Abstract
Signaling by receptor tyrosine kinases regulates pancreatic β cell function. Inactivation of insulin receptor (InsR), IGF1 receptor (Igf1r), or Irs1 in β cells impairs insulin secretion. Conversely, Irs2 ablation impairs β cell replication. In this study, we examined aspects of the Igf1r regulatory signaling cascade in β cells. To examine genetically the involvement of Irs1 and Irs2 in Igf1r signaling, we generated double mutant mice lacking Igf1r specifically in pancreatic β cells in an Irs1- or Irs2-null background. We show that Igf1r/Irs1 double mutants do not differ phenotypically from Irs1 single mutants and exhibit hyperinsulinemia, while maintaining normal β cell mass and glucose tolerance. In contrast, lack of Igf1r function in β cells aggravates the consequences of Irs2 ablation in double mutants and results in lethal diabetes by 6 weeks of age. This additivity of phenotypic manifestations indicates that Irs2 serves a pathway that is largely independent of Igf1r signaling. Consistent with the view that the latter is the InsR pathway, we show that combined β cell-specific knock-out of both Insr and Igf1r results in a phenocopy of double mutants lacking Igf1r and Irs2. We conclude that Igf1r signals primarily through Irs1 and affects insulin secretion, whereas β cell proliferation is mainly regulated by InsR using Irs2 as a downstream signaling effector. The insulin and IGF pathways appear to control β cell functions independently and selectively.
Keywords: Diabetes, Gene Knockout, Growth Factors, Insulin Secretion, Insulin-like Growth Factor (IGF), Receptor Tyrosine Kinase
Introduction
Type 2 diabetes is caused by a combination of insulin resistance and impaired β cell function (1). In prospective studies of individuals at risk of developing (2) or newly diagnosed with type 2 diabetes (3), disease progression is associated with a relatively modest deterioration of insulin resistance, and a steep decrease in β cell function (4). These data have been interpreted to suggest that the progressive defect in glucose control in type 2 diabetes by the combined action of these two parameters (5) primarily reflects the irreversibility of β cell failure.
Defects of β cell function in diabetes are complex, and include reduced insulin secretion (6, 7) and alterations of β cell number (8). There are genetic and acquired (environmental) components to both aspects of β cell failure (9). Thus, an impairment of glucose-stimulated insulin secretion can be detected in non-diabetic first-degree relatives of diabetic patients, consistent with a genetic predisposition (6, 7). But following the onset of diabetes, hyperglycemia itself causes a deterioration of insulin secretion, which can be partly reversed by improved glycemia control (10), demonstrating a role of acquired metabolic abnormalities in β cell failure (11).
It remains unclear whether the two primary components of β cell failure, impaired insulin secretion (7), and reduced β cell mass (12), are mechanistically linked. Turnover studies in mice support the view that β cells are capable of a limited number of replications (13), suggesting that the decrease of β cell mass in humans with diabetes is likely to be the result of uncompensated apoptosis (12). Thus, understanding the mechanism linking β cell renewal and insulin secretion can provide clues to the best therapeutic approaches to β cell failure.
The insulin-like growth factor (IGF)2 system represents an attractive candidate in that regard. We and others (14, 15) have shown that targeted ablation of Igf1r function in pancreatic β cells impairs glucose-induced insulin secretion, without changes in β cell proliferation. Here we have tested the consequences of the same mutation in combination with mutations affecting downstream effectors, more specifically with a model of insulin resistance (Irs1 knock-out) (16, 17) or a model of β cell failure (Irs2 knock-out) (18–20).
EXPERIMENTAL PROCEDURES
Mice
Animals carrying a floxed Igf1r (Igf1rlox) (15) and Insr allele (Insrlox) (21), or null Irs1 (Irs1−/−) and Irs2 alleles (Irs2−/−) (22) and transgenic mice expressing Cre recombinase under the transcriptional control of the rat insulin2 promoter (Rip)-Cre have been described previously. A mating program with Irs1+/−, Irs2+/−, Igf1rlox, Insrlox, and Rip-Cre mice was used to generate progeny of eight genotypes: WT, (Rip)-cre:Igf1rΔlox/Δlox (henceforth, βIgf1r), (Rip)-cre:InsrΔlox/Δlox (βInsr), Irs1−/−, Irs2−/−, βIgf1r:Irs1−/−, βIgf1r:Irs2−/−, and βIgf1r:βInsr. We used PCR analysis for genotyping as described previously (15, 21).
Phenotypic Analysis
We included only male mice in the analyses, as they are more prone to developing diabetes in the strains employed. We measured blood glucose levels using Accucheck meters (Roche Applied Science, Mannheim, Germany), and serum insulin and glucagon by ELISA (Linco Research, St. Charles, MO). We carried out all assays in duplicate (22).
