Abstract
Nucleic acid cytidine deaminases of the activation-induced deaminase (AID)/APOBEC family are critical players in active and innate immune responses, playing roles as target-directed, purposeful mutators. AID specifically deaminates the host immunoglobulin (Ig) locus to evolve antibody specificity, whereas its close relative, APOBEC3G (A3G), lethally mutates the genomes of retroviral pathogens such as HIV. Understanding the basis for the target-specific action of these enzymes is essential, as mistargeting poses significant risks, potentially promoting oncogenesis (AID) or fostering drug resistance (A3G). AID prefers to deaminate cytosine in WRC (W = A/T, R = A/G) motifs, whereas A3G favors deamination of CCC motifs. This specificity is largely dictated by a single, divergent protein loop in the enzyme family that recognizes the DNA sequence. Through grafting of this substrate-recognition loop, we have created enzyme variants of A3G and AID with altered local targeting to directly evaluate the role of sequence specificity on immune function. We find that grafted loops placed in the A3G scaffold all produced efficient restriction of HIV but that foreign loops in the AID scaffold compromised hypermutation and class switch recombination. Local targeting, therefore, appears alterable for innate defense against retroviruses by A3G but important for adaptive antibody maturation catalyzed by AID. Notably, AID targeting within the Ig locus is proportionally correlated to its in vitro ability to target WRC sequences rather than non-WRC sequences. Although other mechanisms may also contribute, our results suggest that local sequence targeting by AID/APOBEC3 enzymes represents an elegant example of co-evolution of enzyme specificity with its target DNA sequence.
Keywords: Antibodies, DNA, Enzymes, HIV, Immunology, Innate Immunity, Protein-DNA Interaction, Class Switch Recombination, Retroviral Restriction, Somatic Hypermutation
Introduction
The activation-induced deaminase (AID)3/APOBEC3 family of nucleic acid cytidine deaminases is used by both the innate and adaptive immune systems to introduce purposeful mutations through the targeted deamination of cytosine bases in DNA. As a part of the innate immune response to retroviruses, the host enzyme APOBEC3G (A3G) is packaged within HIV virions (Fig. 1A) and actively deaminates the (−)-strand cDNA produced by reverse transcription upon infection of a new cell (1, 2). Although non-catalytic mechanisms may also contribute to retroviral restriction, the targeted introduction of deoxyuridine via cytosine deamination plays an important role in the anti-HIV activity of A3G (3–6). The importance of this innate defense factor is evidenced by the inclusion of the viral accessory protein Vif within the parsimonious HIV genome that functions to target A3G for ubiquitination (7, 8).
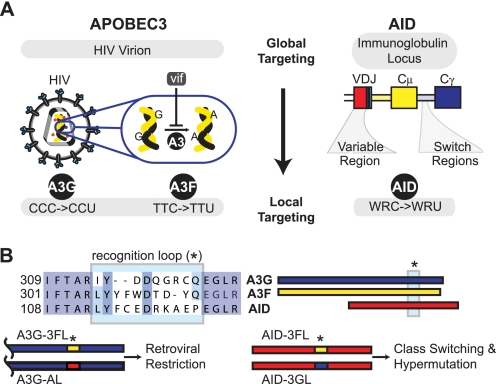
FIGURE 1.
Levels of targeting by AID/APOBEC enzymes. A, global and local targeting. APOBEC3 enzymes, used by the innate immune system for retroviral restriction, are globally targeted to the HIV virion and show a local preference for deamination of cytidine residues following a pyrimidine. The viral accessory protein Vif prevents global targeting by promoting the degradation of the host APOBEC defense factor. AID, the master catalyst governing somatic hypermutation and class switch recombination, is globally targeted to the immunoglobulin locus and locally targeted to WRC sequences within the variable locus and switch regions. B, loop grafting. An alignment of AID, A3G, and A3F demonstrates a region of sequence divergence, the hotspot recognition loop (*), flanked by highly conserved sequences. Grafting of the loop between AID/APOBEC3 family members offers a means to probe the importance of local sequence targeting upon retroviral restriction, somatic hypermutation, and class switch recombination.
The adaptive immune system uses a closely related deaminase family member, AID (9, 10), to introduce targeted mutations into the host genome at the immunoglobulin (Ig) gene locus (Fig. 1A). During somatic hypermutation (SHM), mutations are introduced within the complementarity-determining regions (CDRs) of the antibody variable genes, which can be subsequently selected for enhanced antigen binding through the process of affinity maturation. AID also initiates class switch recombination (CSR), which results from deamination within switch regions located upstream from the various constant genes that encode antibody isotypes. These resulting mutations promote double-stranded breaks, recombination, and ultimately, antibody isotype switching.
Given the mutagenic potential of these deaminase enzymes, an intriguing question is how they are targeted to their intended sites of action as opposed to other genomic sites (11). Mistargeting of these enzymes can have serious consequences for HIV therapy and human cancers. For example, the introduction of mutations within the viral genome by A3G has been suggested to play a role in HIV drug resistance (12–14), and mistargeting by AID has been associated with IgH-myc translocations commonly found in B-cell lymphoma (15, 16).
