Abstract
The DNA binding activity of NF-κB is critical for VCAM-1 expression during inflammation. DNA-dependent protein kinase (DNA-PK) is thought to be involved in NF-κB activation. Here we show that DNA-PK is required for VCAM-1 expression in response to TNF. The phosphorylation and subsequent degradation of I-κBα as well as the serine 536 phosphorylation and nuclear translocation of p65 NF-κB were insufficient for VCAM-1 expression in response to TNF. The requirement for p50 NF-κB in TNF-induced VCAM-1 expression may be associated with its interaction with and phosphorylation by DNA-PK, which appears to be dominant over the requirement for p65 NF-κB activation. p50 NF-κB binding to its consensus sequence increased its susceptibility to phosphorylation by DNA-PK. Additionally, DNA-PK activity appeared to increase the association between p50/p50 and p50/p65 NF-κB dimers upon binding to DNA and after binding of p50 NF-κB to the VCAM-1 promoter. Analyses of the p50 NF-κB protein sequence revealed that both serine 20 and serine 227 at the amino terminus of the protein are putative sites for phosphorylation by DNA-PK. Mutation of serine 20 completely eliminated phosphorylation of p50 NF-κB by DNA-PK, suggesting that serine 20 is the only site in p50 NF-κB for phosphorylation by DNA-PK. Re-establishing wild-type p50 NF-κB, but not its serine 20/alanine mutant, in p50 NF-κB−/− fibroblasts reversed VCAM-1 expression after TNF treatment, demonstrating the importance of the serine 20 phosphorylation site in the induction of VCAM-1 expression. Together, these results elucidate a novel mechanism for the involvement of DNA-PK in the positive regulation of p50 NF-κB to drive VCAM-1 expression.
Keywords: Inflammation, NF-Kappa B, Protein Kinases, siRNA, Transcription Factors, Tumor Necrosis Factor (TNF), DNA-dependent Protein Kinase, TNF, VCAM-1
Introduction
Expression of VCAM-12 on the surface of structural and inflammatory cells, as well as endothelial cells, is critical for the initiation and progression of inflammatory diseases, such as asthma and arthrosclerosis (1, 2), as well as cancer (3). The role of VCAM-1 expression in structural cells is believed to facilitate transmigration, accumulation of leukocytes, and cell-cell interactions at inflammatory sites (4–6). The expression of VCAM-1 and other adhesion molecules plays an important role in both the recruitment of Th2 cells and the accumulation of eosinophils in allergic inflammatory foci (7, 8). Similarly, VCAM-1 is a determining factor in early lesion establishment during atherogenesis (9). Under these pathological conditions, the expression of adhesion molecules like VCAM-1 is induced by cytokines, such as TNF and IL-1β, which are produced by a number of immune and structural cells (1, 10).
DNA-PK is an important factor for repairing double-stranded DNA breaks caused by DNA-damaging agents, such as oxidative stress. DNA-PK is composed of two DNA-binding subunits (Ku70 and Ku86) and one 450-kDa catalytic subunit (DNA-PKcs). Activated DNA-PKcs is a serine/threonine kinase that has been shown to phosphorylate a number of proteins in vitro, including p53, transcription factors, RNA polymerase, and Ku70-Ku86. However, a very limited number of reports have associated such phosphorylation events with actual, functional outcomes (11). Elevated levels of DNA-PK were observed in human atherosclerotic plaques (12), and there is growing evidence demonstrating that DNA damage is extremely important in the pathogenesis of atherosclerosis (13).
NF-κB regulates many genes involved in mammalian immune and inflammatory responses, apoptosis, cell proliferation, and differentiation. Activated NF-κB unmasks the nuclear translocation and binding of NF-κB to specific κB consensus sequences in the chromatin as well as the activation of specific subsets of genes. p50 NF-κB is produced as an inactive p105 precursor protein that lacks transactivation domains. Activated NF-κB is generally composed of p50/p65 or p50/c-Rel heterodimers, whereas homodimeric complexes of p50 or p52 are associated with transcriptional repression. The decisive mechanisms that govern whether p50 NF-κB functions as a suppressor or an activator of NF-κB-dependent genes remain elusive (14).
VCAM-1 is a member of the immunoglobulin gene superfamily that is very important in the human immune system; VCAM-1 also plays a dominant role in the initiation of atherosclerosis and asthma (6, 15). The VCAM-1 promoter harbors two putative κB sites that are required for induction by TNF (16, 17). Many of the studies investigating the role of NF-κB in VCAM-1 expression have focused on the function of the p65 subunit. Studies addressing the mechanism by which p50 NF-κB influences VCAM-1 gene expression are lacking. Accordingly, such a mechanism remains to be elucidated. The objective of the current study was to determine the relationship between DNA-PK and VCAM-1, specifically the involvement of p50 NF-κB in the expression of VCAM-1.
EXPERIMENTAL PROCEDURES
Cell Culture, Treatment Protocols, and Indirect Immunofluorescence
293T cells as well as mouse embryonic fibroblast (MEF) 3T3 and p50−/− NF-κB cells (18) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). M059K (DNA-PK-proficient) and M059J (DNA-PK-deficient) cells were maintained in DMEM/F12 (1:1) medium supplemented with 10% FBS, 1 mm l-glutamine, and 1% nonessential amino acids. All culture supplies were purchased from Invitrogen. Prior to treatment with TNF (10 ng/ml) (Roche Applied Science, Mannheim, Germany) or IL-1β (2 or 10 ng/ml) (Sigma-Aldrich) for the indicated times, cells at 50–80% confluence were starved by incubation in medium with 0.5% cell culture-tested BSA (Sigma-Aldrich) for 5 h. For immunofluorescence, cells grown on chamber slides, treated as indicated in the Fig. 2 legend, were then fixed with 3.7% paraformaldehyde in PBS. Cells were permeabilized and subjected to immunofluorescence as described (19) with antibodies to p50 or p65 NF-κB.
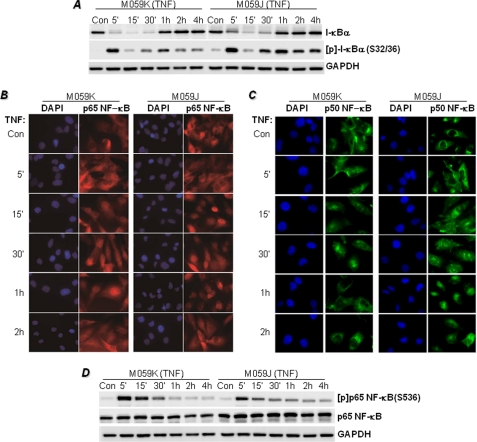
FIGURE 2.