Immunohistochemical and Morphometric Analyses
Pancreata were removed from WT, βIgf1r, Irs1−/−, Irs2−/−, βIgf1r:Irs1−/−, and βIgf1r:Irs2−/− mice at P30, weighed, and fixed overnight in 4% paraformaldehyde. 4-μm-thick sections were immunostained for β cells using mouse anti-insulin antibodies (n = 19–36 per genotype) or rabbit anti-Glut2 polyclonal antibody (n = 9–12 per genotype) (Calbiochem) and for α-cells using mouse anti-glucagon antibodies (n = 19–36 per genotype) (Sigma). For immunohistochemistry with anti-Glut2 antiserum (Calbiochem), Pdx1, and MafA (Bethyl) pancreata were fixed in 4% paraformaldehyde and incubated at a 1:300, 1:10,000 and 1:1,000 dilutions, respectively. Immunoreactivity was detected with the ABC system for Glut2 (DAKO), or with a FITC-conjugated secondary antibody for Pdx1 (23) and MafA (24). For morphometric analysis of β cell mass, 4–7 animals of each genotype were analyzed at P30. For each pancreas, several sections ∼160 μm apart were covered systematically by accumulating images from non-overlapping fields with an Olympus IX-70 inverted fluorescence microscope (Olympus America, Melville, NY), and images were captured using a Spot digital camera (Diagnostic Instruments, Sterling Heights, MI), and analyzed using the NIH Image 1.60 software (22). Results were expressed as percentage of the total surveyed pancreatic area occupied by α and β cells. For qualitative analysis of β cell insulin content, two independent operators scored sections blindly, using the same material prepared for morphometry. Cells were arbitrarily binned as “strongly-positive” or “weakly positive” based on intraislet comparisons to avoid staining artifacts due to comparison of sections processed independently.
Detection and Quantitation of β Cell Replication
We evaluated β cell proliferation using immunohistochemistry with the cell cycle antigen Ki67 (NCL-Ki 67p, Castra). For these experiments, we obtained pancreata from WT, βIgf1r, Irs1−/−, Irs2−/−, βIgf1r:Irs1−/−, and βIgf1r:Irs2−/− mice at P30 (15). For double staining with anti-Ki67 and anti-insulin antibodies, we used guinea pig anti-insulin antibody. Quantitation of β-cell replication was performed by counting Ki67-positive cells in 3–5 sections spaced more than 160 μm apart in each pancreas. The mean value of Ki67-positive cells was multiplied by β cell area as calculated above to obtain an arbitrary Ki67 labeling index.
Statistical Analysis
Descriptive statistics, t test for paired data, and analysis of variance (ANOVA) were performed using the Statsview software.
RESULTS
Experimental Design
Defective glucose-dependent insulin secretion is a common phenotypic feature of β cell-specific Igf1r mutants and globally nullizygous Irs1−/− mice, even though the latter exhibit additional abnormalities (14, 15, 25). Therefore, considering the upstream position of a receptor in a signaling cascade, a prediction of our genetic study was that, if Igf1r signals exclusively or predominantly through Irs1, double mutants βIgf1r:Irs1−/− should not differ phenotypically from single mutants βIgf1r or Irs1−/−. If this prediction were fulfilled, it would be also interpreted as indicating the lack of a signaling relationship between Igf1r and Irs2, as an abnormality in insulin secretion is not a trait of the latter mutants (19). For experimental verification of this conjecture, we generated double βIgf1r:Irs2−/− mutants expecting that, if our overall hypothesis were correct, they would manifest additive phenotypic features, i.e. the lack of Igf1r signaling in β cells would aggravate the diabetic phenotype of single Irs2−/− mutants. To test the hypothesis, we generated mice of six different genotypes (WT, βIgf1r, Irs1−/−, Irs2−/−, βIgf1r:Irs1−/−, and βIgf1r:Irs2−/−), and analyzed their growth and metabolic features.
