Abstract
The expression of a variety of cytoprotective genes is regulated by short cis-acting elements in their promoters, called antioxidant response elements (AREs). A central regulator of ARE-mediated gene expression is the NF-E2-related factor 2 (Nrf2). Human hepatitis B virus (HBV) induces a strong activation of Nrf2/ARE-regulated genes in vitro and in vivo. This is triggered by the HBV-regulatory proteins (HBx and LHBs) via c-Raf and MEK. The Nrf2/ARE-mediated induction of cytoprotective genes by HBV results in a better protection of HBV-positive cells against oxidative damage as compared with control cells. Furthermore, there is a significantly increased expression of the Nrf2/ARE-regulated proteasomal subunit PSMB5 in HBV-positive cells that is associated with a decreased level of the immunoproteasome subunit PSMB5i. In accordance with this finding, HBV-positive cells display a higher constitutive proteasome activity and a decreased activity of the immunoproteasome as compared with control cells even after interferon α/γ treatment. The HBV-dependent induction of Nrf2/ARE-regulated genes might ensure survival of the infected cell, shape the immune response to HBV, and thereby promote establishment of the infection.
Keywords: Gene Expression, Hepatitis Virus, Oxidative Stress, Signal Transduction, Transcription Regulation
Introduction
Infection with human hepatitis B virus (HBV)3 can cause acute or chronic inflammation of the liver. In addition, HBV is considered as a major etiological factor in the development of human hepatocellular carcinoma (1). Almost all HBV-associated HCCs harbor chromosomally integrated HBV DNA (2). The HBV genome encodes two regulatory proteins, the PreS2 activator LHBs and the HBx protein. Both proteins are known to trigger a variety of different intracellular signal transduction cascades (3). The activation of the Raf-MEK-Erk signal transduction pathway plays a central role for the LHBs- or HBx-dependent modulation of transcription factors, leading to the activation of various promoter elements (4). Therefore, the HBV regulatory proteins are discussed as potential factors that confer to the development of HBV-associated HCC (4, 5). Moreover, there is evidence that the function of the HBV regulatory proteins is required to support viral replication (6).
The inflammatory process and the cell stress due to permanent overproduction of viral proteins can result in an increased level of radicals and other reactive oxygen species (7, 8). Crucial players in the defense to oxidative stress are antioxidant proteins and enzymes that are involved in the detoxification of electrophiles and radicals. Examples are NAD(P)H quinone oxidoreductase 1 (NQO1), glutathione peroxidases (GPx), and the glutathione biosynthesis enzymes glutamate-cysteine ligase catalytic subunit and regulatory subunit or glutathione S-transferases (GST) γa and π (9, 10). The expression of these cytoprotective proteins is controlled by cis-acting elements in the promoters of these genes, the antioxidative response elements (AREs) (11). An important factor triggering the expression of ARE-regulated genes is the NF-E2-related factor 2 (Nrf2) (12). Nrf2 is a member of the cap “n” collar family of transcription factors. In its inactive state, Nrf2 is associated with the actin-anchored protein Keap1 and localized within the cytoplasm. Keap1-associated Nrf2 is subjected to rapid proteasomal degradation. However, upon its activation initiated by electrophiles or oxidative molecules, Nrf2 dissociates from Keap1 and escapes proteasomal degradation (13). In the nucleus, Nrf2 binds to ARE sequences and thereby functions in partnership of other nuclear proteins as a strong transcriptional activator of ARE-responsive genes. Recently, a crucial role of Nrf2 was found in liver regeneration (14) and in protection from fibrosis (15). However, in many tumors increased expression of ARE-regulated genes was observed that might protect tumor cells from elimination by increased radical levels or from chemotherapy through up-regulation of multidrug resistance proteins, for example (16).
Here, we analyze the capacity of HBV to modulate the expression of ARE-regulated genes, investigate the mechanisms by which HBV interferes, and study the physiological relevance for the viral life cycle and pathogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture
Human hepatoma-derived cell lines HepG2, Huh7.5, and the HBV-positive stable cell lines HepG2.2.15 (17) and HepAD38 (18) were grown as described previously (17, 18). Primary human hepatocytes were isolated by a modified two-step EGTA/collagenase perfusion procedure as described previously (19, 20) and infected with HepAD38-derived supernatant as described previously (21). Primary mouse hepatocytes were isolated from Nrf2−/− (22) and WT C57BL/6 mice using a two-step collagenase perfusion and cultivated as described previously (23). Generation of recombinant adenovirus and infection with recombinant adenovirus were performed as described previously (24).
Chemicals and Antibodies
Anti-NQO1 (A180), anti-PSMB1 (FL-241), anti-PSMB2 (MCP165), anti-γ-GCS(H-300), anti-sMaf F/G/K (H-100), and anti-LMP2 (H200) antibodies were all purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Anti-PSMB5 was purchased from ABR Affinity BioReagents and anti-proteasomal β5i-subunit (23-223) from Calbiochem; anti-β-actin from Sigma, and anti-GPx (C8C4) from Cell Signaling Technology. MA18/07 (25) was kindly provided by Dr. Glebe, Göttingen, Germany; polyclonal goat anti-Hbs was purchased from DAKO, Denmark, and rabbit anti-HBc was from Dianova, Germany.
Kinase inhibitors PD98059, SB203580, calphostin-C, and Bay 43-9006 and the antioxidant tert-butylhydroquinone (tBHQ) were purchased from Calbiochem/Merck. Bortezomib (PS-341) was purchased from Selleck Chemicals LLC (Houston, TX) and dissolved in PBS. Stimulation with insulin and EGF (both from Sigma) were performed as described previously (14, 26).
Real Time PCR
HBV-genome quantification was done using COBAS® Ampli PREP/COBAS® TaqMan® HBV test (Roche Diagnostics) according to the manufacturer's instructions.
Immunohistochemistry
Consecutive sections of paraffin-embedded liver samples derived from HBV patients with chronic or acute hepatitis were deparaffinized and immunostained with anti-γ-GCS and anti-HBs antibodies using the Vectastain kit (Vector Laboratories, Inc., Burlingame, CA).
Plasmids
Luciferase reporter constructs harboring the antioxidant response elements from NAD(P)H-dependent quinone oxidoreductase 1 (pNQO1luc), γ-glutamylcysteine synthetase (pγ-GCSluc), and gastrointestinal glutathione peroxidase (pGI-GPxluc) were generated using the pGL3-promoter vector (Promega). Double-stranded oligonucleotides were synthesized containing a part of the rat NQO1 promoter (5′-CTCTAGAGTCACAGTGACTTGGCAAAATCTGAC-3′, in case of pNQO1luc) (27), the murine Gclm promoter (5′-CCTGGAAGACAATGACTAAGCAGAAAC-3′, in case of pγ-GCSluc) (28), or the human GI-GPx promoter (5′-CCTGTTTTGCTAAGTCATCCTGGGGACC-3′, in case of pGI-GPxluc) (29), including the antioxidant response elements (underlined). After annealing of the complementary oligonucleotides, the double-stranded oligonucleotides were inserted into the pGL3-promoter vector.
Constitutively active (phcaNrf2) and trans-dominant negative Nrf2 expression constructs (ptdnNrf2) were described recently (30). The PSMB5 reporter constructs were kindly provided by Dr. Kwak, Seoul, Korea (31). A mutated PSMB5 reporter construct (p1.1PSMB5lucΔARE341/52) lacking the two functional ARE sequences (ARE341 and ARE52) was generated by PCR using p1.1PSMB5luc as template and primers containing the mutated ARE341(GCCTGGGCAGTGACCAAAC3GCCTGGGTGGCAACCAAAC) or ARE52 (TGA-CGTCGCGGCGTTGCCA3CAACGTCGCGGCGTTGCTG).
NQO1 and γ-GCS expression constructs fused to yellow fluorescence protein (YFP) were purchased from ImaGenes GmbH, Berlin, Germany. For inhibition of c-Raf, the dominant negative mutant pRafC4 was used (32). For HBV expression, a 1.2-fold HBV-genome ayw (pHBV1.2) was used.