Effects of DNA-PK deficiency on I-κBα phosphorylation and its subsequent degradation and NF-κB phosphorylation and nuclear translocation in response to TNF stimulation. A, M059K and M059J cells were treated with TNF for the indicated times, after which protein extracts were subjected to immunoblot analysis with antibodies to I-κBα, phosphor(Ser-36)-I-κBα ([p]-I-κBα(S32/36)), or GAPDH. M059K and M059J cells grown in chamber slides were treated with TNF for the indicated times. Con, control. B and C, cells were then fixed and subjected to immunofluorescence staining with antibodies to p65 (B) or p50 NF-κB (C) followed by staining with DAPI (nuclei). D, the immunoblots displayed in A were stripped and reprobed with antibodies to the phosphorylated form of p 65 NF-κB at serine 536 ([p]p65NF-κB(S536)) or to antibodies recognizing full-length p65 NF-κB. Note that the immunoblot for GAPDH is the same as displayed in A.
Immunoblot Analysis, Nuclear Extract Preparations, EMSA, and Conventional RT-PCR
After treatment with the indicated reagents, cells were subjected to immunoblot analysis essentially as described (20) with antibodies to I-κBα, VCAM, p65 NF-κB, actin (Santa Cruz Biotechnology, Santa Cruz, CA), DNA-PKcs, p50 NF-κB (Abcam, Cambridge, UK), GAPDH, or GST (Novus Biologicals, Littleton, CO), as well as antibodies to the phosphorylated forms (serine residues 32 and 36) of I-κBα (p-I-κBα) (Cell Signaling Technology, Danvers, MA) or of p65 NF-κB at serine 536 (Abcam). The use of antibodies against actin or GAPDH was related to the availability of the antibodies. Immune complexes were detected with the appropriate secondary antibodies and chemiluminescence reagents (PerkinElmer Life Sciences). The preparation of nuclear extracts was performed using a commercial kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. EMSA analysis of DNA binding activity in prepared nuclear extracts was performed as described (21) using a radiolabeled oligonucleotide containing the NF-κB consensus sequence 5′-TCGACAGAGGGGACTTTCCGAGAGGCTCGA-3′ (Promega, Madison, WI). For RT-PCR, total RNA was extracted from cells using the RNeasy Plus micro kit (Qiagen, Valencia, CA). One microgram of total RNA was used as a template to make first-strand cDNA by random priming using the iScript cDNA synthesis kit (Bio-Rad). Oligonucleotide primers (Integrated DNA Technologies, San Jose, CA) to specifically amplify a fragment of VCAM-1 or β-actin were as follows: VCAM-1, forward primer, 5′-TAAAATGCCTGGGAAGATGG-3′; reverse primer, 5′-AGTTTTATGGCCTCCTCCTGA-3′; and β-actin, forward primer, 5′-ACCGTGAAAAGATGACCCAGATC-3′; reverse primer, 5′-TAGTTTCATGGATGCCACAGG-3′. The amplification program was as follows: 5 min at 94 °C, 30 s at 94 °C, 45 s at 60 °C, and 45 s at 72 °C. The cycle numbers were optimized for each primer pair. The PCR products were then incubated for 15 min at 72 °C. The resulting PCR products were subjected to electrophoresis in a 2% agarose gel and stained with ethidium bromide.
Construction of Wild-type GST-p50 NF-κB and Mutant GST-p50 NF-κB Plasmids, Transfections, and Electroporation
For the construction of the GST-p50 NF-κB expression vector, the p50 NF-κB cDNA was cloned into the Gateway entry vector pENTR/SD/D-TOPO (Invitrogen). The accuracy of the construct was verified by sequence analysis. The expression clone was generated by performing an LR recombination reaction between the entry clone and the destination vector GST-pDEST27 (Invitrogen). Phosphorylation prediction software, NetPhosK 1.0 and NetPhos 2.0, was used to locate possible DNA-PK phosphorylation sites on p50 NF-κB. p50 NF-κB site-directed mutants were prepared using the GeneTailor site-directed mutagenesis system (Invitrogen). All sequences were confirmed by sequencing. 293T cells were transiently transfected with the GST-p50 NF-κB vector using Lipofectamine LTX (Invitrogen). MEF p50−/− NF-κB cells were transfected using the Amaxa MEF1 Nucleofector kit (Lonza, Walkersville, MD). The transfections were performed according to the manufacturer's specifications and instructions.
Short Interfering RNA (siRNA) and Short Hairpin RNA (shRNA)
Knockdown of DNA-PKcs expression in DNA-PK-proficient M059K cells was achieved using DNA-PKcs siRNA (sc-35200, Santa Cruz Biotechnology) or DNA-PKcs shRNA (sc-35200-V, Santa Cruz Biotechnology). Both transfection and transduction were performed according to the manufacturer's specifications.
GST Pulldown and Immunoprecipitation
293T cells were transiently transfected with the GST-p50 NF-κB vector or its mutants using Lipofectamine LTX (Invitrogen). GST-p50 NF-κB was pulled down using glutathione-SepharoseTM 4 fast flow (GE Healthcare). Immunoprecipitation was conducted using nuclear extracts essentially as described (21).
In Vitro Kinase Assay
Purified recombinant wild-type GST-p50, GST-p50 mutants, or His-tagged p50 NF-κB (Promega, Madison, WI or Axxora, San Diego, CA) and DNA-PK kinase (Promega) were incubated in kinase reaction buffer containing 4 mm ATP and 10 μCi of [γ-32P]ATP (PerkinElmer Life Sciences). The samples were resolved using SDS-PAGE followed by autoradiography.
Chromatin Immunoprecipitation Assay (ChIP)
Cells were treated as described in the Fig. 3 and 5 legends and fixed with formaldehyde to cross-link the chromatin and nuclear proteins. The ChIP assay was conducted using a kit (Active Motif) according to the manufacturer's instructions. Briefly, after enzymatic shearing, antibodies against p50 NF-κB were added to precipitate the sheared chromatin. After cross-linking reversal, DNA was isolated. One-tenth of the final precipitated DNA was used in each reaction. The following primers were used. To target one κB site on the human VCAM-1 promoter, 5′-CCAATGGGGGAGATAGACCT-3′ and 5′-ACCGCAAACCCAGTTAAAAA-3′ were used to target the −1015 to −775 region, or to target one site on the murine VCAM-1 promoter, 5′-GAACGGCCATAGGAAAATCA-3′ and 5′-TAATTGCCTGCCTAACTTGGCATCCATTTCTGC-3′ were used to target the −296 to −53 region.
