Abstract
Connexin43 (Cx43) is a transmembrane protein that forms gap junction channels. Regulation of Cx43 turnover is one mechanism to control the level of intercellular communication that occurs through gap junction channels. Proteasomal degradation of Cx43 is regulated in part through CIP75, a ubiquitin-like and ubiquitin-associated domain containing protein. CIP75 interacts with endoplasmic reticulum-localized Cx43, as demonstrated through co-immunoprecipitation and immunofluorescence microscopy experiments. CIP75 also binds to free monoubiquitin and lysine 48-linked tetraubiquitin chains in vitro and binds to ubiquitinated proteins in cellular lysates. However, analysis of Cx43 that immunoprecipitated with CIP75 demonstrated that the Cx43 associated with CIP75 was not ubiquitinated, and a mutant form of Cx43 that lacked lysines capable of ubiquitination retained the capacity to interact with CIP75. These results suggest that although CIP75 can interact with ubiquitinated cellular proteins, its interaction with Cx43 and stimulation of Cx43 proteasomal degradation does not require the ubiquitination of Cx43.
Keywords: Connexin, Endoplasmic Reticulum (ER), Proteasome, Protein Degradation, Ubiquitin, Ubiquitination, CIP75, ERAD
Introduction
Gap junctions are plasma membrane channels that mediate direct cell to cell communication. These channels allow the intercellular transfer of ions and molecules that are less than 1000 daltons, such as secondary messengers and small metabolites. The regulation of gap junctions is critical for the maintenance of normal tissue function. Gap junctions are composed of individual proteins (connexins) that oligomerize to form a six-member multimeric connexon, with two connexons in adjacent cell membranes docking across the intercellular space to form a complete gap junction channel (1). Gap junctional intercellular communication has an important role during development as well as in maintaining normal cell homeostasis (2–4). Misregulation of gap junctional intercellular communication and connexin levels can result in human diseases, such as heart arrhythmias (5–7), developmental diseases including oculodentodigital dysplasia (8), and cancers (9).
Connexins are highly labile proteins, having short half-lives of 1.5–5 h (10–14). Therefore, the mechanisms that control connexin levels and turnover must be tightly regulated. Connexins are known to be degraded through both the lysosomal and proteasomal degradation pathways (14–18). However, the exact mechanisms that regulate these pathways for the most prevalent connexin, Cx43, are unclear.
Connexin-interacting proteins have been studied with a great deal of interest because several have been implicated in aiding Cx43 degradation (19). The actin-binding protein ZO-1 (zonula occludens 1) has been demonstrated to be involved in the internalization of gap junction-associated Cx43 from the cell membrane, presumably leading to lysosomal degradation (20–26). Our laboratory discovered that the RabGAP protein CIP85 is also involved in Cx43 lysosomal degradation (27). Previous studies have also demonstrated Cx43 degradation via the proteasome pathway (14, 17, 18, 28–30). In particular, Cx43 that is translocated out of the ER2 can be degraded by the proteasome via the process of ER-associated degradation (ERAD) (14, 30). However, the mechanism by which this occurs has not been fully elucidated, although most proteins that are degraded by the ERAD pathway are typically marked by ubiquitination (31, 32).
UbL-UBA proteins are a part of the UDP (ubiquitin domain protein) superfamily (33). They contain a N-terminal UbL domain and one or more C-terminal UBA domains. UbL-UBA proteins have been found to be involved in a variety of processes, such as nucleotide excision repair (34) and spindle pole body duplication (35), but have also been implicated in proteasomal degradation (36–40). Multiple UbL-UBA proteins have been demonstrated to bind to components of the 26 S proteasome holoenzyme (40–43). For example, Rad23 is able to interact with Rpn1 and Rpn10, members of the 19 S cap subunit, through the UbL domain (40, 43). The UDP family of proteins is also known to interact with ubiquitinated proteins via the UBA domain (42, 44–48). Because ubiquitination is typically considered the tag that marks proteins for proteasomal degradation (31, 32), this suggests a role for the UbL-UBA proteins in proteasomal degradation, possibly as a shuttle or an adaptor protein to bring target substrates to the proteasome to be degraded. In fact, Rad23 is involved in the degradation of the tumor suppressor p53, the CDK inhibitor Sic1, and the SCFCdc4 substrate Far1 by proteasomes (49–51). Our previous studies identified the UbL-UBA protein CIP75 as a Cx43-interacting protein that is also able to interact with the Rpn1 and Rpn10 proteasomal proteins via its UbL domain (52), suggesting that CIP75 may be involved in Cx43 proteasomal degradation. However, whether ubiquitination is a prerequisite for an interaction between Cx43 and CIP75 is not known.
Because the regulation of connexin levels has a direct correlation with gap junctional intercellular communication, understanding the mechanism by which connexin levels are regulated is critical. In this present work, we have found that CIP75 was a ubiquitin-binding protein that interacts with Cx43 located in the ER and that this interaction increases upon inhibition of proteasomal degradation. However, in contrast with the more typical ubiquitin-dependent pathway for proteasomal degradation, the CIP75-Cx43 interaction was not dependent on Cx43 ubiquitination. These results suggest that CIP75 regulates proteasomal degradation of non-ubiquitinated Cx43, perhaps in concert with mediating the degradation of other ubiquitinated cellular protein substrates.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Human cervical carcinoma HeLa cells without endogenous Cx43 or stably expressing ER-localized Cx43 (HeLa-Cx43HKKSL) (53) were cultured in high glucose DMEM (Invitrogen) supplemented with 10% FBS, 20 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2. Transient transfections were conducted using Lipofectamine 2000 (Invitrogen) or the Nucleofector system (Lonza). Transfected cells were harvested 24–48 h after transfection.
Antibodies
p27 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); CIP75 monoclonal clones A333, M398, and L64 (54); Cx43 monoclonal clone P2D12 (from Paul Lampe, Fred Hutchinson Cancer Research Center, Seattle, WA); HA (Santa Cruz Biotechnology, Inc.); ubiquitin (Santa Cruz Biotechnology, Inc.); GST (Santa Cruz Biotechnology, Inc.); His (Santa Cruz Biotechnology, Inc.); FLAG M2 (Sigma); and calnexin (Thermo) antibodies were used.
Site-directed Mutagenesis/Plasmids
Cx43 lysine to arginine mutants were generated using the QuikChange Lightning multisite-directed mutagenesis kit (Agilent Technologies) using a series of primers (Table 1). After the mutagenesis PCR, DNA was transformed into XL-10 gold-competent cells according to the manufacturer's instructions. DNA was isolated from bacteria using the Qiagen Miniprep kit, sequenced, and then subcloned into the pcDNA3.1 expression vector using the EcoRI and XbaI restriction sites. Mutant 1 was generated with Cx43HKKSL-pcDNA DNA (53) as the template using primers Cx43K265R, Cx43K288R, and Cx43K303R. Mutant 2 was generated using Mutant 1 as the template with primers Cx43K235–242R, Cx43K259,265R, and Cx43K346,347R. Mutant 3 was generated with Mutant 2 as the template using the remaining primers.
TABLE 1.
Sequences of primers used to mutate Cx43 lysines to arginines
| Primer name | Sequence |
|---|---|
| Cx43K9,13R | gcc ttg ggg agg ctt ctg gac agg gtc caa gcc |
| Cx43K23R | gct gga ggg agg gtg tgg ctg tca gtg c |
| Cx43K69R | acg tct gct atg aca ggt cct tcc cc |
| Cx43K103–110R | gtg atg agg agg gaa gag agg cta aac agg aga gaa gag gag c |
| Cx43K115R | gag gag ctc aga gtg gcc cag act gac g |
| Cx43K129–137R | tgc acc tga ggc aga ttg aaa tca gga ggt tca ggt acg gga ttg |
| Cx43K145,147R | gag cac ggc agg gtg aga atg agg ggc g |
| Cx43K163R | gtt cta cac ctg cag gag aga tcc c |
| Cx43K189R | gca tcc tct tca ggt ctg tct tcg agg |
| Cx43K207R | ccc acg gag aga acc atc ttc atc atc ttc atg c |
| Cx43K235–242R | cgt ctt ctt cag agg cgt tag gga tcg cgt gag ggg aag aag c |
| Cx43K259,265R | gag ccc atc aag aga ctg cgg atc tcc aag ata cgc c |
| Cx43K265R | gga tct cca aga tac gcc tac ttc aat ggc |
| Cx43K288R | ctc ctc ctg ggt aca ggc tgg tta ctg g |
| Cx43K303R | gcc gca att aca aca ggc aag cta gcg agc |
| Cx43K346,347R | gac aac cag aat gcc aga aga gtt gct gct gg |
Tagged constructs were produced by PCR amplification with Accuzyme (Bioline) using wild-type Cx43 and Cx43 lysine to arginine Mutants 1–3 as the templates. The ER retention tag RRRRISLS was added by two consecutive rounds of PCR amplification onto the 3′-end of each using the 5′ primer GTA TGG ATC CAT GGG TGA CTG GAG TGC CTT and two different 3′ primers: the 3′ primer TGA AAT TCT TCT TCT TCT AAT CTC CAG GTC ATC AG first and then the 3′ primer ACG AAT TCT TAT GAC AGT GAA ATT CTT CTT CT second. PCR products were subcloned into pcDNA3.1 using the BamHI and EcoRI restriction sites and sequenced to confirm their authenticity.