Mutant HBV genomes lacking PreS2 activator function (pHBVΔPreS2) or functional HBx (pHBVΔHBx) or the activator-deficient double mutant (pHBVΔPreS2/ΔHBx) were described recently (33, 34). For silencing of HBV expression, the following siRNA was used, 5′-AAGACCUAGUCAGUUAUG-3′. Transfection was performed using Lipofectamine (Invitrogen), as described recently (35), using 3 μg/ml of HBV-specific siRNA or scrambled siRNA per well of a 6-well plate. Cells were harvested 72 h after siRNA transfection.
Transient Transfection and Reporter Gene Activity Assay
Huh7.5 and HepG2 cells were transfected using linear polyethyleneimine (PEI) (Polysciences, Inc) as described recently (36). All transfection experiments were performed in 6-well plates. The plasmid amounts refer to one well. Details to the transfection experiments are given in the figure legends. Transfection of peGFP was used to determine the transfection efficiency that was found between 40 and 50%. Stimulation with tBHQ was performed at a concentration of 60 μm for 16 h. Luciferase activity was measured using a luminometer (Berthold Detection Systems, Wildbad, Germany). Activities, shown as multiples of induction, are mean values from three independent experiments. The error bars represent the standard deviations.
SDS-PAGE and Western Blot Analysis
SDS-PAGE and Western blot analysis were performed according to standard procedures (16) Detection of bound secondary antibody was performed by ECL using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Freiburg, Germany). All experiments were performed in triplicate. One representative experiment is shown.
Protein Oxidation Measurement, 8-OHdG Formation
Protein carbonylation by reactive oxygen intermediates was detected by using OxyBlotTM protein oxidation detection kit (Millipore, Germany). All experiments were performed in triplicate. One representative experiment is shown. Formation of the oxidative DNA adduct 8-OHdG was analyzed by ELISA kit (JalCA, Japan). Relative amounts are mean values from three independent experiments.
Analysis of Proteasome Activity
Proteasomes were isolated by differential centrifugation based on the protocol of Robek et al. (37). Analysis of constitutive proteasome function was performed using a commercial assay system (20 S proteasome activity assay kit, Millipore, Germany) measuring release of the fluorophor 7-amino-4-methylcoumarin (AMC) after cleavage from the labeled substrate peptide LLVY-AMC. Analysis of the immunoproteasome activity was performed as described. The peptide Cbz-VVRR-AMC that was derived from the HBV core protein (amino acids 141–151) was used as substrate (38) Stimulation with interferon α or interferon γ was performed for 3 days using 102 and 103 units per ml. Recombinant purified HBx was isolated as described previously (34). All experiments were performed in triplicate.
Indirect Immunofluorescence Analysis
Fixation and staining were performed as described recently (39). Immunofluorescence staining was analyzed using a confocal laser scanning microscope (CLSM 510 Carl Zeiss, Germany).
RESULTS
HBV Induces ARE-dependent Gene Expression in Vitro and in Vivo
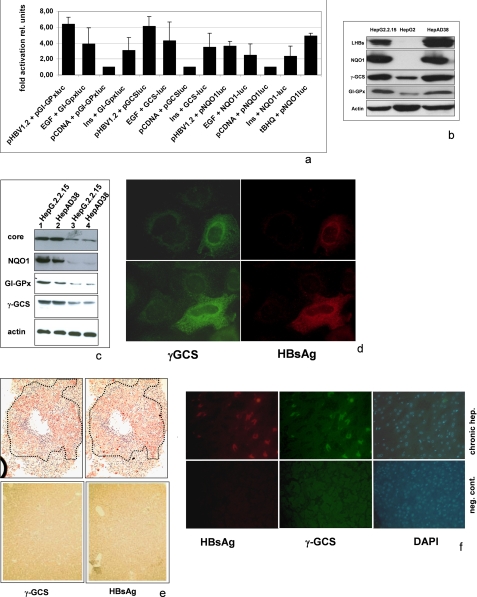
AREs are present in the promoters/enhancers of a variety of cytoprotective genes. To study the effect of HBV on ARE-regulated genes, we performed reporter gene assays. A plasmid harboring a 1.2-fold HBV genome (pHBV1.2) was cotransfected with various reporter constructs harboring a luciferase reporter gene under the control of NQO1, GI-GPx, or γ-GCS-derived ARE sequences. The reporter gene experiments demonstrate that HBV induces a strong expression of the reporter genes (Fig. 1a). This is reflected by increased levels of γ-GCS, GI-GPx, or NQO1 in HBV-producing cells as demonstrated by Western blot analyses of cellular lysates derived from the HBV-positive stable cell lines HepAD38 or HepG2.2.15 compared with HBV-negative HepG2 cells (Fig. 1b). To control whether the observed effect indeed is due to HBV, the expression of HBV was impaired by siRNA. Suppression of HBV gene expression in HepAD38 and HepG2.2.15 cells by siRNA abolishes the induction of NQO1 or γ-GCS (Fig. 1c). Consistent with this result, transient transfection of HuH7.5 or HepG2 with pHBV1.2 resulted in significantly higher amounts of NQO1 or γ-GCS as compared with the pCDNA.3 transfected control (data not shown). Analysis of HBV-infected primary human hepatocytes by double immunofluorescence shows that in HBV-positive cells the amount of γ-GCS is increased as compared with the noninfected cells (Fig. 1d).
FIGURE 1.
HBV-dependent induction of ARE-regulated genes. a, reporter gene assay in HepG2 cells cotransfected with pHBV1.2 (0.8 μg) and luciferase reporter constructs (0.2 μg) harboring the NQO1, GI-GPx, or γ-GCS-derived ARE sequences. Stimulation with tBHQ served as positive control. Moreover, cells were stimulated by the addition of EGF (10 ng/ml) or insulin (50 nm) for 12 h. Cotransfection with pCDNA.3 served as control and was set as 1. The error bars represent the standard deviation. rel units, relative units. b, Western blot analysis of cellular lysates derived from HepG2.215, HepG2, or HepAD38 cells using NQO1-, γ-GCS-, or GI-GPx-specific antisera. Expression of the HBV genome was demonstrated by a LHB-specific antiserum (MA18/7). c, Western blot analysis of cellular lysates from HepG2.215 or HepAD38 cells transfected with HBV-specific siRNA (lanes 3 and 4) or scrambled control siRNA (lanes 1 and 2) using NQO1-, γ-GCS-, or GI-GPx-specific antisera. Expression of the HBV genome was demonstrated by a core-specific antiserum. d, double immunofluorescence microscopy (200-fold magnification) of formaldehyde-fixed HBV-infected primary human hepatocytes stained with anti-LHBs(MA18–7) (red) and anti-γ-GCS (green). e, immunohistochemistry of consecutive sections of paraffin-embedded liver samples derived from an HBV patient with an acute hepatitis (hep.) B infection (upper panels) stained with anti-HBs (right panel) and anti-γ-GCS (left panel). The HBs-positive cells in the consecutive sections are marked by dotted lines. A liver sample from an HBV-negative patient (lower panels) served as negative control (neg. cont). f, double immunofluorescence microscopy of paraffin-embedded liver samples derived from an HBV patient with a chronic hepatitis B infection (upper panels) stained with anti-HBs (red) and anti-γ-GCS (green). The nuclei are visualized with DAPI (blue). A liver sample from an HBV-negative patient (lower panels) served as negative control.
Moreover, immunohistochemistry/double immunofluorescence microscopy of liver samples from patients with chronic or acute HBV infection shows a tight correlation between HBsAg-positive cells and increased amounts of γ-GCS as well (Fig. 1, e and f). Taken together, these data demonstrate that HBV activates the expression of ARE-regulated genes in vitro and most likely also in vivo.