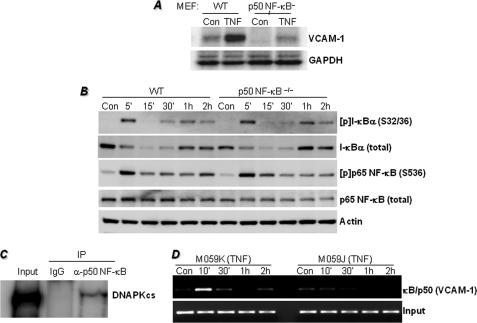
FIGURE 3.
Requirement of p50 NF-κB for TNF-induced VCAM-1 expression is dominant over the requirement for p65 NF-κB activation and may be associated with the interaction between p50 NF-κB and DNA-PK and the requirement for DNA-PK for efficient κB site occupancy on the VCAM-1 promoter by p50 NF-κB. A, MEFs derived from WT or p50−/− NF-κB mice were treated with 10 ng/ml TNF for 18 h. Protein extracts were subjected to immunoblot analysis with antibodies to VCAM-1 or GAPDH. Con, control. B, WT and p50−/− NF-κB MEFs were treated with TNF for the indicated times, after which protein extracts were subjected to immunoblot analysis with antibodies to I-κBα, phospho(Ser-32/36)-I-κBα ([p]-I-κBα(S32/36)), phospho(Ser-536)-p65 NF-κB ([p]p65 NF-κB (S536)), p65 NF-κB, or actin. C, M059K cells were treated with TNF for 1 h, after which nuclear extracts were prepared and subjected to immunoprecipitation (IP) with antibodies to DNA-PKcs or with normal IgG. Immunoprecipitates were subjected to immunoblot analysis with antibodies to DNA-PKcs. Ten percent of the amount used for immunoprecipitation was used as input control (see supplemental Fig. S2 for the original uncropped immunoblot). D, M059K or M059J cells were treated with TNF for the indicated times. Cells were then fixed and subjected to ChIP assay using antibodies to p50 NF-κB. Levels of immunoprecipitated chromatin fragments (−1015 to −775 region) of the human VCAM-1 promoter or input were examined by PCR.
Data Analysis
All data are expressed as the means ± S.E. PRISM software (GraphPad, San Diego, CA) was used to analyze the differences between experimental groups by two-way analysis of variance followed by Bonferroni post tests to compare replicate means by row.
RESULTS
DNA-PK Is Required for VCAM-1 Expression in Response to TNF Treatment
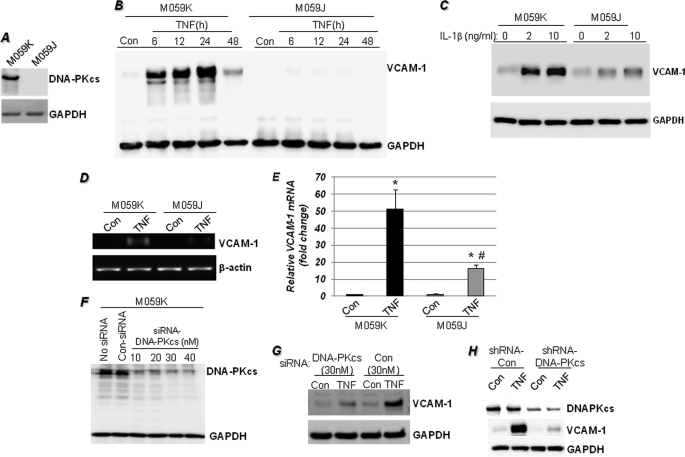
We took advantage of DNA-PK-proficient (M059K) and -deficient (M059J) cells isolated from the same glioblastoma (Fig. 1A) to test whether DNA-PK is critical for VCAM-1 expression in response to TNF. Fig. 1B shows that TNF treatment induced time-dependent expression of VCAM-1 protein in the DNA-PK-proficient (M059K) cells and that such an expression pattern was absent in the DNA-PK-deficient (M059J) cells. This clearly suggests that DNA-PK is required for the expression of VCAM-1 in response to TNF. The requirement of DNA-PK for VCAM-1 expression does not appear to be limited to TNF as the DNA-PK-deficient M059J cells failed to efficiently induce the adhesion molecule in response to IL-1β (Fig. 1C). The requirement of DNA-PK is likely at the mRNA level as assessed by conventional (Fig. 1D) or quantitative PCR (Fig. 1E), suggesting an important role for DNA-PK in the transcription of VCAM-1. Moreover, DNA-PK appears to be required for the transcription of a great number of additional NF-κB-dependent genes in response to TNF, such as inducible nitric-oxide synthase, BCL3, CD40, IL-8, IL-10, TLR7, and TLR9 among others (see supplemental Fig. S1).
FIGURE 1.
DNA-PK is required for VCAM-1 expression in response to TNF treatment. A, immunoblot analysis of extracts from untreated M059K (DNA-PK-proficient) or M059J (DNA-PK-deficient) cells with antibodies to human DNA-PKcs or GAPDH. B, M059K and M059J cells were stimulated with TNF for different times, after which protein extracts were subjected to immunoblot analysis with antibodies to VCAM-1 or GAPDH. Con, control. C, M059K and M059J cells were stimulated with 2 or 10 ng/ml IL-1β for 18 h, after which protein extracts were subjected to immunoblot analysis with antibodies to VCAM-1 or GAPDH. D and E, cells were treated with TNF for 6 h, after which total RNA was prepared and subjected to cDNA generation followed by conventional (D) or quantitative (E) PCR with primers specific to human VCAM-1 or β-actin. *, different from respective untreated cells; #, different from TNF-treated M059K cells; p < 0.01. F, M059K cells were treated with different doses of DNA-PKcs siRNA; 48 h later, protein extracts were prepared and subjected to immunoblot analysis with antibodies to DNA-PKcs or GAPDH. G, M059K cells were treated with siRNA against DNA-PKcs or control siRNA; 48 h later, cells were treated with TNF for 18 h. Protein extracts were subjected to immunoblot analysis with antibodies to VCAM-1 or GAPDH. H, M059K cells were transduced with a lentiviral vector (Santa Cruz Biotechnology) expressing either control shRNA or an shRNA targeting DNA-PKcs. Forty-eight hours later, cells were treated with TNF for 18 h, and the resulting protein extracts were subjected to immunoblot analysis with antibodies to DNA-PKcs, VCAM-1, or GAPDH.