Growth of Mutant Mice
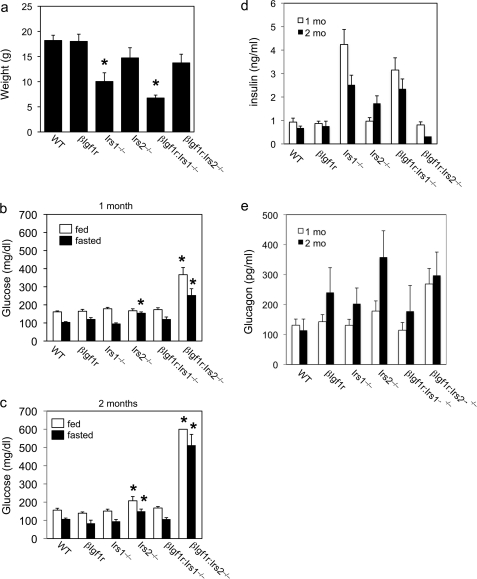
To acquire some indication of whether the examined combinations of mutations were affecting mouse growth, we monitored body weights at selected time points and concluded that this limited analysis was in agreement with previous observations (15). Thus, the βIgf1r mutants did not differ significantly from WT littermates in weight, whereas the Irs1−/− mice (carrying the mutation in a C57BL/6 × 129/Sv genetic background) were ∼50% of normal in body size, as expected (Fig. 1a) (16). The Irs2−/− mutants showed a non-significant trend toward reduced body weight, likely as a consequence of weight loss through polyuria (Fig. 1a). Moreover, we observed that the absence of Igf1r in β cells had no growth consequences for the double mutants. Thus, the weight of βIgf1r:Irs2−/− mice was comparable to normal, while the growth retardation of βIgf1r:Irs1−/− animals was indistinguishable from that of Irs1−/− single mutants (Fig. 1a).
FIGURE 1.
Weights and metabolic parameters in mutant mice. a, body weights of 1-month-old mice (n = >6 per genotype). b and c, fed (empty bars) and fasting glucose levels (full bars) in 1-month (b) and 2-month-old mice (c). n = 15–41 for each genotype and each condition at 1 month, and 8–23 for each genotype and each condition at 2 months, except for βIgf1r (n = 4) and βIgf1r:Irs2−/− (n = 3). d, fed insulin levels in 1- (empty bars) and 2-month-old mice (full bars). e, fasted glucagon levels in 1- (empty bars) and 2-month-old mice (full bars). n = 15–35 for 1-month-old mice, and 5–19 for 2-month-old mice for both insulin and glucagon measurements. An asterisk indicates p < 0.05 by ANOVA versus other genotypes in the same age group.
Metabolic Features of Mutant Mice Lacking Igf1r and Irs1, or Igf1r and Irs2 in β Cells
When we measured glucose levels in 1-month and 2-month-old fasted and fed mice, we observed normal values in βIgf1r, Irs1−/−, and βIgf1r:Irs1−/− mice (Fig. 1, b and c), whereas Irs2−/− showed fasting hyperglycemia starting at 1 month, and both fasting and fed hyperglycemia at 2 months. βIgf1r:Irs2−/− mutants exhibited more pronounced fasting and fed hyperglycemia than any other mutants at all time points examined. The double mutants died at a young age. Thus, 87% of them (20/23) died by 2 months of age, as opposed to 11% (2/18) of Irs2−/− mice during the same time period. By 4 months of age, there were no βIgf1r:Irs2−/− survivors (Table 1).
TABLE 1.
Death records
Deaths among mutant mice were recorded over a 1-year period.
| n (total) | Dead at 2 months | Dead at 4 months | Dead at 6 months | |
|---|---|---|---|---|
| WT | 29 | 0 | 0 | 0 |
| bIgf1r | 35 | 0 | 0 | 0 |
| Irs1−/− | 16 | 4 | 0 | 0 |
| bIgf1r/Irs1−/− | 19 | 1 | 4 | >10 |
| Irs2−/− | 18 | 2 | >10 | |
| bIgf1r/Irs2−/− | 23 | 20 | 3 |
Analyses of insulin levels in the fed state demonstrated hyperinsulinemia in 1-month-old Irs1−/− and βIgf1r:Irs1−/− mutant mice (Fig. 1d). This finding was confirmed in 2-month-old mice and is consistent with a state of compensated systemic insulin resistance (16, 17). There were no differences in insulin levels among 1-month-old WT, Irs2−/−, and βIgf1r:Irs2−/− mice. In contrast, 2-month-old Irs2−/− and βIgf1r:Irs2−/− mice showed opposite changes, with Irs2−/− mice displaying increased insulin levels, and βIgf1r:Irs2−/− mice displaying very low insulin levels (Fig. 1d). These findings indicate that single Irs2−/− mutants are able to mount a transient, limited compensatory response to counter hyperglycemia, whereas double mutants βIgf1r:Irs2−/− have exhausted their β cell compensatory ability.
One-month-old double mutants βIgf1r:Irs2−/− also showed elevated fasting glucagon levels, consistent with impaired insulin secretion (Fig. 1e) (26). By two months of age, the difference was no longer statistically significant, probably reflecting the advanced metabolic deterioration in these mutants.
The metabolic data indicate that the phenotype due to systemic ablation of Irs1 is not modified by β cell-specific deletion of Igf1r, suggesting that the proteins encoded by these two genes act on the same signaling pathway in the β cell, i.e. the two phenotypes are overlapping. In contrast, the diabetic phenotype resulting from systemic Irs2 ablation is significantly worsened by Ifg1r ablation in β cells, suggesting that the signaling pathways mediated by Igf1r and Irs2 differ, at least in part.