Targeting by AID/APOBEC3 enzymes can be thought of as occurring at multiple levels (Fig. 1A). On a global level, APOBEC3 enzymes are physically targeted to the HIV virion (17, 18), and Vif, therefore, functions to thwart global targeting (8). Similarly, AID must be globally targeted to the Ig locus rather than to other genomic loci where its mutagenic activity could result in deleterious mutations. At a more local level, productive mutations are largely confined to the CDRs or switch regions of Ig genes rather than adjacent, but structurally essential, framework regions. At an even finer level, the AID/APOBEC3 enzymes exhibit local DNA sequence targeting, which differs between family members. A3G preferentially targets the third cytosine of a CCC motif in (−)-strand DNA, APOBEC3F (A3F) targets TTC sequences for deamination (19), and AID localizes mutations to WRC motifs (W = A/T, R = A/G) (20).
The importance of local DNA sequence targeting by these enzymes has been subject to numerous hypotheses. The sequence specificity of A3G introduces numerous stop codons into the HIV genome that have been proposed to be relevant for viral restriction (2, 21). Similarly, enrichment of AID hotspots in the CDRs and switch regions immediately suggests the intriguing possibility that AID specificity and the Ig locus DNA sequence have co-evolved to promote productive SHM and CSR (22).
Isolating the determinants of local sequence targeting has been an iterative process. As many APOBEC3 deaminases contained duplicated deaminase domains, initial studies using domain swaps between family members helped to localize the determinants of sequence preference into the catalytically active deaminase domain (23). Subsequently, several studies with point mutations initially identified a divergent protein loop in the AID/APOBEC3 family that could influence the motif specificity (24–26). Building on these observations, we recently demonstrated that local targeting could be predictably altered. By grafting the entire 9–11-amino acid protein loops of A3G or A3F into the scaffold of AID, the resulting chimeric enzymes shifted toward the local sequence targeting preferences of the donor enzyme using either purified oligonucleotide substrates in vitro or by examining their mutagenic profiles in bacteria (27). These findings have subsequently been confirmed by several groups (28, 29). In the most recent studies loop grafting in AID was demonstrated to impact SHM and CSR, although no conclusions could be drawn if this was due to altered local sequence targeting or to altered enzyme activity (29). Our biochemically validated loop swapping approach with kinetically characterized enzyme variants offers a unique opportunity to probe the importance of local sequence targeting on the function of AID/APOBEC family members in immune defense. Here, we utilize reciprocal loop grafting to specifically examine and compare how DNA sequence preferences of enzymes from this family affect retroviral restriction, SHM and CSR (Fig. 1B). Our studies demonstrate that the A3G chimeras all produce efficient retroviral restriction, but the AID chimeras are less effective at SHM and CSR of Ig genes.
EXPERIMENTAL PROCEDURES
Loop Variant and HIV Plasmids
Wild-type APOBEC3G (pcDNA3.1-A3G-V5-His6) was obtained from the National Institutes of Health AIDS Reagent Program from Drs. B. Matija Peterlin and Yong-Hui Zheng (30). AID retroviral vector pMSCV2.2*hAID-eGFP and overexpression vector pMSCV2.2*hAID-puro were generous gifts from Sebastian Fugmann (NIA/NIH) (31). Excision of A3G or AID yielded empty vector controls. Loop graft variant and catalytic mutants were created using the previously described overlap extension method (27) using mutagenic oligonucleotides (supplemental Table S1). Using a previously constructed pNL4–3-ΔVif construct (32) encoding a deletion of Vif residues 29–88, subcloning a NdeI/EcoRI fragment into pNL4–3 ΔE-eGFP (33) yielded pNL4–3 ΔVif-ΔE-eGFP.
Generation of Vif-deficient, APOBEC-packaged HIV Virions
Pseudotyped virus was generated in adherent HEK 293T cells (1.5 × 107 cells) co-transfected with viral vector, APOBEC3G-containing pCDNA vector, and VSV-G envelope expression plasmid (20:10:10 μg) using Lipofectamine 2000 (Invitrogen). Viruses were purified by previously reported protocols (33) and additionally through a 20% sucrose cushion and standardized using p24 enzyme-linked immunoabsorbancy assay (PerkinElmer Life Sciences). A3G packaging was examined with anti-Penta-His antibody (Qiagen) and loading controlled with HIV-1 p24 Gag monoclonal antibody (#24-2) obtained through the AIDS Reagent Program from Dr. Michael H. Malim (34, 35).
Retroviral Infection, Real Time Analysis, and Viral Clone Sequencing
For analysis of infection efficiency, increasing titers of purified virus were used to infect 0.5 × 106 Jurkat cells by spin inoculation at 1200 × g for 2 h at 30 °C. After 48 h, cells were washed and fixed. Productive infection was quantified by detecting GFP-positive cells in the live-cell gate on a FACSCalibur (BD Biosciences). For real time analysis and viral clone sequencing, total DNA was collected (Qiagen) from infections of 2.0 × 106 Jurkat cells (50 ng of total p24) carried out for 24 h under similar conditions and treated with DpnI to minimize plasmid carryover from purified virus. Reverse transcripts were quantified by quantitative PCR using pNL4–3 plasmid for a standard curve (36). For sequencing analysis, nested PCR products (primers, supplemental Table S1) were digested with AgeI and NdeI and cloned into pUC19 (XmaI/NdeI sites), and insert-containing clones were sequenced and analyzed as below.