Other plasmids used were pMT-HA-ubiquitin (from Dirk Bohmann, University of Rochester Medical Center, Rochester, NY) (55), pTrcHis-CIP75FL (54), pET-CIP75ΔUbL (52), pET-CIP75ΔUBA (52), pcDNA-FLAG-CIP75 (52), and pcDNA-Cx43HKKSL (53).
Tissue Blots
For the Northern tissue blot, a region from the CIP75 3′-UTR (bp 1981–2220 based on the sequence for the UBIN mRNA, NCBI accession number AB0400050) was PCR-amplified with Accuzyme using the 5′ primer AGG GGA TCC ATT CGG GTC ACC GTC AAG and 3′ primer TGG GAA TTC AGC AGC AGC AGT CAC TGG. The probe was transcribed using this template and used on the First Choice Northern blot Mouse Blot I (Ambion) using the DIG Northern Starter Kit (Roche Applied Science) according to the manufacturer's instructions. For the Western tissue blot, the Cruz Blot-H (Santa Cruz Biotechnology, Inc.) mouse tissue blot was obtained and immunoblotted using the CIP75 M398 monoclonal antibody.
Co-immunoprecipitations/Pull-downs
For co-immunoprecipitation of proteins in the HeLa-Cx43HKKSL stable cell line, cells were grown to confluence and then lysed in 0.2% Nonidet P-40 lysis buffer (0.2% Nonidet P-40, 150 mm NaCl, 20 mm Tris-HCl, pH 8.0, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 2 mm PMSF, 1 mm benzamidine, 2 mm N-ethylmaleimide). Cells were sonicated on ice 3 times for 5 s each, and the lysates clarified by centrifugation at 14,000 rpm (13,900 × g) for 30 min at 4 °C. Clarified supernatant proteins were precleared with Pierce Protein G-agarose (Thermo) overnight at 4 °C and then immunoprecipitated with the Cx43 P2D12 monoclonal antibody or CIP75 M398 monoclonal antibody for 2–5 h at 4 °C. The immune complexes were collected with Protein G-agarose and washed three times with Nonidet P-40 lysis buffer, and the proteins were released from the agarose by boiling for 5 min in SDS-PAGE sample buffer. The proteins were analyzed by SDS-PAGE and immunoblotting for Cx43 and CIP75. This analysis was repeated using HeLa cells that were transiently transfected with pcDNA-Cx43HKKSL, pcDNA-Cx43RRRRISLS, or each of the Cx43 lysine mutants.
For co-immunoprecipitation of proteins in cells treated with various degradation inhibitors, cells were grown to confluence; treated with the proteasome inhibitor lactacystin (20 μm) (Sigma), the proteasome inhibitor bortezomib (0.5 μm) (LC laboratories), or the lysosome inhibitor NH4Cl (50 mm) (Sigma) for 5 h; and then lysed in Nonidet P-40 lysis buffer for CIP75 or RIPA buffer (150 mm NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 10 mm Tris, pH 7.2, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 2 mm PMSF, 1 mm benzamidine, 2 mm N-ethylmaleimide) for p27 immunoprecipitations. The subsequent steps were performed as described above.
For the sequential immunoprecipitation experiments, complexes immunoprecipitated with the CIP75 M398 antibody for 2 h at 4 °C were incubated in RIPA buffer for 1–2 h at 4 °C to disrupt protein complexes. Released proteins were then immunoprecipitated with either the Cx43 P2D12 or HA antibodies, and the immune complexes were collected with Protein G-agarose overnight at 4 °C. The immune complexes were washed three times with 0.2% Nonidet P-40 lysis buffer, and the proteins were released from the agarose by boiling for 5 min in SDS-PAGE sample buffer. The proteins were analyzed by SDS-PAGE and immunoblotting for HA, Cx43, or CIP75.
For in vitro His pull-down assays, His-CIP75wt, His-CIP75ΔUbL, and His-CIP75ΔUBA were expressed in Escherichia coli BL21 cells grown to log phase. After induction with 2 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h, the cells were harvested, resuspended in PBS supplemented with 2 mm PMSF, and sonicated on ice six times for 25 s each, and lysates were clarified by centrifugation. Lysates with His-CIP75 were incubated with Talon cobalt beads (Agilent) in binding buffer (50 mm NaH2PO4, 500 mm NaCl, 5% glycerol, 0.3% Nonidet P-40) for 2 h at 4 °C. After washing the beads, 1 μg of GST-monoubiquitin or Lys48-linked tetraubiquitin (Enzo) was added and incubated with beads for 2 h at 4 °C. The beads were washed five times with binding buffer before boiling with sample buffer to release proteins. The proteins were resolved on SDS-containing 10% polyacrylamide gels and analyzed by immunoblotting with His, GST, ubiquitin, or CIP75 antibodies. All co-immunoprecipitation and pull-down experiments were performed a minimum of three times, and representative experiments are shown.
Laser-scanning Confocal Microscopy
HeLa cells were transiently transfected with pcDNA-FLAG-CIP75 and/or pcDNA-Cx43HKKSL, pcDNA-Cx43RRRRISLS, and all Cx43 lysine mutants. 24–48 h after transfection, cells were fixed in cold 80% methanol, 20% acetone for 30 min at −20 °C and washed three times with 0.1% Triton X-100 in PBS (PBSTx) for 5 min each. The cells were then blocked with 5% normal goat serum and 1% BSA in PBSTx for 30 min prior to incubation with rabbit Cx43 antibody, mouse IgG2a CIP75 L64 antibody, and mouse IgG1 calnexin antibody in blocking solution for 1 h. After three 5-min washes with PBSTx, the cells were incubated with goat anti-rabbit Alexa 594, goat anti-mouse IgG2a Alexa 488, and goat anti-mouse IgG1 Alexa 647-conjugated secondary antibodies (Invitrogen) for 1 h and then washed three times with PBS. The cells were mounted on slides with Prolong antifade reagent (Invitrogen), and the subcellular localization of Cx43, CIP75, and calnexin was visualized using the Leica ×63 HCX PL Apo oil immersion objective (numerical aperture 1.4) on a Leica TCS SP5 AOBS confocal microscope.
Statistical Analysis
ImageJ, GraphPad Prism 5.0, and SigmaPlot 9.0 were used for Western blot quantification and the statistical analysis of the amounts of Cx43 associated with CIP75, relative to CIP75 immunoprecipitates, using independent Student's t tests. These data are represented as the -fold change in CIP75-associated Cx43 compared with control cells and expressed as the mean ± S.E. of three or more independent experiments.
RESULTS
CIP75 Is Expressed in Multiple Tissues
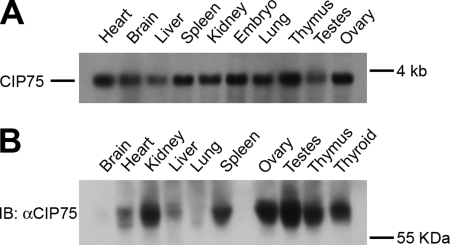
We have previously demonstrated that CIP75 interacts with Cx43 (52). To further characterize CIP75, we examined the expression of CIP75 mRNA in mouse tissues by Northern blot, using a RNA probe containing a region of the CIP75 3′-UTR that has a unique sequence to avoid detection of other members of the UbL-UBA family. Using a blot containing mRNA from various mouse tissues, a 3.5 kb band corresponding to the CIP75 mRNA was detected in all tissues examined to varying degrees, with the highest levels of expression in the heart and thymus; moderate levels of expression in the brain, spleen, kidney, lung, and ovary; and lower levels of expression in the liver and testes (Fig. 1A). CIP75 mRNA was also detected at high levels in whole embryo extracts. We also examined the expression of the CIP75 protein in mouse tissues by Western blot. Using a commercial blot containing protein lysates from various mouse tissues, CIP75 was detected in all tissues examined to varying degrees, with the highest levels of expression in the testes, ovary, thymus, thyroid, and kidney; moderate levels of expression in the spleen, heart, and liver; and low levels of expression in the brain and lung (Fig. 1B). Interestingly, although most of the mouse tissues contained comparable levels of the CIP75 transcript, several tissues did not express the levels of CIP75 protein predicted by the mRNA levels. In particular, brain and lung tissues contained lower amounts of CIP75 protein than the thymus and kidney while having similar CIP75 mRNA levels. Also, the testes contained higher levels of CIP75 protein while expressing lower levels of CIP75 mRNA. However, overall, CIP75 is ubiquitously transcribed and expressed in murine tissues.