Morphometric Analysis of β Cells
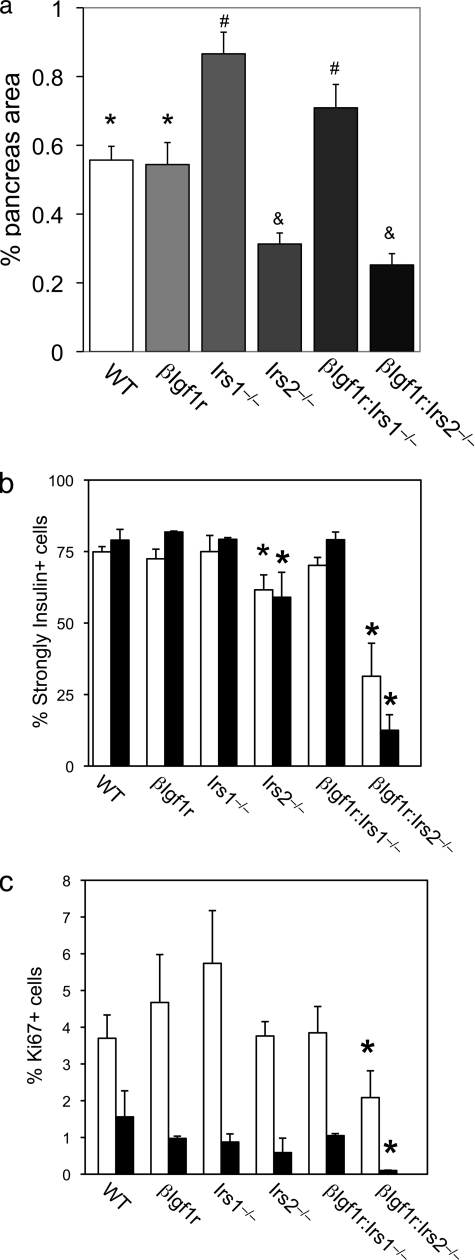
To investigate the mechanism of the observed phenotypes, we determined the percentage of pancreatic area occupied by α and β cells as an index of islet mass and also measured β cell turnover. In 1-month-old mice, we observed a ∼50% reduction of β cell mass in both Irs2−/− and βIgf1r:Irs2−/− genotypes (Fig. 2a). In contrast, in Irs1−/− and βIgf1r:Irs1−/− mice, we detected a ∼40% increase in β cell mass that correlated with the increase in circulating insulin levels. There were no changes in α cell mass (not shown).
FIGURE 2.
Islet morphometry and β cell turnover in mutant mice. a, β cell mass in 1-month-old mice (n = 4–7 mice and 19–36 sections for each genotype). Identical symbols (*, #, and &) denote genotypes that do not differ from one another, but differ from each of the other two groups by ANOVA. b, number of strongly positive β cells, as detected by immunohistochemistry, in 1- and 2-month-old mice, indicated by the empty and filled bars, respectively (n = 4–7 mice and 19–36 sections for each genotype). c, Ki67 labeling indexes in β cells of mutant mice at 1 month (empty bars) and 2 months (full bars). An asterisk indicates p < 0.05 by ANOVA versus other genotypes in the same age group.
In addition to a decreased number of β cells, insulin immunohistochemistry revealed numerous weakly positive β cells in βIgf1r:Irs2−/− mice. In WT, βIgf1r, and Irs1−/− mutants, alone or in combination, nearly 80% of islet cells showed intense immunoreactivity with anti-insulin antiserum. In contrast, the percentage fell to ∼60% in Irs2−/− mutants, and to 30% in βIgf1r:Irs2−/− mutants (Fig. 2b). The defect became more pronounced in 2-month-old βIgf1r:Irs2−/− mice that had only 10% strongly reactive β cells in islets (Fig. 2b).
We next asked whether the decrease in β cell mass was due to decreased proliferation, increased apoptosis, or a combination of both. The percentage of Ki67-labeled, insulin-positive cells was similar between WT, βIgf1r, Irs2−/−, and βIgf1r:Irs1−/− mice, and was slightly increased in Irs1−/− mutants at 1 month, albeit the difference was not statistically significant. In contrast, the Ki67 labeling index decreased by ∼45% in βIgf1r:Irs2−/− mice at both 1 and 2 months (Fig. 2c). The proliferation index of non-β cells was comparable among the six genotypes at both ages (not shown). Caspase-3 immunohistochemistry failed to detect differences in apoptosis among the various genotypes at any of the ages examined (data not shown). These data indicate that the primary defect in βIgf1r:Irs2−/− mice is decreased β cell proliferation (27, 28).
