Abstract
Heparan sulfate (HS) is a structurally complex polysaccharide that interacts with a broad spectrum of extracellular effector ligands and thereby is thought to regulate a diverse array of biologic processes. The specificity of HS-ligand interactions is determined by the arrangement of sulfate groups on HS, which creates distinct binding motifs. Biologically important HS motifs are expected to exhibit regulated expression, yet there is a profound lack of tools to identify such motifs; consequently, little is known of their structures and functions. We have identified a novel phage display-derived antibody (NS4F5) that recognizes a highly regulated HS motif (HSNS4F5), which we have rigorously identified as (GlcNS6S-IdoA2S)3. HSNS4F5 exhibits a restricted expression in healthy adult tissues. Blocking HSNS4F5 on cells in culture resulted in reduced proliferation and enhanced sensitivity to apoptosis. HSNS4F5 is up-regulated in tumor endothelial cells, consistent with a role in endothelial cell activation. Indeed, TNF-α stimulated endothelial expression of HSNS4F5, which contributed to leukocyte adhesion. In a mouse model of severe systemic amyloid protein A amyloidosis, HSNS4F5 was expressed within amyloid deposits, which were successfully detected by microSPECT imaging using NS4F5 as a molecularly targeted probe. Combined, our results demonstrate that NS4F5 is a powerful tool for elucidating the biological function of HSNS4F5 and can be exploited as a probe to detect novel polysaccharide biomarkers of disease processes.
Keywords: Amyloid, Antibodies, Apoptosis, Heparan Sulfate, Inflammation, Sulfation
Introduction
Heparan sulfate proteoglycans (HSPGs),3 major components of the cell surface and the extracellular matrix, are involved in a variety of biological phenomena, including cell adhesion, proliferation, differentiation, and inflammation as well as being associated with pathologic events such as atherosclerosis and amyloidosis. Because of their high negative charge, HS chains interact with a variety of proteins, including growth factors/morphogens and their receptors, the amyloid precursor protein serum amyloid protein A (AA), chemokines, and extracellular matrix proteins. HS-protein interactions vary with regard to specificity and may depend on charge density in addition to strict sequence motifs of HS.
The interaction of heparin and HS with FGFs and their receptors has been characterized in great detail. Specific HS structures are predominantly determined by the regulated positioning of N-, 2-, 6-, and 3-O-sulfate groups along HS chains (1). For example, FGF-2 requires both N- and 2-O-sulfate groups for binding to HS. The 6-O-sulfate group is not essential for binding to FGF-2 but is critical for activation of the FGF receptor (2). In contrast, binding of hepatocyte growth factor, platelet-derived growth factor, lipoprotein lipase, and herpes simplex virus glycoprotein C to HS is dependent on 6-O-sulfation (3). The activation of antithrombin III by HS/heparin is mediated by a specific pentasaccharide in which a 3-O-sulfate group is crucial (4). Thus, the biological functions of HSPGs are controlled by biosynthetic events that define HS structures.
Although it is increasingly recognized that a specific domain structure, typically from a tetra- to a decasaccharide in size, is responsible for HS modulation of biological activity, in only a few cases is there any structural information regarding sequences (3–5). Therefore, accelerating our understanding of structure-function relationships for HS requires the development of rapid yet thorough sequencing methodologies. Antibodies reactive with HS have proven valuable tools to study specific HS motifs in cells and tissues (6–10). From the phage display-derived anti-HS antibodies available, some chemical HS modifications crucial for antibody binding have been identified; however, the exact saccharide sequence(s) of the motif remains elusive (7, 9–11). This impedes the assignment of specific HS motifs/sequences to biological and pathological phenomena.
Here, we report on the selection, characterization, and biological application of a novel phage display-derived antibody, NS4F5. The antibody reacts with a specific stretch of highly sulfated GlcNS6S-IdoA2S disaccharides (HSNS4F5) that is common in heparin but that exhibits a very restricted distribution in HS in healthy tissues. To study the cell biological relevance of this HSNS4F5 motif, we used the antibody as a blocking agent and showed that in vitro it inhibits cell proliferation, sensitizes cells to apoptosis, but does not affect cell attachment to collagen I. Increased expression of HSNS4F5 by activated human umbilical vein endothelial cells (HUVEC) contributed to leukocyte adhesion. The HSNS4F5 motif was highly up-regulated in human ovarian cancer. We further observed in mice the presence of this motif in visceral AA amyloid deposits with little or no reactivity of the antibody in healthy, amyloid-free tissues. Using labeled NS4F5 antibodies, amyloid deposits could be visualized by single photon emission computed tomography. Our studies provide new insight into the distribution and function of a specific HS motif (GlcNS6S-IdoA2S)3 and show that in addition to mediating cellular behavior it represents a novel biomarker of amyloid disease and possibly ovarian tumors. Taken together, the ability to identify specific HS structures with unique antibodies will accelerate our understanding of the relevance of polysaccharide structures to the regulation of biological functions.
EXPERIMENTAL PROCEDURES
Tissues and Cells
Rats (Wistar, male, 8 weeks old) were obtained from the Central Animal Laboratory (Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands). Human lung epithelial cells (A549) were cultured in F-12K nutrient mixture (Kaign's modification), supplemented with 10% FCS. CHO-K1 and CHO-pgsF-17 cells were cultured in Ham's F-12 medium supplemented with 10% FCS. HUVEC and the leukocyte (granulocyte) cell line 32Dcl3 were cultured as described (12, 13). Human ovarian cryosections from ovary, endometrioid, and serous adenocarcinomas were obtained from the archives of the Institute of Pathology of the Radboud University Nijmegen.
ScFv Antibodies
The DNA sequences encoding the antibodies were subcloned into vector pUC119-His-VSV (J. M. H. Raats, Department of Biochemistry, Faculty of Sciences, Radboud University Nijmegen Medical Centre). Periplasmic fractions of infected bacteria were isolated as described (9). Briefly, the bacteria were grown and induced by isopropyl β-d-thiogalactopyranoside to produce antibodies. Periplasmic fractions containing antibodies were isolated, dialyzed against PBS, and stored at −20 °C. The antibodies were purified using protein A-agarose or HIS-cobalt affinity-agarose beads. The antibody concentration was determined at 280 nm, and BSA (1%) and sodium azide (0.02%) were added for stabilization and preservation. The efficiency of purification was assessed by SDS-PAGE and Western blotting as described earlier (10).
Antibody Treatment
The cells were incubated for 24 h after passage before further manipulation. The medium was removed and replaced with 1% FCS or containing antibodies (10 or 50 μg/ml) in medium with 1% FCS. The cells were incubated 16 h for immunohistochemistry. As controls, the cells were incubated with control antibody MPB49, which is >95% identical to most antibodies used but does not bind glycosaminoglycans (GAGs).