Somatic Hypermutation Analysis
DT40 AID−/− UNG−/− cells were cultured in chicken cell media (RPMI 1640, with 10% FBS, 1% chicken serum, 1% penicillin/streptomycin, and 50 μm β-mercaptoethanol). Cells were transfected with 40 μg of linearized DNA using the Gene Pulser (Bio-Rad) at 580–700 V, 25 microfarads. Stably transfected clones were selected using chicken cell media containing 0.5 μg/ml puromycin. 5–12 individual transfectants from each construct were isolated and cultured for 54–99 days in selective media. Clones from the same construct were then pooled and sorted for IgM loss (anti-chicken IgM-FITC antibody, Bethyl Laboratories). Genomic DNA was extracted from sorted cells (lowest 1.5% FITC), and the rearranged light chain variable (Vλ) sequences were amplified, cloned into the NdeI and HindIII sites of pUC19, and sequenced.
Sequencing Analysis
For HIV and DT40 experiments, mutated sequences were catalogued to calculate mutagenesis rates and targeting. Only unique clones contributed to the cataloged mutations, as identical sequences likely represent amplification of the same initial clone. Targeting sequence analysis was also confined to mutated sequences by exclusion of sequences that contained no point mutations, an insertion, deletion, or a DT40 pseudogene sequence (rare events). Relative to the cytosine mutated, the −4 to +4 nucleotides of the HIV (−)-strand cDNA or the cytosine-containing target strand for DT40 were used to calculate a logo representation of enzyme targeting (37). For DT40, tables were constructed that included the number of mutations within CDRs (a), unmutated observations within CDRs (b), mutations outside CDRs (c), and unmutated observations outside CDRs (d). Thus, the odds ratio for mutation of a CDR residue versus a non-CDR residue is given by ((a/(a + b))/(c/(c + d)). Comparison of the tables yielded a χ2 value from a standard test of homogeneity, and the associated p value is reported (Table 2). Complete hypermutated sequences are available upon request.
TABLE 2.
AID loop graft variants impact hypermutation
| DT40 cells transfected with: | Vλ base pairs sequenced | #C/G → T/A mutations | #Non C/G → T/A mutations | C/G → T/A mutation frequency | CDR mutation frequency | Non-CDR mutation frequency | CDR odds ratioa (95% CI) | p valueb |
|---|---|---|---|---|---|---|---|---|
| ×10−3 | ×10−3 | ×10−3 | ||||||
| AID-WT | 19,149 | 113 | 4 | 5.9 | 10.5 | 5.0 | 2.09 (1.34–3.20) | |
| AID-3FL | 17,185 | 121 | 5 | 7.0 | 5.9 | 7.3 | 0.81 (0.44–1.37) | 0.005 |
| AID-3GL | 12,275 | 42 | 4 | 3.3 | 1.0 | 3.9 | 0.26 (0.03–1.02) | 0.004 |
| AID-E58A | 9,820 | 0 | 0 | 0.0 | N/A | N/A | N/A | N/A |
| No AID (pMSCV) | 13,748 | 0 | 0 | 0.0 | N/A | N/A | N/A | N/A |
a Odds ratio for a base being mutated if it resides within the CDR versus if it resides outside of the CDR.
b A test of homogeneity performed on mutations from loop graft variants against AID-WT was used to calculate a χ2 value. The probability associated with that χ2 value is reported representing the likelihood that the CDR/non-CDR mutational pattern of the variant is distinct from that of AID-WT.
Class Switching Analysis
Retroviral particles were generated in HEK 293T cells by co-transfection of the pMSCV2.2*hAID-eGFP variant vectors with packaging vector pCL-Eco (Imgenex) by calcium phosphate precipitation (31). AID−/− mouse splenic B cells were isolated and cultured as described (38). To prompt CSR, cells (106 cells/ml) were incubated for 24 h in the presence of 5 μg/ml LPS (Sigma) and 5 ng/ml IL-4 (BD Biosciences). After 24 h, retroviral particles were added, incubated for an additional 72 h, and analyzed by flow cytometry analysis (phycoerythryin-labeled anti-mouse IgG1 antibody, Southern Biotech). Western blot analysis on 1.6 × 106 GFP-sorted cells was performed with anti-AID antibody, generated against the keyhole limpet hemocyanin conjugated C-terminal peptide Cys-185—Phe-198 (Covance) and monoclonal AC-15 anti-β-actin antibody (Sigma). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the NIA/NIH Animal Care and Use Committee.
Quantitative PCR
Transcription of AID variants was quantified from mouse B-cells sorted for GFP or from transfected DT40 cells selected with puromycin and sorted for IgM loss. Total cellular RNA was isolated by standard procedures (RNeasy mini column, Qiagen). cDNA was synthesized from 50 ng of RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Transcripts were quantified using PerfeCTa SYBR Green SuperMix (Quanta Biosciences) in a MyiQ real-time instrument (Bio-Rad) using primers (supplemental Table S1) described against human AID with β-actin as a control.
RESULTS
Loop Graft Variants Effectively Restrict HIV
Bhagwat and co-workers (28) have recently examined the reciprocity of our loop grafting approach using a highly engineered version of A3G known as CTD-2K3A as their scaffold. This construct consists of the GST-fused monomeric C-terminal domain of A3G, with an additional five-point mutation for improved soluble expression. Using this modified construct, they demonstrated that substitution of the AID loop broadened substrate specificity of mutagenesis upon expression in Escherichia coli. However, the impact of loop substitutions in the scaffold of full-length A3G and the physiologic impact of this targeted change upon retroviral restriction remains unknown.
