Abstract
PCSK9, a target for the treatment of dyslipidemia, enhances the degradation of the LDL receptor (LDLR) in endosomes/lysosomes, up-regulating LDL-cholesterol levels. Whereas the targeting and degradation of the PCSK9-LDLR complex are under scrutiny, the roles of the N- and C-terminal domains of PCSK9 are unknown. Although autocatalytic zymogen processing of PCSK9 occurs at Gln152↓, here we show that human PCSK9 can be further cleaved in its N-terminal prosegment at Arg46↓ by an endogenous enzyme of insect High Five cells and by a cellular mammalian protease, yielding an ∼4-fold enhanced activity. Removal of the prosegment acidic stretch resulted in ∼3-fold higher binding to LDLR in vitro, in ≥4-fold increased activity on cellular LDLR, and faster cellular internalization in endosome/lysosome-like compartments. Finally, swapping the acidic stretch of PCSK9 with a similar one found in the glycosylphosphatidylinositol-anchored heparin-binding protein 1 does not impair PCSK9 autoprocessing, secretion, or activity and confirmed that the acidic stretch acts as an inhibitor of PCSK9 function. We also show that upon short exposure to pH values 6.5 to 5.5, an ∼2.5-fold increase in PCSK9 activity on total and cell surface LDLR occurs, and PCSK9 undergoes a second cleavage at Arg248, generating a two-chain PCSK9-ΔN248. At pH values below 5.5, PCSK9 dissociates from its prosegment and loses its activity. This pH-dependent activation of PCSK9 represents a novel pathway to further activate PCSK9 in acidic endosomes. These data enhance our understanding of the functional role of the acidic prosegment and on the effect of pH in the regulation of PCSK9 activity.
Keywords: Cholesterol, Convertases, Dyslipidemia, Endosomes, Intracellular Processing, Lipoprotein Receptor, Post-translational Modification, furin, PCSK9, Prosegment
Introduction
Complications resulting from cardiovascular disorders are the main cause of death worldwide. High levels of circulating low density lipoprotein-cholesterol represent a major risk factor that leads to coronary heart disease associated with increased death and morbidity worldwide (1). LDL is constantly cleared by internalization into cells by the LDL receptor (LDLR),4 which binds and internalizes LDL via its unique apolipoprotein B (apoB) protein. Mutations in LDLR or APOB genes are major causes for the frequent autosomal dominant genetic disorder known as familial hypercholesterolemia (2, 3). More recently, the PCSK9 gene (4), which is highly expressed in liver and small intestine (5), was identified as the third locus associated with familial hypercholesterolemia (6). PCSK9 binds the LDLR and triggers its intracellular degradation in acidic endosomes/lysosomes, resulting in increased circulating LDL-cholesterol (7–10).
Following its autocatalytic cleavage, PCSK9 is secreted as a stable noncovalent complex with its 122-amino acid (aa 31–152) N-terminal prosegment (5, 7). This cleavage results in a conformational change (11), which favors the binding of PCSK9 to the EGFA domain of the LDLR (12), with much increased affinity at acidic pH values (11).
Overexpression studies in liver suggested that PCSK9 targets the LDLR (9, 13, 14) toward degradation in late endosomes/lysosomes (7–10). The extracellular pathway is defined by the ability of extracellular PCSK9 to target the LDLR for destruction. A recent study presented a clinically relevant monoclonal antibody (mAb) that inhibits PCSK9 interaction with LDLR at the cell surface and results in an ∼80% reduction of LDL-cholesterol that lasted for 2-weeks in cynomolgus monkey (15). However, we recently presented evidence for the presence of both intracellular and extracellular pathways of PCSK9-induced LDLR degradation (16).
The crystal structure of PCSK9 revealed three separate domains as follows: the prosegment (aa 31–152) and catalytic domain (aa 153–404) in tight complex, and the spatially separated hinge region (aa 405–454) (17) and Cys-His rich domain (CHRD; aa 455–692) (11, 18, 19). In all reported crystal structures, aa 31–60 of the prosegment and portions of the CHRD were unresolved, indicating their unstructured nature. Biochemical (20) and co-crystal structure (12) studies revealed that aa 153–156 and 367–381 directly interact with the EGFA domain of the LDLR. The most severe mutation, D374Y associated with hypercholesterolemia (21), is within aa 367–381 and results in an ∼25-fold higher affinity of PCSK9 toward LDLR (11). The shallow binding surface on PCSK9 is distant from its catalytic site, and the EGFA domain of LDLR apparently makes no contact with either the prosegment or the CHRD.
Even though the prosegment does not bind the EGFA domain of LDLR (12), it negatively regulates the binding of PCSK9 to LDLR. Deletion of an N-terminal stretch (aa 31–53), which exhibits Tyr38 sulfation (5) and Ser47 phosphorylation (22), enhances the binding of PCSK9 to soluble LDLR by >7-fold (12). Whether under certain conditions this unstructured acidic stretch (11, 18, 19) binds another domain of PCSK9 or LDLR and/or interacts with another protein have yet to be defined. Interestingly, natural variants of PCSK9 exist in this highly acidic stretch of the prosegment. Of note, the variants E32K and E57K reduce the overall negative charge of this part of the prosegment and result in a gain of function (GOF), whereas the reduced positive charge in the R46L variant results in loss of function (LOF).
In this study, we focused on the N-terminal acidic region of the prosegment and on the effect of pH in the regulation of the activity of PCSK9 toward LDLR. The information presented herein opens new avenues in deciphering the structural components of PCSK9 that dynamically regulate its ability to enhance LDLR degradation.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
Human PCSK9 and its mutant cDNAs were generated and cloned into pIRES2-EGFP (Clontech) as described previously (5). HepG2, HuH7, and HEK293 cells (American Type Culture Collection) were routinely cultivated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Wisent). Low density lipoprotein coupled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (diI-LDL) and lipoprotein-deficient serum (LPDS) were from Biomedical Technologies. Purified recombinant human hPCSK9 was obtained from a baculovirus expression system (see below). The cDNA of GPIHBP1 was a kind gift of Drs. Anne Beigneux and Stephen G. Young (UCLA).
Expression and Purification of Full-length (30–692) and Truncated (47–692) Forms of PCSK9 from Baculovirus/Insect Cells
Human PCSK9 cDNA (residues 30–692) was PCR-amplified and cloned into a pAcHLT vector (Pharmingen) with GP67 secretion signal, confirmed by sequencing to confer expression and secretion of hexahistidine-tagged (C-terminal) PCSK9. Recombinant baculovirus was generated by co-transfection with BaculoGold DNA (Pharmingen) and amplified once. The resulting viral stock (133 ml) was used to inoculate 40 liters of High Five cells (Invitrogen) in ESF921 media (Expression System LLC) at a density of 2 × 106 cells/ml. Cells were grown at 27 °C in two 25-liter WAVE Bioreactors (GE Healthcare) for 65–72 h and harvested when cell viability reached 70%. Culture supernatants were centrifuged at 1,000 × g for 15 min and kept at 20 °C for 30–60 min prior to purification. The supernatants from 40 liters of baculovirus culture were concentrated and exchanged into 1 liter of Buffer A (25 mm HEPES, 200 mm NaCl, pH 7.4). The remaining purification steps were performed at 0–5 °C. The sample was supplemented with 5 mm imidazole, centrifuged, and applied at 10 ml/min to a 9.5 × 2.6-cm (50 ml) nickel-nitrilotriacetic acid column (Qiagen) pre-equilibrated in Buffer A. The column was then washed with 20 column volumes (CV) of Buffer A + 5 mm imidazole, 5 CV of Buffer A + 10 mm imidazole, and 10 CV of Buffer A + 35 mm imidazole. PCSK9 was then eluted in two 5-CV portions of Buffer A + 250 mm imidazole, partially concentrated, and then diluted 15-fold in Buffer B (25 mm Tris-HCl, pH 7.5) immediately prior to loading onto a 9.5 × 2.6-cm (50 ml) SOURCE 15Q column (GE Healthcare) pre-equilibrated in Buffer B containing 10 mm NaCl. Truncated (47–692) and full-length (30–692) forms of PCSK9 were base-line-resolved and eluted in this order using a 12-CV linear gradient from 10 to 600 mm NaCl in Buffer B. Fractions found to contain each form of PCSK9 by SDS-PAGE were separately pooled and concentrated to 5 mg/ml. Each form of PCSK9 was loaded onto a 60 × 2.6-cm (320 ml) HiLoad Superdex 200 column (GE Healthcare) pre-equilibrated and isocratically eluted at 3 ml/min in final storage buffer (25 mm HEPES, pH 7.4, 200 mm NaCl, 5% glycerol). Fractions containing monomeric PCSK9 were pooled, concentrated to 3 mg/ml, and stored at −80 °C. Purified proteins were analyzed by Bradford, SDS-PAGE, dynamic light scattering using a Plate Reader Plus (Wyatt Technologies) with a laser wavelength of 829 nm, and LC/ESI-MS using a LTQ mass spectrometer (Thermo) in positive ion mode. Expression and purification process yields typically ranged from 80 to 110 mg of purified PCSK9(30–692)-His6 and from 5 to 10 mg of purified PCSK9(47–692)-His6 per 40 liters of baculovirus supernatants. Analytical data showed ∼95% purity with hydrodynamic radii of 4.1 nm, consistent with a glycosylated monomer with 11% polydispersity, and mass spectra consistent with heterogeneous N-linked glycosylation of the catalytic CHRD and partial sulfation of the nontruncated prodomain form.
diI-LDL Uptake Cell-based Assay for PCSK9 Activity
A cell culture method to assay PCSK9 functional activity on the LDLR was developed measuring cellular LDLR activity as the uptake of fluorescently labeled LDL. Our method was adapted from the diI-LDL uptake procedure by Teupser et al. (23). HepG2 cells were plated at 20,000 cells/well in a 96-well format in RPMI + 10% FBS, and on the 2nd day the medium was changed to RPMI + 10% LPDS with 100 nm statin (catalog no. 423526, Bristol-Myers Squibb) for 16 h. Cells were then preincubated with recombinant human PCSK9 protein at the levels indicated for 2 h, followed by the addition of 5 μg/ml diI-LDL for a further 2-h incubation at 37 °C. Uptake was stopped by the addition of 4% formaldehyde in 10 μm Hoechst 33342 for 20 min at 20 °C. Cells were washed twice with PBS, and fluorescence measured for Hoechst (DNA content) at excitation/emission 360/460 nm (dichroic mirror = 400 nm) was read. After DNA reading, the cells were lysed in 0.1 n NaOH, 0.1% SDS and shaken for 10 min followed by fluorescence reading for diI-LDL excitation/emission at 530/580 nm (dichroic mirror = 561 nm) on an LJL Analyst instrument. For data analysis, the fluorescence ratio of diI-LDL/Hoechst 33342 was used to normalize diI-LDL uptake value to cell count.