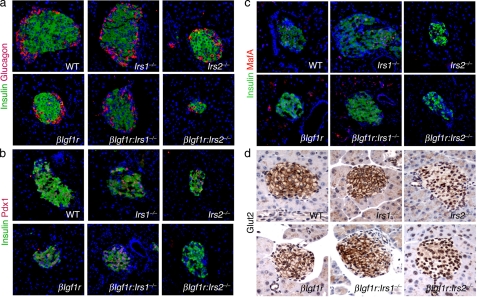
We analyzed islet morphology and expression of genes required for proper β cell function. Islets from double mutants βIgf1r:Irs2−/− showed abnormal morphology, while those from other genotypes showed normal size and distribution of endocrine cells (Fig. 3a). Transcription factor Pdx1 is important in differentiated β cells for maintenance of normal β cell mass (23, 30). Accordingly, we found a marked reduction of Pdx1 in Irs2−/− and βIgf1r:Irs2−/− mice, but normal expression in mice of other genotypes (Fig. 3b). These data link Pdx1 expression with Irs2, but not Irs1 signaling (23, 31, 32). The facilitative glucose transporter Glut2 (encoded by the gene Slc2a2) plays a critical role in coupling glucose sensing to insulin secretion in β cells (33), and was also decreased in double mutant βIgf1r:Irs2−/− mice (Fig. 3c). Finally, transcription factor MafA is expressed in β cells following the “secondary transition”, when cells acquire the ability to secrete insulin in response to variations in glucose levels (34–36), and has been shown to decrease in diabetic mice (24). We found that MafA expression mirrored that of Pdx1, with decrements in both Irs2−/− and βIgf1r:Irs2−/− mice, but not in other genotypes (Fig. 3d).
FIGURE 3.
Immunohistochemical analysis of marker expression in mutant mouse pancreas. a–d, illustrative examples of immunohistochemistry of pancreatic sections from 1-month-old mice of the indicated genotypes stained with anti-insulin (green) and anti-glucagon (a) (n = >9 per genotype), anti-Pdx1 (b), anti-MafA (c) (all in red) (n = 4–7 per genotype) or with anti-Glut2 antibodies (brown) (n = 9–12 per genotype) (d).
Phenotypic Characterization of Double Mutant Mice Lacking Igf1r and InsR in β Cells
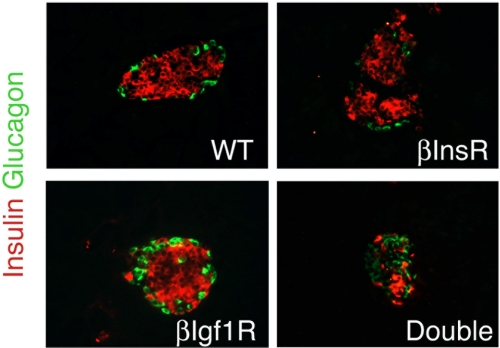
The data described support a model in which Igf1r signals primarily through Irs1, while Irs2 signaling is distinct. To ascertain whether our model accounted for all possible signaling through the InsR/Igf1r system, we generated mice lacking both receptors in β cells. Double mutants lacking βIgf1r and βInsR both were born at term in the expected Mendelian ratios. Unlike single mutants, double mutants βIgf1r:βInsR showed marked hyperglycemia at 3 weeks (>500 mg/dl) and died of diabetic ketoacidosis within 4–8 weeks. Islet immunohistochemistry revealed only scattered remnants of β cells, with normal numbers of α cells (Fig. 4) (37).
FIGURE 4.
Islet morphology in mice with single and combined mutations of InsR and Igf1r. Examples of immunohistochemistry of pancreatic sections from 1-month-old mice of the indicated genotypes stained with anti-insulin (red) and anti-glucagon antibodies (green) (n = 4 per genotype).
DISCUSSION
The discovery that signaling by receptor tyrosine kinases of the insulin/IGF family affects pancreatic β cell function marked a watershed in diabetes research. There is now a substantial body of work indicating that the two receptors and their main substrates, Irs1 and Irs2, can regulate insulin secretion and β cell turnover (14, 15, 23, 38–41). Moreover, activation of InsR appears to be required to mediate β cell hyperplasia in response to peripheral insulin resistance (28, 42). Not only do InsR and Igf1r act in terminally differentiated β cells, but their combined ablation also results in altered cell fate specification in the embryonic pancreas (43), highlighting the overarching role of this pathway in the maturation and maintenance of pancreatic endocrine function.