HBV-dependent Activation of ARE-regulated Genes Requires Integrity of Nrf2
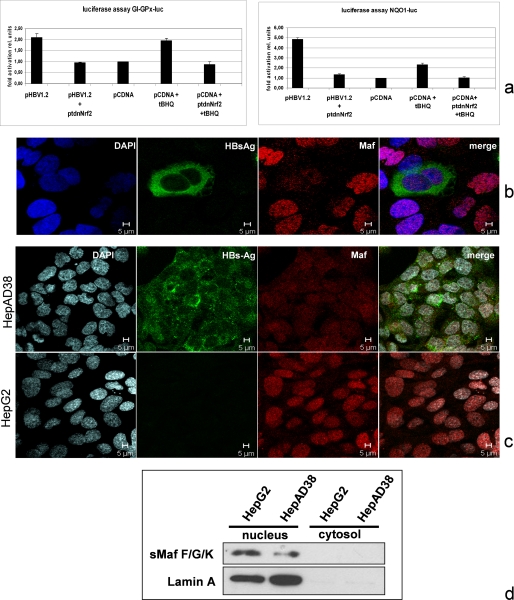
The transcription factor Nrf2 plays a crucial role in the stimulation of ARE-regulated genes. To investigate the relevance of Nrf2 for the HBV-dependent induction of ARE-regulated genes, we cotransfected HepG2 cells with pHBV1.2 and the ARE-driven luc reporter constructs described above and inhibited Nrf2 activity by coexpression of a transdominant negative (tdn) mutant of Nrf2 (ptdnNrf2). The reporter gene assays show that inhibition of Nrf2 by coexpression of the tdn mutant completely abolished the HBV-dependent induction of the reporter genes (Fig. 2a). Although Nrf2 heterodimerizes in the nucleus with small Maf proteins, it was found that increased amounts of small Mafs impair Nrf2-dependent activation of ARE sequences (40).
FIGURE 2.
Integrity of Nrf2 is required for the HBV-dependent induction of ARE-regulated genes. a, reporter gene assay in HepG2 cells cotransfected with pHBV1.2 (0.8 μg) and pNQO1luc (0.2 μg) luciferase reporter construct or pHBV1.2 (0.4 μg) and pGI-GPxluc (0.2 μg). Nrf2 activity was inhibited by cotransfection of ptdnNrf2 (0.4 μg); cotransfection of pCDNA.3 (0.4 μg) served as control and was set as 1. Stimulation with tBHQ served as a positive control. Activities, shown as multiples of induction, are mean values from three independent experiments. The error bars represent the standard deviation. rel units, relative units. b and c, confocal laser scanning immunofluorescence microscopy (630-fold magnification) of formaldehyde-fixed Huh7.5 cells that were transfected with pHBV1.2 (b) or of HepAD38 and HepG2 cells (c). The immunofluorescence staining was performed using the polyclonal small Maf-specific serum (red fluorescence) and the LHBs-specific antibody MA18/7; nuclei were stained with DAPI. d, Western blot analysis of the nuclear and cytosolic fraction derived from HepG2 or HepAD38 cells using a sMaf-specific antiserum. Lamin A was used as a nuclear marker.
In light of the HBV-dependent activation of ARE-dependent genes and the relevance of Nrf2 for this process, we asked whether there is a difference in the amount of small Mafs in HBV-positive and -negative cells. Confocal immunofluorescence microscopy of pHBV1.2-transfected HuH7.5 cells showed a decreased level of small Mafs in the case of HBV-positive cells as compared with the HBV-negative cells (Fig. 2b). Comparable results were obtained when HepG2 cells were compared with HepAD38 cells (Fig. 2c). Western blot analysis of cellular lysates derived from HepG2 and HepAD38 cells confirms a decreased amount of small Mafs in the HBV-positive cells HepAD38 cells as compared with HepG2 cells (Fig. 2d).
Activation of ARE-dependent Genes by HBV Is Not Affected by N-Acetyl-l-cysteine
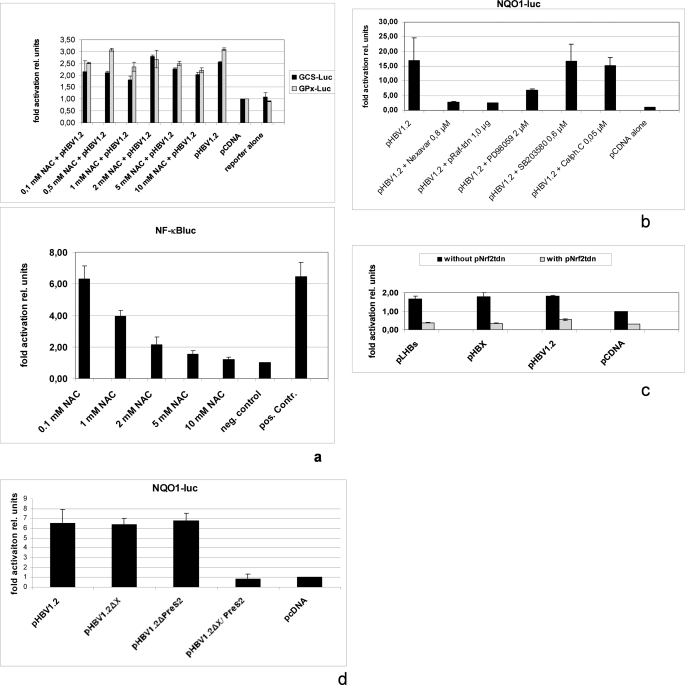
Activation of ARE-regulated genes can be induced by increased radical levels. To study the potential relevance of increased ROS levels for induction of ARE-regulated genes by HBV, we performed cotransfection experiments of HepG2 cells with pHBV1.2 and pγ-GCSluc or pGI-GPxluc and incubated the cells in the presence of the radical scavenger N-acetylcysteine (NAC). The reporter gene assay shows that even high concentrations of NAC result only in a small decrease of HBV-dependent induction of ARE-regulated genes (Fig. 3a). Functionality of NAC was shown by inhibition of H2O2-dependent induction of NF-κB (Fig. 3a). On the other hand, activation of Nrf2 can be triggered by c-Raf (41, 42). HBV is a well known activator of the c-Raf signal transduction cascade (43) Cotransfection experiments of HepG2 cells with pHBV1.2 and the reporter construct pNQO1luc revealed that inhibition of c-Raf by coexpression of the tdn mutant RafC-4 or by the small molecule inhibitor Bay 43-9006/Nexavar® caused a complete loss of HBV-dependent induction of the reporter gene (Fig. 3b). Moreover, inhibition of MEK with PD98059 caused a significant reduction, whereas inhibition of p38 MAPK with SB203580 or inhibition of protein kinase C with calphostin-C failed to exert any effect on HBV-mediated activation of the reporter gene.
FIGURE 3.
HBV-dependent activation of Nrf2/ARE-regulated genes requires c-Raf. a, reporter gene assay in HepG2 cells cotransfected with pHBV1.2 (0.4 μg) and luciferase reporter constructs pγ-GCSluc (0.2 μg) or pGI-GPxluc (0.2 μg). The vector pCDNA.3 (0.4 μg) served as control and was set as 1. N-acetyl-l-cysteine (NAC) was added to the medium and served as radical scavenger. In the lower panel, functionality of NAC was shown by inhibition of the H2O2-dependent activation of the NF-κB reporter construct. HepG2 cells were transfected with a luciferase reporter construct 2×NF-κBluc (0.2 μg), and grown in the presence of increasing concentrations of NAC. 6 h prior harvest, cells were stimulated with 150 μm H2O2. Cells stimulated with 150 μm H2O2 for 6 h in the absence of NAC served as positive control (pos. Contr.), and unstimulated cells served as negative (neg.) control. Activities, shown as multiples of induction, are mean values from three independent experiments. The error bars represent the standard deviation. rel units, relative units. b, reporter gene assay of HepG2 cells cotransfected with pHBV1.2 (1.0 μg), pNQO1luc (0.2 μg), and pCDNA.3 (1.0 μg) to adjust the total DNA concentration. Cotransfection of pRafC4 (1.0 μg) was used to inhibit c-Raf, the small molecule inhibitors PD98095 for MEK, SB203580 for p38 MAPK, and calphostin C as a PKC inhibitor. Activities, shown as multiples of induction, are mean values from three independent experiments. The error bars represent the standard deviation. c, HepG2 cells were cotransfected with pHBV1.2 (0.4 μg), pHBx (0.4 μg), or pSVLM−S− (0.4 μg) and the luciferase reporter construct pγ-GCSluc (0.2 μg). The vector pCDNA.3 (0.8 μg) served as control and was set as 1. For inhibition of Nrf2 activity, ptdnNrf2 (0.4 μg) was cotransfected. In the other samples, the equal amount of pCDNA.3 (0.4 μg) was added to the transfection mixture. Activities, shown as multiples of induction, are mean values from three independent experiments. The error bars represent the standard deviation. d, HepG2 cells were cotransfected with pHBV1.2 (0.8 μg), pHBV1.2ΔHBx (0.8 μg), pHBV1.2Δ PreS2 (0.8 μg), or pHBV1.2ΔHBx/PreS2 (0.8 μg) and the luciferase reporter construct pNQO1luc (0.2 μg). The vector pCDNA.3 (0.8 μg) served as control and was set as 1. Activities, shown as multiples of induction, are mean values from three independent experiments. The error bars represent the standard deviation.