FRET-based Assays for PCSK9:EGFA and PCSK9:LDLR Binding
Time-resolved fluorescence resonance energy transfer (FRET)-based assays were developed to determine the binding affinity of PCSK9 with LDLR, adapted from the general method described previously by Miao et al. (24). The PCSK9:EGFA FRET assay measures PCSK9 binding to the LDLR epidermal growth factor precursor homology domain (EGFA domain) using recombinant human PCSK9 expressed in baculovirus and a synthetic 40-mer EGFA peptide (biotinylated) produced by solid-phase chemistry. EGFA has been shown to represent the key interacting domain of LDLR with PCSK9 (12). This assay uses a PCSK9 C-terminal domain binding mAb produced in Bristol-Myers Squibb (mAb-4H5) labeled with europium-chelate to provide FRET interaction with biotinyl-EGFA through the streptavidin/allophycocyanin fluorophore complex. A second assay measured PCSK9 binding to the full-length LDLR ectodomain using biotinyl-LDLR-His6 and the same mAb-4H5 system described above.
Transfections, Biosynthetic Analyses, and Immunoprecipitations of PCSK9
Transfections were done with 3 × 105 HEK293 cells using Effectene (Qiagen) and a total of 0.4 μg of cDNAs. Alternatively, 5 × 105 HuH7 or 6 × 105 HepG2 cells were transfected with a total of 4 μg of cDNAs in Lipofectamine 2000 (Invitrogen). In some cases, 2-day post-transfection, HEK293 cells were washed and then incubated for various times with either 250 μCi/ml [35S]Met or [35S]Cys (PerkinElmer Life Sciences). The cells were lysed in modified RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and a protease inhibitor mixture (Roche Applied Science), after which the lysates and media were prepared for immunoprecipitation (25). The antibodies used were the anti-V5 mAb (Invitrogen, 1:500), goat anti-prosegment PCSK9 (Imx-3786, 1:200), and rabbit anti-PCSK9 31–454 (A-03) (10). Immunoprecipitates were resolved by SDS-PAGE on 8% Tricine gels and autoradiographed. Most of these experiments were repeated at least three times. Quantitation was performed on a Storm Imager (Amersham Biosciences) by using the ImageQuant version 5.2 software.
PCSK9 Incubations at Different pH Values
Full-length and various PCSK9 constructs secreted from HEK293 cells in media composed of RPMI in 5% LPDS and 1% sodium pyruvate, pH 7.2, were incubated at 37 °C for 1–4 h at various pH values from 7.2 to 4.5. The pH was then immediately neutralized with NaOH to pH 7.2. The samples were incubated for 4 h with HuH7 cells for FACS analyses and then 4 h or overnight with either HepG2 or HuH7 cells for Western blots of PCSK9 and LDLR.
LDLR Activity Assay
HepG2 cells were plated in a 96-well poly-d-lysine-coated plate (BD Biosciences) at a density of 15,000 cells/well in RPMI 1640 medium (Invitrogen), containing 5% lipoprotein-deficient serum (Intracel). After 18 h, purified PCSK9 protein was added to the cells in 30 μl of medium (RPMI 1640 containing 5% LPDS). After 2 or 24 h of incubation at 37 °C, 5 μg/ml diI-LDL (Biomedical Technologies) were added to the cell media, and cells were returned to the tissue culture incubator for another 2 h. Cells were then fixed in the presence of a 2% paraformaldehyde solution containing 4 μg/ml Hoechst (Molecular Probes). After three washes of the cell layer, plates were scanned on Cellomics Array-scan (Thermo). diI-LDL uptake measured as the mean total intensity of fluorescence per cell was obtained from quadruplicate wells with an average read of 500–700 cells/well for each treatment condition. Data were analyzed using GraphPad Prism 4.
Western Blot Analysis
Cells were washed three times in phosphate-buffered saline (PBS) and lysed in complete RIPA buffer (50 mm Tris-HCl, pH 8.0, 1% (v/v) Nonidet P-40, 0.5% sodium deoxycholate, 150 mm NaCl, and 0.1% (v/v) SDS) supplemented with 1× Complete Protease Inhibitor Mixture (Roche Applied Science). Proteins were separated by 8% SDS-PAGE and blotted on polyvinylidene difluoride (PVDF, PerkinElmer Life Sciences) membranes (GE Healthcare), which were blocked for 1 h in TBS-T (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Tween 20) containing 5% nonfat dry milk. Membranes were then incubated for 3 h in 1% nonfat milk with a polyclonal hPCSK9 antibody (1:2500) (10) and human LDLR antibody (1:1,000, R & D Systems). Appropriate horseradish peroxidase-conjugated secondary antibody (1:10,000, Sigma) was used for detection with enhanced chemiluminescence using the ECL Plus kit (GE Healthcare).
FACS
HuH7 cells were incubated for 1–4 h at 37 °C with various PCSK9 constructs and then washed three times with calcium/magnesium-free Dulbecco's PBS containing 0.5% bovine serum albumin (Sigma) and 1 g/liter glucose (solution A). Cells were then incubated 5 min at 37 °C with 500 μl of 1× Versene solution (Invitrogen) and layered on 5 ml of solution A. Cells were then centrifuged for 5 min at 1,000 rpm and resuspended in 1 ml of solution A containing 1:100 of monoclonal LDLR antibody C7 directed against human LDLR (mAb-C7, Santa Cruz Biotechnology) for 40 min. Cells were washed once with 5 ml of solution A, centrifuged, and resuspended for 20 min in 1 ml of PBS containing 1:250 of Alexa Fluor 647 donkey anti-mouse (Molecular Probes). Cells were washed and resuspended in 300 μl of PBS 0.2% of propidium iodide. Viable cells (propidium iodide-negative) were then analyzed by FACS for both propidium iodide and Alexa Fluor 647 using the FACS BD LSR (BD Biosciences).
Immunocytochemistry
Cells were washed three times with PBS, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100/PBS for 10 min, and incubated with 150 mm glycine to stabilize the aldehydes. The cells were then incubated for 30 min with 1% BSA (Fraction V, Sigma) containing 0.1% Triton X-100, followed by overnight incubation at 4 °C with selected antibodies (1:100 goat polyclonal anti-LDLR, R & D Systems; 1:500 mouse anti-V5, Invitrogen). The cells were then incubated for 60 min with corresponding Alexa Fluor-conjugated secondary antibodies (Molecular Probes) and mounted in 90% glycerol containing 1% 1,4-diazabicyclo-[2.2.2]octane (Sigma). Immunofluorescence analyses were performed with a Zeiss LSM-510 confocal microscope coupled with a Nikon Eclipse TE2000-U laser-scanning microscope with 408-, 488-, and 543-nm laser lines. Images were processed with Adobe Photoshop CS2, version 9.0 (Adobe Systems). For internalization experiments, HepG2 cells were incubated for 60 min at 4 °C with conditioned media from HEK293 cells overexpressing PCSK9-V5 or the deletion mutant Δpro-33–58-PCSK9-V5. Cells were then incubated at 37 °C for 0, 30, and 60 min before immunofluorescence as described above.
RESULTS
Effect of Prosegment on the PCSK9-induced LDLR Degradation
We first concentrated on the very acidic sequence (aa 31–60) at the N terminus of the prosegment of PCSK9 (aa 31–152), which exhibits ∼50% Asp and Glu residues, as well as Tyr38 sulfation (5) and Ser47 phosphorylation (22). This segment is not seen in any of the crystal structures analyzed at both acidic and neutral pH values in the presence or absence of the EGFA domain of LDLR (11, 12, 26), suggesting high flexibility, or the need for another protein to stabilize it.
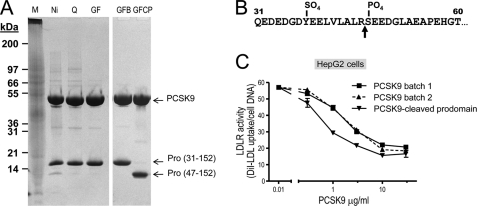
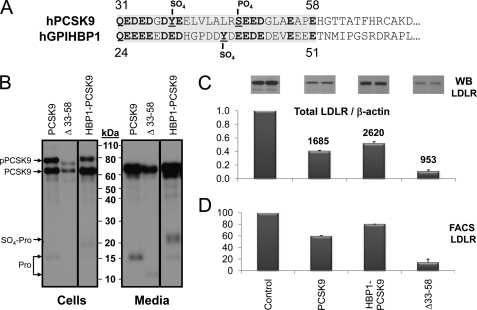
Upon purification of full-length PCSK9 produced by the baculovirus expression system in High Five insect cells, we noticed the presence of a shorter prosegment form likely resulting from processing by an endogenous protease(s) during expression/purification (Fig. 1A). The SOURCE 15Q column employed in purification effectively resolved two forms of PCSK9, with a truncated form of PCSK9(47–692) eluting between 100 and 150 mm NaCl, and a full-length form of PCSK9(30–692) eluting between 250 and 300 mm NaCl. N-terminal sequence analysis revealed that the shorter form started at Ser47 (data not shown), demonstrating that it represents a PCSK9 prosegment cleaved after Arg46↓ and missing residues AQEDEDGDYEELVLALR46 (Fig. 1B).
FIGURE 1.