Mechanistically, it remains unclear how these signaling pathways can function effectively, when large amounts of insulin released by the β cell should theoretically cause ligand-induced receptor internalization, and attendant desensitization. Cell polarity, pulsatile release of tightly packed insulin crystals that may have poor access to receptor binding sites, anatomy of vascular flow, and capillary permeability in the islet's portal system, or formation of low-affinity, desensitization-resistant hybrid receptors composed of an InsR monomer and an Igf1r monomer (44) are reasonable potential explanations for the preservation of feedback control of β cell function by InsR or Igf1r in the face of overwhelming insulin concentrations (1).
The present study was undertaken to examine the overlap between InsR and Igf1r signaling in β cells, and thus shed light on mechanisms coupling insulin secretion with β cell proliferation, two processes that lie at the core of β cell dysfunction in type 2 diabetes. We have suggested that excessive β cell replication, especially in the metabolically unfavorable environment caused by hyperglycemia, is detrimental to β cell preservation (24), and have proposed that induction of β cell quiescence should be a goal of diabetes treatment (45), a concept loosely related to “β cell rest” (46, 47). Our analyses indicate that Igf1r is coupled to Irs1 signaling in β cells to regulate insulin secretion, whereas it is surprisingly uncoupled from its traditional proliferative role. In β cells, this function appears to be the purview of Irs2, acting to relay primarily InsR-dependent signals.
We have previously shown that peripheral insulin resistance due to InsR haploinsufficiency accelerates β cell failure in Irs2 knock-out mice (23, 27). Others have shown that Igf1r haploinsufficiency precipitates diabetes in Irs2 knock-out mice, suggesting that the two proteins mediate different pathways (41). But those studies could not distinguish between direct effects on β cells from indirect effects due to changes in peripheral insulin sensitivity. The assignment of specific roles to Irs1 and Irs2 in response to InsR and Igf1r, respectively, is noteworthy in several ways: (i) it indicates that signal transduction in β cells occurs differently from liver, where Irs1 and Irs2 have overlapping roles (29, 48); (ii) it provides a potential mechanistic explanation for the heterogeneity of β cell failure, which in some instances could be due to impaired secretion (i.e. an Irs1-dependent pathway), and in some to defects in β cell proliferation (i.e. an Irs2-dependent pathway); (iii) it suggests that the two systems could be exploited separately for therapeutic ends.
Our data in combined β cell-specific knock-out of InsR and Igf1r are largely consistent with those of Ueki et al. (37). Unlike Ueki et al., we show that decreased β cell proliferation trumps increased apoptosis as the main cause of islet demise, a discrepancy that can possibly be due to different time points examined in the two studies (2 weeks versus 4 weeks). It is interesting to note that, although the combined knock-out of InsR and Igf1r has a more severe phenotype than the Irs2 knock-out, neither InsR nor Igf1r individual knockouts are associated with decreased β cell proliferation. Our interpretation of this peculiar finding is that the “proliferative” effects of InsR and Igf1r signaling are secondary to their effects on insulin secretion.
In conclusion, we show that InsR and Igf1r signaling affect pancreatic β cell function in largely distinct ways through their actions on Irs1 and Irs2. The unmet challenge for this field is to demonstrate how these signaling pathways can be modulated in vivo, overriding the alleged paracrine effects of large amounts of insulin, and how these processes play out in the unfolding of β cell failure in diabetes.
Acknowledgments
We thank members of the Efstratiadis and Accili Laboratories for useful discussions, C. Wright for the gift of anti-Pdx1 antiserum, and R. Stein for the gift of anti-MafA antiserum.
This work was supported, in whole or in part, by National Institutes of Health Grants DK58282 and DK63608 (Columbia University Diabetes & Endocrinology Research Center) and a research grant from the Russell Berrie Foundation (to A. E. and D. A.).
- IGF
- insulin-like growth factor
- InsR
- insulin receptor
- ANOVA
- analysis of variance.