FIGURE 1.
The expression of CIP75 in mouse tissues. A, a DIG-labeled RNA probe to the 3′-UTR of CIP75 was used to probe a commercial membrane containing 2 μg/lane of poly(A) RNA isolated from mouse tissues. A 3.5-kb CIP75 mRNA is expressed in all tissues, to varying degrees. B, a commercial membrane containing 50 μg of protein lysates isolated from mouse tissues was immunoblotted (IB) for CIP75. The CIP75 protein was expressed in all tissues, to varying degrees.
CIP75 Interacts with ER-localized Cx43
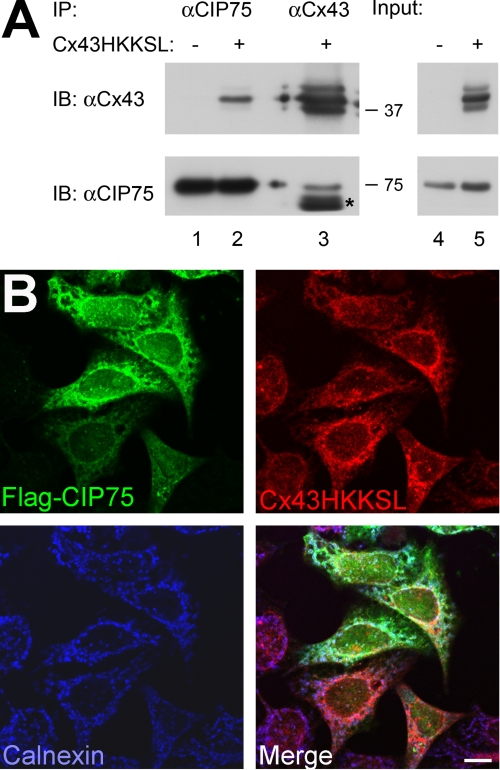
CIP75 has a role in the degradation of Cx43, possibly through ERAD. We have previously demonstrated that CIP75 co-localizes with Cx43 in or near the ER (52). To determine whether CIP75 does in fact interact with Cx43 located in the ER, the Cx43HKKSL protein was used. HKKSL is an ER retention motif that has previously been used to study the proper folding and oligomerization of Cx43 and its trafficking through the ER and Golgi (53, 56, 57). A HeLa cell line stably expressing Cx43HKKSL was used to assess whether this would enhance the interaction between Cx43 and CIP75. Whole cell lysates were immunoprecipitated with either CIP75 or Cx43 antibodies. The CIP75 immunoprecipitation revealed the presence of Cx43HKKSL (Fig. 2, lane 2), and the reciprocal Cx43 immunoprecipitation contained CIP75 (Fig. 2, lane 3). These results demonstrate that CIP75 interacted with the ER-retained Cx43HKKSL. Immunoprecipitation of CIP75 from a non-Cx43-expressing HeLa cell line was used as the negative control (Fig. 2, lane 1).
FIGURE 2.
CIP75 interacts with ER-localized Cx43. A, co-immunoprecipitation of CIP75 and Cx43 in HeLa and HeLa-Cx43HKKSL cells. Immunoprecipitated CIP75 interacts with ER-retained Cx43 (A, lane 2), which is not seen in cells that do not express Cx43 (A, lane 1). Immunoprecipitated Cx43 containing an ER retention motif (HKKSL) is able to interact with CIP75 (A, lane 3). The presence of Cx43 does not affect CIP75 expression. Input levels represent 2% of the amount of lysates used for immunoprecipitations. Migration positions of the molecular mass markers in kilodaltons are indicated at the right of the co-immunoprecipitation panels. *, IgG heavy chain. B, HeLa-Cx43HKKSL cells were transiently transfected with FLAG-CIP75. The subcellular localization of CIP75 (green) and Cx43HKKSL (red) with the ER marker calnexin (blue) was visualized by LSCM. Cx43HKKSL localizes to the ER, together with the ER marker calnexin, and CIP75 co-localizes with Cx43HKKSL. Scale bar, 10 μm. IP, immunoprecipitation; IB, immunoblot.
To confirm these results, HeLa-Cx43HKKSL cells were transiently transfected with FLAG-CIP75 and labeled with antibodies against Cx43, CIP75, and the ER-resident protein calnexin. Subcellular co-localization was examined using laser-scanning confocal microscopy. Cx43HKKSL was observed to localize to the ER, as previously published (53, 56, 57), co-localizing with calnexin (Fig. 2, C and D). Overexpressed CIP75 was found to co-localize with the ER-localized Cx43HKKSL (Fig. 2, B, C, and E), supporting the interaction of CIP75 and Cx43HKKSL demonstrated by the biochemical co-immunoprecipitation assays. Thus, CIP75 interacted with Cx43 that is located in the ER compartment.
CIP75 Interacts with Mono- and Tetraubiquitin in Vitro
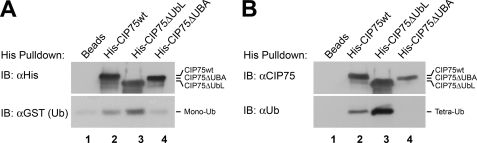
The UbL-UBA family of proteins has been shown to be a part of the ubiquitin-binding protein superfamily. The UBA domain, in particular, is a known ubiquitin-binding domain (42, 44, 45). To determine whether the CIP75 UBA domain is also able to bind to ubiquitin, in vitro binding studies utilizing purified ubiquitin were conducted using either GST-tagged monoubiquitin or Lys48-linked tetraubiquitin chains. Mono- and tetraubiquitin serve different functions by targeting proteins to different pathways. Monoubiquitination is primarily associated with protein endocytosis and targeting to multivesicular bodies, as has been demonstrated for the internalization of Cx43 from the plasma membrane (58, 59). By contrast, Lys48-linked tetraubiquitin is more typically associated with substrate proteins that are marked for proteasomal degradation (31, 32). Members of the UbL-UBA family have been demonstrated to interact with mono- and/or tetraubiquitin to varying degrees (46–48). We used an in vitro assay to assess the ability of CIP75 to interact with mono- and tetraubiquitin. His-tagged wild-type CIP75 and CIP75 mutants deleted for either the UbL domain (CIP75ΔUbL) or the UBA domain (CIP75ΔUBA) were expressed in bacteria and then bound to cobalt beads. The ability of purified monoubiquitin or tetraubiquitin to bind to the immobilized wild-type CIP75 and/or the CIP75 deletion mutants was assessed by a co-precipitation assay, which was analyzed by immunoblot. Wild-type CIP75 interacted with both monoubiquitin (Fig. 3A, lane 2) and the Lys48-linked tetraubiquitin chain (Fig. 3B, lane 2). This interaction was dependent on the UBA domain because the CIP75ΔUbL deletion mutant also bound both monoubiquitin (Fig. 3A, lane 3) and Lys48-tetraubiquitin (Fig. 3B, lane 3), whereas the CIP75ΔUBA deletion mutant failed to bind to either type of ubiquitin (Fig. 3, A (lane 4) and B (lane 4)). Thus, as is characteristic of other members of the UbL-UBA protein family, CIP75 was able to bind to both monoubiquitin and Lys48-linked tetraubiquitin via its UBA domain.
FIGURE 3.
CIP75 interacts with monoubiquitin and Lys48-linked tetraubiquitin chain in vitro. A and B, pull-down of His-tagged wild-type CIP75 (CIP75wt), CIP75 deleted for the N-terminal UbL domain (CIP75ΔUbL), and CIP75 deleted for the C-terminal UBA domain (CIP75ΔUBA), with beads alone as a negative control. Interaction of CIP75 and the deletion mutants with GST-tagged monoubiquitin (A) and untagged tetraubiquitin (B) were examined. CIP75wt and CIP75ΔUbL were able to interact with both mono- (A, lanes 2 and 3) and tetraubiquitin (Tetra-UB) (B, lanes 2 and 3), whereas CIP75ΔUBA was not (A and B, lane 4). IB, immunoblot.