Comparative activity of PCSK9 and its shortened prosegment isoform. PCSK9-(His)6 was produced in large quantities from 35-liter baculovirus High Five cells and subjected to multiple steps of purification including Ni2+-affinity followed by Mono Q anion exchange and finally gel filtration chromatography. The anion exchange (SOURCE 15Q) column effectively resolved two forms of PCSK9. The truncated form of PCSK9(47–692) eluted between 100 and 150 mm NaCl, whereas the full-length form of PCSK9(30–692) eluted between 250 and 300 mm NaCl. A, Coomassie Blue staining of 18 μg of total proteins from each preparation separated on a 4–12% gradient in MES buffer, SDS-PAGE under nonreducing conditions are shown. The position of the molecular size markers, total proteins in the High Five supernatant before purification, purification by Ni2+-affinity chromatography (Ni), SOURCE 15Q anion exchange (Q), and Superdex 200 gel filtration (GF) are shown. Notice the presence of a small amount of cleaved prosegment associated with PCSK9 apparent after the nickel purification step, and the two PCSK9 complexes with normal and shorted prosegments were finally resolved and purified to homogeneity after the anion exchange and gel filtration columns in two different preparations (right panels). M, Mark 12 (25 μl); S, supernatant (10 μl); Ni, nickel-nitrilotriacetic acid pool (18 μg); Q, SOURCE Q pool (18 μg); GF, gel filtration pool of batch 7 (18 μg); GFB, gel filtration pool of batch 7B (18 μg); GFCP, gel filtration pool of batch 7CP (18 μg). B, microsequencing analysis of the prosegments revealed an 47SEEDG … sequence demonstrating that the prosegment was cleaved Arg46 (arrow). C, concentration dependence for PCSK9 effect on LDLR of HepG2 cells using diI-LDL incorporation (16). PCSK9(31–692) and PCSK9(47–692) produced and purified from baculovirus/insect cell expression as shown in A were tested for biological activity on the LDLR in the HepG2 cell diI-LDL uptake assay as described under “Experimental Procedures.” Notice that the PCSK9 associated with the truncated 47–152 prosegment is ∼4-fold more active than the WT protein in two independent preparations (batches 1 and 2).
Separation and complete purification of both forms of the PCSK9 heterodimer with either the prosegment 30–152 or 47–152 allowed us to test side by side their functional activity on HepG2 cells based on diI-LDL incorporation (16). Clearly, the loss of aa 30–46 resulted in increased PCSK9 functional activity on the LDLR, with an ∼4-fold reduction in the PCSK9 concentration needed to reduce diI-LDL incorporation by 50%: EC50 ∼1 μg/ml for the truncated prosegment form and 3.5 μg/ml for the full-length form (Fig. 1C). We conclude that the N-terminal acidic domain of the prosegment naturally inhibits the activity of PCSK9 on LDLR.
Finally, we had previously reported that the mammalian basic amino acid-specific proprotein convertase PC7 (27) was the only the proprotein convertase that can partially process the prosegment of PCSK9 into a shorter fragment (28). We now further demonstrate that this fragment is also most likely generated by cleavage after Arg46↓, because the natural hypocholesterolemic R46L variant is no longer cleaved by PC7 (data not shown). Therefore, both an endogenous convertase in insect High Five cells and mammalian PC7 can process the prosegment at Arg46↓ resulting in a more active PCSK9 complex.
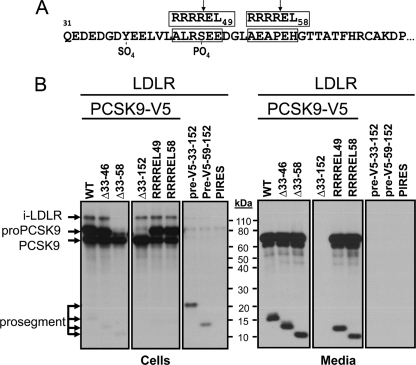
We next wanted to define the boundaries of this inhibitory domain. Because all reported crystal structures of PCSK9 did not detect aa 31–60 (11, 12, 26), most of our present constructs concentrated on this part of the prosegment. For this, we generated V5-tagged PCSK9 constructs (Fig. 2) either lacking aa 33–46 (Δ33–46), Δ33–58, or the whole prosegment Δ33–152. We kept aa 31 and 32 to make sure the signal peptide was correctly cleaved at the Ala30↓ (5). We also generated RRRR↓EL mutants (Fig. 2A) that are optimal for cleavage (↓) by furin in the trans-Golgi network/cell surface (28). These were strategically placed at RRRREL49 and RRRREL58 (Fig. 2A). Finally, we also generated (signal peptide-V5-prosegment) constructs containing the V5 tag at the N terminus of either aa 33–152 (pre-V5–33-152) or 59–152 (pre-V5–59-152) of the prosegment. All the above C-terminally V5-tagged constructs were co-expressed with LDLR. The autoradiographic data obtained after pulse labeling of HEK293 cells for 4 h with [35S]Met and -Cys followed by immunoprecipitation of cell lysates and media with either mAb-V5 or an LDLR mAb-C7, as reported previously (10), and SDS-PAGE analysis (28) are shown in Fig. 2B. The data demonstrate that processing and secretion of most of these constructs were as expected, resulting in secretory proteins consistent with their modifications or cleavage by endogenous furin-like enzymes in HEK293 cells (28). Exceptions are PCSK9 lacking the whole prosegment (Δ33–152-PCSK9) (29), as well as the prosegments alone pre-V5–33-152 and pre-V5–59-152, which likely remain in the endoplasmic reticulum (ER), as previously observed with other prosegment mutants (7).
FIGURE 2.
Biosynthetic analysis of PCSK9 constructs. A, sequence representation of the prosegment, its Tyr38 sulfation and Ser47 phosphorylation, and the location of the two RRRREL mutants. The segment 31–60 was not seen in any of the crystal structures analyzed. B, various PCSK9-V5 constructs produced were co-expressed with the LDLR and analyzed by SDS-PAGE separation of mAb-V5 immunoprecipitated materials obtained following a 4-h pulse labeling with [35S]Met + -Cys of HEK293 cells transiently overexpressing each construct.
As reported before, only pro-PCSK9 co-immunoprecipitated with the immature ER-associated form of LDLR (∼110 kDa) (10). This was further confirmed in immunoprecipitates using the LDLR mAb-C7 (data not shown). Interestingly, except for Δ33–58-PCSK9, all other secretable constructs that contained the PCSK9 catalytic subunit co-immunoprecipitated with immature LDLR in the ER. It is likely that the fast processing of the Δ33–58-pro-PCSK9 into Δ33–58-PCSK9 (Fig. 2B, cells) resulted in too low ER-associated zymogen levels for detection of the complex. The fact that the Δ33–152-PCSK9 construct that is unable to leave the ER (29) still binds immature form of LDLR strongly argues that this interaction does not require the prosegment and likely occurs via the catalytic subunit of PCSK9, as found at the cell surface for the mature forms of PCSK9 and LDLR (12).
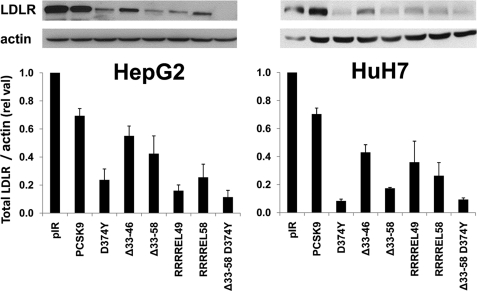
Upon overexpression of the PCSK9 prosegment deletants in HepG2 and HuH7 cells, we analyzed by Western blot their effect on total cellular LDLR levels. Thus, WT PCSK9 and its D374Y GOF mutant caused an overall decrease in total LDLR by ∼30 and 75% in HepG2 cells and by ∼30 and 92% in HuH7 cells, respectively (Fig. 3). In contrast, Δ33–46 and Δ33–58 decreased LDLR levels by ∼45 and ∼60% in HepG2 cells and by ∼65 and ∼80% in HuH7 cells, respectively, confirming that removal of aa 33–58 enhances by ∼3–4-fold the PCSK9 activity on total LDLR. Finally, the most active constructs were PCSK9-D374Y and D374Y lacking aa 33–58 (Δ33–58-D374Y), and the latter results in an ∼90% decrease in total LDLR levels in both cell lines (Fig. 3).
FIGURE 3.
Effect of extracellular PCSK9 and its GOF mutants on total LDLR levels in HepG2 cells. Western blot analysis of LDLR in HepG2 (left panel) and HuH7 (right panel) cells transfected with PCSK9 and its gain of function mutants D374Y, Δ33–46, Δ33–58, and D374Y-Δ33–58. The estimated % decrease in total LDLR was normalized to β-actin levels. These data are representative of three independent experiments.
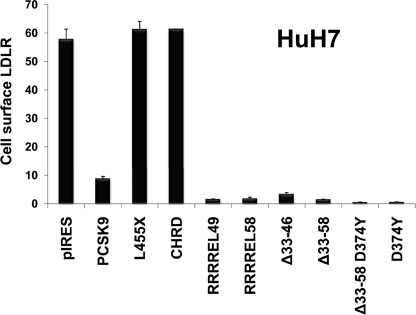
FACS analysis was also performed to test the effect of the best constructs on the level of cell surface LDLR. Thus, ∼0.7 μg/ml of each protein construct secreted from HEK293 cells, estimated by ELISA directed against aa 31–454 (17) (not recognizing the CHRD), was incubated for 4 h with HuH7 cells. The cells were then detached and immediately incubated with the mAb-C7 and then analyzed by FACS for LDLR levels (Fig. 4). The data show that removal of the acidic prosegment (Δ33–58) results in an ∼6-fold increase in the activity of PCSK9 on cell surface LDLR and that the most active extracellular PCSK9 constructs were the D374Y and (Δ33–58)-D374Y (Fig. 4), similar to what was observed in HepG2 and HuH7 cells (Fig. 3).
FIGURE 4.
Effect of extracellular PCSK9 and its mutants on HuH7 cell surface LDLR. PCSK9 and its gain of function mutants D374Y, Δ33–46, Δ33–58, and D374Y-Δ33–58 obtained from HEK293 cells (0.7 μg/ml) were incubated with HuH7 cells for 4 h. The cells were then detached, and the levels of cell surface LDLR were measured by FACS analysis. As control, we used either a construct lacking the CHRD (L455X) or only a secreted form of the CHRD, which had no effect on the LDLR. These data are representative of at least four independent experiments.
The data also show that a secretory CHRD construct (SP-CHRD) or one lacking the CHRD (aa 1–454; L455X) has no effect on LDLR degradation (Fig. 4), even though L455X can bind the LDLR (30). This reinforces the notion that the CHRD, although not binding the LDLR (11), is critical for the internalization and/or sorting of the PCSK9-LDLR complex into endosomes/lysosomes for degradation.