REFERENCES
- 1.Accili D. (2004) Diabetes 53, 1633–1642 [DOI] [PubMed] [Google Scholar]
- 2.Weyer C., Bogardus C., Mott D. M., Pratley R. E. (1999) J. Clin. Invest. 104, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy J., Atkinson A. B., Bell P. M., McCance D. R., Hadden D. R. (1998) Diabet. Med. 15, 290–296 [DOI] [PubMed] [Google Scholar]
- 4.Kahn S. E. (2003) Diabetologia 46, 3–19 [DOI] [PubMed] [Google Scholar]
- 5.Monnier L., Colette C., Dunseath G. J., Owens D. R. (2007) Diabetes Care 30, 263–269 [DOI] [PubMed] [Google Scholar]
- 6.O'Rahilly S., Turner R. C., Matthews D. R. (1988) N. Engl. J. Med. 318, 1225–1230 [DOI] [PubMed] [Google Scholar]
- 7.Polonsky K. S., Given B. D., Hirsch L. J., Tillil H., Shapiro E. T., Beebe C., Frank B. H., Galloway J. A., Van Cauter E. (1988) N. Engl. J. Med. 318, 1231–1239 [DOI] [PubMed] [Google Scholar]
- 8.Rahier J., Guiot Y., Goebbels R. M., Sempoux C., Henquin J. C. (2008) Diabetes Obes. Metab. 10, Suppl. 4, 32–42 [DOI] [PubMed] [Google Scholar]
- 9.Bell G. I., Polonsky K. S. (2001) Nature 414, 788–791 [DOI] [PubMed] [Google Scholar]
- 10.Leahy J. L., Bonner-Weir S., Weir G. C. (1992) Diabetes Care. 15, 442–455 [DOI] [PubMed] [Google Scholar]
- 11.Ferrannini E. (1998) Endocrine Rev. 19, 477–490 [DOI] [PubMed] [Google Scholar]
- 12.Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. (2003) Diabetes 52, 102–110 [DOI] [PubMed] [Google Scholar]
- 13.Teta M., Long S. Y., Wartschow L. M., Rankin M. M., Kushner J. A. (2005) Diabetes 54, 2557–2567 [DOI] [PubMed] [Google Scholar]
- 14.Kulkarni R. N., Holzenberger M., Shih D. Q., Ozcan U., Stoffel M., Magnuson M. A., Kahn C. R. (2002) Nat. Genet. 31, 111–115 [DOI] [PubMed] [Google Scholar]
- 15.Xuan S., Kitamura T., Nakae J., Politi K., Kido Y., Fisher P. E., Morroni M., Cinti S., White M. F., Herrera P. L., Accili D., Efstratiadis A. (2002) J. Clin. Invest. 110, 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araki E., Lipes M. A., Patti M. E., Brüning J. C., Haag B., 3rd, Johnson R. S., Kahn C. R. (1994) Nature 372, 186–190 [DOI] [PubMed] [Google Scholar]
- 17.Tamemoto H., Kadowaki T., Tobe K., Yagi T., Sakura H., Hayakawa T., Terauchi Y., Ueki K., Kaburagi Y., Satoh S., Sekihara H., Yoshioka S., Horikoshi H., Furuta Y., Ikawa Y., Kasuga M., Yazaki Y., Aizawa S. (1994) Nature 372, 182–186 [DOI] [PubMed] [Google Scholar]
- 18.Withers D. J., Sanchez-Gutierrez J., Towery H., Burks D. J., Ren J. M., Previs S., Zhang Y., Bernal D., Pons S., Shulman G. I., Bonner-Weir S., White M. F. (1998) Nature 391, 900–904 [DOI] [PubMed] [Google Scholar]
- 19.Kubota N., Tobe K., Terauchi Y., Eto K., Yamauchi T., Suzuki R., Tsubamoto Y., Komeda K., Nakano R., Miki H., Satoh S., Sekihara H., Sciacchitano S., Lesniak M., Aizawa S., Nagai R., Kimura S., Akanuma Y., Taylor S. I., Kadowaki T. (2000) Diabetes 49, 1880–1889 [DOI] [PubMed] [Google Scholar]
- 20.Terauchi Y., Matsui J., Suzuki R., Kubota N., Komeda K., Aizawa S., Eto K., Kimura S., Nagai R., Tobe K., Lienhard G. E., Kadowaki T. (2003) J. Biol. Chem. 278, 14284–14290 [DOI] [PubMed] [Google Scholar]
- 21.Kitamura T., Kitamura Y., Nakae J., Giordano A., Cinti S., Kahn C. R., Efstratiadis A., Accili D. (2004) J. Clin. Invest. 113, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kido Y., Burks D. J., Withers D., Bruning J. C., Kahn C. R., White M. F., Accili D. (2000) J. Clin. Invest. 105, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura T., Nakae J., Kitamura Y., Kido Y., Biggs W. H., 3rd, Wright C. V., White M. F., Arden K. C., Accili D. (2002) J. Clin. Invest. 110, 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura Y. I., Kitamura T., Kruse J. P., Raum J. C., Stein R., Gu W., Accili D. (2005) Cell. Metab. 2, 153–163 [DOI] [PubMed] [Google Scholar]
- 25.Kulkarni R. N., Winnay J. N., Daniels M., Brüning J. C., Flier S. N., Hanahan D., Kahn C. R. (1999) J. Clin. Invest. 104, R69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gromada J., Franklin I., Wollheim C. B. (2007) Endocr. Rev. 28, 84–116 [DOI] [PubMed] [Google Scholar]