CIP75 Interacts with Ubiquitinated Proteins Bound for Proteasomal Degradation
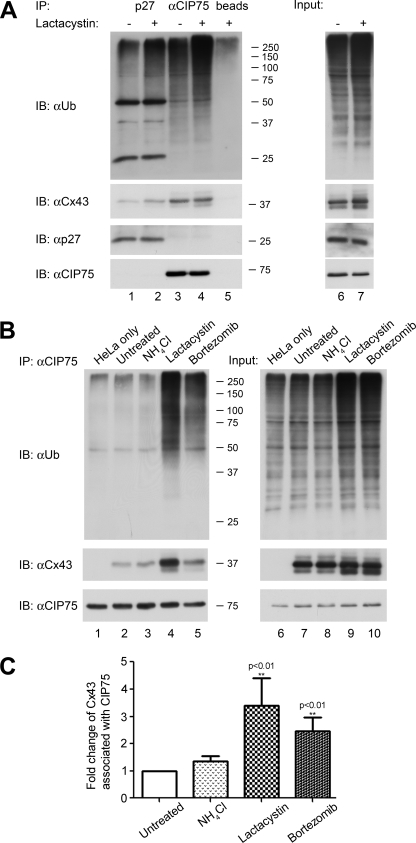
To determine whether the CIP75-ubiquitin interaction, demonstrated in vitro, could be extended to an interaction with ubiquitinated proteins, immunoprecipitation of whole cell lysates with the CIP75 antibody was performed. Using HeLa-Cx43HKKSL cells with or without a 5-h treatment with the proteasome inhibitor lactacystin, CIP75 was immunoprecipitated, and the associated proteins were examined by immunoblot for Cx43 or ubiquitinated proteins using the Cx43 or ubiquitin antibodies, respectively. As expected, lactacystin treatment increased the total level of ubiquitinated proteins compared with the levels in untreated control cells (Fig. 4A, top, lanes 6 and 7). As a positive control to ensure that we were able to specifically detect ubiquitinated proteins, p27 was studied because its ubiquitination has been well documented (60–63). p27 was immunoprecipitated, and these immune complexes were also probed with the ubiquitin antibody on the immunoblot. There was an increase in the ubiquitin signal in the proteins from lactacystin-treated cells, which was more readily apparent in the slower migrating polyubiquitinated proteins near the top of the blot. Moreover, p27 itself was also detected by the ubiquitin antibody (Fig. 4A, top, lanes 1 and 2). CIP75 interacted with ubiquitinated proteins as indicated by the multitude of proteins of varying sizes detected with the ubiquitin antibody (Fig. 4A, top, lane 3), and this interaction increased with lactacystin treatment (Fig. 4A, top, lane 4). The level of ubiquitinated proteins detected exceeded the nonspecific binding found in the immunoprecipitation control using the agarose beads only (Fig. 4A, top, lane 5), which indicated that the ubiquitin signal associated with the CIP75 immunoprecipitates was authentic. Interestingly, the p27 immunoprecipitate appeared to contain a protein migrating similarly to Cx43 (Fig. 4A, middle, lanes 1 and 2). However, this was not a reproducible result because use of a different Cx43 antibody failed to detect co-immunoprecipitated Cx43, indicating that this signal was not a specific interaction (data not shown). These results were replicated using cells treated with another proteasome inhibitor, bortezomib, for 5 h (data not shown). Thus, these combined results indicated that CIP75 can interact with both ER-localized Cx43 and ubiquitinated proteins. The observation that blocking proteasomal degradation increased the interaction of CIP75 with ubiquitinated proteins underscored a possible role for CIP75 in the proteasomal degradation of cellular proteins.
FIGURE 4.
CIP75 interacts with ubiquitinated proteins. A, immunoprecipitation of p27 and CIP75 in untreated or lactacystin-treated HeLa-Cx43HKKSL cells. Lactacystin treatment increases the levels of immunoprecipitated ubiquitinated proteins (A, top, lanes 1 and 3 compared with lanes 2 and 4). CIP75 is not affected by lactacystin treatment (A, bottom, lanes 3 and 4). B, immunoprecipitation of CIP75 in untreated HeLa or HeLa-Cx43HKKSL cells and HeLa-Cx43HKKSL cells that were treated with NH4Cl, lactacystin, or bortezomib. Immunoprecipitated CIP75 is able to interact with ubiquitinated proteins regardless of Cx43 expression (B, top, lanes 1–5). Treatment with proteasomal inhibitors increases this interaction (B, top, lanes 4 and 5) compared with untreated cells or treatment with a lysosomal inhibitor (B, top, lanes 2 and 3). CIP75 interaction with Cx43 is also increased in cells treated with the proteasomal inhibitors (B, middle, lanes 4 and 5), but CIP75 is not affected by the various treatments (B, bottom, lanes 2–5). Input levels represent 2% of the amount of lysates used for immunoprecipitations. Migration positions of the molecular mass markers in kilodaltons are indicated at the right of the co-immunoprecipitation panels. The amount of proteins in the input material is shown in the right panels of A and B. C, Cx43 levels found in the co-immunoprecipitation assays were quantified with ImageJ and analyzed relative to the immunoprecipitated CIP75 levels with GraphPad Prism 5.0 and SigmaPlot 9.0 software. The data represent the mean of at least four independent experiments ± S.E. (n = 10 for untreated cells, n = 4 for NH4Cl-treated cells, n = 6 for lactacystin-treated cells, n = 8 for bortezomib-treated cells). **, -fold change in Cx43 levels relative to CIP75 with statistically significant difference (p < 0.01) compared with the untreated control cells. IP, immunoprecipitation; IB, immunoblot.
To confirm that CIP75 is acting specifically to regulate proteasomal degradation versus lysosomal degradation, changes in the interaction between CIP75 and ubiquitinated proteins were examined in HeLa-Cx43HKKSL cells treated with either a proteasome inhibitor (lactacystin or bortezomib) or a lysosomal inhibitor (NH4Cl) for 5 h. CIP75 was immunoprecipitated from treated whole cell lysates and compared with immunoprecipitates from untreated cells as well as untreated, non-Cx43-expressing HeLa cells. As previously demonstrated, inhibiting proteasome degradation with lactacystin or bortezomib increased the amount of ubiquitinated proteins that interacted with CIP75 (Fig. 4B, top, lanes 4 and 5). This stimulation was specific to proteasome inhibitors because inhibiting lysosomal degradation with NH4Cl did not exhibit the same effect. CIP75 appeared to have the same level of association with ubiquitinated proteins in cells expressing Cx43HKKSL versus cells that were not expressing Cx43 (Fig. 4B, top, lanes 1 and 2), which confirmed the previous observations that CIP75 is probably interacting with ubiquitinated proteins beyond Cx43.
The amount of Cx43 interacting with CIP75 also appeared to increase after treatment with lactacystin and bortezomib. Statistical analysis was conducted on the amount of Cx43 associated with CIP75, compared with the relative amount of CIP75, and the -fold change in this association compared with untreated cells was calculated in the treated cells. Using Student's t test to determine significance, proteasomal inhibition with both lactacystin and bortezomib resulted in statistically significant increases in Cx43HKKSL association with CIP75 (p < 0.01), whereas lysosomal inhibition with NH4Cl did not result in a statistically significant change (Fig. 4C). This quantitative analysis supports our previous finding that CIP75 is involved in the proteasomal degradation of Cx43 (52).