Polysaccharides
For characteristics of polysaccharides, see supplemental Table S1.
Oligosaccharides
The specific activity of size-defined 3H end-labeled heparin oligosaccharides was ∼7.5 × 106 cpm/nmol. Heparin was chemically O-desulfated to eliminate all 6-O-sulfate and approximately half of the 2-O-sulfate groups and was subjected to partial deaminative cleavage at low pH followed by end labeling through reduction with NaB3H4. 3H end-labeled 8-mer (∼1.4 × 106 cpm/nmol) was recovered and separated into subfractions with defined numbers of 2-O-sulfates. The subfractions were separately incubated with mastocytoma microsomal enzymes in the presence of (unlabeled) adenosine 3′-phosphate 5′-phosphosulfate, primarily de novo attaching sulfate groups in the 6-O position (14).
Binding Assays
Affinity chromatography was performed on columns with 1 mg of antibody coupled to 0.5 mg of protein A beads in 50 mm Tris/HCl, pH 7.4, as indicated. Radiolabeled polysaccharide samples were eluted with a stepwise gradient of 0.15–2 m NaCl in 50 mm Tris/HCl, pH 7.4. Polysaccharide samples compared for antibody binding were applied in similar amounts, based on specific radioactivity determined by colorimetric analysis. Size-defined heparin oligosaccharides and oligosaccharide libraries were separated by affinity chromatography.
Reactivity of antibodies with K5 polysaccharide derivatives and modified heparin preparations (supplemental Table S1) was evaluated by ELISA (15). Wells of microtiter plates (Greiner, Frickenhausen, Germany) were coated as described (11). After blocking with PBS containing 3% (w/v) BSA and 1% (v/v) Tween 20 for 1 h, anti-HS antibodies (in PBS containing 1% (w/v) BSA and 0.1% (v/v) Tween 20) were added for 1.5 h. As a control, antibody MPB49 was used. Bound antibodies were detected using mouse IgG anti-VSV tag antibody P5D4 (1:10; Roche Applied Science, Mannheim, Germany), followed by incubation with alkaline phosphatase-conjugated rabbit anti-mouse IgG (1:2000; Dako, Glostrup, Denmark), both for 1 h. Enzyme activity was detected using 100 μl of 1 mg of p-nitrophenyl phosphate (ICN, Aurora, OH)/ml of 1 m diethanolamine, 0.5 mm MgCl2, pH 9.8, as a substrate. Absorbance was read at 405 nm.
Characterization of the HSNS4F5 Motifs
For sequencing of HSNS4F5 motifs, selected library octasaccharides were affinity-purified on an NS4F5 column. Binding and nonbinding fragments were separately pooled, desalted, dried, and sequenced (16). The samples were subjected to partial HNO2 cleavage by 2 mm NaNO2 in 20 mm HCl and stopped after different time points (0–60 min). Aliquots were pooled, and a fraction of the pool was analyzed by anion exchange chromatography directly, whereas a fraction was subjected to enzymatic digestion with recombinant IdoA2S-sulfatase. Digests were analyzed on a Propac column, and sequence information was obtained by detecting shifts in the elution positions of the enzyme-treated fragments.
Immunohistochemistry
The sections (4 μm) were air-dried and blocked with PBS containing 2% (w/v) BSA and 0.05% (v/v) Tween 20 (blocking buffer) for 20 min. The sections were incubated with anti-HS antibodies in blocking buffer for 1 h. As a control, antibody MPB49 was used. Bound antibodies were detected by incubation with mouse anti-VSV tag antibody P5D4 (1:10), followed by Alexa 488-conjugated goat anti-mouse IgG (1:200; Molecular Probes, Eugene, OR), both for 45 min. After each incubation, the sections were washed three times for 5 min with PBS containing 0.1% (v/v) Tween 20. The sections were fixed in 100% (v/v) ethanol for ∼10 s, air-dried, and embedded in 10% (w/v) Mowiol.
To evaluate antibody specificity, the sections were digested with the glycosidases heparin lyase III (digests HS) 0.04 IU/ml in 50 mm NaAc, 50 mm Ca(Ac)2, pH 7.0, or 0.02 unit/ml chondroitinase ABC (digests chondroitin sulfate and dermatan sulfate) in 25 mm Tris/HCl, pH 8.0 (2 h at 37 °C, refreshing the enzyme after 1 h). As a control, the sections were incubated in reaction buffer without enzyme. After washing three times with PBS and blocking for 30 min, the sections were incubated with anti-HS antibodies and processed for immunohistochemistry as above. The efficiency of heparin lyase III and chondroitinase ABC treatment was evaluated by incubation of sections with antibodies against GAG “stubs” generated by the glycosidases. For HS stubs, the antibody 3G10 (Seikagaku, Tokyo, Japan) was used. For chondroitin sulfate stubs the antibody 2B6 (Seikagaku) was used. All of the tests were performed at least three times. The sections were examined using a Leica CTR6000 microscope.
Immunoelectron Microscopy
Tissue was fixed for 3 h in Somogyi solution containing 4% (v/v) formaldehyde and 0.05% (v/v) glutaraldehyde in 0.1 m phosphate buffer. The sections (200 μm) were cut and incubated in increasing amounts of glycerol (10, 20, or 30%) in phosphate buffer for 30 min. The sections were oriented on Thermanox (LAB-TEK DVI, Miles Laboratories Inc., Naperville) and frozen in liquid propane. Freeze substitution was performed as described (17). Ultra-thin lowicryl HM20 resin sections were cut on a Reichert Ultracut-E and mounted on one-hole nickel grids coated with a Formvar film. The sections were preincubated in PBS containing 0.2% BSA and 0.05% cold fish skin gelatin (PBG). The sections were incubated overnight at 4 °C in drops of PBG containing antibodies and washed for 20 min in PBG. The bound antibodies were visualized using anti-VSV tag antibody P5D4 and goat anti-mouse IgG-labeled with gold spheres (10 nm; Aurion, Wageningen, Netherlands). The sections were washed in PBS and post-fixed with 2.5% (v/v) glutaraldehyde in PBS for 5 min. After washing with distilled water, the sections were contrasted with uranyl acetate and studied using a Jeol TEM 1010 electron microscope.
Cell Proliferation Assay
Human lung epithelial cells were incubated at 37 °C with or without purified scFv antibodies. Incubation was for 4 days in a humidified atmosphere. Cell proliferation was determined with the cell proliferation reagent WST-1 (Boehringer Mannheim), a tetrazolium salt cleaved by the mitochondria of viable cells to yield a soluble formazan chromophore. Relative cell density was determined according to the instructions provided by the manufacturer.