To confirm that the observed dependence of VCAM-1 expression on DNA-PK was due to DNA-PK and not to any difference that might have developed between the two cell lines over time, we examined the effects of DNA-PK knockdown with specific siRNAs (Fig. 1F) on the expression of VCAM-1 following TNF treatment. Indeed, knockdown of DNA-PKcs severely reduced VCAM-1 expression in TNF-treated M059K cells when compared with cells that were transfected with control siRNA (Fig. 1G). This was further confirmed using a lentiviral vector-expressing shRNA, targeting DNA-PKcs (Fig. 1H). Together, these results establish a strong relationship between the expression of DNA-PK and VCAM-1 in response to TNF. The next question to answer was how DNA-PK influences transcription of VCAM-1 upon TNF stimulation.
I-κBα Phosphorylation and Its Subsequent Degradation as Well as p65 NF-κB Phosphorylation and Nuclear Translocation Are Insufficient for the Induction of VCAM-1 Expression in Response to TNF Stimulation
To begin examining the mechanism by which DNA-PK regulates VCAM-1 expression, we examined the possibility that DNA-PK influences TNF-induced, NF-κB-dependent signal transduction. Surprisingly, I-κBα phosphorylation and subsequent degradation were very similar in both TNF-treated DNA-PK-proficient cells (M059K) and TNF-treated DNA-PK-deficient (M059J) cells (Fig. 2A), preserving the transient nature of I-κBα phosphorylation and degradation. Interestingly, a moderate level of I-κBα phosphorylation persisted in TNF-treated M059J cells, which does not explain the complete absence of VCAM-1 expression. These results strongly suggest that the NF-κB signal transduction machinery after TNF stimulation was intact in the DNA-PK-deficient cells and that the defect may reside in the actual translocation of the transcription factor to the nucleus rather than in its association with I-κBα. Fig. 2, B and C, show that DNA-PK deficiency does not induce any noticeable alteration in the translocation patterns of p65 or p50 NF-κB to the nuclei of TNF-treated cells. A quantitative assessment of NF-κB subcellular localization did not reveal any significant changes between the translocation patterns of NF-κB to the nuclei of M059K and M059J cells upon TNF treatment (data not shown).
The release of NF-κB from inhibition by I-κBα phosphorylation of the p65 subunit at serines 276, 529, and 536 is decisive for its interaction with transcriptional co-activators, such as CREB-binding protein (CBP)/p300, and for binding to the κB sequences of target genes (22). TNF induced robust phosphorylation of p65 NF-κB at the serine 536 site in DNA-PK-proficient cells, and the effect was slightly reduced in DNA-PK-deficient cells (Fig. 2D). Similar to the effects of DNA-PK deficiency on I-κBα phosphorylation, such modest changes do not explain the severe reduction of VCAM-1 in TNF-treated DNA-PK-deficient cells. These results suggest that DNA-PK does not critically regulate I-κBα phosphorylation or degradation or p65 NF-κB phosphorylation and nuclear translocation. Similar results were attained in TNF-treated M059K cells that were subjected to DNA-PKcs knockdown (data not shown). These results suggest that the regulatory step affected by DNA-PK may reside after the actual translocation of the transcription factor to the nuclei of TNF-treated cells.
Requirement of p50 NF-κB for VCAM-1 Expression in Response to TNF Is Dominant over the Requirement for p65 NF-κB Activation and May Be Associated with the Interaction between p50 NF-κB and DNA-PK and the Requirement of DNA-PK for Efficient κB Site Occupancy by p50 NF-κB on the VCAM-1 Promoter
As shown above, DNA-PK deficiency exerted little effect on p65 NF-κB activation, and what effect it had did not explain the drastic reduction in VCAM-1 expression in DNA-PK-deficient cells following TNF treatment. Accordingly, we surmised that the relationship between the DNA-PK and VCAM-1 was potentially associated with the regulation of the p50 NF-κB subunit. Initially, we examined whether VCAM-1 expression could be affected by p50 NF-κB deficiency. TNF induced robust VCAM-1 expression in wild-type MEFs that was severely compromised in p50 NF-κB−/− cells (Fig. 3A). Such effects were independent of the fate of I-κBα as the phosphorylation and subsequent degradation of the inhibitor were very similar in both TNF-treated wild-type (WT) and TNF-treated p50 NF-κB−/− cells (Fig. 3B). Although p50 NF-κB deficiency promoted a moderate reduction in the expression of p65 NF-κB, the extent of its phosphorylation upon TNF treatment was virtually identical to that observed in the WT cells. These results strongly suggest that the TNF-induced NF-κB signal transduction machinery is not critically altered by p50 NF-κB deficiency. These results also suggest that p65 NF-κB activation is insufficient to induce VCAM-1 expression and that p50 NF-κB may play a dominant role in determining the expression of the adhesion molecule.
Given that p50 NF-κB is generally regarded as either an assisting partner of the p65 subunit or a suppressor of transcription when present as a homodimer, we surmised that DNA-PK might influence p50 NF-κB to function as a gene expression-promoting factor. Our initial effort was aimed at determining whether there was a physical interaction between DNA-PK and p50 NF-κB. Using an immunoprecipitation strategy with antibodies to DNA-PKcs and nuclei extracts prepared from TNF-treated cells, we showed that DNA-PK and p50 NF-κB could interact in the nuclei of TNF-treated DNA-PK-proficient cells, suggesting a potential direct relationship (Fig. 3C). These results suggest that a physical interaction between DNA-PK and p50 NF-κB may result in the phosphorylation of p50 NF-κB.
We next examined whether expression of DNA-PK influenced p50 NF-κB occupancy of the κB site on the VCAM-1 promoter using a ChIP assay with antibodies against the transcription factor. Fig. 3D shows that in DNA-PK-proficient (M059K) cells, TNF treatment induced a rapid and transient binding of p50 NF-κB to the −1015- to −775-region of the human VCAM-1 promoter. Such a pattern of κB binding was largely absent in the DNA-PK-deficient (M059J) cells except for minimal binding after 10 min of TNF treatment, suggesting that the kinase may influence the binding of the transcription factor to the VCAM-1 promoter.