CIP75 Interacts with Non-ubiquitinated Cx43
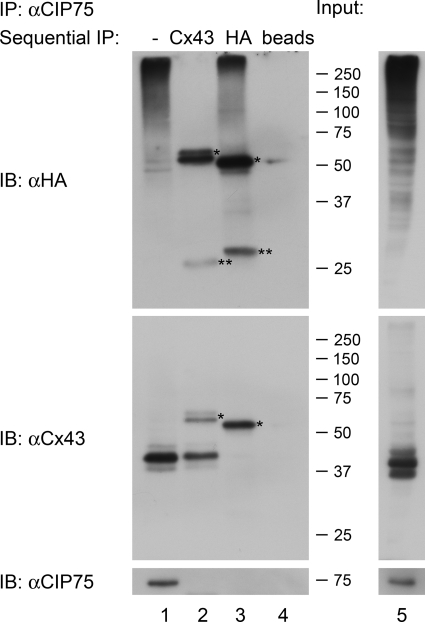
The previous results indicated that CIP75 interacts with ubiquitinated cellular proteins and with Cx43. However, the ubiquitination state of Cx43 specifically interacting with CIP75 was uncertain. Previous studies have demonstrated that Cx43 is ubiquitinated and that this occurs in various stages of its life cycle (17, 18, 58, 64, 65). Recently, ubiquitination of Cx43 was found to occur when plasma membrane-localized Cx43 is internalized in response to 12-O-tetradecanoylphorbol-13-acetate treatment (58, 59). To establish if CIP75-interacting Cx43 is ubiquitinated, a sequential immunoprecipitation assay was performed. In this assay, HeLa-Cx43HKKSL cells were transiently transfected with HA-tagged ubiquitin for 48 h and then treated with lactacystin for 5 h to increase the levels of associated ubiquitinated protein. CIP75 was first immunoprecipitated from the transfected HeLa-Cx43HKKSL whole cell lysates after lactacystin treatment. This first immunoprecipitation was performed in a less stringent Nonidet P-40 lysis buffer to preserve interactions of proteins associated with CIP75, as performed in earlier co-immunoprecipitation experiments. After the first immunoprecipitation, the Protein G-agarose beads containing the CIP75 antibody and CIP75-protein complexes were subjected to the RIPA buffer containing SDS, which would disrupt protein-protein interactions. After releasing proteins from any CIP75-protein complexes, the proteins were then immunoprecipitated a second time, using either the Cx43 antibody or the HA antibody to extract ubiquitinated proteins. Both sets of immunoprecipitations were then analyzed by immunoblot with Cx43 or HA antibodies. As controls, the agarose beads with the first immunoprecipitate incubated in RIPA buffer as well as a beads alone negative control for the second immunoprecipitation were used. Incubation of protein complexes in RIPA buffer released almost all proteins (data not shown), whereas the agarose beads alone negative control did not isolate any proteins (Fig. 5, lane 4). The CIP75 immunoprecipitation contained Cx43 and various ubiquitinated proteins (Fig. 5, top and middle, lane 1). The Cx43 sequential immunoprecipitation revealed Cx43 (Fig. 5, middle, lane 2); however, significantly, ubiquitinated protein was not detected (Fig. 5, top, lane 2). Conversely, the HA sequential immunoprecipitation revealed ubiquitinated proteins (Fig. 5, top, lane 3); however, this immunoprecipitation did not isolate Cx43 (Fig. 5, middle, lane 3). The only signal detected with the HA immunoprecipitation in the Cx43 immunoblot was the IgG heavy chain (Fig. 5, middle, lane 3). Likewise, the only signal detected with the Cx43 immunoprecipitation in the HA immunoblot was the IgG heavy and light chains (Fig. 5, top panel, lane 2). Together, these results suggested that although CIP75 does interact with ubiquitinated proteins, the subset of ER-localized Cx43 that interacts with CIP75 is not ubiquitinated.
FIGURE 5.
CIP75 interacts with non-ubiquitinated Cx43. Sequential immunoprecipitation of ubiquitinated protein and Cx43HKKSL that interacts with CIP75 in lactacystin-treated HeLa-Cx43HKKSL cells. CIP75 was first immunoprecipitated from cellular lysates. Immune complexes were subsequently disrupted using RIPA buffer and subjected to a second immunoprecipitation of Cx43 or HA-tagged ubiquitinated proteins to isolate the subset of proteins that interact with CIP75. CIP75 interacts with Cx43HKKSL and HA-ubiquitin-tagged proteins (top and middle, lane 1). Cx43HKKSL that was initially co-immunoprecipitated with CIP75 is not ubiquitinated (top and middle, lanes 2 and 3), whereas ubiquitinated proteins that were initially co-immunoprecipitated with CIP75 are separately recovered (top, lane 3). CIP75 is not recovered after the sequential immunoprecipitation (bottom, lanes 2 and 3). *, IgG heavy chain; **, light chain. Migration positions of the molecular mass markers in kilodaltons are indicated at the right of the co-immunoprecipitation panels. Protein inputs are shown in the right panels and represent 2% of the amount of lysates used for immunoprecipitations. IP, immunoprecipitation; IB, immunoblot.
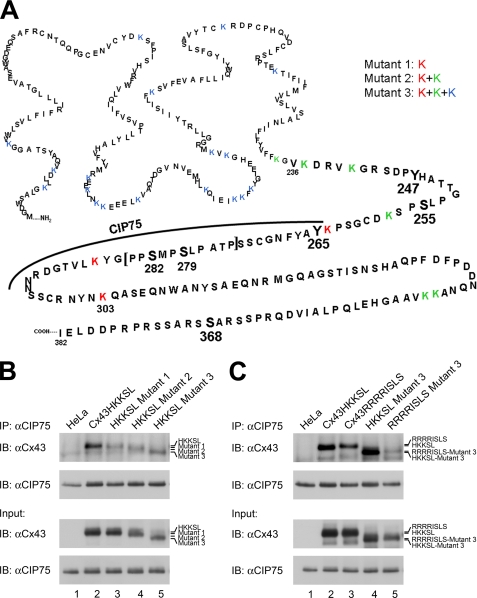
To validate this critical and surprising finding, Cx43 mutants that lacked lysine ubiquitination sites were generated. In the process of ubiquitination, ubiquitin moieties are covalently bound to lysine residues of target substrates. A substitution of arginine in the place of lysine blocks the attachment of ubiquitin. Because we do not know which residues in Cx43 are ubiquitinated, three separate sets of Cx43HKKSL arginine mutants were generated (Fig. 6A). We previously identified the region of Cx43 that interacted with CIP75 to be within the Lys264–Asn302 region in the C-terminal tail (52, 66). Thus, Mutant 1 contained three point mutations, K264R, K287R, and K303R, to residues that are located in or immediately adjacent to the CIP75 interaction domain. Mutant 2 contained five additional mutations, resulting in the mutation of all lysines to arginines in the Cx43 C-terminal tail. In Mutant 3, every lysine residue in Cx43 was mutated to arginine.
FIGURE 6.
CIP75 interacts with Cx43 that lacks lysine ubiquitination sites. A, site-directed mutagenesis was performed to mutate lysine residues to arginine to eliminate potential ubiquitination sites. Mutant 1 consists of mutations K264R, K287R, and K303R (red) because these wild-type residues appear to be involved or near those that are involved in CIP75 binding. Mutant 2 consists of the same mutations in Mutant 1 plus the additional mutations K236R, K239R, K243R, K345R, and K346R (green). Mutant 2 represents mutations of all lysine residues in the C-terminal tail. In Mutant 3, all lysine residues in Cx43 are mutated (blue). B and C, co-immunoprecipitation of transiently transfected HeLa cells. B, HeLa cells were transfected with either Cx43HKKSL, Mutant 1, Mutant 2, or Mutant 3. Untransfected HeLa cells were used as the negative control. Immunoprecipitated CIP75 interacted with Cx43HKKSL (B, lane 2), Mutant 1 (B, lane 3), Mutant 2 (B, lane 4), and Mutant 3 (B, lane 5), suggesting that ubiquitination of ER-localized Cx43 is not necessary for the interaction with CIP75. B, HeLa cells were transfected with either Cx43HKKSL, Cx43RRRRISLS, Cx43HKKSL Mutant 3, or Cx43RRRRISLS Mutant 3. CIP75 can also interact with the ER-retained wild-type Cx43RRRRISLS (C, lane 3) and Cx43RRRRISLS Mutant 3 (C, lane 5), which does not contain any lysines for ubiquitination. Input levels represent 2% of the amount of lysates used for immunoprecipitations. IP, immunoprecipitation; IB, immunoblot.