To study whether activation of MEK per se is sufficient to trigger induction of ARE-dependent genes, HepG2 cells were transfected with pNQO1-luc, pγ-GCS-luc, or pGI-GPxluc and treated with insulin or EGF. The reporter gene assays demonstrate that under these conditions a significant induction of the reporter genes could be observed (Fig. 1a).
Because the two regulatory proteins of HBV, HBx and LHBs, are known to activate c-Raf (3, 4, 44), we asked whether HBx or LHBs are able to induce expression of ARE-regulated genes. Expression vectors for HBx (pHBx) or LHBs (pSVLM-S-) (45) were cotransfected with pNQO1luc. The reporter gene assay shows that both regulatory proteins activate the reporter gene to a similar extent as complete HBV. Moreover, the HBx- or LHBs-dependent activation of the reporter constructs depends on the integrity of Nrf2; coexpression of tdnNrf2 completely abolishes the HBx- or LHBs-dependent activation of the reporter gene (Fig. 3c).
To confirm the relevance of HBx and LHBs for the HBV-dependent induction of ARE-regulated genes, HepG2 cells were cotransfected with a complete HBV genome or mutant genomes lacking a functional PreS2 activator (HBVΔPreS2), or HBx (HBVΔHBx), or a double mutant lacking both regulatory functions (HBVΔHBx/PreS2) and the pNQO1luc reporter construct. The reporter gene assay shows that lack of one regulatory protein does not result in a significant reduction of the reporter gene activation, whereas the double mutant failed to induce the reporter gene (Fig. 3d). This is in accordance with previous results that have demonstrated that loss of one regulatory protein can be compensated by the other one (33, 34, 45). Taken together, these data indicate that HBV via its regulatory proteins HBx and LHBs induces an activation of Nrf2/ARE-regulated genes that depends on the functionality of c-Raf.
HBV Replication Does Not Depend on Nrf2
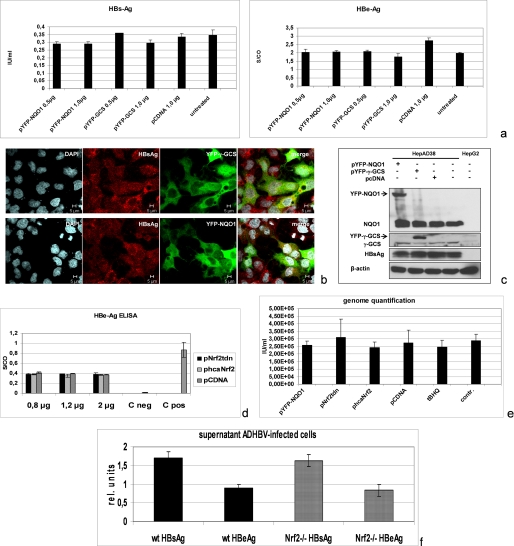
The data described above demonstrate the capacity of HBV to activate ARE-regulated genes via Nrf2. In the next set of experiments, we asked whether the expression level of ARE-regulated genes directly affects HBV. To investigate this, the HBV-positive stable cell line HepAD38 was cotransfected with expression vectors encoding NQO1 or γ-GCS or the empty control vector pCDNA.3. The effect on HBV replication was analyzed by HBeAg or HBsAg-ELISA, Western blot analysis, confocal double fluorescence microscopy, or real time PCR for quantification of the secreted viral particles. All approaches demonstrate that overexpression of NQO1 or γ-GCS does not affect HBV replication (Fig. 4, a–c). In the next set of experiments, a general overexpression of ARE-regulated genes was achieved by transfection of HepAD38 with a constitutively active mutant of Nrf2. The effect on HBV replication was analyzed by HBeAg-specific ELISA (Fig. 4d) and by real time PCR (Fig. 4e). The ELISA and the real time PCR demonstrate that activation of ARE-regulated gene expression does not affect HBV replication. Functionality of constitutively active Nrf2 was demonstrated by induction of NQO1 expression after transient transfection (data not shown).
FIGURE 4.
HBV expression is not affected by enhanced or reduced expression of ARE-regulated genes. HepAD38 were cotransfected with expression vectors encoding YFP-NQO1 or YFP-γ-GCS-fusion proteins. Cotransfection with YFP served as control. a, secretion of HBsAg and of HBeAg was quantified by ELISA. b, confocal laser scanning immunofluorescence microscopy (630-fold magnification). LHBs was detected by MA18/7 (red fluorescence) and YFP-NQO1 or YFP-γ-GCS (green fluorescence); nuclei were stained with DAPI. c, Western blot analysis of cellular lysates using an LHB-specific antiserum (MA18/7). Expression of the YFP-NQO1 or YFP-γ-GCS fusion proteins was demonstrated by NQO1- or γ-GCS-specific antibodies. d and e, HepAD38 cells were cotransfected with pcaNrf2 or ptdnNrf2. Transfection with pCDNA.3 served as control. The amount of secreted HBeAg (d) was determined by ELISA. The amount of secreted viral particles was quantified by real time PCR (e). f, primary mouse hepatocytes isolated from Nrf2−/− mice or from the corresponding WT mice were infected with AdHBV. The expression of HBV was quantified by HBsAg- and HBeAg-specific ELISA.
To study the effect of Nrf2 inhibition on HBV replication, HepAD38 cells were transfected with ptdnNrf2. HBeAg-specific ELISAs as well as real time PCR revealed that inhibition of Nrf2 does not impair HBV replication (Fig. 4, d and e).
To confirm the independence of HBV replication from Nrf2, primary mouse hepatocytes were isolated from Nrf2−/− mice (22) or the corresponding wild-type mice and infected with recombinant adenovirus encoding HBV. HBsAg- and HBeAg-specific ELISA revealed that Nrf2-deficiency does not affect HBV gene expression (Fig. 4f).
Taken together these data indicate that HBV replication does not depend on Nrf2. Neither inhibition of Nrf2 nor constitutive activation affects expression of HBV genes.