DNA-PK Phosphorylates p50 NF-κB, and Binding of the Transcription Factor to a κB-containing DNA Sequence Greatly Enhances Its Susceptibility to Such Phosphorylation
DNA-PK has been shown to phosphorylate a number of proteins, including itself, p53, several transcription factors, and RNA polymerase (11). To test the above mentioned possibilities, we utilized a cell-free system using purified recombinant proteins in a DNA-PK phosphorylation reaction. Fig. 4A shows that, as expected, DNA-PK phosphorylated its known substrates, p53 and itself. The affinity of DNA-PK to its substrates, such as p53, is much higher than that to itself, which explains the reduction in DNA-PK autophosphorylation when combined with p53. Fig. 4B shows that p50 NF-κB is an excellent substrate for the kinase. The substantially lower levels of autophosphorylation of DNA-PK are indicative of its higher affinity toward p50 NF-κB. To further support the finding that DNA-PK could phosphorylate p50 NF-κB, we subjected p50 NF-κB to a DNA-PK kinase reaction with non-radioactive ATP. The proteins were then separated by high density SDS-PAGE followed by immunoblot analysis. The addition of DNA-PK and sonicated DNA generated a lower mobility band that was recognized by antibodies against p50 NF-κB (Fig. 4, C and D), suggestive of a phosphorylated form of the protein. Altogether, these results demonstrate that p50 NF-κB can be influenced by DNA-PK. However, these data are insufficient to unequivocally show that such influence plays a critical role in VCAM-1 expression.
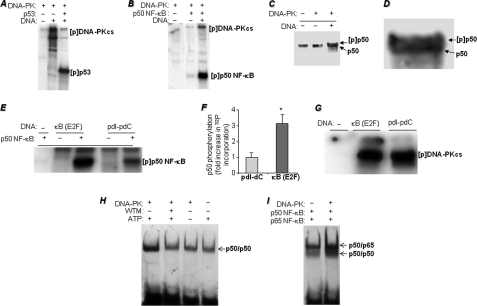
FIGURE 4.
Phosphorylation of p50 NF-κB by DNA-PK, effects of p50 NF-κB binding to a κB-containing DNA sequence on phosphorylation susceptibility, and consequences of p50 NF-κB phosphorylation on its DNA binding activity. A and B, in vitro kinase assay was performed using [32P]ATP and purified DNA-PK complex (Promega) with pure recombinant p53 (A), a positive control, or recombinant p50 NF-κB (B). The reactions were terminated by the addition of sample buffer and subjected to SDS-PAGE followed by autoradiography. [p] indicates phospho. C, the kinase reaction was repeated with cold ATP, and the reaction was subjected to immunoblot analysis with antibodies to p50 NF-κB; D represents a larger image of the double band for better visualization. E, recombinant p50 NF-κB was incubated in the presence of either a 500-bp κB-containing fragment of the E2F gene promoter or synthetic poly(dI-dC) for 15 min before the addition of DNA-PK and [32P]ATP in the kinase reactions. The reactions were terminated and subjected to SDS-PAGE followed by autoradiography. 32P incorporation in the p50 NF-κB bands was assessed using a Storm PhosphorImager system (GE Healthcare). pdI-pdC, poly(dI-dC). F, data are the means ± S.D. of values from three separate reactions. *, different from p50 NF-κB phosphorylated in a reaction containing poly(dI-dC); p < 0.01. G, autophosphorylation of DNA-PK in response to activation by either poly(dI-dC) or the 500-bp κB-containing fragment of the E2F gene promoter. H, p50 NF-κB was incubated in a kinase reaction in the presence or absence of DNA-PK, ATP (cold), and the DNA-PK inhibitor wortmannin (WTM) as shown in the figure. All kinase reactions were terminated by the addition of wortmannin. The reactions were then subjected to EMSA with a 32P end-labeled oligonucleotide containing the NF-κB-binding site. Gels were dried and subjected to autoradiography. I, p50 NF-κB was incubated in the presence or absence of DNA-PK in a kinase reaction. After reaction termination by wortmannin, p65 NF-κB was added prior to incubation with the 32P-labeled NF-κB oligonucleotide probe.
To determine whether p50 NF-κB binding to its consensus sequence influences its susceptibility to phosphorylation by DNA-PK, we incubated the transcription factor with either a 500-bp κB-containing dsDNA sequence isolated from the E2F promoter (23, 24) or a poly(dI-dC) synthetic polynucleotide prior to the kinase reaction. Fig. 4, E and F, show that incubation of p50 NF-κB with the κB-containing E2F promoter significantly increased its phosphorylation by DNA-PK by more than 3-fold when compared with levels achieved using the synthetic, nonspecific poly(dI-dC) sequence. Such differences in p50 NF-κB phosphorylation were not the result of an increase in DNA-PK kinase activity as assessed by autophosphorylation in the absence of p50 NF-κB (Fig. 4G).
Phosphorylation of p50 NF-κB by DNA-PK Promotes a Higher Level of Binding to the κB Sequence and an Elevation in the Formation of p50/p50 and p50/p65 Dimers
We next investigated the potential consequence(s) of DNA-PK phosphorylation of p50 NF-κB by examining the effects of such a modification on the interaction of p50 NF-κB with its target κB sequence in vitro. When p50 NF-κB was incubated with DNA-PK in the presence of ATP, its binding to the κB sequence was markedly increased (Fig. 4H). The increase in DNA binding activity was eliminated by either the absence of ATP or the addition of the DNA-PK inhibitor wortmannin. The addition of DNA-PK and ATP also enhanced the binding of the p50 and p65 NF-κB subunits to the κB sequence (Fig. 4I). These results suggest that DNA-PK-mediated modification of p50 NF-κB enhances its binding to its target sequence as a homodimer or as a heterodimer with p65 NF-κB, which may indicate a more robust binding to the promoters of target genes, resulting in more pronounced gene expression.