Cx43HKKSL and lysine Mutants 1–3 were transiently transfected into non-Cx43-expressing HeLa cells. Whole cell lysates from transfected cells were immunoprecipitated using the CIP75 antibody and subsequently immunoblotted for Cx43 to determine whether CIP75 could still interact with the various lysine mutants. Immunoprecipitated CIP75 interacted with Cx43HKKSL (Fig. 6B, lane 2), lysine Mutant 1 (Fig. 6B, lane 3), lysine Mutant 2 (Fig. 6B, lane 4), and lysine Mutant 3 (Fig. 6B, lane 5). Untransfected HeLa cells were used as a negative control (Fig. 6B, lane 1). These results confirmed that Cx43 does not require ubiquitin modification(s) in order to interact with CIP75. However, the ER retention tag HKKSL contains two additional lysine residues. To ensure that ubiquitination of these two remaining lysine residues was not contributing to the interaction of the ER-localized Cx43 with CIP75, an alternate ER retention tag, RRRRISLS, was substituted for the HKKSL tag (67). Wild-type Cx43 and lysine Mutant 3 were both generated containing the alternate RRRRISLS tag. Cx43HKKSL, Cx43HKKSL Mutant 3, Cx43RRRRISLS, and Cx43RRRRISLS Mutant 3 were transiently transfected into non-Cx43-expressing HeLa cells. CIP75 was again immunoprecipitated from whole cell lysates, with untransfected HeLa cells used as a negative control (Fig. 6C, lane 1). Compared with Cx43HKKSL (Fig. 6C, lane 2) and the Cx43HKKSL Mutant 3 (Fig. 6C, lane 4), Cx43RRRRISLS (Fig. 6C, lane 3) and the Cx43RRRRISLS Mutant 3 (Fig. 6C, lane 5) were still able to interact with CIP75, indicating that the HKKSL tag was not aberrantly mediating interaction with CIP75. These data further supported the conclusion that CIP75 interacts with non-ubiquitinated Cx43. Inspection of the Cx43 immunoblots appeared to suggest possible differences in the association of CIP75 with the lysine mutants. However, differences in band intensities on the immunoblots were probably due to variations in transfection efficiencies of the transient transfections. To establish this, quantitative analyses were performed on the levels of Cx43 associated with immunoprecipitated CIP75, normalized to the relative levels of Cx43, similar to the statistical analysis previously performed for the Cx43-CIP75 association after degradation inhibitor treatment. We performed Student's t test on the difference in association of the various Cx43 proteins (wild-type Cx43HKKSL or Cx43RRRRISLS or the lysine mutants) from CIP75 and found that in all cases, the p values were greater than 0.1. Thus, there was no significant difference in the binding between CIP75 and the wild-type Cx43HKKSL, compared with any of the lysine mutants, and no significant difference in binding between CIP75 and wild-type Cx43RRRRISLS, compared with the corresponding Mutant 3.
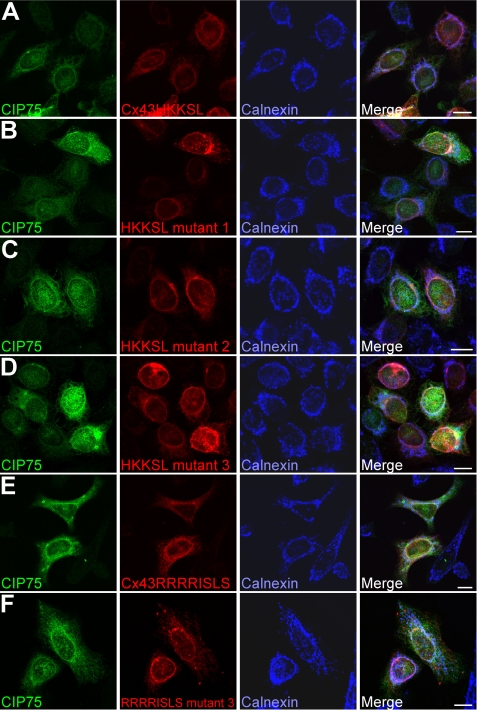
Finally, non-Cx43-expressing HeLa cells were transiently co-transfected with FLAG-CIP75 and either Cx43HKKSL, each of the three corresponding lysine mutants, Cx43RRRRISLS, or the corresponding lysine Mutant 3. These cells were labeled with antibodies against CIP75, Cx43, and calnexin and visualized by laser scanning confocal microscopy. As expected, Cx43HKKSL (Fig. 7A), Cx43HKKSL Mutant 1 (Fig. 7B), Cx43HKKSL Mutant 2 (Fig. 7C), Cx43HKKSL Mutant 3 (Fig. 7D), Cx43RRRRISLS (Fig. 7E), and Cx43RRRRISLS Mutant 3 (Fig. 7F) localized to the ER, along with the ER marker, calnexin. Transfected FLAG-CIP75 was found partially co-localized with Cx43HKKSL (Fig. 7A), Cx43RRRISLS (Fig. 7E), and all of the lysine mutants (Fig. 7, B–D and F). These combined results further indicated that the ubiquitination of Cx43 is not a requirement for its interaction with CIP75.
FIGURE 7.
CIP75 co-localizes with wild-type and mutant Cx43. HeLa cells were transiently co-transfected with FLAG-CIP75 and ER-retained Cx43 (wild-type (A and E) or lysine mutants (B–D and F)). The subcellular localization of CIP75 (green) and Cx43 (red) with the ER marker calnexin (blue) was visualized by LSCM. CIP75 co-localizes with Cx43, regardless of the extent of lysine to arginine mutations. CIP75 and Cx43 co-localization occurs in an ER-like pattern. Scale bar, 10 μm.
DISCUSSION
In this work, we found that CIP75 interacted specifically with ER-localized Cx43 through biochemical assays and immunofluorescence microscopy studies. Our previous data had suggested a role for CIP75 in Cx43 proteasomal degradation (52). We have now demonstrated an increased association of CIP75 and ER-localized Cx43 after inhibition of proteasomal degradation but not after inhibition of lysosomal degradation. This observation provides additional support for a role for CIP75 in the ERAD of Cx43. Proteasomal degradation typically requires a ubiquitin tag (thought to specifically be at least a Lys48-linked tetraubiquitin chain) covalently attached to target substrates, which marks these proteins for degradation via the proteasome (31, 32). However, our sequential immunoprecipitation studies demonstrated that although CIP75 is a ubiquitin-binding protein, it did not appear to interact with ubiquitinated Cx43. Furthermore, the lack of ubiquitination of the Cx43 lysine to arginine mutants did not affect their ability to interact with CIP75 in the biochemical co-immunoprecipitation and immunofluorescence microscopy experiments. These results thus present a rare case of a non-ubiquitinated protein that is targeted for proteasomal degradation. There are only a few proteins known to be degraded by the proteasome in the absence of ubiquitination (68). The most conclusively demonstrated example is the enzyme ornithine decarboxylase, which is not ubiquitinated and yet undergoes proteasomal degradation (69–71). Our results are surprising because Cx43 ubiquitination has been previously well documented (17, 58, 64, 65). However, we cannot exclude other mechanisms that might facilitate the proteasomal degradation of ubiquitinated Cx43 that do not involve CIP75. The precise subcellular location(s) of Cx43 ubiquitination has not been thoroughly resolved. However, previous studies on this issue have suggested that the ubiquitination of Cx43 occurs primarily as Cx43 is trafficked from the plasma membrane (58, 59, 65). Thus, it is conceivable that Cx43 that is in the ER and subjected to ERAD is not ubiquitinated, which would be consistent with our results.
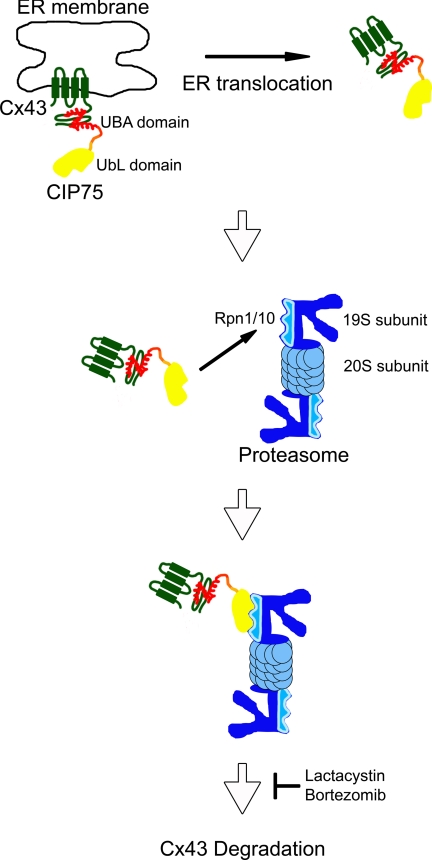
A “shuttle hypothesis” has been proposed for other members of the UbL-UBA protein family, such as Rad23, where these proteins may act to shuttle target substrates to the proteasome for degradation (33, 72). Based on this and our previous and current results, we propose a model for the role of CIP75 in facilitating Cx43 degradation where CIP75 interacts with non-ubiquitinated Cx43 in the ER and then acts as a shuttle to transport Cx43 from the ER to the proteasome for degradation, through ERAD (Fig. 8). CIP75 would bind to the C-terminal tail of ER-localized Cx43 with its UBA domain, perhaps aiding the translocation of Cx43 out of the ER membrane and then facilitating the transport of Cx43 to the proteasome, where the UbL domain of CIP75 interacts with the 19 S proteasome cap subunits, Rpn1 and Rpn10, leading to the proteasomal degradation of Cx43. Interestingly, no direct interaction has yet been reported between Cx43 and any proteasomal proteins. However, we have previously shown that CIP75 can interact with the proteasome proteins, Rpn1 and Rpn10, subunits of the 19 S cap subunit of the 26 S proteasome (52). The Rpn1 and Rpn10 19 S subunits have been reported to be involved in the recognition of proteasomal substrates (73–75). Thus, CIP75 may mediate the interaction between Cx43 and the proteasome and may represent a “missing link” in this process.