Acidic Region of the Prosegment Dampens the Binding PCSK9 to Either EGFA or LDLR
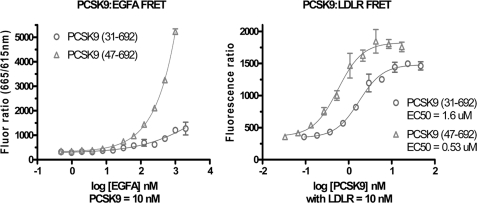
Two FRET-based assays were used to examine the binding affinity of the full-length versus N-terminal truncated forms of PCSK9 purified from the baculovirus/High Five system. The binding of PCSK9 to the LDLR epidermal growth factor precursor homology domain peptide (EGFA domain) was markedly increased for truncated PCSK9(47–692) compared with full-length PCSK9(30–692), as seen by the higher FRET signal with increasing levels of EGFA peptide added (Fig. 5, left panel). Using full-length LDLR ectodomain with the two forms of PCSK9 in a second FRET assay revealed that truncated PCSK9 bound LDLR with >3 times higher affinity (lower EC50) than full-length PCSK9 (Fig. 5, right panel). These findings indicate that loss of the acidic N terminus of the prodomain resulted in a marked increase in PCSK9 binding affinity to the LDLR EGFA domain consistent with the increased functional activity observed in the cell-based assays.
FIGURE 5.
Binding affinities by FRET assays. Left panel, binding EGFA to full-length or truncated prodomain forms of PCSK9. Either form of PCSK9 (10 nm) was incubated with increasing levels of biotinyl-EGFA 40-mer peptide with 1 nm mAb 4H5-europium and 1 nm streptavidin-allophycocyanin conjugated at pH 7.4. Right panel, binding affinity by FRET assay for LDLR ectodomain binding to full-length or truncated prodomain forms of PCSK9. Biotinyl-PCSK9 (10 nm) was incubated with increasing levels of LDLR-His along with 1 nm anti-His-mAb-europium and 1 nm streptavidin-allophycocyanin conjugated.
Internalization Rate of PCSK9 and Its Gain of Function Deletant
By using mAb-V5, we assessed by immunocytochemistry the rate of internalization of PCSK9 and PCSK9(Δ33–58) (supplemental Fig. S1). Incubation of HepG2 cells at 4 °C for 1 h with ∼0.7 μg/ml of these constructs was followed by 0-, 30-, and 60-min incubations at 37 °C. Immunocytochemical analysis of the V5 and endogenous LDLR immunoreactivities revealed a much faster rate of internalization of PCSK9(Δ33–58) as compared with the wild type proteins. This is especially clear at the 30-min (supplemental Fig. S1B) and 60-min (supplemental Fig. S1C) time points, where the punctate-like structures, likely representing endosomes/lysosomes where PCSK9 and LDLR co-localize (10), are much more evident. In conclusion, removal of the 33–58 segment of PCSK9 leads to a >4-fold higher activity toward cell surface LDLR and results in a higher cellular internalization rate of the PCSK9-LDLR complex into endosomal/lysosomal-like structures.
Functional Similarity between the Acidic Stretch of the Prosegment of PCSK9 and That of GPIHBP1
Could the inhibitory effect of aa 31–58 of PCSK9 be due to the highly acidic nature of this segment? Literature search of secretory proteins that exhibit a similar acidic stretch playing a functional role in regulating their activity singled out GPIHBP1. Thus, we noted a significant similarity in acidity and length of the Tyr38-sulfated, Ser47-phosphorylated aa 31–58, 31QEDEDGDYEELVLALRSEEDGLAEAPEH58, of PCSK9 to the heparin-like, Tyr-sulfated, acidic stretch, including aa 24–51, 24QEEEEEDEDHGPDDYDEEDEDEVEEEET51, at the N terminus of the glycosylphosphatidylinositol-anchored protein GPIHBP1. The latter domain binds lipoprotein lipase and apolipoprotein AV on chylomicrons, thereby enhancing lipoprotein lipase activity on triglycerides (31–33). Note that 13/14 acidic residues of PCSK9 (including the Tyr sulfation and Ser phosphorylation sites) align with similar residues in human GPIHBP1 (Fig. 6A).
FIGURE 6.
Biosynthetic analysis and comparative activity of PCSK9 and its HBP1 chimera. A, alignment of the acidic N-terminal domains of human PCSK9 and GPIHBP1. The shaded regions were swapped in the HBP1-PCSK9 chimera. Boldface residues emphasize the homology of acidic residues, including the Tyr sulfation and Ser phosphorylation. B, SDS-PAGE analysis of mAb-V5 immunoprecipitated materials obtained following a 3-h pulse labeling with [35S]Met + -Cys of HEK293 cells transiently overexpressing each construct, including wild type PCSK9, its Δ33–58 construct, and HBP1-PCSK9 chimera. The slower migrating prosegment associated with the HBP1-PCSK9 chimera is likely to be the Tyr-sulfated form, as predicted (31). C, Western blot analysis of total LDLR levels of HuH7 cells exposed to various constructs for 4 h. The bar graph represents the average values of two independent experiments shown above each bar. The numbers above each bar represent the levels of PCSK9 in ng/ml measured by ELISA. D, FACS analysis for LDLR expression following a 4-h incubation of HuH7 cells with 0.7 μg/ml of the represented constructs, estimated by ELISA.
Accordingly, using PCR mutagenesis we swapped the above sequences, and the chimeric HBP1-PCSK9 with aa 24–51 of GPIHBP1 replacing aa 31–58 of PCSK9 was analyzed. Biosynthetic (Fig. 6B), Western blot (Fig. 6C), and FACS (Fig. 6D) analyses revealed that the HBP1-PCSK9 chimeric construct behaved similar to WT PCSK9, in terms of its autocatalytic activation, secretion from HEK293 cells (∼1.6-fold enhanced, estimated by ELISA) (Fig. 6B), and activity on total (Fig. 6C) and cell surface (Fig. 6D) LDLR in HuH7 cells (∼20% less efficient). This suggested that the acidic stretch of the prosegment of PCSK9 can be replaced by that of GPIHBP1 without overt deleterious effects on its activity on LDLR.
pH Dependence of the PCSK9 Association with Cells
To investigate a possible role of pH on the activity of extracellular PCSK9 on cell surface LDLR, we first defined the best conditions to analyze this process on two human hepatocyte-derived HepG2 and HuH7 cell lines. Incubation of each cell line with 0.7 μg/ml of PCSK9 at 37 °C for 30 min up to 4 h led to a gradual decrease of cell surface LDLR, as measured by FACS analysis of the cells after each incubation period (supplemental Fig. S2A). We noticed that even though both naive cell lines start with similar amounts of cell surface LDLR, HepG2 cells are ∼3-fold more sensitive to PCSK9 within the first 30 min of incubation. Thus, in 30 min, LDLR levels decreased by ∼1.7%/min in HepG2 cells and only by ∼0.5%/min in HuH7 cells (supplemental Fig. S2A). However, following the first 30 min, the loss of cell surface LDLR slowed down in both cell lines, reaching a linear value of ∼0.1%/min (supplemental Fig. S2B). The difference between the two cell lines may in part be due to the presence of the PCSK9 inhibitor annexin A2 at the plasma membrane of HuH7 cells, but not HepG2 cells (34), thus slowing down the initial effect of PCSK9. After 4 h of incubation with PCSK9, ∼51 and ∼24% of LDLR remained at the cell surface of HuH7 and HepG2 cells, respectively.
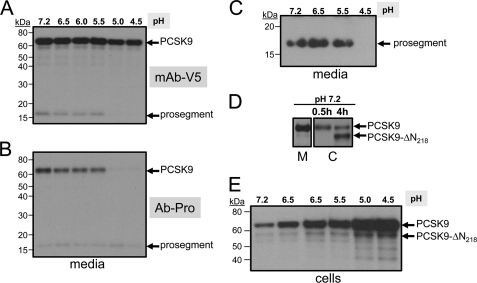
Because PCSK9 is known to better bind the EGFA domain of the LDLR at acidic pH values (11, 20), we sought to test the effect of pH on its domain structure and its ability to bind and enhance the degradation of the LDLR. 35S-PCSK9-V5, obtained from the media of HEK293 cells pulse-labeled for 4 h with [35S]Cys + -Met, was incubated with HepG2 cells for 30 min in DMEM containing 0.2% BSA, of which the pH values were adjusted to 7.2, 6.5, 6.0, 5.5, 5.0, and 4.5. The media were then harvested and immediately neutralized to pH 7.2, and the proteins were immunoprecipitated with the mAb-V5, and the supernatants were immunoprecipitated with an in-house polyclonal prosegment antibody Ab-Pro47–67 generated in goat. SDS-PAGE analysis revealed that although mature PCSK9 and its prosegment formed a heterodimer (5) at pH values 7.2 to 5.5, the prosegment dissociated from the catalytic subunit of PCSK9 at pH values 5.0 and 4.5, as it is no longer immunoprecipitated with the mAb-V5 (Fig. 7A), but it is still immunoprecipitated with the prosegment antibody as a single entity of ∼17 kDa (Fig. 7B). To further prove that the prosegment dissociates from mature PCSK9 at pH 4.5, immunoprecipitation of the media with mAb-V5, followed by immunoprecipitation of the supernatant with the prosegment antibody Ab-Pro47–67 and SDS-PAGE analysis, failed to reveal the ∼17-kDa prosegment only when the media were incubated for 30 min at pH 4.5 but not at pH values 7.2, 6.5, or 5.5 (Fig. 7C).
FIGURE 7.
pH dependence of the PCSK9-prosegment complex and its association with HepG2 cells. HepG2 cells were incubated with 35S-PCSK9 (obtained from a 4-h pulse labeling of HEK293 cells overexpressing PCSK9-V5 (28)) for 30 min at 37 °C; the media were harvested and immediately neutralized at pH 7.2. The latter were then sequentially immunoprecipitated with mAb-V5 (A) and their supernatants (B) re-immunoprecipitated with the prosegment polyclonal antibody (Ab-Pro47–67). The immunoprecipitates were analyzed on 8% SDS-PAGE in Tricine buffer, and their autoradiograms were depicted with 2-h (A) or 20-h (B) exposures. C, following immunoprecipitations with mAb-V5, the supernatants were then immunoprecipitated with the prosegment antibody Ab-Pro and then analyzed by SDS-PAGE. SDS-PAGE analysis of the immunoprecipitations of the 35S-PCSK9-V5 bound/internalized to/into HepG2 cells after the 30-min or 4-h incubation period at pH 7.2 (D) and exposed for 30 min at various pH values (E). Notice the formation of the furin-cleaved form (PCSK9-ΔN218) at 4 h of incubation at pH 7.2 and 30-min ones at acidic pH values.