We aimed to directly examine the reciprocity of loop grafting in the scaffold of full-length native A3G and evaluated the impact on restriction of HIV. Prior studies have firmly established that the C-terminal domain of A3G is the catalytically active domain (4, 23, 24), whereas global targeting of A3G into virions has been mapped to interaction of the N-terminal domain with nucleocapsid proteins and viral RNA (17, 18, 24). We, therefore, specifically targeted the loop (residues 314–322) in the C terminus of A3G, postulating that global targeting would be preserved, whereas local sequence targeting could be altered. To examine this hypothesis, we generated A3G variants containing either the A3F or AID loop (A3G-3FL or A3G-AL) or a mutation of a catalytically essential active site residue (A3G-E259A).
VSV-G HIV pseudovirions were generated by transfecting HEK 293T cells with a plasmid containing the HIV genome (NL4–3 ΔVif-ΔE-eGFP) and the VSV-G envelope in the presence of A3G transient expression. The HIV plasmid contains a non-functional partial deletion of vif to permit A3G packaging. Replacement of the envelope protein with GFP serves the dual purpose of limiting the virus to a single cycle of infection and allows for infectivity to be monitored by flow cytometry (33). As predicted, although minor differences in packaging efficiency were apparent within purified virions, packaging of all A3G loop variants was observed and is, therefore, independent of the recognition loop located in the C-terminal domain (Fig. 2A).
FIGURE 2.
Loop graft variants of A3G effectively restrict HIV despite altered local sequence targeting. A, global targeting is preserved with loop graft variants. After SDS-PAGE on purified pseudoviruses, blots were probed with monoclonal anti-His and anti-p24 antibodies demonstrating that A3G-WT, A3G-3FL, A3G-AL, and A3G-E259A are all packaged similarly into virus particles. B, viral cDNA production is modestly impacted by A3G incorporation. Real time PCR performed with primers specific for late cDNA transcripts was used to quantify the total viral cDNA after infection of Jurkat cells. Measurements from 5–7 replicate infections were combined to obtain mean copies and S.E. C, loop graft variants are effective in retroviral restriction. Increasing amounts of virus were used to infect Jurkat cells, which were assessed for GFP expression. The fraction of GFP-positive cells and S.E. from infections with three independent virus preparations is shown. D, grafting a foreign loop alters local sequence targeting. Local sequences in the region from −4 to +4 relative to the target mutation were catalogued. The probability logos in the (−)-strand cDNA target are shown (C → U transition at position 0). The most probable residue at each position is indicated along the top. The height of each residue corresponds to the probability of its occurrence at a given position, with the exception of position 0.
Inhibition of retroviral replication by A3G is at least partially attributable to the targeted deamination of cytosine in the retroviral cDNA (3, 39), which has multiple potential effects upon viral infection. First, it may subject the cDNA to DNA repair pathways that could ultimately degrade the viral genome, although it is unclear if classic base excision repair pathways play a role (40). Second, the introduction of deoxyuridine can compromise integration into the host genome (41). Finally, cytosine deamination can introduce missense or nonsense mutations that render the essential viral proteins non-functional and abort the virus lifecycle (2).
To probe different stages of the viral life cycle that might be affected by deamination, we used two complementary measurements, quantitative PCR and GFP expression, that reflect the total amount of viral cDNA produced and the amount of productively integrated virus capable of protein expression, respectively. A validated set of primers internal to the viral cDNA were selected that allow quantification of complete reverse transcripts (36). A3G-WT confers a 6-fold decrease in complete reverse transcripts relative to transcripts in the absence of A3G (Fig. 2B). The loop variants A3G-3FL, A3G-AL, and the catalytic mutant A3G-E259A all demonstrated smaller reductions in overall transcripts (1.5–2-fold).
Although the impact of A3G and the loop variants on total cDNA copies is modest, all deamination proficient A3G variants caused significant retroviral restriction (Figs. 2C and supplemental Fig. S1). Infection reached saturating levels with virions that lacked A3G or that contained the packaged catalytic mutant A3G-E259A. By contrast, A3G-WT and the loop variants A3G-3FL and A3G-AL all efficiently restricted GFP expression and showed similar potency. The amplified restrictive effect in this single cell infectivity assay at a step after reverse transcription likely results from the additional requirement for integration and subsequent expression of functional GFP (33). Although it is possible that the A3G-WT might more efficiently restrict infection as compared with the loop graft variants under conditions of high viral titers, it is clear that the loop graft variants are all able to reduce GFP expression by 4 orders of magnitude, establishing that they are highly competent in retroviral restriction.
Loop Graft Variants Target Different Sequences in Viral cDNA
We then further explored two possibilities for the robust restriction of HIV by the loop graft variants A3G-3FL and A3G-AL. First, it was possible that the loop graft variants retain the A3G-WT local sequence targeting preferences against HIV. Alternatively, if loop graft variants did alter the mutagenesis targeting in vivo, this would suggest that the sequence targeting preference of A3G-WT is a relatively minor contributor to retroviral restriction. To examine these possibilities, we isolated total DNA from infected cells and amplified viral DNA by nested PCR in the region of the virus known to be highly targeted by A3G-WT (21). In the absence of A3G or with the catalytic mutant, mutations were negligible within the viral genome (Table 1). In contrast, for the viral DNA recovered from cells infected with pseudovirions containing A3G-WT or loop graft variants, extensive mutations were observed (supplemental Fig. S2). Almost all of the mutations are G → A transitions, indicating that they do not arise from error-prone reverse transcription or from PCR amplification.
TABLE 1.