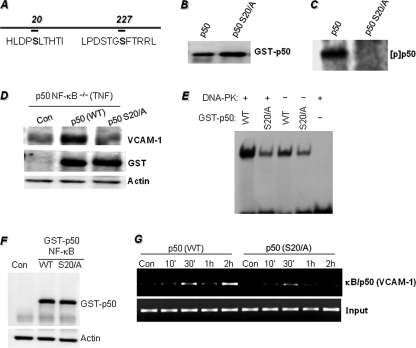
DNA-PK Phosphorylates a Single Serine Residue, Ser-20, at the Amino Terminus of p50 NF-κB, and This Phosphorylation Is Required for TNF-induced VCAM-1 Expression
A critical issue was to determine the site(s) of DNA-PK-mediated phosphorylation of p50 NF-κB. We used NetPhosK 1.0 and NetPhos 2.0 software (Center for Biological Sequence Analysis) (25) to identify the sites at which DNA-PK might phosphorylate p50 NF-κB. The analysis revealed serines 20 and 227 of the amino terminus of p50 NF-κB as potential phosphorylation sites (Fig. 5A). When Ser-20 was substituted with an alanine (S20A), DNA-PK no longer phosphorylated p50 NF-κB (Fig. 5B), suggesting that this site was the sole site at which DNA-PK could modify the transcription factor. Mutation of Ser-227 did not abolish DNA-PK-mediated phosphorylation (data not shown), strengthening the conclusion that Ser-20 is the sole phosphorylation site of p50 NF-κB. To determine the importance of the Ser-20 phosphorylation site in VCAM-1 expression, p50 NF-κB−/− MEFs were transfected with expression vectors encoding either GST-tagged WT p50 NF-κB or S20A mutant p50 NF-κB (Fig. 5C). Transfected cells were then treated with TNF for 18 h, after which total cellular extracts were subjected to immunoblot analysis with antibodies against VCAM-1, GST, or GAPDH. Expression of the WT, but not the S20A mutant, p50 NF-κB induced expression of VCAM-1 following TNF treatment (Fig. 5D). The expression levels of the WT and mutant p50 proteins were relatively the same in the transfected cells. These results demonstrate the importance of the Ser-20 phosphorylation site on p50 NF-κB for the expression of VCAM-1 in response to TNF.
FIGURE 5.
Phosphorylation of p50 NF-κB on a single serine residue (serine 20) at the amino terminus and requirement of this phosphorylation site for TNF-induced VCAM-1 expression. A, using NetPhosK 1.0 and NetPhos 2.0 software, two DNA-PK phosphorylation sites on p50 NF-κB at serines 20 and 227 were predicted. B, p50 NF-κB-tagged with GST was subjected to site-directed mutagenesis to achieve the substitution of alanine for serine 20. The generated expression vectors were transiently transfected into 293T cells. Purified WT or S20A mutant GST-tagged p50 NF-κB proteins were subjected to immunoblot analysis with antibodies to GST. C, purified WT or S20A mutant GST-tagged p50 NF-κB proteins were subjected to a kinase reaction with DNA-PK and [32P]ATP; the reactions were terminated by the addition of sample buffer and subjected to SDS-PAGE followed by autoradiography. [p] indicates phospho. D, p50−/− NF-κB MEFs were transfected with plasmids expressing WT or S20A mutant p50 (p50 S20/A) NF-κB or a control vector (Con) by electroporation. Cells were then treated with TNF for 18 h. The resulting protein extracts were subjected to immunoblot analysis with antibodies to VCAM-1, GST, or actin. E, purified WT or S20A mutant GST-tagged p50 NF-κB proteins were incubated in a kinase reaction in the presence or absence DNA-PK and ATP (cold). All kinase reactions were terminated by the addition of wortmannin. The reactions were then subjected to EMSA with a 32P end-labeled oligonucleotide containing the NF-κB-binding site. Gels were dried and subjected to autoradiography. F, p50−/− NF-κB MEFs were transfected with plasmids expressing WT or S20A mutant p50 NF-κB or a control vector (Con) by electroporation. Protein extracts from untreated cells were subjected to immunoblot with antibodies to GST or actin. G, a ChIP assay was performed using antibodies to p50 NF-κB in WT or S20A mutant p50-expressing p50−/− NF-κB MEFs treated with TNF for the indicated times. Levels of immunoprecipitated chromatin fragments (the −296 to −53 region) of the mouse VCAM-1 promoter or input were examined by PCR.
To further establish the correlation between Ser-20 phosphorylation and p50 NF-κB DNA binding activity, GST-tagged WT or mutant recombinant proteins were subjected to a kinase reaction with DNA-PK followed by an assessment of their respective DNA binding activity by EMSA. Fig. 5E shows that although incubation of WT p50 NF-κB with DNA-PK markedly increased its DNA binding activity to the κB sequence, incubating the mutant protein with the kinase did not improve its binding activity to the DNA sequence. These results further exemplify the importance of the Ser-20 site and its phosphorylation by DNA-PK for the DNA binding activity of p50 NF-κB.
Using the ChIP assay, we next examined whether the Ser-20 phosphorylation site on p50 NF-κB influenced the occupancy of the transcription factor of its binding site on the VCAM-1 promoter in p50 NF-κB−/− MEFs transfected with either the WT or the S20A mutant p50 NF-κB-encoding plasmids. Fig. 5G shows that in p50 NF-κB−/− MEFs transfected with WT p50-encoding plasmid, TNF treatment induced a rapid and biphasic binding of p50 NF-κB to the −296- to −53-region of the mouse VCAM-1 promoter. In p50 NF-κB−/− MEFs transfected with the S20A mutant p50-encoding plasmid, TNF treatment induced a short and weak binding of the mutant p50 NF-κB to the same region of the mouse VCAM-1 promoter. These results strongly support the importance of the Ser-20 phosphorylation site for the efficient occupancy of the transcription factor to its binding site on the VCAM-1 promoter, ultimately affecting the expression of the adhesion molecule.