Cell Attachment Assay
Flat-bottomed 96-well plates (Greiner, Frickenhausen, Germany) were coated with 100 μl of type I collagen (Symatese, Chaponost, France) for 60 min and washed three times with PBS. The cells were labeled with calcein AM (Molecular Probes) in PBS for 30 min at 37 °C in the dark while shaking. The labeled cells were incubated with scFv antibodies (50 μg/ml) in medium supplemented with 1% FCS for 30 min and allowed to adhere for 45 min. Nonadherent cells were removed. The attached cells were lysed, and fluorescence was quantified in a cytofluorometer (Perseptive Biosystems). Adhesion was expressed as the mean percentage (± S.D.) of bound cells from four wells relative to untreated cells.
Detection of Apoptosis in Human Lung Epithelial Cells after Treatment with scFv Antibodies
The cells were treated with scFv antibodies (50 μg/ml) for 16 h, washed with PBS, fixed in ice-cold methanol for 10 min, washed three times with PBS, and blocked with blocking buffer for 10 min. Permeabilization and TUNEL labeling were performed according to the manufacturer's instructions (In Situ Cell Death Detection kit; Roche Applied Science), and the cells were incubated with 0.5 μg/ml DAPI for 1 min. The cells were washed with PBS, rinsed with ice-cold methanol, air-dried, and embedded in Mowiol. In addition to the TUNEL assay, apoptosis was detected using an M30 CytoDEATH antibody that detects a caspase cleavage site in cytokeratin 18 and labels apoptotic epithelial cells instead of viable or necrotic cells (18). To detect caspase cleavage, the cells were treated with scFv antibodies (50 μg/ml) for 16 h. After blocking, the cells were incubated with M30 antibody for 1 h. The cells were washed with PBS and incubated with goat anti-mouse IgG Alexa 488 for 1 h. The cells were washed with PBS, incubated with 0.5 μg/ml DAPI, then washed with PBS, air-dried, fixed with ice-cold methanol, and embedded with Mowiol.
Flow Cytometry
HUVEC cells were detached with 10 mm EDTA and washed with PBS. The cells were incubated with purified anti-HS antibodies (25 μg/ml) in PBA (PBS containing 0.5% (w/v) BSA) for 30 min and subsequently washed in PBA. Bound antibodies were detected by incubation with mouse anti-VSV tag antibody P5D4 (1:10), followed by Alexa 488-conjugated goat anti-mouse IgG (1:200; Molecular Probes), both in PBA for 30 min. The cells were washed and resuspended in PBA. Fluorescence was measured using a FACScan (Becton Dickinson, San Jose, CA).
Static Leukocyte Adhesion Assay
HUVEC was cultured in 96-well plates until confluence. 32Dcl3 cells (6 × 105 cells/ml) were labeled with calcein AM (25 μg/ml) in PBS for 30 min at 37 °C, washed in PBS, and resuspended in medium. HUVEC and 32Dcl3 cells were preincubated with purified anti-HS antibodies as indicated or the control MPB49 antibody (25 μg/ml) for 30 min at 37 °C. Standard curves of adhesion of different numbers of labeled leukocytes to HUVEC were always included. In general, 60,000 labeled leukocytes were added in one well and incubated for 30 min at 37 °C. After incubation, the plates were filled with medium and centrifuged upside down for 5 min at 300 × g. This washing step was repeated once; then subsequently cells were lysed in 50 mm Tris, pH 8.3, with 0.1% (w/v) SDS, and fluorescence was measured.
Single Photon Emission Computed Tomographic (SPECT) Imaging of Radiolabeled NS4F5 in Mice
AA amyloidosis was induced in 8-week-old female H2-Ld-huIL-6 Tg Balb/c (C) transgenic mice by administering intravenously in the lateral tail vein 100 μg of amyloid enhancing factor in 100 μl of PBS (19). The mice were used 6 weeks after the injection of amyloid enhancing factor, at which time AA deposits are widespread in the almost all visceral organs and tissues. Antibody NS4F5 was labeled with iodine-125 (125I) via production of iodotyrosine using chloramine T as the oxidizing agent (20). Free iodide was removed from the preparation chromatographically by using an Ultrogel AcA 34 column (Amersham Biosciences). Groups of three AA mice or healthy age-matched controls received ∼200 μCi of 125I-NS4F5 antibody (∼20 μg) intravenously in ∼200 μl of sterile PBS, and 4 h thereafter the animals were sacrificed by isoflurane inhalation overdose. SPECT and x-ray computed tomographic images were acquired using a microCAT II + SPECT imaging system (Siemens Preclinical Solutions, Knoxville, TN) as described previously (20–22).
Microautoradiography and Histology
Immediately following SPECT/computed tomographic imaging, the tissues were harvested and fixed in 10% formalin for 24 h, before being paraffin-embedded. 6-μm-thick sections were cut and placed on Probond microscope slides (Fisher Scientific). For autoradiography, the tissue sections were dipped in NTB-2 emulsion (Eastman Kodak), stored in the dark, developed after a 72-h exposure, and then counter-stained with hematoxylin and eosin. Consecutive tissue sections were stained for the presence of amyloid using alkaline Congo red. All of the sections were coverslipped using Permount (Fisher) and examined by light or polarizing microscopy. Digital images were acquired using a cooled CCD camera (Diagnostic Instruments, Sterling Heights, MI).
Statistical Analysis
Four independent experiments were subjected to statistical analysis. The results were expressed as the means ± S.D. The data were analyzed using Student's t test, as appropriate. p < 0.05 was considered significant for all tests.
RESULTS
Selection of scFv Antibody NS4F5
Four rounds of panning were performed against GAGs isolated from human lung tissue, the semi-synthetic scFv library 1 (23). Panning resulted in an increase of phage titer from 3 × 104 colony-forming units in the first round to 2 × 108 colony-forming units in the fourth round. Supernatants containing antibodies of clones obtained after the third and fourth selection were initially tested for reactivity with various GAGs. Of the clones reactive with heparin/HS, one clone produced an antibody (NS4F5) with a very selective reactivity toward (modified) heparin preparations and a restricted tissue staining pattern (see below). Therefore, this antibody was selected for further analysis. Antibody NS4F5 belongs to the VH3 family, has a DP53 germ line gene segment, and contains a heavy chain complementarity-determining region 3-amino acid sequence of SGRKGRMR.