APOBEC3G loop graft variants promote HIV mutation
N/A, not applicable.
| HIV virion with packaged: | HIV base pairs sequenced | #G → A mutations | G → A mutation frequency | #Non G → A mutations | % Nonsense mutations | % Missense mutations |
|---|---|---|---|---|---|---|
| ×10−3 | ||||||
| A3G-WT | 7,589 | 109 | 14.4 | 1 | 22 | 63 |
| A3G-3FL | 11,451 | 64 | 5.6 | 3 | 9 | 67 |
| A3G-AL | 10,252 | 57 | 5.6 | 1 | 12 | 72 |
| A3G-E259A | 13,993 | 0 | 0.0 | 0 | N/A | N/A |
| No A3G (pCDNA) | 16,660 | 1 | 0.1 | 0 | N/A | N/A |
When the sequence context of the mutations was examined, as predicted, the loop graft variants were found to target different local sequences than A3G-WT (Fig. 2D). Relative to the target cytosine (position 0), A3G-WT shows strong preference for a cytosine base at the −1 position (∼80% of deaminated sequences), and modest sequence preferences extend from the −3 to +1 positions. With A3G-3FL, sequence preferences are altered substantially, with the majority of deaminated sequences now containing a −1 and −2 thymidine, as predicted from the natural sequence preference of A3F (23, 24). For the AID-loop construct A3G-AL, the most highly targeted sequence is unchanged from that of A3G-WT. However, tolerance for a purine residue at the −1 position is more than twice that of A3G-WT, suggesting a partial shift to a more AID-like sequence preference.
Local Sequence Targeting by AID Localizes Somatic Hypermutation to CDRs
To determine the impact of altered local sequence targeting on SHM, we grafted the A3G and A3F loops into the wild-type, full-length AID scaffold using an expression vector with a puromycin selectable marker. The AID variants were transfected into AID−/−UNG−/− DT40 cells, a chicken B-cell line that undergoes SHM in the presence of AID but has limited gene conversion (42). These cells originally express surface IgM and can lose the marker after SHM in the heavy and light chains. After selecting for individual transfectants and culturing the populations for 8–14 weeks, the rearranged light chain variable gene was amplified from genomic DNA obtained from IgM−-sorted cells, and individual clones were sequenced. We confirmed that AID transcripts were comparable across the transfected cell lines, as judged by quantitative PCR (supplemental Fig. S3A).
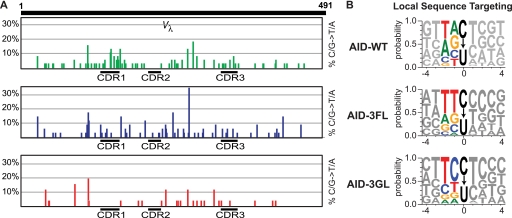
In the absence of AID or with the catalytic mutant AID-E58A, mutations are absent in the sorted cells (Table 2). Expression of AID-WT, AID-3FL, or AID-3GL all restore hypermutation with pronounced C/G → T/A transitions (Fig. 3A). Although levels were judged to be comparable across cell lines, we focused our analysis on two outcomes that are independent of overall mutation rate; that is, the local DNA sequences targeted by each enzyme and the frequency of targeting the CDR regions as opposed to adjacent scaffold regions. For AID-WT, the local sequence preferences were in complete agreement with the canonical WRC hotspot preference (Fig. 3B), and these in vivo results correlate well with our prior investigations on in vitro sequence preferences (27). Expression of the AID loop variants generates C/G → T/A transition mutations with the predicted local sequence preference of the loop donor. For AID-3FL, a shift toward the A3F spectrum TTC is notable, whereas AID-3GL demonstrates a preference for a −1 cytosine (Fig. 3B). Additionally, a preference for a pyrimidine at the +1 position is conserved across all constructs, suggesting this aspect of local targeting is inherent to AID and not perturbed by loop grafting. Despite the scaffolds being distinct for A3G-3FL and AID-3FL, the in vivo sequence targeting is remarkably similar for these two chimeric enzymes that share a grafted loop from A3F (Figs. 2D and 3B). Furthermore, the reciprocal variants A3G-AL and AID-3GL take on similar sequence preferences that are intermediate between the AID and A3G wild-type preferences (Figs. 2D and 3B). As neither A3G-AL nor AID-3GL entirely take on the sequence preference of the donor loop, it is clear that the loop, although a significant determinant, is not the sole influence upon sequence targeting. Other regions of the enzyme or second-shell mutations are likely to further refine sequence preferences in the AID/A3 family, as recently demonstrated by several other studies of local sequence targeting (28, 43).
FIGURE 3.
Targeted hypermutation at CDRs is perturbed by loop graft mutants. A, targeting of CDRs is influenced by loop grafting. From transfected AID−/− UNG−/− DT40 B-cells propagated under selection and sorted for loss of IgM expression, the Vλregion was amplified and sequenced. The frequency of mutations found at each position in Vλ is shown, with the CDRs highlighted. B, AID variants alter the local sequence targeting of hypermutation. The local DNA sequence in the region from −4 to +4 relative to the target mutations were catalogued. The sequences were read relative to the sense strand for C → T mutations and in the antisense strand for G → A mutations. The probability logo is shown (target C → U at position 0). The most probable residue at each position is indicated along the top. The height of each residue corresponds to the probability of its occurrence at a given position, with the exception of position 0.