HBV-positive Cells Are More Protected against Oxidative Damage Than Control Cells
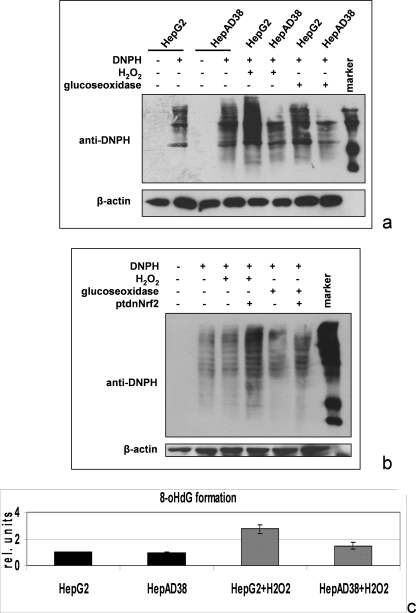
During the infection process, HBV-positive cells encounter increased ROS levels generated by inflammatory cells as part of the immune response. Therefore, it may well be that induction of ARE-regulated genes protects HBV-positive cells from oxidative damage. To test this hypothesis, we incubated the HBV-positive cell line HepAD38 with H2O2 or glucose oxidase and analyzed the formation of oxidized proteins using OxyBlot analysis. Comparable signals were obtained in the OxyBlot in case of untreated HepG2 and HepAD38 cells. However, after H2O2 or glucose oxidase treatment of the cells, the OxyBlot shows that significantly less oxidative damage of proteins occurred in HBV-positive cells as compared with the HepG2 control cells (Fig. 5a). Inhibition of the HBV-dependent induction of ARE-regulated genes by coexpression of the tdnNrf2 mutant abolished the protection of the HBV-positive cells against H2O2-dependent protein oxidation (Fig. 5b). Comparable results were obtained by analyzing the formation of the oxidative DNA adduct 8-OHdG by ELISA. In HBV-positive cells, significantly less 8-OHdG was formed after H2O2 stimulation as compared with the control cells (Fig. 5c) Taken together, these findings indicate that HBV-positive cells are better protected against oxidative damage than the corresponding control cells through Nrf2-mediated induction of ARE-regulated genes.
FIGURE 5.
HBV-dependent induction of the ARE-regulated genes protects HBV-positive cells from oxidative damage. a, OxyBlot analysis of cellular lysates derived from HepG2 or HepAD38 cells. In case of the 5th and 6th lanes, cells were treated for 30 min with 2 mm H2O2 and in case of the 7th and 8th lanes for 4 h with 20 milliunits of glucose oxidase. Protein oxidation was analyzed using 2,4-dinitrophenylhydrazine (DNPH) (2nd, 4th, and 5th to 8th lanes) for covalent modification of oxidized proteins. 2,4-Dinitrophenylhydrazine was omitted in the 1st and 3rd lanes to demonstrate the specificity of the observed signals. b, OxyBlot analysis of cellular lysates derived from ptdnNrf2- or control plasmid-transfected HepAD38 cells treated for 30 min with 2 mm H2O2 in case of the 3rd and 4th lanes or for 4 h with 20 milliunits of glucose oxidase (5th and 6th lanes). c, 8-OHdG-specific ELISA of hydrolyzed chromosomal DNA isolated from untreated or H2O2-treated HepG2 or HepAD38. Stimulation with H2O2 was performed as described above (a and b).
Decreased Activity of the Immunoproteasome in HBV-positive Cells
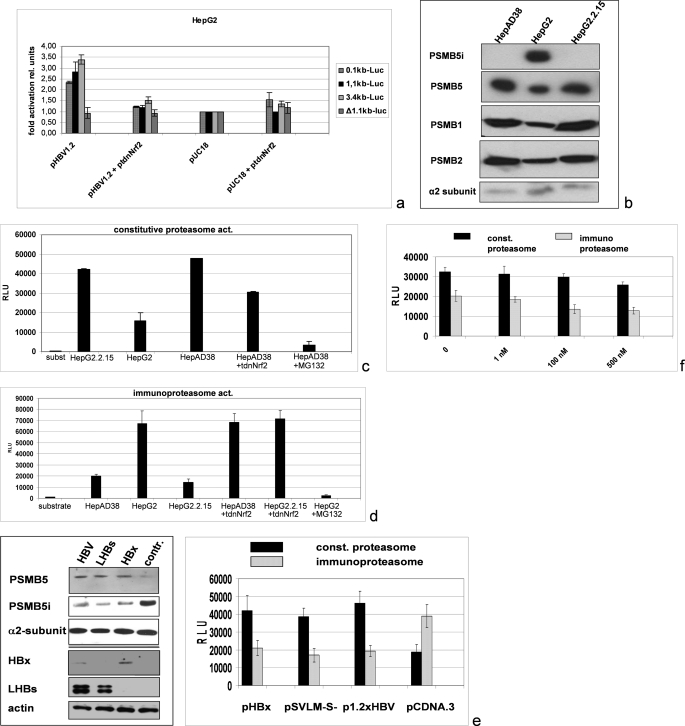
A crucial factor for the establishment of the viral infection and for the viral pathogenesis is the antigen processing. Recent reports described that Nrf2/ARE is involved in the expression control of the proteasomal β-subunits 1, 3, and 5 (31). Cotransfection of the HBV expression vector pHBV1.2 with luciferase reporter construct harboring the complete promoter of the proteasomal subunit PSMB5 (pPSMB5luc) showed a significant activation of the reporter gene (Fig. 6a). The PSMB5 promoter encompasses close to its 5′ end three XRE sequences and close to its 3′ end two ARE sequences (31, 46). Cotransfection of pHBV1.2 with a 5′ end deleted reporter construct that lacks all XRE sequences but still possesses the two ARE sequences (p1.1PSMB5luc) revealed a significant activation of this reporter construct by HBV comparable with the activation of the complete promoter (Fig. 6a). To analyze the relevance of the ARE sites for the HBV-dependent induction of the reporter construct, the two ARE sites were destroyed by site-directed mutagenesis. The reporter gene assay shows that cotransfection of pHBV1.2 with the mutated construct (p1.1PSMB5lucΔARE341/52) does not result in a significant induction of the reporter construct (Fig. 6a). This demonstrates the relevance of the ARE sequence for the HBV-dependent induction of the PSMB5 expression.
FIGURE 6.
Increased activity of the constitutive and decreased activity of the immunoproteasome in HBV-positive cell lines as compared with HepG2 cells. a, reporter gene assay of HepG2 cells cotransfected with pHBV1.2 (0.4 μg) and luciferase (luc) reporter constructs (0.2 μg) harboring different fragments of the promoter of the proteasomal subunit PSMB5 as follows: the complete promoter in case of 3.4-kb luciferase encompassing three XRE and two ARE sequences; the 1.1-kb fragment lacking the three XRE sequences but still harboring two ARE sequences; the 0.1-kb fragment that still harbors one ARE sequence; or the mutated 1.1-kb fragment harboring two mutated ARE sequences (p1.1PSMB5lucΔARE341/52). ptdnNrf2 (0.4 μg) was used to inhibit Nrf2 activity. pUC 18 (0.4 μg) was used as negative control and to adjust equal DNA amounts in all samples. Activities, shown as multiples of induction, are mean values from three independent experiments. The error bars represent the standard deviation. b, Western blot analyses of purified proteasomes from HepAD38, HepG2.2.15, and HepG2 cells using PSMB5- and PSMB5i (=LMP7)-specific antisera. Moreover PSMB1- and PSMB2-specific antisera were used. Detection of the α2 subunit served as loading control. c and d, analysis of constitutive (const.) (c) or immune (d) proteasomal activity (act.) by measuring the cleavage of the substrate peptides LLVY-AMC (c) or Cbz-VVRR-AMC (d). Proteasomes were isolated from HepG2 cells (control (contr.)) or from the HBV-positive cell lines HepG2.2.15 or HepAD38. Nrf2 was inhibited by coexpression of ptdnNrf2. MG132 was used to inhibit proteasomal activity. e, Western blot analyses (left panel) of proteasomes purified from pHBx, pSVLM-S-, pHBV1.2, or pCDNA.3-transfected HepG2 cells. For Western blotting PSMB5-, PSMB5i (=LMP7)-, HBx-, and PreS1/LHBs-specific antisera were used. Detection of the α2 subunit (upper panel) and of β-actin (lower panel) served as loading controls. Analysis of constitutive and immunoproteasomal activities was performed as above (right panel). f, proteasomal activities of HepG2 cell-derived proteasomes were determined in the presence of the indicated concentrations of HBx that was added to the reaction mixture. RLU, relative light units.