Characterization of HSNS4F5-HSNS4F5 Has a Defined Saccharide Sequence: (GlcNS6S-IdoA2S)3
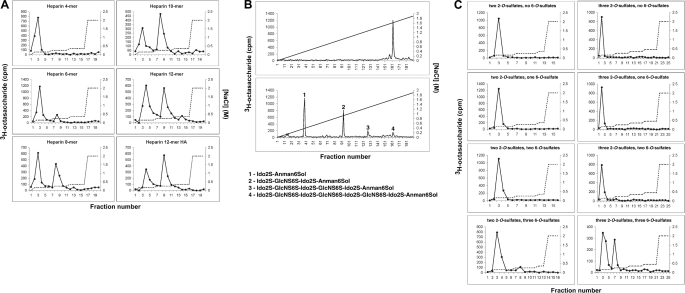
To identify HSNS4F5 and characterize its structure in detail, different approaches were taken. Initially, a large set of (modified) heparin and Escherichia coli cell wall polysaccharide K5 and other GAG preparations (supplemental Table S1) was analyzed for reactivity with NS4F5 (supplemental Fig. S1). To establish the saccharide sequence, we turned to heparin-derived oligosaccharide libraries (Fig. 1). Using a library containing 3H end-labeled heparin fragments, it was established that the minimal length for significant binding was an 8-mer (Fig. 1A). The binding heparin 8-mer eluted as an almost homogeneous late eluting (highly sulfated) oligosaccharide from an anion exchange column (Fig. 1B, upper panel). Cleavage of the bound, end-labeled 8-mer resulted in three additional fragments corresponding to end-labeled di-, tetra-, and hexasaccharides. Comparison of the elution characteristics of these saccharides with those of standards elucidated the saccharide sequence of the fragments and, by combination, the parent oligosaccharide (24). The disaccharide corresponded to IdoA2S-anManol6S (2-O-sulfated iduronic acid-6-O-sulfated anhydromannitol). The tetrasaccharide corresponded to IdoA2S-GlcNS6S-IdoA2S-anManol6S (GlcNS6S = N- and 6-O-sulfated glucosamine), the hexasaccharide to IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-anManol6S, and the octasaccharide, i.e. the original octasaccharide bound by the antibody, to IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-anManol6S. To substantiate these data, a biosynthetic heparin library was used, consisting of fully N-sulfated, epimerized octasaccharides with various numbers of 2-O- and/or 6-O-sulfation (Fig. 1C). Only the highest sulfated octasaccharide, harboring three 2-O-sulfated iduronic acids and three N- and 6-O-sulfated glucosamines, was able to bind NS4F5. Given the notion that IdoA2S residues are clustered in the heparin octasaccharide library, the binding octasaccharide had two monosaccharide-sequence possibilities: IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA-anManol and IdoA-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-anManol. The latter sequence is more likely because the library has been constructed by partial nitrous acid treatment that preferentially cleaves at sites of nonsulfated iduronic acid residues. Thus, HSNS4F5 contains three N-sulfates, three 2-O-sulfates, and three 6-O-sulfate groups. Taking into account the data obtained from the oligosaccharide libraries, it can be deduced that NS4F5 defines the sequence (GlcNS6S-IdoA2S)3. 3-O-Sulfation is of no importance because 3-O-sulfate-containing 12-mers (the antithrombin III high affinity fraction; Fig. 1A, Heparin 12-mer HA) do not bind any better than oligosaccharides without this modification.
FIGURE 1.
Sequence analysis of HSNS4F5. A, affinity chromatography on NS4F5-loaded columns using size-defined, 3H end-labeled heparin oligosaccharides (4–12-mer) and a 12-mer oligosaccharide preparation affinity selected on antithrombin III (12-mer HA). B, fractionation of NS4F5 affinity-purified octasaccharides (as in A) before (upper panel) or after (lower panel) partial deaminative cleavage by anion exchange HPLC on a Propac column, using a linear salt gradient for elution (diagonal line). C, reactivity of immobilized NS4F5 with heparin-derived, 3H end-labeled octasaccharide libraries with defined numbers of 2-O-sulfate groups and variable numbers of 6-O-sulfate groups using a stepwise increasing salt gradient as indicated (dotted line). The left column shows affinity separation of oligosaccharides with two 2-O-sulfate groups and zero, one, two, or three added 6-O-sulfate groups, respectively. The right panel displays corresponding patterns for octasaccharides containing three 2-O-sulfate groups (n = 4).
HSNS4F5 Is Present in HS and Has a Restricted Tissue Distribution
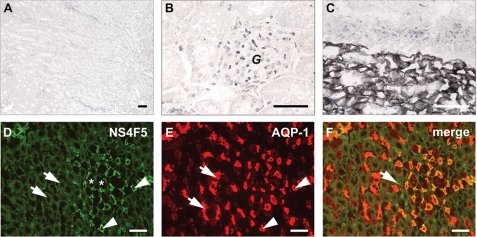
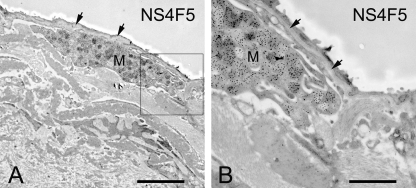
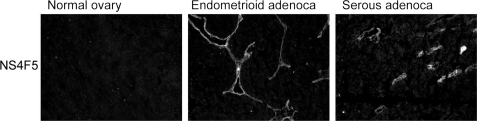
In rat renal tissue, NS4F5 stains the medullar region and not the cortex (Fig. 2). This is highly unusual because all of the anti-HS antibodies studied to date reacted strongly with the cortical region (25, 26). Treatment of renal sections with heparin lyase III, which removes HS, abolished staining, indicating that (GlcNS6S-IdoA2S)3 is present in HS. To further characterize HSNS4F5 in renal tubules, we performed co-localization studies with an antibody against aquaporin-1. NS4F5 staining was restricted to the descending thin limbs (Fig. 2D, asterisk) and small blood vessels (Fig. 2D, arrows). In the descending thin limbs, water is passively absorbed from the tubular lumen, and highly sulfated HSNS4F5 may play a role in this process; it could function by binding sodium ions, thereby providing conditions that favor water uptake. In the lung, only mast cells are positive (Fig. 3, M), staining being confined to the heparin-containing granules. Strikingly, basement membranes, a rich source of HS, were completely negative (Fig. 3, arrows). A remarkable staining pattern was also observed in human ovarian tissue; HSNS4F5 was up-regulated in endometrioid and serous adenocarcinomas (Fig. 4). Not all of the tumors examined showed expression of HSNS4F5, but when up-regulated, a strong staining confined to blood vessels alone or blood vessels and extracellular matrix was observed, which implicates a role for HSNS4F5 in angiogenesis. None of the normal ovaries showed HSNS4F5 expression. Combined, these data show that HSNS4F5 is present in heparin and a restricted subset of HS.
FIGURE 2.
Restricted location of the HSNS4F5 motif in rat kidney. A and B, cortical area. C, cortical-medullary area. D–F, medullary area; identification of the HSNS4F5 motif by co-localization. Medullary areas were incubated with antibody NS4F5 (D, green) and with an antibody against aquaporin-1 (E, AQP-1), a marker for descending thin limbs of the loops of Henle and proximal tubules (red). F, overlay (n = 4). Bars, 50 μm; G, glomerulus; asterisks in D, ascending thin limbs of the loops of Henle; arrows in D, ascending thick limbs of the loops of Henle; arrowheads in D, descending thin limbs of the loops of Henle; arrows in E, proximal tubules; arrowheads in E, descending thin limbs of the loops of Henle; arrow in F, small blood vessel.