It has been hypothesized that WRC sequence motifs are enriched in the CDRs in order to promote hypermutation at the antigen-antibody interface while leaving the antibody framework regions outside of the CDRs relatively intact. Indeed, for AID-WT, the probability of enzymatic deamination of a base located in the CDR was twice that of a base located in a non-CDR region (Table 2). Relative to AID-WT, both AID-3FL and AID-3GL are altered in their CDR to non-CDR targeting (p ≤ 0.05) and have increasing preference for deamination outside of the CDRs. The propensity for regional targeting of each construct correlates remarkably with the previously reported (27) preference for deamination of oligonucleotides with WRC/non-WRC sequences in vitro (supplemental Fig. S4A). These results support the hypothesis that the sequence specificity of AID and the DNA sequence of the Ig locus have co-evolved to allow for improved function of SHM to increase antibody affinity.
Altering AID Motif Recognition Is Significant for Class Switch Recombination
During the process of CSR, the introduction of multiple deoxyuridines on both DNA strands results in the formation of double-strand breaks. It has been hypothesized that switch regions have evolved to contain frequent AGCT sequences, which contains WRC hotspots on both strands, to promote a high level of deamination (22). These potential hotspots are interspersed in otherwise G-rich clusters in the template strands that have been demonstrated to promote R-loop formation, exposing a single-stranded DNA substrate (44, 45).
Variants were introduced into AID−/− mouse B-cells, and each demonstrated similar expression (Fig. 4A and supplemental Fig. S3B). When stimulated to undergo class switching from IgM to IgG1, AID-3FL retained 50% switching ability, whereas AID-3GL had only 13% switching compared with AID-WT (Fig. 4, B and C). Notably, the decreased switching by loop graft variants correlates with the deamination activity of each variant on WRC motifs in vitro (supplemental Fig. S4B), where AID-3FL has low activity, but AID-3GL has markedly less activity. Thus, the efficiency of CSR, unlike viral restriction, is quantitatively linked to the ability to deaminate specifically at the AID hotspot, WRC.
FIGURE 4.
Frequency of class switch recombination is proportional to AID hotspot targeting. A, expression of AID variants by Western blot. After SDS-PAGE on protein extracts from AID−/− splenic B-cells infected with retroviruses expressing AID variants, the blots were probed with polyclonal anti-AID and anti-β-actin antibodies to demonstrate that AID-WT, AID-3FL, AID-3GL, and AID-E58A are expressed similarly. B, class switching is influenced by loop grafting. After infection, B-cells were stimulated to promote CSR from IgM to IgG1. Shown is a representative dataset gating on GFP expression as a marker of infection and IgG1 expression as a measure of class switching from IgM. C, AID variants that less efficiently target WRC sequences compromise CSR. Switching was calculated as the IgG1+ GFP+ population compared with the total GFP+ population with S.E. from three replicates shown.
DISCUSSION
The targeted deamination of cytosine by the AID/APOBEC3 enzymes provides a key weapon in the battle between the immune system and pathogens. Correct targeting involves both global and local contributions. In this work we have explored the contribution of local sequence targeting in the AID/APOBEC3 family to their respective function in retroviral restriction, SHM and CSR.
As of yet, no nucleic acid-bound structure of an AID/APOBEC3 family deaminase exists. Available structures of the unliganded enzymes have led to debate over their DNA binding mode as well as a general deficit in our understanding of how they recognize cytosine in different sequence contexts (25, 46). Building upon observations that identified a protein loop within the catalytic deaminase domain that appeared relevant for local sequence targeting (23–26, 47), we previously demonstrated that grafting this loop could transfer the sequence specificity from the A3 enzymes to AID (27). In other varied scaffolds, using either CTD-2K3A variant of A3G or “upmutant” variants of AID, the importance of this loop in local DNA sequence recognition has also been consistently seen (28, 29). In this work we demonstrate that loop grafting in two wild-type, full-length scaffolds (A3G and AID) can alter local sequence targeting in vivo. Taken together, these studies demonstrate that loop grafting changes the local DNA sequence preferences with a shift from the wild-type motif to that of the loop donor. However, for each of the loop graft variants, the extent of the shift and the impact on overall catalytic function are distinct. Some of these differences may arise from disruption of the optimal structural requirements of the loop, which we have found could result if grafting is performed in a suboptimal position of the loop sequence (supplemental Fig. S5) (29).
For A3G, we found that even though the A3G-3FL and A3G-AL loop variants targeted different sequences within the viral genome relative to A3G-WT, they were all effective in restricting productive HIV infection. In earlier studies hinting at similar findings, forced incorporation of packaging-deficient A3G variants or fusion of the N-terminal domain of A3G with the entire catalytically active domains from other A3 enzymes (domain swaps) can potentiate effective retroviral restriction (24, 48, 49). Collectively, our results and these prior findings together indicate that retroviral restriction can be achieved independent of hotspot recognition as long as a catalytically competent deaminase domain is packaged within the virion via efficient global targeting.
It has been postulated that local sequence targeting by A3G is particularly relevant for restriction because its sequence specificity introduces stop codons in place of tryptophan codons (2). As tryptophan is only encoded by the codon TGG, deamination of either cytosine in the corresponding viral (−)-strand cDNA (CCA) would yield a stop codon. Our A3G-3FL and A3G-AL loop variants introduce ∼2-fold fewer stop codons than A3G-WT (Table 1) yet still yield a 4-order of magnitude reduction in GFP expression. Therefore, the generation of stop codons does not appear to be a major determinant of retroviral restriction. This is further supported by the fact that other A3 enzymes, such as A3F, can restrict HIV despite having different local sequence targets than A3G.