In accordance to this, Western blot analysis of purified proteasomes derived from the HBV-positive cell lines HepAD38 or HepG2.2.15 compared with HepG2-derived proteasomes clearly demonstrates an increased level of PSMB5 in HBV-positive cells as compared with HBV-negative HepG2 cells (Fig. 6b). Comparable results were obtained for the β1- and β2-subunits (PSMB1 and PSMB2) (Fig. 6b).
In the immunoproteasome, the β1-, β2-, and β5-subunits are replaced by their interferon-γ inducible forms β1i, β2i, and β5i, resulting in an altered proteasomal specificity. The products of the immunoproteasome with an average length of 8–10 amino acids seem to be optimized for the presentation by MHC-1 on the cell surface. The amounts of PSMB5 and of PSMB5i (= LMP7) that is part of the immunoproteasome are inversely regulated (47). Western blot analysis of purified proteasomes or cellular lysates derived from HepG2.2.15 or HepAD38 indeed revealed significantly lower levels of PSMB5i/LMP7 in these cells as compared with HepG2 cells (Fig. 6b).
The resulting question was whether these changes are reflected by a higher constitutive proteasome activity in HBV-positive cells compared with HBV-negative cells. Analyses of proteasomal activity indeed show a significantly increased constitutive proteasome activity in the case of HepG2.2.15 or HepAD38 cells as compared with HepG2. Interestingly, coexpression of tdnNrf2 caused a decrease in the constitutive proteasome activity of HBV-positive cells, confirming the relevance of Nrf2 for the HBV-dependent induction of the constitutive proteasome activity (Fig. 6c).
Analysis of the immunoproteasome activity using the HBV core(141–151)-derived peptide Cbz-VVRR-AMC (38) revealed that the immunoproteasome activity in HBV-positive cells is decreased as compared with the HepG2 control cells (Fig. 6d). Here, coexpression of the tdnNrf2 mutant resulted in an increased activity of the immunoproteasome in HBV-positive cells (Fig. 6d). Comparable results were obtained for the HBV-pol(803–811)-derived peptide substrate Cbz-SPSV-AMC (37) (data not shown). To clarify the relevance of the viral regulatory proteins for the deregulation of proteasomal activity and to verify the results described above by a transient expression system, HepG2 cells were transfected with pHBx, pSVLM-S-, or with pHBV1.2 as positive control and transfection with pCDNA.3 as negative control.
Western blot analysis of the purified proteasomes revealed that both HBx and LHBs trigger the increased formation of PSMB5 that is associated with a decreased formation of PSMB5i (Fig. 6e). In accordance with this finding, we found increased activity of the constitutive proteasome and decreased activity of the immunoproteasome in LHBs- or HBx-expressing cells as compared with the negative control (Fig. 6e). The decrease of the immunoproteasome activity was stronger in HBx-producing cells than in LHBs-producing cells. Recent reports demonstrated that HBx could impair proteasomal activity (48–50). To study this, we added increasing amounts of purified HBx to the proteasomal fraction isolated from HepG2 cells and measured the activities of the immunoproteasome and of the constitutive proteasome. Although the constitutive proteasomal activity measured by the cleavage of the fluorophor-coupled substrate peptide was only impaired by higher concentrations, a stronger inhibitory effect was observed for the activity of the immunoproteasome (Fig. 6f).
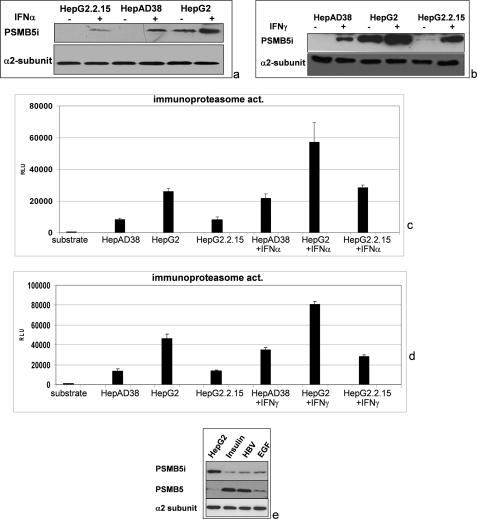
IFNα and IFNγ are strongly expressed in the HBV-infected liver. It was found that LMP7 and LMP2 subunits affect the magnitude of CD8 T-cell response in HLA-A2 transgenic mice (37). Because of the relevance of the interferon-inducible subunits for the HBV-specific CD8 T-cell response and thereby for the progression of the infection, we analyzed whether stimulation of HBV-replicating cells with IFNα or IFNγ overcomes the inhibitory effect of HBV on the activity of the immunoproteasome. Western blot analysis shows that both IFNα and IFNγ increase the amount of PSMB5i in HBV-replicating cells, but the level of PSMB5i observed for HepG2 cells was not achieved (Fig. 7, a and b). Comparable results were obtained upon analysis of the immunoproteasomal activity. For both HBV-replicating cell lines, an increase in the activity of the immunoproteasome was observed after IFNα or IFNγ stimulation. However, the immunoproteasomal activity remained significantly lower as compared with the HBV-negative HepG2 control cells (Fig. 7, c and d).
FIGURE 7.
Decreased immunoproteasome activity in HBV-expressing cells is not fully rescued by interferon α or interferon γ. HepAD38, HepG2.2.15, and HepG2 cells were stimulated for 3 days with IFNα (a and c) or IFNγ (b and d) prior to isolation of the proteasomes. a and b, Western blot analyses of purified proteasomes using PSMB5i-specific antisera. Detection of the α2 subunit served as loading control. c and d, analysis of immunoproteasomal activity by measuring the cleavage of the substrate peptide Cbz-VVRR-AMC. e, Western blot analyses of proteasomes purified from pHBV1.2- or pCDNA.3-transfected HepG2 cells. Moreover, control transfected cells were stimulated by the addition of EGF (10 ng/ml) or insulin (50 nm) for 48 h. For Western blotting, PSMB5- and PSMB5i-specific antisera were used. Detection of the α2 subunit served as loading control. RLU, relative light units.
In accordance to the data described above that have demonstrated that stimulation of MEK by EGF or insulin MEK per se is sufficient to trigger induction of ARE-dependent reporter genes in HepG2 cells (Fig. 1a), we observed that stimulation with EGF or insulin for 48 h resulted in an increased amount of PSMB5 and a decreased amount of PSMB5i (Fig. 7e). This demonstrates that constitutive activation of c-Raf/MEK signal transduction cascade affects the expression of PSMB5 and PSMB5i. Taken together, these data indicate that the activity of the constitutive proteasome is increased in HBV-positive cells, whereas the activity of the immunoproteasome is decreased.
DISCUSSION
In this study we demonstrate the capacity of HBV to stimulate the expression of a variety of cytoprotective genes that are Nrf2/ARE-regulated. The HBV-dependent induction of these genes is primarily initiated by the two regulatory proteins of HBV, HBx and LHBs, and is mediated by c-Raf. In accordance to this, stimulation of the c-Raf/MEK signal transduction cascade by insulin or EGF induces expression of Nrf2/ARE-dependent genes in cell culture. This seems to argue against an HBV-specific component triggering the expression of Nrf2/ARE-regulated genes in vitro. However, regarding long term effects in an organism, there might be a difference between a growth factor-dependent stimulation and an induction triggered by a chronic HBV infection. The latter one represents a process based on multiple factors besides the induction of this signaling cascade by the two regulatory proteins. Our data argue against a prominent role of ROS as mediators of the HBV-dependent induction of Nrf2/ARE-regulated genes (51). However, there are many reports discussing ROS as mediators of the regulatory protein function or as important factors for HBV-dependent induction of HCCs (52). Indeed, endoplasmic reticulum overload due to increased production of the HBV envelope proteins could enhance the formation of intracellular radicals, together with the immune system that tries to eliminate HBV-positive cells. In light of this, the induction of cytoprotective genes that mediate the inactivation of ROS might confer to ensure the survival of the host cell and to maintain the genetic integrity of the host and viral genomes. In contrast to reports discussing radical formation as a causative factor for HBV-associated HCC (7), our data suggest (in accordance with a previous report (53)) that HBV can induce protection against oxidative damage by triggering the expression of Nrf2/ARE-regulated genes.