FIGURE 3.
Immunoelectron microscopical localization of HSNS4F5 in human lung. For experimental details, see text. Only heparin-containing granules in mast cells (M) are clearly positive. Basement membranes (arrows), a rich source of HS, are negative (n = 4). Bar in A, 15 μm; bar in B, 5 μm.
FIGURE 4.
Immunolocalization of HSNS4F5 in ovarian tumor tissue. Cryosections of human ovary, ovarian endometrioid adenocarcinoma, and serous cystadenocarcinoma were stained with NS4F5. Bound antibodies were visualized by mouse monoclonal antibody P5D4 followed by Alexa 488-conjugated goat anti-mouse IgG (n = 3).
Blocking HSNS4F5 Decreased Cell Proliferation, Stimulated Apoptosis but Had No Influence on Cell Attachment
HSPGs have many roles in cell physiology (27). To study the effect of blockage of HSNS4F5 on cellular behavior, human lung epithelial cells (A549) were used. In culture these cells produce HSNS4F5 (supplemental Fig. S2A), which can be removed by heparin lyase III treatment (supplemental Fig. S2B). This indicates that HSNS4F5 is present in HS produced by these cells. Heparin lyase III has no effect on chondroitin sulfate (supplemental Fig. S2, C and D).
Dose-dependent Effect of NS4F5 on Cell Proliferation
To study the effect of HSNS4F5 on cell proliferation, A549 cells were treated with NS4F5, and proliferation was analyzed. Treatment of cells with 10 μg of NS4F5/ml resulted in a reduced cell proliferation to 76.6 ± 7.7% (supplemental Fig. S3). Treatment with 50 μg/ml further reduced proliferation to 51.4 ± 7.8%. MPB49 had no effect on proliferation.
Effect of Antibody NS4F5 on Apoptosis
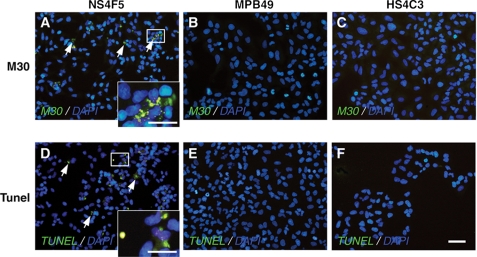
To determine whether HSNS4F5 plays a role in apoptosis, A549 cells were treated with NS4F5. MPB49 and the anti-HS antibody HS4C3, which has a different specificity than NS4F5 (10, 25), were also studied. After treatment with NS4F5, a number of cells were positive for immunostaining with antibody M30 (Fig. 5A, arrows), a marker for apoptosis. NS4F5-treated cells were also positive for TUNEL staining (Fig. 5D), which detects 3′-OH DNA ends within nuclei, further indicating that treatment with NS4F5 induced apoptosis. Cells treated with MPB49 (Fig. 5, B and E) and HS4C3 (Fig. 5, C and F) did not show signs of apoptosis.
FIGURE 5.
HSNS4F5 immunoblocking induces morphologic changes consistent with apoptosis. Human lung epithelial cells were treated with purified NS4F5 (A and D), control antibody MPB49 (B and E), or anti-HS antibody HS4C3 (C and F). The cells were fixed and concomitantly stained with M30 (A–C) or with TUNEL (D–F). DAPI was used to identify nuclei. The arrows in A identify caspase-cleaved cytokeratin 18, whereas the arrows in D identify TUNEL-positive cells (n = 4). Bars, 50 μm.
Effect of NS4F5 on Cell Attachment to Type I Collagen
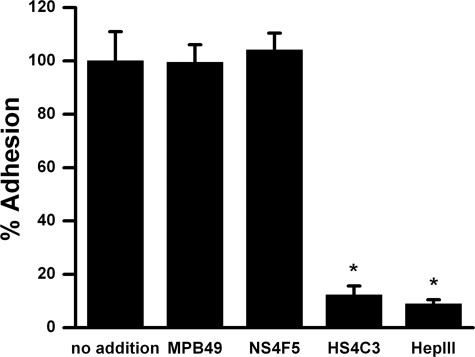
To study the effect of HSNS4F5 on cellular attachment, calcein AM-labeled human lung epithelial cells were treated without and with heparin lyase III or with purified NS4F5 and allowed to attach to type I collagen-coated wells. As controls, the cells were incubated with MPB49 or HS4C3. Digestion of HS decreased the number of adherent cells to 8.9 ± 1.6% (Fig. 6). HS4C3 had a similar effect (12.3 ± 3.4%). NS4F5, however, did not have any effect on cell attachment to type I collagen. These data indicated that, although cell attachment is HS-dependent, HSNS4F5 does not play a major role in this process.
FIGURE 6.
HSNS4F5 immunoblocking has no effect on cell attachment. Calcein AM-labeled human lung epithelial cells were incubated with or without purified NS4F5, MPB49, or HS4C3 (50 μg/ml) or heparin lyase III (HepIII) and plated in wells precoated with type I collagen. After washing off nonadherent cells, phase contrast images of the remaining adherent cells were taken (without heparin lyase III or with heparin lyase III), or cells were lysed and fluorescence was quantified. The values are expressed as the means ± S.D. (n = 5). *, p < 0.05.
HSNS4F5 Is Present on Activated HUVEC and Contributes to Firm Adhesion of Leukocytes
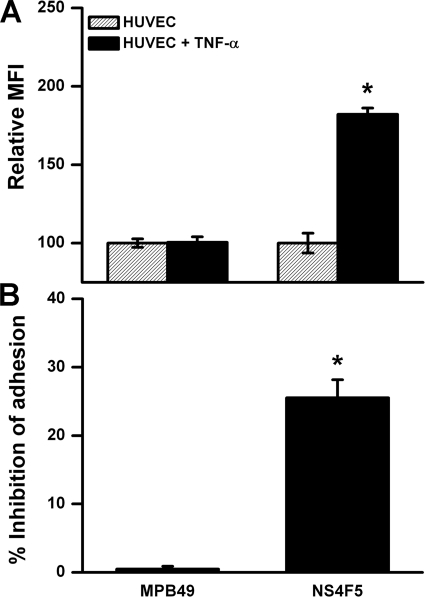
Endothelial inflammatory HS motifs appear to be important mediators in leukocyte trafficking in vitro and also have been detected in vivo on endothelium in inflamed renal tissue (26, 28). Expression of HSNS4F5 was significantly up-regulated after activation of HUVEC with TNF-α (Fig. 7A). In addition, blocking of HSNS4F5 on activated HUVEC revealed a significant reduction in firm leukocyte adhesion (25.5 ± 2.6%) (Fig. 7B). Thus, upon activation HSNS4F5 appears on endothelium and contributes to firm leukocyte adhesion.