It remains plausible that A3G hotspot targeting is important for reasons other than retroviral restriction. Through their enzymatic action, A3 enzymes may also introduce mutations that confer a selective advantage to virions by increasing diversity (12–14) without crossing the threshold of lethal mutagenesis (50). Stop codons may be less likely than missense mutations to contribute to drug resistance or immune escape mutations. Furthermore, clustering of mutations at C-rich loci may be important for tipping the balance toward lethal mutagenesis, in line with the error catastrophe model.
With AID, we find strong evidence to support the significance of local hotspot targeting in both SHM and CSR. Loop grafting altered the spectrum of SHM with respect to CDR and adjacent non-CDR regions. The preference for AID-WT to deaminate in WRC motifs localized mutations to CDRs, which contain many WRC sequences, at twice the frequency of non-CDR regions. In contrast, AID-3GL mutated non-CDR regions at four times the rate of the CDR regions (Table 2). Similarly, the switch regions are enriched for WRC sequences, particularly within AGCT palindromic repeats, and accordingly, the level of switching was reduced with the loop variants (Fig. 4). These results correlate with qualitative observations on SHM and CSR in a recent study by Wang et al. (29) and provide unique new insights into Ig targeting. Given our prior kinetic characterization of AID chimeras in vitro, we now provide evidence that for SHM, the ratio of CDR/non-CDR targeting is proportional to the ratio of WRC/non-WRC deamination by AID, whereas for CSR, the rate of switching is proportional to the rate of WRC targeting (supplemental Fig. S4). SHM and CSR are potentially the product of multiple catalytic events in vivo. It is, therefore, intriguing and informative that a correlation occurs between biological and biochemical measures of AID action, as it is also feasible that enzymatic variants could have demonstrated amplified impacts on SHM and CSR.
Our conclusions shed light on the possible evolution of viral and host DNA sequences to combat or facilitate the action of DNA cytosine deaminases. A study of APOBEC protein evolution suggests that many counteracting genetic changes have occurred over the course of the battle between the virus and the host (51). For instance, the biased A-rich nucleotide composition of HIV, including a ∼60% incidence of A at the third base of degenerate codons, has been proposed to result in part from A3G-mediated mutation (2, 52). In stark contrast, the Ig locus has evolved its sequence to promote specific AID targeting. For example, CDRs show a preference for coding Ser with AGY codons (a WRC hotspot) over TCN codons, whereas this preference is reversed in the nearby framework regions where mutations could disrupt antibody structural integrity (53). Even more notable is the enrichment of hotspots within switch regions, where the AGCT sequence is represented 16× more frequently in the IgM locus than would be predicted by chance (22). AID and the Ig locus appear to represent an intriguing example where an enzyme and its preferred substrate DNA sequence have co-evolved for optimization of biological function.
Supplementary Material
Acknowledgments
We express thanks to Patti Longo, Sebastian Fugmann, Stuart Ray, and Meenakshi Bewtra for advice and Robert Wersto, Tonya Wolf, and Coung Nguyen for cell sorting.
This work was supported, in whole or in part, by NIA, National Institutes of Health (Intramural Research Program), T32 Grant GM06669, and Grants GM056834 (to J. T. S.) and KAI089242A (to R. M. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–5.
- AID
- activation-induced deaminase
- A3G
- APOBEC3G
- SHM
- somatic hypermutation
- CSR
- class switch recombination
- CDR
- complementarity-determining region
- A3F
- APOBEC3F
- A3G-3FL
- A3G-APOBEC3F loop variant
- A3G-AL
- A3G-AID loop variant
- AID-3FL
- AID-APOBEC3F loop variant
- AID-3GL
- AID-A3G loop variant
- UNG
- uracil DNA N-glycosylase
- WRC
- W = A/T, R = A/G
- Ig
- immunoglobulin.
REFERENCES
- 1.Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) Cell 113, 803–809 [DOI] [PubMed] [Google Scholar]
- 2.Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J. M., Landau N. R. (2004) Nat. Struct. Mol. Biol. 11, 435–442 [DOI] [PubMed] [Google Scholar]
- 3.Browne E. P., Allers C., Landau N. R. (2009) Virology 387, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro F., Bollman B., Chen H., König R., Yu Q., Chiles K., Landau N. R. (2005) Virology 333, 374–386 [DOI] [PubMed] [Google Scholar]
- 5.Schumacher A. J., Haché G., Macduff D. A., Brown W. L., Harris R. S. (2008) J. Virol. 82, 2652–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyagi E., Opi S., Takeuchi H., Khan M., Goila-Gaur R., Kao S., Strebel K. (2007) J. Virol. 81, 13346–13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 8.Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 9.Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., Tezcan I., Ersoy F., Kayserili H., Ugazio A. G., Brousse N., Muramatsu M., Notarangelo L. D., Kinoshita K., Honjo T., Fischer A., Durandy A. (2000) Cell 102, 565–575 [DOI] [PubMed] [Google Scholar]
- 10.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 11.Odegard V. H., Schatz D. G. (2006) Nat. Rev. Immunol. 6, 573–583 [DOI] [PubMed] [Google Scholar]