We also analyzed the 35S-PCSK9-V5 bound/internalized to/into HepG2 cells after 30 min or 4 h of preincubation at pH 7.2 (Fig. 7D) or at various pH values (Fig. 7E). The data revealed that PCSK9 association with HepG2 cells increased gradually as a function of lowering the pH, reaching a maximum at pH 4.5 (Fig. 7E). The strong cell association of PCSK9 that lacks the prosegment at pH 4.5 may be related to its cell surface aggregation at this pH (see below). We also noticed in cell lysates the increased generation of the furin-cleaved form at Arg218 (PCSK9-ΔN218) (28) with time (4 h versus 0.5 h; Fig. 7D). This suggests that this processing, which results in the loss of function of PCSK9, is achieved by cell surface furin (35).
pH Dependence of the PCSK9-induced Reduction of Cell Surface LDLR Levels
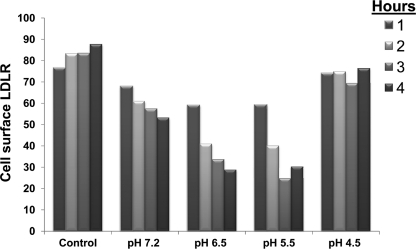
We next assessed whether short exposure of PCSK9 to acidic pH values affects its ability to enhance the degradation of cell surface LDLR. Accordingly, we incubated PCSK9 at the above pH values for 1 h and then immediately neutralized the media. These were then incubated for 1–4 h with HuH7 cells at pH 7.2, and cell surface LDLR levels were measured by FACS, with a representative example (out of three independent ones) shown in Fig. 8. To our surprise, the data revealed an irreversible pH-dependent increased activity of PCSK9 at all incubation times (1–4 h) from pH 7.2 to 5.5, with a maximal ∼2.5-fold higher activity after 3 h of incubation at pH 5.5. Interestingly, incubation of PCSK9 for 1 h at pH 4.5, followed by neutralization, resulted in an irreversible loss of its activity toward cell surface LDLR in HuH7 cells at all incubation times studied (Fig. 8), likely reflecting a profound conformational change of PCSK9 resulting in the dissociation of its prosegment (Fig. 7).
FIGURE 8.
FACS analysis of HuH7 cells for LDLR expression. PCSK9 obtained from HEK293 cells (0.7 μg/ml) was incubated at 37 °C for 1 h at pH 7.2, 6.5, 6.0, 5.5, 5.0, and 4.5. The media were then neutralized to pH 7.2 and incubated with HuH7 cells for 1–4 h. The cells were then extensively washed, treated with Versene for 5 min to suspend them, and then were immediately analyzed by FACS analysis for LDLR expression at the cell surface. Cell viability was higher than 70% in all cases, as judged by FACS analysis. The levels of measured cell surface LDLR are plotted (in arbitrary units) against pH and compared with control HuH7 cells not exposed to PCSK9 but subjected to similar manipulations using media from naive HEK293 cells. Similar data were obtained in more than three separate experiments.
We next tested the pH-dependent extracellular activity of the various PCSK9 constructs lacking or not the acidic stretch of the prosegment (Fig. 2). The level of all proteins secreted from HEK293 cells were quantified by an ELISA (17). Equal amounts of PCSK9 (0.7 μg/ml) from each preparation were then incubated at 37 °C at various pH values for 1 h, neutralized to pH 7.2, and then incubated with HuH7 cells for 4 h, and the cell surface LDLR was quantified by FACS (supplemental Fig. S3). The data show that the natural mutants R46L and A53V do not significantly modify the PCSK9 activity or its pH dependence. However, the constructs lacking aa 33–46 and especially 33–58 are about 2- and >4-fold more active than WT PCSK9, at all pH values, approaching the activity of the most potent PCSK9 D374Y mutant (11). These data clearly show that removal of aa 33–58 results in a significantly enhanced activity on cell surface LDLR of HuH7 cells. Because it was reported that co-expression of the prosegment and PCSK9 lacking the latter results in a secreted bioactive complex (29, 36, 37), we also used this approach to verify whether the lack of aa 33–58 would also result in an enhancement of the pH-dependent activity of PCSK9 on LDLR. Although the high level of activity achieved upon removal of the acidic stretch of the prosegment masked the pH-dependent effect, we did see that it still remained when we co-expressed PCSK9 lacking the prosegment (PCSK9-Δpro) with either the full-length prosegment or its Δ33–58 mutant (supplemental Fig. S3, right histograms). Although in trans co-expression of the prosegment and mature PCSK9 results in a less active protein that the native one, it is still sensitive to the irreversible pH 5.5 activation. This suggests that the determinant responsible for the pH-dependent activation of PCSK9 does not reside in aa 33–58 of the prosegment and more specifically that the conserved His58 is not involved.
pH-dependent Internalization of PCSK9 and Its Co-localization with LDLR
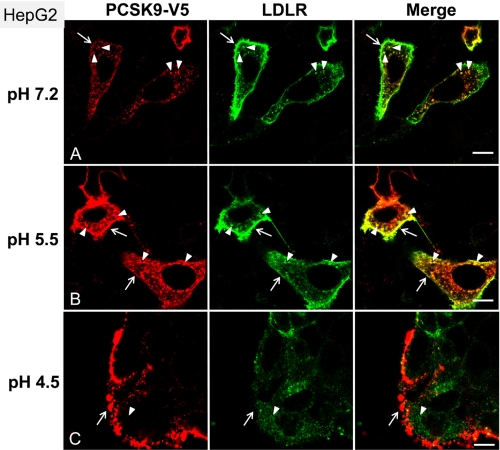
HepG2 cells were incubated with ∼0.7 μg/ml of PCSK9 that were preincubated for 1 h at pH values 7.2, 5.5, and 4.5, followed by neutralization at pH 7.2. Immunocytochemistry at the confocal level revealed a stronger intensity of labeling and degree of co-localization of PCSK9 and LDLR at the cell surface and in endosome/lysosome-like punctate structures upon preincubation at pH 5.5 versus pH 7.2 (Fig. 9). In contrast, preincubation at pH 4.5 resulted in an aggregated PCSK9 that stuck nonspecifically to cells at areas mostly devoid of LDLR.
FIGURE 9.
Increased internalization of PCSK9 after exposure to pH 5.5. HepG2 cells were incubated for 60 min at 37 °C with condition media, from HEK293 cells overexpressing PCSK9-V5, that have been preincubated at the indicated pH (A–C), as described under “Experimental Procedures.” After fixation and permeabilization of the cells, surface binding and internalization of PCSK9 as well as the localization of LDLR were detected by immunofluorescence with anti-V5 (red) and anti-LDLR antibodies (green). Arrows point to the cell surface labeling of PCSK9 and LDLR, and arrowheads indicate the internalized PCSK9 and its co-localization with LDLR. Bars, 10 μm.
Novel Cleavage of PCSK9 at pH 5.5
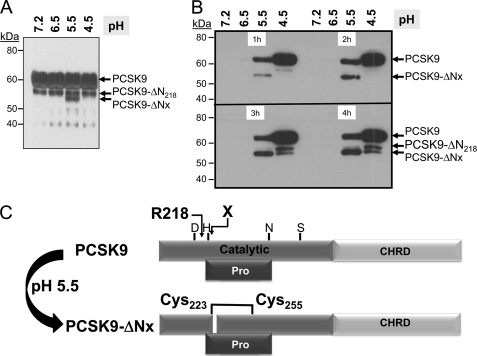
To probe the mechanism behind the irreversible activation of PCSK9 following a 1-h incubation at pH 5.5, we examined the molecular forms of PCSK9 by Western blot. Upon incubation of PCSK9-V5 secreted from HEK293 cells, which contains both full-length and the furin-cleaved PCSK9-ΔN218, only at pH 5.5, we clearly observed the formation of a novel V5-positive fragment that is ∼3 kDa N-terminally shorter than PCSK9-ΔN218, hereafter called PCSK9-ΔNx (Fig. 10A). HuH7 cells were then incubated with the above neutralized media for 1–4 h and washed extensively, and the PCSK9-bound/internalized forms were analyzed by Western blotting (Fig. 10B). Clearly, a gradual increase in PCSK9-ΔNx was seen in cell lysates of the samples originating from the pH 5.5 incubations. We observed only small amounts of cell-associated PCSK9 at higher pH values possibly because they are rapidly hydrolyzed. Notice that the inactive furin-cleaved form PCSK9-ΔN218 (28) does not associate with cells. The novel cleavage site is believed to occur after the active site His226, based on its molecular mass relative to PCSK9-ΔN218. From the crystal structure data (11), this would predict that PCSK9-ΔNx is composed of two disulfide-bonded subunits held by Cys223–Cys255 (Fig. 10C). Whether this form is more active than the full-length PCSK9 or is actually an inactivated form remain to be defined, but it is produced coincidentally at pH 5.5, and much less so at pH 6.5, which coincides with the observed maximal PCSK9 activity on LDLR. Interestingly, PCSK9 arising from the pH 4.5 preincubation, which resulted in dissociation of the prosegment (Fig. 7), was strongly bound to HuH7 cells (Fig. 10B) but seemingly was not able to enhance the degradation of the LDLR (Fig. 8). This might be due to its propensity to aggregate at pH 4.5, as seen on the surface of HepG2 cells (Fig. 9).
FIGURE 10.
Generation of a novel PCSK9-ΔNx product at pH 5.5. A, SDS-PAGE analysis of the media containing 35S-PCSK9 exposed to various pH values for 1 h. Notice the appearance of the novel PCSK9-ΔNx product at pH 5.5, migrating below the furin-cleaved PCSK9-ΔN218. B, SDS-PAGE analysis of the lysates of HuH7 cells exposed to 35S-PCSK9 preincubated at various pH values, neutralized, and then incubated for 1–4 h with the cells. Notice that very little PCSK9 can be detected in cells at pH values below 5.5, likely due to its rapid uptake and degradation. The level of the PCSK9-ΔNx product gradually increases with time. Notice that the furin-cleaved PCSK9-ΔN218 is not taken up by the cells, in accordance with its loss of aa 152–218 and the prosegment (28). C, proposed model for the generation at pH 5.5 of a two-chain disulfide-linked PCSK9-ΔN218 still bound to the prosegment.