FIGURE 8.
Model of CIP75 facilitating the proteasomal degradation of Cx43. CIP75 may act a shuttle to transport non-ubiquitinated Cx43 from the ER to the proteasome for degradation. CIP75 would bind to the C-terminal tail of ER-localized Cx43 via the UBA domain, perhaps aid in translocating Cx43 out of the ER membrane into the cytosol, and then transport Cx43 to the proteasome, where CIP75 binds to the 19 S proteasome cap subunits Rpn1 and Rpn10 via the CIP75 UbL domain, and Cx43 is subsequently degraded.
Cx43 has been observed to be subject to ERAD. Studies from the laboratory of L. Musil (14, 30) have examined Cx43 degradation through ERAD. They have shown that the specific blockade of proteasome degradation resulted in an increase in Cx43 levels, an increase in gap junction assembly, and the corresponding increase in intercellular communication (14, 30). These effects were not observed with lysosomal inhibitors. These studies suggested that a portion of the newly synthesized Cx43 is degraded through ERAD while the rest is trafficked to the membrane and contributes to increased gap junction formation and intercellular communication. It has also been demonstrated that different types of cellular stresses result in different consequences in the trafficking of Cx43. Specific ER stresses, such as that induced by DTT treatment, result in increased translocation of Cx43 out of the ER and increased proteasomal degradation. However, cytosolic stresses, such as hyperthermia or sodium arsenite treatment, have the opposite effect, resulting in decreased Cx43 ER translocation, increased trafficking to the plasma membrane, and gap junction communication (30). Thus, the regulation of ERAD can result in changes in the level of Cx43, which may alter gap junctional communication. The discovery of a mechanism involving CIP75 in facilitating ER-localized Cx43 proteasomal degradation suggests that regulation of CIP75 activity, and hence Cx43 levels, may have a significant effect on gap junctional intercellular communication.
We also found that CIP75 can interact with a variety of unidentified ubiquitinated cellular proteins, and in vitro pull-down experiments demonstrated that CIP75 binds to both monoubiquitin and Lys48-linked tetraubiquitin chains via its UBA domain. Because Lys48-linked tetraubiquitin is the prevalent ubiquitin marker for proteasome degradation (31, 32) and because the interaction of CIP75 with ubiquitinated proteins in cells increases upon inhibition of proteasomal degradation, it is possible that CIP75 may regulate the proteasomal degradation of substrates other than Cx43. We have recently demonstrated that the amino acid residues that interact with Cx43 are not the same as the residues that interact with free ubiquitin (66), further supporting the notion that CIP75 may interact with other ubiquitinated substrates bound for proteasome degradation. Other members of the UbL-UBA family interact with multiple substrates (33), suggesting that this family of proteins may have a widespread role in cellular homeostasis, specifically through the regulation of proteasomal degradation. CIP75 is ubiquitously transcribed and expressed in all murine tissues we examined via Northern and Western blots, albeit to varying degrees, supporting a possible ubiquitous activity of CIP75. Interestingly, the levels of CIP75 mRNA did not always correlate with the levels of CIP75 protein found in the various tissues, such as in the brain and lung tissues, suggesting a perhaps more complex mechanism that regulates the translation of the CIP75 protein from the mRNA. We also observed that CIP75 is able to bind to free monoubiquitin. Different forms of ubiquitination are thought to have distinct consequences. Whereas Lys48-linked tetraubiquitin typically marks a protein for proteasomal degradation, Lys63-linked ubiquitin chains are thought to be involved in signaling, and monoubiquitin has been reported to have a role in endocytosis (76). Given the ability of CIP75 to interact with monoubiquitin, CIP75 may have another function that is unrelated to its role in proteasomal degradation.
It is clear that there is still much to be discovered about CIP75 and the functional aspects of its interaction with Cx43 and, in general, the family of UbL-UBA proteins. Elucidating the mechanisms that regulate the function of CIP75 as a mediator of protein degradation via the proteasome will provide insights into how the cell is able to maintain proper levels of proteins employing the members of this UDP subfamily of proteins. In the case of Cx43, uncovering how CIP75 is regulating Cx43 levels may help to extend the understanding of the mechanisms that regulate Cx43 expression and therefore gap junction formation and intercellular communication. Because it is crucial for cell to cell communication to be tightly regulated, a better understanding of the various mechanisms that control different aspects of the Cx43 life cycle can lead to a better understanding of normal cellular function as well as the diseases caused by the misregulation of Cx43.
Acknowledgments
We thank Dirk Bohmann and Paul Lampe for providing molecular and immunological reagents. We would also like to thank Linda Musil, Wendy Kurata, Courtney Luke, Ken Takeuchi, and Dana-Lynn Koomoa for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grant CA052098 (to A. F. L.).
- ER
- endoplasmic reticulum
- UbL
- ubiquitin-like
- UBA
- ubiquitin-associated
- ERAD
- ER-associated degradation
- RIPA
- radioimmune precipitation assay.
REFERENCES
- 1.Goodenough D. A., Goliger J. A., Paul D. L. (1996) Annu. Rev. Biochem. 65, 475–502 [DOI] [PubMed] [Google Scholar]
- 2.Vinken M., Vanhaecke T., Papeleu P., Snykers S., Henkens T., Rogiers V. (2006) Cell. Signal. 18, 592–600 [DOI] [PubMed] [Google Scholar]
- 3.Wei C. J., Xu X., Lo C. W. (2004) Annu. Rev. Cell Dev. Biol. 20, 811–838 [DOI] [PubMed] [Google Scholar]
- 4.White T. W., Paul D. L. (1999) Annu. Rev. Physiol. 61, 283–310 [DOI] [PubMed] [Google Scholar]
- 5.Martin P. E., Evans W. H. (2004) Cardiovasc. Res. 62, 378–387 [DOI] [PubMed] [Google Scholar]
- 6.Severs N. J., Bruce A. F., Dupont E., Rothery S. (2008) Cardiovasc. Res. 80, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Veen A. A., van Rijen H. V., Opthof T. (2001) Cardiovasc. Res. 51, 217–229 [DOI] [PubMed] [Google Scholar]
- 8.Laird D. W. (2008) J. Biol. Chem. 283, 2997–3001 [DOI] [PubMed] [Google Scholar]
- 9.Naus C. C., Laird D. W. (2010) Nat. Rev. Cancer 10, 435–441 [DOI] [PubMed] [Google Scholar]
- 10.Beardslee M. A., Laing J. G., Beyer E. C., Saffitz J. E. (1998) Circ. Res. 83, 629–635 [DOI] [PubMed] [Google Scholar]
- 11.Fallon R. F., Goodenough D. A. (1981) J. Cell Biol. 90, 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darrow B. J., Laing J. G., Lampe P. D., Saffitz J. E., Beyer E. C. (1995) Circ. Res. 76, 381–387 [DOI] [PubMed] [Google Scholar]
- 13.Laird D. W., Puranam K. L., Revel J. P. (1991) Biochem. J. 273, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musil L. S., Le A. C., VanSlyke J. K., Roberts L. M. (2000) J. Biol. Chem. 275, 25207–25215 [DOI] [PubMed] [Google Scholar]
