem. 254, 781–788 [PubMed] [Google Scholar]
- 47.Cutler M. W. (1995) Ph.D. Thesis, Rutgers University, New Brunswick, NJ [Google Scholar]
- 48.Deleted in proof
- 49.Deleted in proof
- 50.Morin J. G., Hastings J. W. (1971) J. Cell Physiol. 77, 313–318 [DOI] [PubMed] [Google Scholar]
- 51.Cutler M. W., Ward W. W. (1997) in Bioluminescence and Chemiluminescence: Molecular Reporting with Photons (Hastings J. W., Kricka L. J., Stanley P. E. eds), pp. 596–599, Wiley-Liss, New York [Google Scholar]
- 52.Wampler J. E., Hori K., Lee J. W., Cormier M. J. (1971) Biochemistry 10, 2903–2909 [DOI] [PubMed] [Google Scholar]
- 53.Wampler J. E., Karkhanis Y. D., Morin J. G., Cormier M. J. (1973) Biochim. Biophys. Acta 314, 104–109 [DOI] [PubMed] [Google Scholar]
- 54.Anderson J. M., Cormier M. J. (1973) J. Biol. Chem. 248, 2937–2943 [PubMed] [Google Scholar]
- 55.Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turnbull W. B., Daranas A. H. (2003) J. Am. Chem. Soc. 125, 14859–14866 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.