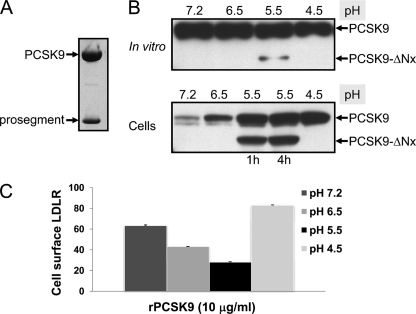
The above data suggested that at pH values 6.5–5.5, PCSK9 could irreversibly adopt a more active structure. Whether or not this is counterbalanced or associated with the production of PCSK9-ΔNx is still not clear. We next incubated in vitro affinity-purified PCSK9 obtained from baculovirus expression (Fig. 11A) for 1 h at various pH values. The data show that here again PCSK9-ΔNx is only produced at pH 5.5, albeit at a very low level. However, further incubation of this reaction pool with HuH7 cells for 1 or 4 h showed much enhanced cleavage and the preferential cellular binding/internalization of the cleaved versus full-length form, as the levels of the two cell-associated forms are now almost equal (Fig. 11B). Finally, FACS analysis of the LDLR levels at the surface of HuH7 cells exposed to this reaction mixture also showed a similar maximal ∼2.5-fold increased activity of the sample preincubated at pH 5.5 (Fig. 11C). Thus, purified PCSK9, which may be contaminated with traces of a protease activity from High Five cells, behaves the same as the one originating from HEK293 cells overexpressing it, and its cleavage at site X is maximal at pH 5.5. Collectively, these data suggest that PCSK9-ΔNx is likely generated by cleavage of PCSK9 by a soluble protease and that this form has a strong affinity to bind, and likely internalize, into cells.
FIGURE 11.
Pure PCSK9 can generate PCSK9-ΔNx at pH 5.5 with increased activity. A, Coomassie staining of pure PCSK9 purified from the media of baculovirus High Five cells, attesting for its complex formation with the prosegment and its purity as confirmed by protein sequence analysis (data not shown). B, Western blot analysis using our PCSK9 polyclonal antibody (10) of pure PCSK9 (10 μg/ml) incubated in vitro at 37 °C at various pH values for 1 h. Notice the production of PCSK9-ΔNx only at pH 5.5. Following neutralization and incubation with HuH7 cells for 4 h, the cells were lysed, and their content was analyzed by Western blot. C, FACS analysis of cell surface LDLR of HuH7 cells exposed to 10 μg/ml of pure PCSK9 preincubated in vitro at 37 °C at various pH values for 1 h, neutralized, and then incubated with the cells for 4 h.
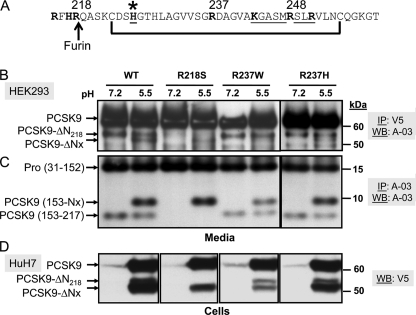
Finally, we wished define the cleavage site resulting in PCSK9-ΔNx. Based on its ∼3-kDa size smaller than the furin-cleaved form at Arg218, and the presence of a disulfide bond, cleavage likely occurs between aa 244 and 254 (Fig. 12A). Accordingly, although cleavage of the natural GOF R218S and LOF R237W mutants still occurred at pH 5.5 (Fig. 12B), the pH dependence of the less active R237W mutant was somewhat blunted (Fig. 12C). This suggested that the degree of pH dependence is sensitive to one or more residue(s) close to Arg237. It seems that the positive charge of Arg237 is important, because its replacement with Trp decreased the extent of the pH 5.5 cleavage, whereas the R237H mutant, which is positively charged at acidic pH values, was almost as well cleaved as the WT. Notably, production of the PCSK9-ΔNx form and its PCSK9(153–217) counterpart (the Arg218 is expected to be removed by a basic carboxypeptidase, e.g. CPD (38)) was not affected by the loss of the furin cleavage at Arg218↓ (R218S; Fig. 12C), providing evidence that its generation is independent from that of the furin cleavage at Arg218↓. Finally, incubation of HuH7 cells with neutralized media expressing either PCSK9 or its R218S, R237W, or R237H mutants, which had been pre-exposed to pH 5.5 for 1 h, also revealed that PCSK9-ΔNx is robustly associated with cells. From the small amounts of PCSK9-ΔNx seen in the media compared with PCSK9 (Fig. 12B), we can deduce that PCSK9-ΔNx is relatively more strongly associated with cells than the uncleaved WT PCSK9 (Fig. 12D).
FIGURE 12.
Evidence that the natural variants R218S and R237W can still generate PCSK9-ΔNx at pH 5.5. A, amino acid sequence of the segment, including aa 215–260 of PCSK9. The various Arg and His residues, including the potential KGASMRSLR↓VL253 furin-like sites, and the disulfide bridge are emphasized. B, SDS-PAGE analysis of the media of HEK293 cells overexpressing WT or R218S, R237W, or R237H. The media of the above cells were incubated for 1 h at 37 °C at either pH 7.2 or 5.5 and then immediately neutralized to pH 7.2 and immunoprecipitated (IP) with mAb-V5 and Western-blotted (WB) with the PCSK9 antibody A-03, recognizing aa 31–454 (10), or immunoprecipitated and Western-blotted with A-03 (C). D, same neutralized media were then incubated for 4 h at 37 °C with HuH7 cells. The cells were extensively washed three times and the lysates analyzed by Western blot with mAb-V5. The emphasized migration positions are those of PCSK9 and its furin-processed form PCSK9-ΔN218, as well as the novel PCSK9-ΔNx product and the corresponding prosegment (aa 31–152) and N-terminal aa 153-Nx and 153–217.
Identification of Arg248 as the Cleavage Site of PCSK9 at pH 5.5
Scrutiny of the PCSK9 sequence revealed two other potential furin-like sites following Arg218, i.e. at either Arg248 or Arg251 (39) in the sequence KGASMR248-SLR251-VL253. However, preincubation of PCSK9 at pH 5.5 for 2 h, followed by overnight incubation with pure furin at neutral pH, did not enhance this cleavage (supplemental Fig. S6). In fact, this resulted in a novel cleavage, likely at REIEGR199↓VM, even in the absence of pH 5.5 preincubation (supplemental Fig. S6). Furthermore, we believe that cleavage does not occur autocatalytically because a similar incubation of the complex prosegment PCSK9-H226A with the active site His226 mutated to Ala (5, 29) also generated the same PCSK9(153-Nx) (supplemental Fig. S6). Interestingly, the H226A mutant is resistant to this novel furin cleavage (supplemental Fig. S6). Thus, at neutral pH the cell surface furin inactivates PCSK9 by cleavage at Arg218↓. In contrast, the conformational changes occurring at pH 5.5 favor activation of PCSK9 with the concomitant production of a two-chain PCSK9-ΔNx through cleavage by another protease, likely within the acidic environment of endosomal compartments.
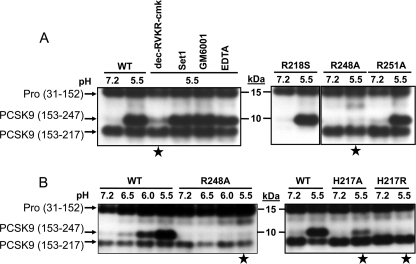
We also noted that such cleavage occurs rapidly, because incubation of PCSK9 secreted from HEK293 cells at pH 5.5 for as short as 10 min generated most of the observed cleavage. However, the presence of 50 μm RVKR-chloromethyl ketone, a furin-like protease inhibitor (40), during the incubation period prevented the observed cleavage (Fig. 13A). In contrast, incubation of PCSK9 with either a mixture of serine and cysteine protease (500 μm 4-(2-aminoethyl)benzenesulfonyl fluoride + 150 nm aprotinin + 1 μm E-64 + 0.5 mm EDTA + 1 μm leupeptin) or metalloprotease (150 μm GM6001 or 15 mm EDTA) inhibitors did not prevent cleavage. Upon mutagenesis of the likely PC-like sites, we observed that only R248A, but not R251A, abolished cleavage at pH 5.5 (Fig. 13A). This suggested that processing of PCSK9 at pH 5.5 occurs at Arg248, which exhibits an unusual PC-like site with a Lys243 at P6 (39), within the sequence KGASMR248-SL. However, the pH-dependent generation of PCSK9-ΔN248, which is maximal at pH 5.5 (Fig. 13, A and B, left panel; aa 153–247, as Arg248 would be removed by a carboxypeptidase), is not autocatalytic (supplemental Fig. S6) nor is it enhanced in the presence of soluble furin, PACE4 or PC5/6 (data not shown), suggesting that a rapidly cleaving enzyme, very active at pH 5.5 and distinct from the PCs, is responsible for this cleavage. The observed inhibition by the furin-like inhibitor RVKR-chloromethyl ketone is likely due to the ability of this inhibitor to bind and inactivate PCSK9 itself by an unusual mechanism (data not shown).
FIGURE 13.
Identification of Arg248 as the site of cleavage and implication of His217. SDS-PAGE analysis of the media of HEK293 cells overexpressing WT PCSK9 at pH 7.2 or 5.5 The media of the above cells were incubated for 1 h at 37 °C at either pH 7.2 or 5.5 and then immediately neutralized to pH 7.2 and immunoprecipitated (IP) with mAb-V5 and Western-blotted (WB) with the PCSK9 antibody A-03, as in Fig. 12. A, effect of protease inhibitors on this pH 5.5 processing. This includes incubation of PCSK9 at pH 5.5 in the presence of either 50 μm of the furin-like inhibitor decanoyl-RVKR-chloromethyl ketone (RVKR-cmk), the serine and cysteine protease inhibitor mixture (Set1: 500 μm 4-(2-aminoethyl)benzenesulfonyl fluoride + 150 nm aprotinin + 1 μm E-64 + 0.5 mm EDTA + 1 μm leupeptin) or the metalloprotease inhibitors 150 μm GM6001 or 15 mm EDTA. Processing of the mutants R218S, R248A, and R251A at pH values 7.2 and 5.5 is also shown. B, left panel, pH-dependent processing of PCSK9 into PCSK9-ΔN247. Note that the mutant R248A is not processed. Right panel, influence of His217 (WT) and its mutants H217A and H217R on the processing at Arg248. The stars point to the conditions that block cleavage at Arg248 at pH 5.5.