- 15.Beardslee M. A., Lerner D. L., Tadros P. N., Laing J. G., Beyer E. C., Yamada K. A., Kléber A. G., Schuessler R. B., Saffitz J. E. (2000) Circ. Res. 87, 656–662 [DOI] [PubMed] [Google Scholar]
- 16.Girão H., Pereira P. (2003) Mol. Vis. 9, 24–30 [PubMed] [Google Scholar]
- 17.Laing J. G., Beyer E. C. (1995) J. Biol. Chem. 270, 26399–26403 [DOI] [PubMed] [Google Scholar]
- 18.Laing J. G., Tadros P. N., Westphale E. M., Beyer E. C. (1997) Exp. Cell Res. 236, 482–492 [DOI] [PubMed] [Google Scholar]
- 19.Laird D. W. (2010) Trends Cell Biol. 20, 92–101 [DOI] [PubMed] [Google Scholar]
- 20.Barker R. J., Price R. L., Gourdie R. G. (2002) Circ. Res. 90, 317–324 [DOI] [PubMed] [Google Scholar]
- 21.Toyofuku T., Akamatsu Y., Zhang H., Kuzuya T., Tada M., Hori M. (2001) J. Biol. Chem. 276, 1780–1788 [DOI] [PubMed] [Google Scholar]
- 22.Toyofuku T., Yabuki M., Otsu K., Kuzuya T., Hori M., Tada M. (1998) J. Biol. Chem. 273, 12725–12731 [DOI] [PubMed] [Google Scholar]
- 23.Duffy H. S., Ashton A. W., O'Donnell P., Coombs W., Taffet S. M., Delmar M., Spray D. C. (2004) Circ. Res. 94, 215–222 [DOI] [PubMed] [Google Scholar]
- 24.Piehl M., Lehmann C., Gumpert A., Denizot J. P., Segretain D., Falk M. M. (2007) Mol. Biol. Cell 18, 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segretain D., Falk M. M. (2004) Biochim. Biophys. Acta 1662, 3–21 [DOI] [PubMed] [Google Scholar]
- 26.Bruce A. F., Rothery S., Dupont E., Severs N. J. (2008) Cardiovasc. Res. 77, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan Z., Kurata W. E., Martyn K. D., Jin C., Lau A. F. (2005) Biochemistry 44, 2385–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laing J. G., Tadros P. N., Green K., Saffitz J. E., Beyer E. C. (1998) Cardiovasc. Res. 38, 711–718 [DOI] [PubMed] [Google Scholar]
- 29.Qin H., Shao Q., Igdoura S. A., Alaoui-Jamali M. A., Laird D. W. (2003) J. Biol. Chem. 278, 30005–30014 [DOI] [PubMed] [Google Scholar]
- 30.VanSlyke J. K., Musil L. S. (2002) J. Cell Biol. 157, 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang S., Weissman A. M. (2004) Cell Mol. Life Sci. 61, 1546–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su V., Lau A. F. (2009) Cell Mol. Life Sci. 66, 2819–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. (2001) J. Mol. Biol. 313, 955–963 [DOI] [PubMed] [Google Scholar]
- 35.Biggins S., Ivanovska I., Rose M. D. (1996) J. Cell Biol. 133, 1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamitani T., Kito K., Fukuda-Kamitani T., Yeh E. T. (2001) J. Biol. Chem. 276, 46655–46660 [DOI] [PubMed] [Google Scholar]
- 37.Kim I., Mi K., Rao H. (2004) Mol. Biol. Cell 15, 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raasi S., Pickart C. M. (2003) J. Biol. Chem. 278, 8951–8959 [DOI] [PubMed] [Google Scholar]
- 39.Rao H., Sastry A. (2002) J. Biol. Chem. 277, 11691–11695 [DOI] [PubMed] [Google Scholar]
- 40.Schauber C., Chen L., Tongaonkar P., Vega I., Lambertson D., Potts W., Madura K. (1998) Nature 391, 715–718 [DOI] [PubMed] [Google Scholar]
- 41.Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., Finley D. (2002) Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. (2001) Nat. Cell Biol. 3, 939–943 [DOI] [PubMed] [Google Scholar]
- 43.Fujiwara K., Tenno T., Sugasawa K., Jee J. G., Ohki I., Kojima C., Tochio H., Hiroaki H., Hanaoka F., Shirakawa M. (2004) J. Biol. Chem. 279, 4760–4767 [DOI] [PubMed] [Google Scholar]
- 44.Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. (2001) Nat. Struct. Biol. 8, 417–422 [DOI] [PubMed] [Google Scholar]
- 45.Chen L., Madura K. (2002) Mol. Cell. Biol. 22, 4902–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raasi S., Orlov I., Fleming K. G., Pickart C. M. (2004) J. Mol. Biol. 341, 1367–1379 [DOI] [PubMed] [Google Scholar]
- 47.Raasi S., Varadan R., Fushman D., Pickart C. M. (2005) Nat. Struct. Mol. Biol. 12, 708–714 [DOI] [PubMed] [Google Scholar]
- 48.Zhang D., Raasi S., Fushman D. (2008) J. Mol. Biol. 377, 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brignone C., Bradley K. E., Kisselev A. F., Grossman S. R. (2004) Oncogene 23, 4121–4129 [DOI] [PubMed] [Google Scholar]
- 50.Glockzin S., Ogi F. X., Hengstermann A., Scheffner M., Blattner C. (2003) Mol. Cell. Biol. 23, 8960–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 52.Li X., Su V., Kurata W. E., Jin C., Lau A. F. (2008) J. Biol. Chem. 283, 5748–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Das Sarma J., Wang F., Koval M. (2002) J. Biol. Chem. 277, 20911–20918 [DOI] [PubMed] [Google Scholar]
- 54.Su V., Knutson A., Lau K., Kurata W., Berestecky J., Lau A. F. (2009) Hybridoma 28, 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Treier M., Staszewski L. M., Bohmann D. (1994) Cell 78, 787–798 [DOI] [PubMed] [Google Scholar]
- 56.Das S., Smith T. D., Sarma J. D., Ritzenthaler J. D., Maza J., Kaplan B. E., Cunningham L. A., Suaud L., Hubbard M. J., Rubenstein R. C., Koval M. (2009) Mol. Biol. Cell 20, 2593–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maza J., Das Sarma J., Koval M. (2005) J. Biol. Chem. 280, 21115–21121 [DOI] [PubMed] [Google Scholar]
- 58.Leithe E., Rivedal E. (2004) J. Biol. Chem. 279, 50089–50096 [DOI] [PubMed] [Google Scholar]
- 59.Leithe E., Kjenseth A., Sirnes S., Stenmark H., Brech A., Rivedal E. (2009) J. Cell Sci. 122, 3883–3893 [DOI] [PubMed] [Google Scholar]
- 60.Ganoth D., Bornstein G., Ko T. K., Larsen B., Tyers M., Pagano M., Hershko A. (2001) Nat. Cell Biol. 3, 321–324 [DOI] [PubMed] [Google Scholar]
- 61.Mishra A., Godavarthi S. K., Jana N. R. (2009) Neurobiol. Dis. 36, 26–34 [DOI] [PubMed] [Google Scholar]
- 62.Montagnoli A., Fiore F., Eytan E., Carrano A. C., Draetta G. F., Hershko A., Pagano M. (1999) Genes Dev. 13, 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagano M., Tam S. W., Theodoras A. M., Beer-Romero P., Del Sal G., Chau V., Yew P. R., Draetta G. F., Rolfe M. (1995) Science 269, 682–685 [DOI] [PubMed] [Google Scholar]
- 64.Rütz M. L., Hülser D. F. (2001) Eur. J. Cell Biol. 80, 20–30 [DOI] [PubMed] [Google Scholar]
- 65.Leithe E., Rivedal E. (2004) J. Cell Sci. 117, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 66.Kieken F., Spagnol G., Su V., Lau A. F., Sorgen P. L. (2010) J. Biomol. NMR 46, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zerangue N., Malan M. J., Fried S. R., Dazin P. F., Jan Y. N., Jan L. Y., Schwappach B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2431–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verma R., Deshaies R. J. (2000) Cell 101, 341–344 [DOI] [PubMed] [Google Scholar]
- 69.Bercovich Z., Rosenberg-Hasson Y., Ciechanover A., Kahana C. (1989) J. Biol. Chem. 264, 15949–15952 [PubMed] [Google Scholar]
- 70.Glass J. R., Gerner E. W. (1987) J. Cell. Physiol. 130, 133–141 [DOI] [PubMed] [Google Scholar]
- 71.Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 72.Goh A. M., Walters K. J., Elsasser S., Verma R., Deshaies R. J., Finley D., Howley P. M. (2008) BMC Biochem. 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004) J. Biol. Chem. 279, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 74.Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartmann-Petersen R., Seeger M., Gordon C. (2003) Trends Biochem. Sci. 28, 26–31 [DOI] [PubMed] [Google Scholar]
- 76.d'Azzo A., Bongiovanni A., Nastasi T. (2005) Traffic 6, 429–441 [DOI] [PubMed] [Google Scholar]