DISCUSSION
Expression of VCAM-1 is critical for the initiation and progression of numerous inflammatory diseases, including atherosclerosis and asthma, as well as for the metastasis of a number of different types of cancer (6, 15, 26). The involvement of DNA-PK in both inflammation and the regulation of inflammatory gene transcription has yet to be addressed. Most studies that have investigated the potential involvement of DNA-PK in gene regulation have focused primarily on the DNA damage response (27–29). In the current study, we demonstrated that DNA-PK expression is crucial for the expression of the adhesion molecule VCAM-1 in response to TNF. We demonstrated that p50 NF-κB is an excellent substrate for DNA-PK and that the consequences of such phosphorylation are directly related to the ability of p50 NF-κB to drive the transcription of VCAM-1 as well as a number of additional NF-κB-dependent genes. Phosphorylation of p50 NF-κB appears to enhance its binding to DNA as either a homodimer or a heterodimer with p65 NF-κB. DNA-PK expression appears to be required for the efficient occupancy of the κB site on the VCAM-1 promoter by p50 NF-κB. The phosphorylation of p50 NF-κB on serine 20 to regulate VCAM-1 expression is supported by the finding that re-establishing the phosphorylation of p50 NF-κB, but not of the serine 20/alanine mutant, allowed p50 NF-κB−/− cells to express VCAM-1 upon TNF treatment. Further, a mutation of this phosphorylation site severely hampered κB site occupancy on the promoter of the VCAM-1 gene. The results of the current study not only present a novel mechanism by which VCAM-1 expression is regulated but also elucidate a completely new mechanism by which p50 NF-κB can play a dominant, positive role in the expression of VCAM-1 and in which the function of DNA-PK appears to be crucial.
The role of DNA-PK in NF-κB-associated signal transduction has been addressed by several studies; some of these analyses have determined that the connection between DNA-PK and NF-κB is exclusively limited to DNA damage. In fact, Basu et al. (27) showed that DNA-PK is required for both NF-κB activation, as assessed by EMSA, and phosphorylation of I-κBα in response to DNA damage but not in response to TNF. This positive function of DNA-PK was recently corroborated in a study using the DNA-damaging agent N-benzyladriamycin, a catalytic inhibitor of topoisomerase II (29). Conversely and interestingly, Liu et al. (28) reported a modulating function of DNA-PK in NF-κB signaling. DNA-PK was shown to phosphorylate both serine 36 and threonine 273 but not serine 32 of I-κBα; this modification rendered I-κBα a more potent inhibitor when compared with the unmodified protein. These conflicting results demonstrate the complexity of the involvement of DNA-PK in NF-κB signaling. Cellular responses to DNA damage differ greatly according to the nature of the insult as well as to the type and level of the resulting damage, which may explain the discrepancies in the above mentioned studies. Such results also require the differentiation of various inducers of DNA damage in terms of the role of DNA-PK activation in the regulation of gene transcription. In addition to in vitro experiments, our study focused on the cellular responses to TNF treatment and the involvement of DNA-PK in the regulation of VCAM-1 expression. It is noteworthy that upon TNF treatment, the phosphorylation and subsequent degradation patterns of I-κBα in both DNA-PK-proficient and DNA-PK-deficient cells were relatively similar, arguing against the possible involvement of DNA-PK in the direct phosphorylation of the NF-κB inhibitor in response to TNF stimulation.
TNF has been shown to increase the production of H2O2 in treated cells, constituting an oxidant signal and ultimately causing DNA damage (30, 31). A number of reports, including those from our laboratory, showed that pyrrolidine dithiocarbamate, an iron-chelating agent, could block TNF-induced VCAM-1 expression, in part, by inhibiting NF-κB activation (32, 33). Pyrrolidine dithiocarbamate prevents superoxide formation by chelating the iron required for the Fenton reaction, thereby blocking the generation of DNA breaks. Whether TNF treatment induces the generation of H2O2 that causes the DNA damage responsible for the activation of DNA-PK is unlikely given the observation that DNA-PK-proficient M059J cells do not express VCAM-1 upon direct treatment with H2O2 (data not shown). Thus, it is unclear what the triggering factor(s) responsible for both DNA-PK activation and VCAM-1 expression is in our experimental model. Currently, we do not know the underlying reason for the failure of M059K cells to respond to H2O2 treatment. Ko and Chin (34) reported that DNA-PK could be activated by thyroid hormone receptor-binding protein (TRBP) in the absence of DNA ends. Furthermore, Soutoglou and Misteli (35) recently showed that activation of the DNA damage response, including its induction by DNA-PK, could occur in the absence of DNA damage, suggesting the potential existence of mechanisms that govern DNA-PK activation without accompanying DNA damage. A great deal of experimentation is required to establish the exact mechanism by which DNA-PK is activated independently of DNA damage.
The members of the NF-κB family can form, theoretically, all possible combinations of homo- and heterodimers as dimerization is an obligatory regimen for DNA binding (36). The p50/p65 NF-κB complex is present in nearly all cells and is usually the most abundant. p50 homodimers are weak or inert gene activators and, in many instances, are considered suppressors because of competition with the transcriptionally active p50/p65 heterodimer (36). Our results suggest that the phosphorylation of p50 NF-κB on serine 20 by DNA-PK affects the formation of p50/p50 NF-κB and p50/p65 NF-κB dimers. Using p50 NF-κB−/− MEFs, we confirmed the requirement for serine 20 phosphorylation in VCAM-1 expression. We also showed that this phosphorylation is enhanced upon the binding of p50 NF-κB to DNA and appears to be a determinant for the interaction of NF-κB with and phosphorylation by DNA-PK. This observation also suggests that NF-κB phosphorylation must take place within the nucleus. It is important to note that these results were achieved using a cell-free system and that the findings must be confirmed in a cell culture system. We do know, however, that binding of p50 NF-κB to the VCAM-1 promoter in a cell culture system is highly dependent on both DNA-PK expression and the Ser-20 phosphorylation site of p50 NF-κB as revealed by ChIP analysis with antibodies to the transcription factor. What renders p50 NF-κB most susceptible to phosphorylation by DNA-PK appears to involve structural changes to the protein, thus rendering the site of phosphorylation more accessible to DNA-PK.