In agreement with the crystal structure of PCSK9, which suggested that His217 can form contacts with residues close to Arg248 (11), the mutants H217A and much more so H217R abolished the cleavage of PCSK9 at pH 5.5 (Fig. 13B, right panel). Thus, the pH-dependent processing of PCSK9 at Arg248 is at least in part regulated by His217, which would become positively charged at pH 5.5, but such an effect is not just due to charge because it is not mimicked by a bulkier Arg residue.
DISCUSSION
The discovery and characterization of PCSK9 in 2003 (5) and the genetic evidence for its implication in LDL-cholesterol regulation (6) were a stepping stone for a major research effort by many laboratories to define the mechanism behind the observed PCSK9 activity (4, 41–43). Very soon it became clear that the main hepatic target of PCSK9 is the LDLR (13) and that the degradation of the latter in acidic endosome/lysosome-like compartments is enhanced by PCSK9 (7–10), and it does not seem to require the PCSK9 proteolytic activity (36, 37). This was rationalized by structural and biochemical studies that revealed that the spatially separate catalytic domain of PCSK9 (11) binds the EGFA domain of LDLR (20) and that this interaction is enhanced at acidic pH values (11), likely due in part to the protonation of His306 of EGFA (12, 44). It also became apparent that the inhibitory N-terminal prosegment, which remains noncovalently complexed to PCSK9 (5) and is not easily dislodged (11), and the C-terminal CHRD play critical roles in regulating the activity of PCSK9 (12) and the cellular trafficking of the PCSK9-LDLR complex (10, 30). Indeed, specific mutations in the prosegment and CHRD (4, 45), as well as the hinge region connecting the catalytic domain and the CHRD (17), were reported to influence the PCSK9 activity likely by modulating its ability to drag the LDLR toward subcellular degradative compartments.
In this study, we examined more closely the boundary of the prosegment that negatively regulates intracellular and extracellular activity of PCSK9 on the LDLR. Our data pointed out that deletion of aa 31–58 of the prosegment results in a >4-fold higher activity of PCSK9 toward cellular LDLR. In addition, a construct combining the lack of this segment and the GOF mutation D374Y results in a >15-fold enhanced activity of PCSK9 (Figs. 3 and 4). The existence of endogenous proteases that can process the prosegment at Arg46↓ was shown both in baculovirus expressing High Five cells (Fig. 1) and by the ability of PC7 to cleave the prosegment of PCSK9 (28). Interestingly, the LOF mutation R46L that mostly affects Caucasians (45), and is associated with hypocholesterolemia (46), is no longer processed by PC7 (data not shown), and it may result in resistance to cleavage in vivo. In addition, another E32K mutant observed in a Japanese population, which reduces the acidic nature of the prosegment, was associated with a GOF of PCSK9 (47).
What is the mechanism behind the inhibitory activity of the acidic aa 31–58? This may be due in part to its inferred effect on the overall conformation of PCSK9, because the removal of aa 31–53 enhances >7-fold the in vitro binding of PCSK9 and EGFA (12). In our hands, removal of aa 30–46 results in a 3-fold better binding to LDLR (Fig. 5) in vitro, and the lack of aa 33–58 significantly enhances the internalization rate of the PCSK9-LDLR complex in HepG2 cells (supplemental Fig. S1). However, this segment was not seen in any of the crystal structures published at both acidic and basic pH values and is believed to be flexible and/or unstructured (11, 18, 19). An alternative, but not mutually exclusive, possibility would be that this acidic prosegment may transiently bind positive charges in a basic domain of PCSK9 (maybe within the CHRD) or in another protein in vivo, which would regulate its inhibitory effect.
A positive correlation between PCSK9 levels and circulating triglycerides was reported in human plasma (17, 48). The similarity of the N-terminal acidic sequence of PCSK9 to that of GPIHBP1, which enhances lipase hydrolysis of triglycerides in chylomicrons (31–33), led us to show that these domains could be swapped without a deleterious effect on the PCSK9 function (Fig. 6). Thus, it is plausible that like GPIHBP1 (31–33), it is the acidic nature of the prosegment that regulates its activity. Through its acidic segment, could PCSK9 up-regulate triglyceride levels by sequestering a limiting lipase such as lipoprotein lipase and/or endothelial lipase or apoAV on lipoprotein particles, thereby reducing triglyceride hydrolysis to free fatty acids? Whether PCSK9 can compete with the negatively charged N-terminal segment of GPIHBP1 for this activity needs to be analyzed. Thus, much more work is required to confirm this hypothesis and to explain the observed positive effect of PCSK9 on circulating triglyceride levels in human.
Even though His306 of the EGFA domain contributes to the pH-dependent interaction of PCSK9 with the LDLR (12, 44), there is currently no clear structural explanation for the increased PCSK9 binding to LDLR at low pH (11). Could aa 534–607 within the second repeat domain of the CHRD that contains 9 His residues, all localized within a groove (18, 19), undergo electrostatic repulsion due to their positive charge at low pH? Is the LDLR degradation favored by changes in the CHRD conformation and/or through binding to another protein?
To begin to answer such questions, we first studied in more detail the effect of pH on the activity of extracellular PCSK9 in HuH7 cells. To our surprise, we found that a short exposure of PCSK9 to acidic pH values (pH values 6.5 or 5.5) followed by neutralization of the solution resulted in an irreversible and significantly enhanced activity of PCSK9 on cell surface LDLR, reaching a maximal ∼2.5-fold activation at pH 5.5 (Fig. 8). This pH dependence was still present in the absence of aa 33–58 (supplemental Fig. S3), suggesting that the critical aa is/are present elsewhere within the molecule. Interestingly, at the most effective pH 5.5, we also observed a novel cleavage resulting in the generation of a likely two-chain disulfide-linked PCSK9-ΔN248 that still retains the prosegment (Figs. 10 and 13). The generation of this product seems to be the result of a protease possibly present at the cell surface of cells (that may also be shed into the medium), because purified PCSK9 can undergo such cleavage and activation in the media and when incubated with cells (Figs. 11 and 13), and it is produced from PCSK9 isolated from either High Five or HEK293 cells. It is conceivable that such cleavage would produce a more flexible two-chain PCSK9 that can better interact with the LDLR and/or the sorting machinery. It is still possible that the observed enhanced activity of PCSK9 at pH 5.5 may be due to a pH-dependent conformational change of full-length PCSK9 and that the generation of PCSK9-ΔN248 rather results in loss of function. However, preliminary FACS data revealed that the activity of the PCSK9-R248A mutant on LDLR is much less sensitive to pH, especially between pH values 6.0 and 5.5, suggesting that cleavage is necessary for maximal activity. However, the ultimate proof of functionality will require the future purification of the cleaved protein at Arg248. What is clear however is that preincubation of PCSK9 at pH 5.5 results in a more active molecule that internalizes faster into endosomes together with cell surface LDLR (Fig. 9). It is noted that although Arg248 in PCSK9 is conserved between various primates and Xenopus laevis, it is replaced by a His in the rodent rat and mouse orthologues. Because the R248A mutant is not processed (Fig. 13), and neither soluble PC5/6A, PACE4, nor furin can cleave at this residue in human PCSK9, it is plausible that the cognate processing enzyme may not be a proprotein convertase nor exclusively Arg-specific. Alternatively, cleavage at Arg248 may be specific to primate and Xenopus laevis PCSK9. Coincidentally, the best pH for PCSK9 activation is close to that of late endosomes (49), suggesting that this process may actually take place within the acidifying endosomes en route to their fusion with lysosomes and delivery of cargo for degradation. The enzymatic activity of PCSK9 is not necessary for LDLR (36, 37) or the VLDL receptor and apoER2 (29) receptor degradation nor is it necessary for the generation of PCSK9-ΔN248 (Figs. 12 and 13 and supplemental Fig. S6). In that context, preincubation of PCSK9 at any pH value from 7.5 to 4.5 did not yield an active protease that would degrade the ectodomain of LDLR, because it was unable to digest soluble LDLR (supplemental Fig. S4). Thus, at least in vitro, the activation of PCSK9 observed at pH 5.5 does not seem to generate a protease activity that would degrade the LDLR. Interestingly, SDS-PAGE analysis of the molecular forms generated at pH 7.2 and 5.5 revealed that at acidic pH values mostly the cleaved forms of PCSK9, but not the CHRD, exhibit the highest propensity to oligomerize under nonreducing conditions (supplemental Fig. S5). This is in accord with a report suggesting that the self-association of PCSK9, especially observed at acidic pH values, correlates with its LDLR degrading activity (50).
In conclusion, this study sheds new light on the importance of the acidic N-terminal sequence of the prosegment and its effect on the activity of PCSK9. We also present a novel mechanism for fine-tuning the activity of PCSK9, which is enhanced at acidic pH values close to those of late endosomes.
Supplementary Material
Acknowledgments
We thank all the members of the Seidah laboratory for helpful discussions and Brigitte Mary for efficacious editorial assistance. We also thank Brian Carpenter, Yaqun Zhang, Deepa Calambur, Mian Gao, and Mark Witmer of Bristol-Myers Squibb for PCSK9 cloning/baculovirus expression and production. We also thank the reviewers of this article who gave us valuable advice that enhanced the content of this manuscript.
This work was supported by Canadian Institutes of Health Research Grants MOP-102741 and CTP-82946 (to N. G. S.), Strauss Foundation grant (to N. G. S.), Bristol-Myers Squibb collaborative research grant, and Canada Chair 216684 (to N. G. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- LDLR
- low density lipoprotein receptor
- CHRD
- Cys-His-rich domain
- EGFA domain
- epidermal growth factor precursor homology domain
- ER
- endoplasmic reticulum
- GOF
- gain of function
- LPDS
- lipoprotein-deficient serum
- LOF
- loss of function
- aa
- amino acid
- diI
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CV
- column volume.