FIGURE 7.
A, expression of HSNS4F5 by HUVEC is increased after activation with TNF-α. Expression of HSNS4F5 was measured by flow cytometry in nonactivated and TNF-α-activated HUVEC. Relative mean fluorescence intensity (MFI) is shown. The mean fluorescence intensity of nonactivated HUVEC is set at 100. The values are expressed as the means ± S.D. (n = 5). *, p < 0.05. B, HSNS4F5 motifs on activated HUVEC contribute to firm leukocyte adhesion. Monolayers of HUVEC activated with TNF-α were coincubated with labeled 32Dcl3 (leukocyte) cells and antibody NS4F5. In addition, the effect of control antibody MPB49 was evaluated. The percentage of inhibition of adhesion is calculated by the formula [(100 − X)/100 × 100%], in which X is the relative adhesion compared with the control without addition (adhesion of 32Dcl3 cells without addition to activated HUVEC is set at 100%). The antibodies were used at 25 μg/ml. The values are expressed as the means ± S.D. (n = 5). *, p < 0.05 versus control without antibody.
Imaging the Biodistribution of HSNS4F5 in Mice: Co-localization with Systemic AA Amyloid
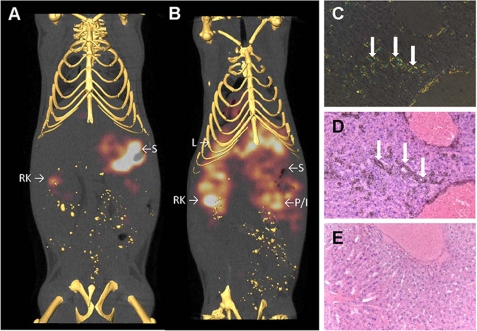
HSPGs are major constituents of all amyloid deposits regardless of the chemical nature of the precursor protein. We therefore investigated the binding in vivo of radioiodinated NS4F5 in healthy mice and those with experimentally induced systemic peripheral AA amyloidosis. The distribution of HSNS4F5 in the healthy animals was limited to kidney (Fig. 8) and stomach. Radioactivity in the stomach is likely due to accumulation of free radioiodide liberated from NS4F5 during catabolism. In contrast, when administered to mice with AA amyloidosis, NS4F5 was retained in sites of amyloid deposition such as liver, kidneys, and intestine and/or pancreas (Fig. 8B). Micro-autoradiography confirmed the precise co-localization of NS4F5 with amyloid deposits as visualized in Congo red-stained adjacent tissue sections (Fig. 8, C and D). In marked contrast radiolabeled NS4F5 was not observed in tissue sections devoid of amyloid (Fig. 8E). These data indicate that HSNS4F5 is present in tissue amyloid and essentially absent in the surrounding healthy tissue, rendering it a potent and selective biomarker of these diseases.
FIGURE 8.
Imaging, and specific co-localization of NS4F5 with AA amyloid deposits in vivo. The distribution of 125I-labeled NS4F5 was visualized in healthy (A) and amyloid-laden (B) mice by microSPECT imaging. The antibody accumulated in amyloid deposits in the liver (L) as well as the pancreas (or intestine, P/I) and kidney (RK for right kidney). In amyloid-free mice, liberated radioiodide was observed in the stomach (S) with some activity also associated with renal clearance of the antibody (RK). Specific uptake of 125I-labeled NS4F5 by hepatic amyloid deposits was confirmed microscopically, where Congo-philic amyloid (C) was co-stained by black grains in the autoradiograph (D). There was no accumulation of 125I-labeled NS4F5 in a healthy liver (E) (n = 4).
DISCUSSION
In this study, we describe the selection, characterization, and application of a novel phage display-derived anti-HS antibody NS4F5. NS4F5 defines HSNS4F5, a specific stretch of highly sulfated disaccharides (GlcNS6S-IdoA2S)3, which is common in heparin (29) and very rare in HS, and has a restricted distribution in healthy tissues. NS4F5 was used as a blocking agent to study the biological relevance of HSNS4F5, and it was demonstrated that binding of NS4F5 to HSNS4F5 reduced cell proliferation and induced apoptosis but did not alter cell attachment. Moreover, we showed an important contribution of HSNS4F5 on endothelium to leukocytes adhesion. These findings provide new insights into the function of a specific HS motif and reiterate the importance of HS sequences (or more specifically, HS with very high charge densities (30)) in cellular behavior.
The chemical heterogeneity of HS, coupled with the lack of appropriate tools to study the fine structure of HS, has seriously limited the investigations into the roles of HS in biological phenomena. Using phage display technology, we have generated a large panel of epitope-specific antibodies against HS (6, 7, 9, 11). Although these antibodies are unique in their preference for specific modification patterns on HS molecules (i.e. 2-O, 6-O, and 3-O sulfation and/or epimerization) (6, 7, 9–11), the exact saccharide sequence remains elusive (31). NS4F5 is the first antibody for which the saccharide sequence motif has been determined precisely. Knowing the specificity of NS4F5 renders it a powerful tool to investigate the expression, localization, and functional role of this specific HSNS4F5 structure in cell biological phenomena.
HS-protein interactions have recently attracted much interest, and it is now widely recognized that HS is a modulator of certain protein functions (32). HS-protein interactions vary with regard to specificity and often seem to depend on specific HS motifs, the degree of sulfation being of major importance (10). For instance, the binding of both FGF-2 and VEGF164 depends on the sulfation state of HS, as does, for example, the process of tumor cell proliferation (33).
The finding that HSNS4F5 is up-regulated in tumors indicates that, in contrast to healthy tissue, a hypersulfated HS structure is present in the tumors. An enormous diversity of HS sequences is possible (34), and considerable variation has been documented in normal adult tissues (7). HS chains are dynamically regulated during development and tumorigenesis (35, 36), consistent with regulatory changes in the binding of signaling molecules. Considerable interest therefore exists in the mechanisms that generate diversity in HS chains. Up-regulation of HSNS4F5 in ovarian tumors could be mechanistically critical for the growth of tumors. Because a myriad of extracellular events are influenced by HS interactions with growth factors, morphogens, cell adhesion molecules, and so forth (37), we can foresee other mechanisms by which cancer cells could exploit the activity of HSNS4F5. Given that expression of HSNS4F5 is undetectable in normal ovarian surface epithelium, compared with ovarian tumor tissue, these observations raise the possibility that ovarian HS serves different roles in normal epithelium compared with tumor cells. In this light, it is highly possible that HSNS4F5 could be used as a biomarker for ovarian tumor cells. Further study of HSNS4F5 and its role in tumor progression is clearly warranted.