- 12.Sadler H. A., Stenglein M. D., Harris R. S., Mansky L. M. (2010) J. Virol. 84, 7396–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E. Y., Bhattacharya T., Kunstman K., Swantek P., Koning F. A., Malim M. H., Wolinsky S. M. (2010) J. Virol. 84, 10402–10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulder L. C., Harari A., Simon V. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki I. M., Kotani A., Honjo T. (2007) Adv. Immunol. 94, 245–273 [DOI] [PubMed] [Google Scholar]
- 16.Ramiro A. R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., Nussenzweig M. C. (2004) Cell 118, 431–438 [DOI] [PubMed] [Google Scholar]
- 17.Huthoff H., Autore F., Gallois-Montbrun S., Fraternali F., Malim M. H. (2009) PLoS Pathog. 5, e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogerd H. P., Cullen B. R. (2008) RNA 14, 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S. (2004) Curr. Biol. 14, 1385–1391 [DOI] [PubMed] [Google Scholar]
- 20.Bransteitter R., Pham P., Calabrese P., Goodman M. F. (2004) J. Biol. Chem. 279, 51612–51621 [DOI] [PubMed] [Google Scholar]
- 21.Armitage A. E., Katzourakis A., de Oliveira T., Welch J. J., Belshaw R., Bishop K. N., Kramer B., McMichael A. J., Rambaut A., Iversen A. K. (2008) J. Virol. 82, 8743–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackney J. A., Misaghi S., Senger K., Garris C., Sun Y., Lorenzo M. N., Zarrin A. A. (2009) Adv. Immunol. 101, 163–189 [DOI] [PubMed] [Google Scholar]
- 23.Haché G., Liddament M. T., Harris R. S. (2005) J. Biol. Chem. 280, 10920–10924 [DOI] [PubMed] [Google Scholar]
- 24.Langlois M. A., Beale R. C., Conticello S. G., Neuberger M. S. (2005) Nucleic Acids Res. 33, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conticello S. G., Langlois M. A., Neuberger M. S. (2007) Nat. Struct. Mol. Biol. 14, 7–9 [DOI] [PubMed] [Google Scholar]
- 27.Kohli R. M., Abrams S. R., Gajula K. S., Maul R. W., Gearhart P. J., Stivers J. T. (2009) J. Biol. Chem. 284, 22898–22904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carpenter M. A., Rajagurubandara E., Wijesinghe P., Bhagwat A. S. (2010) DNA Repair 9, 579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M., Rada C., Neuberger M. S. (2010) J. Exp. Med. 207, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y. H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B. M. (2004) J. Virol. 78, 6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fugmann S. D., Rush J. S., Schatz D. G. (2004) Eur. J. Immunol. 34, 844–849 [DOI] [PubMed] [Google Scholar]
- 32.Opi S., Kao S., Goila-Gaur R., Khan M. A., Miyagi E., Takeuchi H., Strebel K. (2007) J. Virol. 81, 8236–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Zhou Y., Alcock C., Kiefer T., Monie D., Siliciano J., Li Q., Pham P., Cofrancesco J., Persaud D., Siliciano R. F. (2004) J. Virol. 78, 1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fouchier R. A., Meyer B. E., Simon J. H., Fischer U., Malim M. H. (1997) EMBO J. 16, 4531–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon J. H., Fouchier R. A., Southerling T. E., Guerra C. B., Grant C. K., Malim M. H. (1997) J. Virol. 71, 5259–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler S. L., Hansen M. S., Bushman F. D. (2001) Nat. Med. 7, 631–634 [DOI] [PubMed] [Google Scholar]
- 37.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopal D., Maul R. W., Ghosh A., Chakraborty T., Khamlichi A. A., Sen R., Gearhart P. J. (2009) J. Exp. Med. 206, 1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell R. A., Moore M. D., Hu W. S., Pathak V. K. (2009) Retrovirology 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langlois M. A., Neuberger M. S. (2008) J. Virol. 82, 4660–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mbisa J. L., Bu W., Pathak V. K. (2010) J. Virol. 84, 5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saribasak H., Saribasak N. N., Ipek F. M., Ellwart J. W., Arakawa H., Buerstedde J. M. (2006) J. Immunol. 176, 365–371 [DOI] [PubMed] [Google Scholar]
- 43.Wang M., Yang Z., Rada C., Neuberger M. S. (2009) Nat. Struct. Mol. Biol. 16, 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy D., Yu K., Lieber M. R. (2008) Mol. Cell. Biol. 28, 50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu K., Chedin F., Hsieh C. L., Wilson T. E., Lieber M. R. (2003) Nat. Immunol. 4, 442–451 [DOI] [PubMed] [Google Scholar]
- 46.Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008) Nature 452, 116–119 [DOI] [PubMed] [Google Scholar]
- 47.Rausch J. W., Chelico L., Goodman M. F., Le Grice S. F. (2009) J. Biol. Chem. 284, 7047–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goila-Gaur R., Khan M. A., Miyagi E., Kao S., Strebel K. (2007) Retrovirology 4, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bulliard Y., Turelli P., Röhrig U. F., Zoete V., Mangeat B., Michielin O., Trono D. (2009) J. Virol. 83, 12611–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith R. A., Loeb L. A., Preston B. D. (2005) Virus Res. 107, 215–228 [DOI] [PubMed] [Google Scholar]
- 51.Zhang J., Webb D. M. (2004) Hum. Mol. Genet. 13, 1785–1791 [DOI] [PubMed] [Google Scholar]
- 52.Berkhout B., Grigoriev A., Bakker M., Lukashov V. V. (2002) AIDS Res. Hum. Retroviruses 18, 133–141 [DOI] [PubMed] [Google Scholar]
- 53.Wagner S. D., Milstein C., Neuberger M. S. (1995) Nature 376, 732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.