Chronic HBV infection can result in liver fibrosis, cirrhosis, or HCC. A recent report (15, 25) demonstrated that Nrf2 protects from toxin-induced liver injury and fibrosis. At first glance, these observations seem to be contradictory. However, during chronic infection a permanent inflammatory process occurs, which is characterized by an increased ROS level triggered by the insufficient immune response. The induction of Nrf2/ARE-regulated genes by HBV protects HBV-positive cells and thereby ensures viral replication. One could speculate that the lack of the HBV-dependent induction of Nrf2/ARE-regulated genes might result in an earlier onset of fibrosis. Therefore, it will be worthwhile to explore if defects in Nrf2 activation affect the HBV-associated pathogenesis.
On the other hand, the overexpression of ARE-regulated genes in HCCs confers a growth advantage to these tumors (16). Consistent with these findings, we observed an increased expression of ARE-regulated genes in HBV-associated HCCs.4 The overexpression of cytoprotective proteins is likely to protect the tumor from elimination by mechanisms that are based on increased levels of electrophiles or radicals.
In HBV-positive cells, the increased amount of PSMB5 and the higher constitutive proteasomal activity are associated with a decreased amount of PSMB5i (LMP7) and a decreased immunoproteasome activity as compared with the HBV-negative cells. Inhibition of Nrf2 abolished these effects, demonstrating the relevance of Nrf2/ARE for this. The reprogramming of cellular protein synthesis in case of HBV-infected cells and the resulting strong overproduction of viral proteins might be associated with an increased amount of misfolded proteins. The rapid elimination of misfolded proteins by the proteasome prevents their accumulation and thereby the cellular stress (54). Therefore, proteasome activation seems to be a valuable strategy of the infected cells to reduce the stress resulting from misfolded proteins. In accordance to this, an inhibitory effect of proteasome inhibition by bortezomib on HBV replication can be observed in cell culture (see supplemental material) and in HBV transgenic mice (55).
In the immunoproteasome, PSMB5 is replaced by PSMB5i=LMP7 (47). In the mouse model, it was shown that incorporation of LMP7 into the proteasome enhances the production of the immunodominant HBV-pol(803–811) epitope and alters the magnitude and specificity of the CD8 T-cell response to HBV envelope protein (37). Based on this result, it can be speculated that the reduced immunoproteasome activity even after interferon stimulation in HBV-replicating hepatocytes reduces the capability of antigen processing, finally conferring to an impaired elimination of HBV-positive hepatocytes. This would represent a cross-talk between HBV-triggered signal transduction pathways and the immune response.
In contrast to our data, there are reports based on recombinant adenoviruses encoding HBV or HBx overexpression that describe an interference of HBx with the proteasomal complex, resulting in inhibition of the proteasomal activity (48–50). Our in vitro data based on the addition of purified HBx to isolated proteasomes demonstrate that high amounts of HBx can exert an inhibitory effect. However, in the case of proteasomes isolated from HBV-producing cells, a significant activation of the constitutive proteasome is observed. One reason for the differences might be the significantly lower expression level of HBx that exists in the HBV-replicating systems used in our study as compared with a strong overexpression system used in the studies above (48–50). Moreover, the previous reports demonstrated that HBx must be part of the proteasomal complex to exert its inhibitory effect. Therefore, differences in the isolation protocol for the proteasomal complex might explain the observed differences. We could not detect HBx in Western blot analyses of the purified proteasomal fractions from HBV-replicating cells, suggesting that too small amounts of HBx were present in the proteasomal fraction or that HBx was lost during the isolation process and therefore could not exert an inhibitory effect.
However, it should be considered that even if the cells were transfected with a strong expression vector, only a very small amount of HBx molecules is found per cell. For this, the amount was calculated as 8000–24,000 molecules per cell (56). In contrast to this, 0.6–1% of the soluble protein in hepatocytes accounts for proteasomes (57). For L929 cells (which are much smaller compared with hepatocytes), it was calculated that these cells contain 5 × 105 proteasomes (58). In light of the fact that hepatocytes have about 10-fold higher volume (59), it can be calculated that, supposing that almost all of the HBx molecules would bind exclusively to the proteasome, less than 1% of the proteasomes are complexed with HBx. This might not be sufficient for a strong inhibition of the proteasomal activity based on a direct interaction of HBx with the proteasome (50).
Moreover, recent data demonstrated that inhibition of the proteasomal activity impair HBV replication, arguing against an inhibitory effect of HBx on the proteasome (55). In addition, it was reported recently that HBx induces the degradation of insulin receptor substrate 1 via the ubiquitin-proteasome pathway (60).
Taken together, the activation of ARE-regulated genes could be advantageous for HBV for several reasons. (i) The HBV-positive cells are protected from oxidative damage induced by the immune system or by endoplasmic reticulum overload. (ii) The integrity of the Nrf2/ARE system is essential for efficient liver regeneration (14) and thereby ensures survival of the host tissue. (iii) The up-regulation of the constitutive proteasome activity ensures efficient removal of misfolded proteins. (iv) The decreased immunoproteasome activity results in reduced antigen processing. In the future, it will be interesting to determine whether inhibition of Nrf2 activity can be therapeutically explored for the treatment of HBV infections.
Supplementary Material
Acknowledgments
We thank the Human Tissue and Cell Research Foundation, Regensburg, Germany, for providing us with the primary human hepatocytes.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft Excellence Cluster “Inflammation at Interfaces” (to E. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
E. Hildt, unpublished results.
- HBV
- hepatitis B virus
- ARE
- antioxidant response element
- GPx
- glutathione peroxidase
- tBHQ
- tert-butylhydroquinone
- 8-OHdG
- 8-hydroxy-2′-deoxyguanosine
- AMC
- 7-amino-4-methylcoumarin
- tdn
- trans-dominant negative
- NAC
- N-acetyl-l-cysteine
- Cbz
- benzyloxycarbonyl
- HCC
- hepatocellular carcinoma
- LHBs
- large hepatitis B virus surface protein
- HBx
- hepatitis B virus X protein
- ROS
- reactive oxygen species
- XRE
- xenobiotic response element.