HSPGs are intimately associated with pathological amyloid deposits such as those found in the brains of patients with Alzheimer disease and peripherally in patients with chronic inflammatory disorders and plasma cell dyscrasias (38, 39). The deposition of amyloid fibrils is preceded by the up-regulation of HSPG synthesis; indeed, in the absence of sulfated GAGs, amyloid deposition does not occur (40). The serum AA precursor protein has an HS binding motif, and following binding the serum AA protein undergoes a conformational change that results in an increase in the β-sheet content of the protein (41). This interaction therefore renders the serum AA protein in a pro-amyloid form and is a critical step in the process of amyloid deposition. The precise structures of the saccharides in the amyloid-associated HS are not known, but it has been shown that they are structurally distinct from HS isolated from healthy tissues (42). Binding of NS4F5 to amyloid deposits indicates that amyloid contains hypersulfated HSNS4F5, confirming previous observations that HS in amyloid is distinct from most HS in healthy tissues (19). The restricted distribution of HSNS4F5 renders it a potential biomarker of amyloidosis, which can be targeted using novel imaging and therapeutic reagents. The observation that, in vivo, phage display-derived anti-HS antibodies specifically home to their saccharide epitope holds great promise for their diagnostic and therapeutic applications.
The biological function of HSNS4F5 is in line with previous work on the effect of sulfation on cellular behavior. HS 6-O-endosulfatase, an enzyme that removes 6-O-sulfate groups from sulfated HS motifs, resulted in decreased proliferation and increased apoptosis of breast cancer cells (43). Likewise, highly sulfated HS fragments, derived from a melanoma cell line and prepared by heparin lyase III digestion, decreased cell proliferation and increased apoptosis, whereas low sulfated HS fragments, prepared by heparin lyase I had the opposite effect (44). Furthermore, recently we have identified other, distinct HS motifs that also contribute to leukocyte trafficking (28). In this context, HSNS4F5 can also be classified as a functional inflammatory HS motif, which may have therapeutic implications. Finally, apoptosis was noticed in blood vessels of mice with a targeted deletion of the HS biosynthetic enzyme N-deacetylase/N-sulfotransferase-1, which results in a lower degree of HS sulfation (45). Thus, highly sulfated HS motifs generally promote cell proliferation and inhibit apoptosis. This study pinpoints these effects to a specific HSNS4F5 motif (IdoA2S-GlcNS6S)3 and strongly suggests a role for (IdoA2S-GlcNS6S)3 structures in critically important pathways of cellular growth control. Such specific HS motifs may be key targets for the development of novel therapeutic approaches for amyloid-associated disorders such as Alzheimer disease. Some analogy may be drawn here with the drug fondaparinux, a synthetic heparin-based pentasaccharide that binds and activates antithrombin III, thereby reducing blood clot formation. The development of fondaparinux was based on the identification of the active motif within heparin responsible for its anticoagulant characteristics (46). Our findings may be generalizable to pathologies that are dependent on HS/heparin-mediated pathways and may have implications for the development of novel diagnostic reagents and therapeutic interventions. Sequence analysis of other antibody-defined HS motifs and analysis of their cell biological function may thus provide a window for the development of defined oligosaccharides with therapeutic potential.
Supplementary Material
Acknowledgments
We thank Dr. P. Oreste (Glycores, Milan, Italy) for the modified K5 polysaccharides, Dr. A. Naggi (G. Ronzoni Institute for Chemical and Biochemical Research, Milan, Italy) for various modified heparin preparations, Dr. J. Esko for providing wild type and mutant CHO cells, Dr. J. Gallagher for providing modified heparin oligosaccharides, Dr. J. M. H. Raats for providing the pUC119 His-VSV vector, and IBEX Technologies (Montreal, Canada) for providing recombinant heparin lyase III derived from Flavobacterium heparinum. We give our thanks to P. H. Jap for advice in the field of tissue morphology and Gunilla Pettersson for expert technical assistance.
The work was supported by International Human Frontier Science Program Organization Grant RGP0062/2004-C101, Dutch Cancer Society Grant nr 2008-4058, Netherlands Organization for Scientific Research Grant 902-27-292 (to A. L. R.), Dutch Kidney Foundation Grants C05.2152 and KJPB09.010 (to A. L. R.), Swedish Foundation for Strategic Research Grant A303:156e, Swedish Research Council Grant 32X-15023, Swedish Cancer Society Grant 4708-B02-01XAA, and Polysackaridforskning AB (Uppsala, Sweden).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- HSPG
- heparan sulfate proteoglycan
- GAG
- glycosaminoglycan
- HS
- heparan sulfate
- HUVEC
- human umbilical vein endothelial cells
- AA
- amyloid protein A
- SPECT
- single photon emission computed tomographic.