- 27.Kim J. J., Kido Y., Scherer P. E., White M. F., Accili D. (2007) Am. J. Physiol. Endocrinol Metab. 292, E1694–E1701 [DOI] [PubMed] [Google Scholar]
- 28.Okamoto H., Hribal M. L., Lin H. V., Bennett W. R., Ward A., Accili D. (2006) J. Clin. Invest. 116, 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubota N., Kubota T., Itoh S., Kumagai H., Kozono H., Takamoto I., Mineyama T., Ogata H., Tokuyama K., Ohsugi M., Sasako T., Moroi M., Sugi K., Kakuta S., Iwakura Y., Noda T., Ohnishi S., Nagai R., Tobe K., Terauchi Y., Ueki K., Kadowaki T. (2008) Cell. Metab. 8, 49–64 [DOI] [PubMed] [Google Scholar]
- 30.Johnson J. D., Ahmed N. T., Luciani D. S., Han Z., Tran H., Fujita J., Misler S., Edlund H., Polonsky K. S. (2003) J. Clin. Invest. 111, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kushner J. A., Ye J., Schubert M., Burks D. J., Dow M. A., Flint C. L., Dutta S., Wright C. V., Montminy M. R., White M. F. (2002) J. Clin. Invest. 109, 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin X., Taguchi A., Park S., Kushner J. A., Li F., Li Y., White M. F. (2004) J. Clin. Invest. 114, 908–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillam M. T., Hümmler E., Schaerer E., Yeh J. I., Birnbaum M. J., Beermann F., Schmidt A., Dériaz N., Thorens B., Wu J. Y. (1997) Nat. Genet. 17, 327–330 [DOI] [PubMed] [Google Scholar]
- 34.Olbrot M., Rud J., Moss L. G., Sharma A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuoka T. A., Zhao L., Artner I., Jarrett H. W., Friedman D., Means A., Stein R. (2003) Mol. Cell. Biol. 23, 6049–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuoka T. A., Artner I., Henderson E., Means A., Sander M., Stein R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2930–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueki K., Okada T., Hu J., Liew C. W., Assmann A., Dahlgren G. M., Peters J. L., Shackman J. G., Zhang M., Artner I., Satin L. S., Stein R., Holzenberger M., Kennedy R. T., Kahn C. R., Kulkarni R. N. (2006) Nat. Genet. 38, 583–588 [DOI] [PubMed] [Google Scholar]
- 38.Nakae J., Biggs W. H., 3rd, Kitamura T., Cavenee W. K., Wright C. V., Arden K. C., Accili D. (2002) Nat. Genet. 32, 245–253 [DOI] [PubMed] [Google Scholar]
- 39.Hennige A. M., Burks D. J., Ozcan U., Kulkarni R. N., Ye J., Park S., Schubert M., Fisher T. L., Dow M. A., Leshan R., Zakaria M., Mossa-Basha M., White M. F. (2003) J. Clin. Invest. 112, 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulkarni R. N., Brüning J. C., Winnay J. N., Postic C., Magnuson M. A., Kahn C. R. (1999) Cell 96, 329–339 [DOI] [PubMed] [Google Scholar]
- 41.Withers D. J., Burks D. J., Towery H. H., Altamuro S. L., Flint C. L., White M. F. (1999) Nat. Genet. 23, 32–40 [DOI] [PubMed] [Google Scholar]
- 42.Okada T., Liew C. W., Hu J., Hinault C., Michael M. D., Krtzfeldt J., Yin C., Holzenberger M., Stoffel M., Kulkarni R. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8977–8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kido Y., Nakae J., Hribal M. L., Xuan S., Efstratiadis A., Accili D. (2002) J. Biol. Chem. 277, 36740–36747 [DOI] [PubMed] [Google Scholar]
- 44.Frattali A. L., Pessin J. E. (1993) J. Biol. Chem. 268, 7393–7400 [PubMed] [Google Scholar]
- 45.Buteau J., Shlien A., Foisy S., Accili D. (2007) J. Biol. Chem. 282, 287–293 [DOI] [PubMed] [Google Scholar]
- 46.Weng J., Li Y., Xu W., Shi L., Zhang Q., Zhu D., Hu Y., Zhou Z., Yan X., Tian H., Ran X., Luo Z., Xian J., Yan L., Li F., Zeng L., Chen Y., Yang L., Yan S., Liu J., Li M., Fu Z., Cheng H. (2008) Lancet 371, 1753–1760 [DOI] [PubMed] [Google Scholar]
- 47.Greenwood R. H., Mahler R. F., Hales C. N. (1976) Lancet 1, 444–447 [DOI] [PubMed] [Google Scholar]
- 48.Dong X. C., Copps K. D., Guo S., Li Y., Kollipara R., DePinho R. A., White M. F. (2008) Cell. Metab. 8, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]