REFERENCES
- 1.Lloyd-Jones D., Adams R., Carnethon M., De Simone G., Ferguson T. B., Flegal K., Ford E., Furie K., Go A., Greenlund K., Haase N., Hailpern S., Ho M., Howard V., Kissela B., Kittner S., Lackland D., Lisabeth L., Marelli A., McDermott M., Meigs J., Mozaffarian D., Nichol G., O'Donnell C., Roger V., Rosamond W., Sacco R., Sorlie P., Stafford R., Steinberger J., Thom T., Wasserthiel-Smoller S., Wong N., Wylie-Rosett J., Hong Y. (2009) Circulation 119, e21–181 [DOI] [PubMed] [Google Scholar]
- 2.Brown M. S., Goldstein J. L. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maxfield F. R., Tabas I. (2005) Nature 438, 612–621 [DOI] [PubMed] [Google Scholar]
- 4.Seidah N. G., Prat A. (2007) J. Mol. Med. 85, 685–696 [DOI] [PubMed] [Google Scholar]
- 5.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S. B., Stifani S., Basak A., Prat A., Chretien M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abifadel M., Varret M., Rabès J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derré A., Villéger L., Farnier M., Beucler I., Bruckert E., Chambaz J., Chanu B., Lecerf J. M., Luc G., Moulin P., Weissenbach J., Prat A., Krempf M., Junien C., Seidah N. G., Boileau C. (2003) Nat. Genet. 34, 154–156 [DOI] [PubMed] [Google Scholar]
- 7.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., Trillard M., Abifadel M., Tebon A., Attie A. D., Rader D. J., Boileau C., Brissette L., Chrétien M., Prat A., Seidah N. G. (2004) J. Biol. Chem. 279, 48865–48875 [DOI] [PubMed] [Google Scholar]
- 8.Maxwell K. N., Fisher E. A., Breslow J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2069–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S. W., Moon Y. A., Horton J. D. (2004) J. Biol. Chem. 279, 50630–50638 [DOI] [PubMed] [Google Scholar]
- 10.Nassoury N., Blasiole D. A., Tebon Oler A., Benjannet S., Hamelin J., Poupon V., McPherson P. S., Attie A. D., Prat A., Seidah N. G. (2007) Traffic 8, 718–732 [DOI] [PubMed] [Google Scholar]
- 11.Cunningham D., Danley D. E., Geoghegan K. F., Griffor M. C., Hawkins J. L., Subashi T. A., Varghese A. H., Ammirati M. J., Culp J. S., Hoth L. R., Mansour M. N., McGrath K. M., Seddon A. P., Shenolikar S., Stutzman-Engwall K. J., Warren L. C., Xia D., Qiu X. (2007) Nat. Struct. Mol. Biol. 14, 413–419 [DOI] [PubMed] [Google Scholar]
- 12.Kwon H. J., Lagace T. A., McNutt M. C., Horton J. D., Deisenhofer J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maxwell K. N., Breslow J. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. (2006) J. Clin. Invest. 116, 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan J. C., Piper D. E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., Di Y., Shetterly S., Arimura Z., Salomonis H., Romanow W. G., Thibault S. T., Zhang R., Cao P., Yang X. P., Yu T., Lu M., Retter M. W., Kwon G., Henne K., Pan O., Tsai M. M., Fuchslocher B., Yang E., Zhou L., Lee K. J., Daris M., Sheng J., Wang Y., Shen W. D., Yeh W. C., Emery M., Walker N. P., Shan B., Schwarz M., Jackson S. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9820–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirier S., Mayer G., Poupon V., McPherson P. S., Desjardins R., Ly K., Asselin M. C., Day R., Duclos F. J., Witmer M., Parker R., Prat A., Seidah N. G. (2009) J. Biol. Chem. 284, 28856–28864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubuc G., Tremblay M., Paré G., Jacques H., Hamelin J., Benjannet S., Boulet L., Genest J., Bernier L., Seidah N. G., Davignon J. (2010) J. Lipid Res. 51, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampton E. N., Knuth M. W., Li J., Harris J. L., Lesley S. A., Spraggon G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14604–14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piper D. E., Jackson S., Liu Q., Romanow W. G., Shetterly S., Thibault S. T., Shan B., Walker N. P. (2007) Structure 15, 545–552 [DOI] [PubMed] [Google Scholar]
- 20.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. (2007) J. Biol. Chem. 282, 18602–18612 [DOI] [PubMed] [Google Scholar]
- 21.Timms K. M., Wagner S., Samuels M. E., Forbey K., Goldfine H., Jammulapati S., Skolnick M. H., Hopkins P. N., Hunt S. C., Shattuck D. M. (2004) Hum. Genet. 114, 349–353 [DOI] [PubMed] [Google Scholar]
- 22.Dewpura T., Raymond A., Hamelin J., Seidah N. G., Mbikay M., Chrétien M., Mayne J. (2008) FEBS J. 275, 3480–3493 [DOI] [PubMed] [Google Scholar]
- 23.Teupser D., Thiery J., Walli A. K., Seidel D. (1996) Biochim. Biophys. Acta 1303, 193–198 [DOI] [PubMed] [Google Scholar]
- 24.Miao B., Sun S., Santomenna L. D., Zhang J. H., Young P., Mukherjee R. (2002) Methods Enzymol. 357, 180–188 [DOI] [PubMed] [Google Scholar]
- 25.Benjannet S., Elagoz A., Wickham L., Mamarbachi M., Munzer J. S., Basak A., Lazure C., Cromlish J. A., Sisodia S., Checler F., Chrétien M., Seidah N. G. (2001) J. Biol. Chem. 276, 10879–10887 [DOI] [PubMed] [Google Scholar]
- 26.Bottomley M. J., Cirillo A., Orsatti L., Ruggeri L., Fisher T. S., Santoro J. C., Cummings R. T., Cubbon R. M., Lo Surdo P., Calzetta A., Noto A., Baysarowich J., Mattu M., Talamo F., De Francesco R., Sparrow C. P., Sitlani A., Carfí A. (2009) J. Biol. Chem. 284, 1313–1323 [DOI] [PubMed] [Google Scholar]
- 27.Seidah N. G., Hamelin J., Mamarbachi M., Dong W., Tardos H., Mbikay M., Chretien M., Day R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3388–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjannet S., Rhainds D., Hamelin J., Nassoury N., Seidah N. G. (2006) J. Biol. Chem. 281, 30561–30572 [DOI] [PubMed] [Google Scholar]
- 29.Poirier S., Mayer G., Benjannet S., Bergeron E., Marcinkiewicz J., Nassoury N., Mayer H., Nimpf J., Prat A., Seidah N. G. (2008) J. Biol. Chem. 283, 2363–2372 [DOI] [PubMed] [Google Scholar]
- 30.Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13045–13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beigneux A. P., Davies B. S., Gin P., Weinstein M. M., Farber E., Qiao X., Peale F., Bunting S., Walzem R. L., Wong J. S., Blaner W. S., Ding Z. M., Melford K., Wongsiriroj N., Shu X., de Sauvage F., Ryan R. O., Fong L. G., Bensadoun A., Young S. G. (2007) Cell Metab. 5, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gin P., Yin L., Davies B. S., Weinstein M. M., Ryan R. O., Bensadoun A., Fong L. G., Young S. G., Beigneux A. P. (2008) J. Biol. Chem. 283, 29554–29562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beigneux A. P., Davies B. S., Bensadoun A., Fong L. G., Young S. G. (2009) J. Lipid Res. 50, S57–S62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer G., Poirier S., Seidah N. G. (2008) J. Biol. Chem. 283, 31791–31801 [DOI] [PubMed] [Google Scholar]
- 35.Thomas G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNutt M. C., Lagace T. A., Horton J. D. (2007) J. Biol. Chem. 282, 20799–20803 [DOI] [PubMed] [Google Scholar]
- 37.Li J., Tumanut C., Gavigan J. A., Huang W. J., Hampton E. N., Tumanut R., Suen K. F., Trauger J. W., Spraggon G., Lesley S. A., Liau G., Yowe D., Harris J. L. (2007) Biochem. J. 406, 203–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arolas J. L., Vendrell J., Aviles F. X., Fricker L. D. (2007) Curr. Pharm. Des. 13, 349–366 [DOI] [PubMed] [Google Scholar]
- 39.Seidah N. G., Chrétien M. (1999) Brain Res. 848, 45–62 [DOI] [PubMed] [Google Scholar]
- 40.Ozden S., Lucas-Hourani M., Ceccaldi P. E., Basak A., Valentine M., Benjannet S., Hamelin J., Jacob Y., Mamchaoui K., Mouly V., Desprès P., Gessain A., Butler-Browne G., Chrétien M., Tangy F., Vidalain P. O., Seidah N. G. (2008) J. Biol. Chem. 283, 21899–21908 [DOI] [PubMed] [Google Scholar]
- 41.Horton J. D., Cohen J. C., Hobbs H. H. (2007) Trends Biochem. Sci. 32, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidah N. G. (2009) Expert Opin. Ther. Targets 13, 19–28 [DOI] [PubMed] [Google Scholar]
- 43.Horton J. D., Cohen J. C., Hobbs H. H. (2009) J. Lipid Res. 50, S172–S177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., Lagace T. A. (2009) J. Biol. Chem. 284, 10561–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kotowski I. K., Pertsemlidis A., Luke A., Cooper R. S., Vega G. L., Cohen J. C., Hobbs H. H. (2006) Am. J. Hum. Genet. 78, 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berge K. E., Ose L., Leren T. P. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1094–10100 [DOI] [PubMed] [Google Scholar]
- 47.Noguchi T., Katsuda S., Kawashiri M. A., Tada H., Nohara A., Inazu A., Yamagishi M., Kobayashi J., Mabuchi H. (2010) Atherosclerosis 210, 166–172 [DOI] [PubMed] [Google Scholar]
- 48.Lakoski S. G., Lagace T. A., Cohen J. C., Horton J. D., Hobbs H. H. (2009) J. Clin. Endocrinol. Metab. 94, 2537–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czekay R. P., Orlando R. A., Woodward L., Lundstrom M., Farquhar M. G. (1997) Mol. Biol. Cell 8, 517–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan D., Yancey P. G., Qiu S., Ding L., Weeber E. J., Linton M. F., Fazio S. (2008) Biochemistry 47, 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.