REFERENCES
- 1.Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 2.Nakato H., Kimata K. (2002) Biochim. Biophys. Acta 1573, 312–318 [DOI] [PubMed] [Google Scholar]
- 3.Lindahl U., Kusche-Gullberg M., Kjellén L. (1998) J. Biol. Chem. 273, 24979–24982 [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg R. D., Shworak N. W., Liu J., Schwartz J. J., Zhang L. (1997) J. Clin. Invest. 99, 2062–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waksman G., Herr A. B. (1998) Nat. Struct. Biol. 5, 527–530 [DOI] [PubMed] [Google Scholar]
- 6.Jenniskens G. J., Oosterhof A., Brandwijk R., Veerkamp J. H., van Kuppevelt T. H. (2000) J. Neurosci. 20, 4099–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennissen M. A., Jenniskens G. J., Pieffers M., Versteeg E. M., Petitou M., Veerkamp J. H., van Kuppevelt T. H. (2002) J. Biol. Chem. 277, 10982–10986 [DOI] [PubMed] [Google Scholar]
- 8.ten Dam G. B., Hafmans T., Veerkamp J. H., van Kuppevelt T. H. (2003) J. Histochem Cytochem. 51, 727–739 [DOI] [PubMed] [Google Scholar]
- 9.Smits N. C., Robbesom A. A., Versteeg E. M., van de Westerlo E. M., Dekhuijzen P. N., van Kuppevelt T. H. (2004) Am. J. Respir. Cell Mol. Biol. 30, 166–173 [DOI] [PubMed] [Google Scholar]
- 10.Ten Dam G. B., Kurup S., van de Westerlo E. M., Versteeg E. M., Lindahl U., Spillmann D., van Kuppevelt T. H. (2006) J. Biol. Chem. 281, 4654–4662 [DOI] [PubMed] [Google Scholar]
- 11.van Kuppevelt T. H., Dennissen M. A., van Venrooij W. J., Hoet R. M., Veerkamp J. H. (1998) J. Biol. Chem. 273, 12960–12966 [DOI] [PubMed] [Google Scholar]
- 12.van Setten P. A., van Hinsbergh V. W., van der Velden T. J., van de Kar N. C., Vermeer M., Mahan J. D., Assmann K. J., van den Heuvel L. P., Monnens L. A. (1997) Kidney Int. 51, 1245–1256 [DOI] [PubMed] [Google Scholar]
- 13.Roghani M., Mansukhani A., Dell'Era P., Bellosta P., Basilico C., Rifkin D. B., Moscatelli D. (1994) J. Biol. Chem. 269, 3976–3984 [PubMed] [Google Scholar]
- 14.Kurup S., Wijnhoven T. J., Jenniskens G. J., Kimata K., Habuchi H., Li J. P., Lindahl U., van Kuppevelt T. H., Spillmann D. (2007) J. Biol. Chem. 282, 21032–21042 [DOI] [PubMed] [Google Scholar]
- 15.Leali D., Belleri M., Urbinati C., Coltrini D., Oreste P., Zoppetti G., Ribatti D., Rusnati M., Presta M. (2001) J. Biol. Chem. 276, 37900–37908 [DOI] [PubMed] [Google Scholar]
- 16.Kreuger J., Salmivirta M., Sturiale L., Giménez-Gallego G., Lindahl U. (2001) J. Biol. Chem. 276, 30744–30752 [DOI] [PubMed] [Google Scholar]
- 17.Muller M., Marti T., Kriz S. (1980) Proceedings of the 7th European Congress on Electron Microscopy2, 720–721 [Google Scholar]
- 18.Leers M. P., Kölgen W., Björklund V., Bergman T., Tribbick G., Persson B., Björklund P., Ramaekers F. C., Björklund B., Nap M., Jörnvall H., Schutte B. (1999) J. Pathol. 187, 567–572 [DOI] [PubMed] [Google Scholar]
- 19.Solomon A., Weiss D. T., Schell M., Hrncic R., Murphy C. L., Wall J., McGavin M. D., Pan H. J., Kabalka G. W., Paulus M. J. (1999) Am. J. Pathol. 154, 1267–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall J. S., Paulus M. J., Gleason S., Gregor J., Solomon A., Kennel S. J. (2006) Methods Enzymol. 412, 161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wall J. S., Kennel S. J., Paulus M. J., Gleason S., Gregor J., Baba J., Schell M., Richey T., O'Nuallain B., Donnell R., Hawkins P. N., Weiss D. T., Solomon A. (2005) Amyloid 12, 149–156 [DOI] [PubMed] [Google Scholar]
- 22.Wall J. S., Kennel S. J., Paulus M., Gregor J., Richey T., Avenell J., Yap J., Townsend D., Weiss D. T., Solomon A. (2006) J. Nucl. Med. 47, 2016–2024 [PMC free article] [PubMed] [Google Scholar]
- 23.Nissim A., Hoogenboom H. R., Tomlinson I. M., Flynn G., Midgley C., Lane D., Winter G. (1994) EMBO J. 13, 692–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnbull J. E., Hopwood J. J., Gallagher J. T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2698–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smits N. C., Lensen J. F., Wijnhoven T. J., ten Dam G. T., Jenniskens G. J., van Kuppevelt T. H. (2006) Methods Enzymol. 416, 61–87 [DOI] [PubMed] [Google Scholar]
- 26.Rops A. L., van den Hoven M. J., Bakker M. A., Lensen J. F., Wijnhoven T. J., van den Heuvel L. P., van Kuppevelt T. H., van der Vlag J., Berden J. H. (2007) Nephrol. Dial. Transplant 22, 1891–1902 [DOI] [PubMed] [Google Scholar]
- 27.Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 28.Rops A. L., van den Hoven M. J., Baselmans M. M., Lensen J. F., Wijnhoven T. J., van den Heuvel L. P., van Kuppevelt T. H., Berden J. H., van der Vlag J. (2008) Kidney Int. 73, 52–62 [DOI] [PubMed] [Google Scholar]
- 29.Conrad H. E. (1998) in Heparin-binding Proteins, pp. 1–5, Academic Press, San Diego [Google Scholar]
- 30.Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson S. M., Fernig D. G., Jesudason E. C., Losty P. D., van de Westerlo E. M., van Kuppevelt T. H., Turnbull J. E. (2009) J. Biol. Chem. 284, 35621–35631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindahl U. (2007) Thromb. Haemostasis 98, 109–115 [PubMed] [Google Scholar]
- 33.Fuster M. M., Esko J. D. (2005) Nat. Rev. Cancer 5, 526–542 [DOI] [PubMed] [Google Scholar]
- 34.Gallagher J. T. (2001) J. Clin. Invest. 108, 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brickman Y. G., Ford M. D., Gallagher J. T., Nurcombe V., Bartlett P. F., Turnbull J. E. (1998) J. Biol. Chem. 273, 4350–4359 [DOI] [PubMed] [Google Scholar]
- 36.Jayson G. C., Vives C., Paraskeva C., Schofield K., Coutts J., Fleetwood A., Gallagher J. T. (1999) Int. J. Cancer 82, 298–304 [DOI] [PubMed] [Google Scholar]
- 37.Esko J. D., Lindahl U. (2001) J. Clin. Invest. 108, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kisilevsky R., Ancsin J. B., Szarek W. A., Petanceska S. (2007) Amyloid 14, 21–32 [DOI] [PubMed] [Google Scholar]
- 39.Ancsin J. B. (2003) Amyloid 10, 67–79 [DOI] [PubMed] [Google Scholar]
- 40.Li J. P., Galvis M. L., Gong F., Zhang X., Zcharia E., Metzger S., Vlodavsky I., Kisilevsky R., Lindahl U. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6473–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCubbin W. D., Kay C. M., Narindrasorasak S., Kisilevsky R. (1988) Biochem. J. 256, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindahl B., Lindahl U. (1997) J. Biol. Chem. 272, 26091–26094 [DOI] [PubMed] [Google Scholar]
- 43.Hossain M. M., Hosono-Fukao T., Tang R., Sugaya N., van Kuppevelt T. H., Jenniskens G. J., Kimata K., Rosen S. D., Uchimura K. (2010) Glycobiology 20, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu D., Shriver Z., Venkataraman G., El Shabrawi Y., Sasisekharan R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pallerla S. R., Pan Y., Zhang X., Esko J. D., Grobe K. (2007) Dev. Dyn. 236, 556–563 [DOI] [PubMed] [Google Scholar]
- 46.Petitou M., Hérault J. P., Bernat A., Driguez P. A., Duchaussoy P., Lormeau J. C., Herbert J. M. (1999) Nature 398, 417–422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.