Taken together, the results of our study present a novel mechanism for the involvement of NF-κB in the regulation of VCAM-1 expression such that p50 NF-κB is not only a positive inducer of gene transcription but plays a dominant role over that of p65 NF-κB. It is important to mention that p65 NF-κB activation, nuclear translocation, and phosphorylation at sites that are critical for its activity are not sufficient to induce VCAM-1 expression in the absence of either DNA-PK or p50 NF-κB. The data from this investigation also support the possibility that DNA-PK contributes to inflammatory responses in which TNF or IL-1β is a major player, such as asthma and atherosclerosis, as well as cancer. Accordingly, inhibition of DNA-PK kinase activity may represent a novel approach to block such inflammation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL072889 (to A. H. B.). This work was also supported by Grant RSG-116608 from the American Cancer Society and funds from the Louisiana Cancer Research Consortium (to A. H. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- VCAM-1
- vascular cell adhesion molecule-1
- DNA-PK
- DNA-dependent protein kinase
- MEF
- mouse embryonic fibroblast
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1.Braun M., Pietsch P., Schrör K., Baumann G., Felix S. B. (1999) Cardiovasc Res. 41, 395–401 [DOI] [PubMed] [Google Scholar]
- 2.Hughes J. M., Arthur C. A., Baracho S., Carlin S. M., Hawker K. M., Johnson P. R., Armour C. L. (2000) Mediators Inflamm. 9, 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makrilia N., Kollias A., Manolopoulos L., Syrigos K. (2009) Cancer Invest. 27, 1023–1037 [DOI] [PubMed] [Google Scholar]
- 4.Rojas A. I., Ahmed A. R. (1999) Crit. Rev. Oral Biol. Med. 10, 337–358 [DOI] [PubMed] [Google Scholar]
- 5.Huo Y., Ley K. (2001) Acta. Physiol. Scand. 173, 35–43 [DOI] [PubMed] [Google Scholar]
- 6.Cybulsky M. I., Iiyama K., Li H., Zhu S., Chen M., Iiyama M., Davis V., Gutierrez-Ramos J. C., Connelly P. W., Milstone D. S. (2001) J. Clin. Invest. 107, 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazaar A. L., Albelda S. M., Pilewski J. M., Brennan B., Puré E., Panettieri R. A., Jr. (1994) J. Exp. Med. 180, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazaar A. L., Panettieri R. A., Jr. (2005) J. Allergy Clin. Immunol. 116, 488–495; quiz 496 [DOI] [PubMed] [Google Scholar]
- 9.Kasper H. U., Schmidt A., Roessner A. (1996) Gen. Diagn Pathol. 141, 289–294 [PubMed] [Google Scholar]
- 10.Shin W. S., Szuba A., Rockson S. G. (2002) Atherosclerosis 160, 91–102 [DOI] [PubMed] [Google Scholar]
- 11.Collis S. J., DeWeese T. L., Jeggo P. A., Parker A. R. (2005) Oncogene 24, 949–961 [DOI] [PubMed] [Google Scholar]
- 12.Martinet W., Knaapen M. W., De Meyer G. R., Herman A. G., Kockx M. M. (2002) Circulation 106, 927–932 [DOI] [PubMed] [Google Scholar]
- 13.Andreassi M. G. (2008) J. Mol. Med. 86, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 14.Vallabhapurapu S., Karin M. (2009) Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 15.ten Hacken N. H., Postma D. S., Bosma F., Drok G., Rutgers B., Kraan J., Timens W. (1998) Clin. Exp. Allergy 28, 1518–1525 [DOI] [PubMed] [Google Scholar]
- 16.Neish A. S., Williams A. J., Palmer H. J., Whitley M. Z., Collins T. (1992) J. Exp. Med. 176, 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C. W., Lin W. N., Lin C. C., Luo S. F., Wang J. S., Pouyssegur J., Yang C. M. (2006) J. Cell. Physiol. 207, 174–186 [DOI] [PubMed] [Google Scholar]
- 18.Beg A. A., Baltimore D. (1996) Science 274, 782–784 [DOI] [PubMed] [Google Scholar]
- 19.Zerfaoui M., Errami Y., Naura A. S., Suzuki Y., Kim H., Ju J., Liu T., Hans C. P., Kim J. G., Abd Elmageed Z. Y., Koochekpour S., Catling A., Boulares A. H. (2010) J. Immunol. 185, 1894–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulares A. H., Yakovlev A. G., Ivanova V., Stoica B. A., Wang G., Iyer S., Smulson M. (1999) J. Biol. Chem. 274, 22932–22940 [DOI] [PubMed] [Google Scholar]
- 21.Boulares H. A., Giardina C., Navarro C. L., Khairallah E. A., Cohen S. D. (1999) Toxicol. Sci. 48, 264–274 [DOI] [PubMed] [Google Scholar]
- 22.Li Q., Verma I. M. (2002) Nat. Rev. Immunol. 2, 725–734 [DOI] [PubMed] [Google Scholar]
- 23.Simbulan-Rosenthal C. M., Rosenthal D. S., Luo R., Smulson M. E. (1999) Oncogene 18, 5015–5023 [DOI] [PubMed] [Google Scholar]
- 24.Smulson M. E., Simbulan-Rosenthal C. M., Boulares A. H., Yakovlev A., Stoica B., Iyer S., Luo R., Haddad B., Wang Z. Q., Pang T., Jung M., Dritschilo A., Rosenthal D. S. (2000) Adv. Enzyme Regul. 40, 183–215 [DOI] [PubMed] [Google Scholar]
- 25.Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. (2004) Proteomics 4, 1633–1649 [DOI] [PubMed] [Google Scholar]
- 26.Ding Y. B., Chen G. Y., Xia J. G., Zang X. W., Yang H. Y., Yang L. (2003) World J. Gastroenterol. 9, 1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu S., Rosenzweig K. R., Youmell M., Price B. D. (1998) Biochem. Biophys. Res. Commun. 247, 79–83 [DOI] [PubMed] [Google Scholar]
- 28.Liu L., Kwak Y. T., Bex F., García-Martínez L. F., Li X. H., Meek K., Lane W. S., Gaynor R. B. (1998) Mol. Cell. Biol. 18, 4221–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panta G. R., Kaur S., Cavin L. G., Cortés M. L., Mercurio F., Lothstein L., Sweatman T. W., Israel M., Arsura M. (2004) Mol. Cell. Biol. 24, 1823–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das K. C., Lewis-Molock Y., White C. W. (1995) Mol. Cell. Biochem. 148, 45–57 [DOI] [PubMed] [Google Scholar]
- 31.Wheelhouse N. M., Chan Y. S., Gillies S. E., Caldwell H., Ross J. A., Harrison D. J., Prost S. (2003) Int. J. Mol. Med. 12, 889–894 [PubMed] [Google Scholar]
- 32.Satriano J., Schlondorff D. (1994) J. Clin. Invest. 94, 1629–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zerfaoui M., Suzuki Y., Naura A. S., Hans C. P., Nichols C., Boulares A. H. (2008) Cell. Signal. 20, 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko L., Chin W. W. (2003) J. Biol. Chem. 278, 11471–11479 [DOI] [PubMed] [Google Scholar]
- 35.Soutoglou E., Misteli T. (2008) Science 320, 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown K. D., Claudio E., Siebenlist U. (2008) Arthritis Res. Ther. 10, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.