REFERENCES
- 1.Lupberger J., Hildt E. (2007) World J. Gastroenterol. 13, 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beasley R. P., Hwang L. Y., Lin C. C., Chien C. S. (1981) Lancet 2, 1129–1133 [DOI] [PubMed] [Google Scholar]
- 3.Benn J., Schneider R. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10350–10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hildt E., Munz B., Saher G., Reifenberg K., Hofschneider P. H. (2002) EMBO J. 21, 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu H., Wang Y., Chen J., Cheng G., Xue J. (2004) Exp. Mol. Pathol. 76, 44–50 [DOI] [PubMed] [Google Scholar]
- 6.Melegari M., Wolf S. K., Schneider R. J. (2005) J. Virol. 79, 9810–9820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagen T. M., Huang S., Curnutte J., Fowler P., Martinez V., Wehr C. M., Ames B. N., Chisari F. V. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12808–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehermann B., Nascimbeni M. (2005) Nat. Rev. Immunol. 5, 215–229 [DOI] [PubMed] [Google Scholar]
- 9.Jaiswal A. K. (2004) Free Radic. Biol. Med. 36, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 10.Vasiliou V., Ross D., Nebert D. W. (2006) Hum. Genomics 2, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasserman W. W., Fahl W. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aleksunes L. M., Manautou J. E. (2007) Toxicol. Pathol. 35, 459–473 [DOI] [PubMed] [Google Scholar]
- 13.Li W., Kong A. N. (2009) Mol. Carcinog. 48, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyer T. A., Xu W., Teupser D., auf dem Keller U., Bugnon P., Hildt E., Thiery J., Kan Y. W., Werner S. (2008) EMBO J. 27, 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W., Hellerbrand C., Köhler U. A., Bugnon P., Kan Y. W., Werner S., Beyer T. A. (2008) Lab. Invest. 88, 1068–1078 [DOI] [PubMed] [Google Scholar]
- 16.Lau A., Villeneuve N. F., Sun Z., Wong P. K., Zhang D. D. (2008) Pharmacol. Res. 58, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sells M. A., Chen M. L., Acs G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladner S. K., Otto M. J., Barker C. S., Zaifert K., Wang G. H., Guo J. T., Seeger C., King R. W. (1997) Antimicrob. Agents Chemother. 41, 1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dayoub R., Thasler W. E., Bosserhoff A. K., Singer T., Jauch K. W., Schlitt H. J., Weiss T. S. (2006) Biochem. Biophys. Res. Commun. 345, 181–187 [DOI] [PubMed] [Google Scholar]
- 20.Thasler W. E., Dayoub R., Mühlbauer M., Hellerbrand C., Singer T., Gräbe A., Jauch K. W., Schlitt H. J., Weiss T. S. (2006) J. Pharmacol. Exp. Ther. 316, 822–829 [DOI] [PubMed] [Google Scholar]
- 21.Lupberger J., Mund A., Kock J., Hildt E. (2006) J. Hepatol. 45, 547–552 [DOI] [PubMed] [Google Scholar]
- 22.Chan K., Kan Y. W. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12731–12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klingmuller U., Bauer A., Bohl S., Nickel P. J., Breitkopf K., Dooley S., Zellmer S., Kern C., Merfort I., Sparna T., Donauer J., Walz G., Geyer M., Kreutz C., Hermes M., Gotschel F., Hecht A., Walter D., Egger L., Neubert K., Borner C., Brulport M., Schormann W., Sauer C., Baumann F., Preiss R., MacNelly S., Godoy P., Wiercinska E., Ciuclan L., Edelmann J., Zeilinger K., Heinrich M., Zanger U. M., Gebhardt R., Maiwald T., Heinrich R., Timmer J., von Weizsacker F., Hengstler J. G. (2006) Syst. Biol. 153, 433–447 [DOI] [PubMed] [Google Scholar]
- 24.Sprinzl M. F., Oberwinkler H., Schaller H., Protzer U. (2001) J. Virol. 75, 5108–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heermann K. H., Goldmann U., Schwartz W., Seyffarth T., Baumgarten H., Gerlich W. H. (1984) J. Virol. 52, 396–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich B., Wollersheim M., Brandenburg B., Foerste R., Will H., Hildt E. (2005) J. Hepatol. 43, 696–703 [DOI] [PubMed] [Google Scholar]
- 27.Favreau L. V., Pickett C. B. (1995) J. Biol. Chem. 270, 24468–24474 [DOI] [PubMed] [Google Scholar]
- 28.Bea F., Hudson F. N., Chait A., Kavanagh T. J., Rosenfeld M. E. (2003) Circ. Res. 92, 386–393 [DOI] [PubMed] [Google Scholar]
- 29.Banning A., Deubel S., Kluth D., Zhou Z., Brigelius-Flohé R. (2005) Mol. Cell. Biol. 25, 4914–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.auf dem Keller U., Huber M., Beyer T. A., Kümin A., Siemes C., Braun S., Bugnon P., Mitropoulos V., Johnson D. A., Johnson J. A., Hohl D., Werner S. (2006) Mol. Cell. Biol. 26, 3773–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwak M. K., Kensler T. W. (2006) Biochem. Biophys. Res. Commun. 345, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 32.Bruder J. T., Heidecker G., Rapp U. R. (1992) Genes Dev. 6, 545–556 [DOI] [PubMed] [Google Scholar]
- 33.Stöckl L., Berting A., Malkowski B., Foerste R., Hofschneider P. H., Hildt E. (2003) Oncogene 22, 2604–2610 [DOI] [PubMed] [Google Scholar]
- 34.Hafner A., Brandenburg B., Hildt E. (2003) EMBO Rep. 4, 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying C., De Clercq E., Neyts J. (2003) Biochem. Biophys. Res. Commun. 309, 482–484 [DOI] [PubMed] [Google Scholar]
- 36.Ehrhardt C. S., Schmolke M., Matzke A., Knoblauch A., Will C., Wixler V., Ludwig S. (2006) Signal Transduction, Vol. 6, pp. 179–184, Weinheim, Germany [Google Scholar]
- 37.Robek M. D., Garcia M. L., Boyd B. S., Chisari F. V. (2007) J. Virol. 81, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sijts A. J., Ruppert T., Rehermann B., Schmidt M., Koszinowski U., Kloetzel P. M. (2000) J. Exp. Med. 191, 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandenburg B., Stockl L., Gutzeit C., Roos M., Lupberger J., Schwartlander R., Gelderblom H., Sauer I. M., Hofschneider P. H., Hildt E. (2005) Hepatology 42, 1300–1309 [DOI] [PubMed] [Google Scholar]
- 40.Dhakshinamoorthy S., Jaiswal A. K. (2000) J. Biol. Chem. 275, 40134–40141 [DOI] [PubMed] [Google Scholar]
- 41.Kensler T. W., Wakabayashi N., Biswal S. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 42.Shen G., Hebbar V., Nair S., Xu C., Li W., Lin W., Keum Y. S., Han J., Gallo M. A., Kong A. N. (2004) J. Biol. Chem. 279, 23052–23060 [DOI] [PubMed] [Google Scholar]
- 43.Benn J., Schneider R. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11215–11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hildt E., Saher G., Bruss V., Hofschneider P. H. (1996) Virology 225, 235–239 [DOI] [PubMed] [Google Scholar]
- 45.Bruss V., Vieluf K. (1995) J. Virol. 69, 6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwak M. K., Wakabayashi N., Greenlaw J. L., Yamamoto M., Kensler T. W. (2003) Mol. Cell. Biol. 23, 8786–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung T., Catalgol B., Grune T. (2009) Mol. Aspects Med. 30, 191–296 [DOI] [PubMed] [Google Scholar]
- 48.Hu Z., Zhang Z., Doo E., Coux O., Goldberg A. L., Liang T. J. (1999) J. Virol. 73, 7231–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z., Protzer U., Hu Z., Jacob J., Liang T. J. (2004) J. Virol. 78, 4566–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z., Torii N., Furusaka A., Malayaman N., Hu Z., Liang T. J. (2000) J. Biol. Chem. 275, 15157–15165 [DOI] [PubMed] [Google Scholar]
- 51.Durchdewald M., Beyer T. A., Johnson D. A., Johnson J. A., Werner S., auf dem Keller U. (2007) J. Invest. Dermatol. 127, 646–653 [DOI] [PubMed] [Google Scholar]
- 52.Chemin I., Zoulim F. (2009) Cancer Lett. 286, 52–59 [DOI] [PubMed] [Google Scholar]
- 53.Severi T., Vander Borght S., Libbrecht L., VanAelst L., Nevens F., Roskams T., Cassiman D., Fevery J., Verslype C., van Pelt J. F. (2007) Chem. Biol. Interact. 168, 128–134 [DOI] [PubMed] [Google Scholar]
- 54.Pahl H. L., Baeuerle P. A. (1997) Trends Biochem. Sci. 22, 63–67 [DOI] [PubMed] [Google Scholar]
- 55.Bandi P., Garcia M. L., Booth C. J., Chisari F. V., Robek M. D. (2010) Antimicrob. Agents Chemother. 54, 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su F., Schneider R. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka K., Ii K., Ichihara A., Waxman L., Goldberg A. L. (1986) J. Biol. Chem. 261, 15197–15203 [PubMed] [Google Scholar]
- 58.Princiotta M. F., Finzi D., Qian S. B., Gibbs J., Schuchmann S., Buttgereit F., Bennink J. R., Yewdell J. W. (2003) Immunity 18, 343–354 [DOI] [PubMed] [Google Scholar]
- 59.Higuchi A., Tsukamoto Y. (2004) J. Biomed. Mater. Res. A 71, 470–479 [DOI] [PubMed] [Google Scholar]
- 60.Kim K., Kim K. H., Cheong J. (2010) PLoS One 5, e8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.