Abstract
Hypertension is a serious risk factor for cardiovascular disease, and the angiotensinogen (AGT) gene locus is associated with human essential hypertension. The human AGT (hAGT) gene has an A/G polymorphism at −6, and the −6A allele is associated with increased blood pressure. However, transgenic mice containing 1.2 kb of the promoter with −6A of the hAGT gene show neither increased plasma AGT level nor increased blood pressure compared with −6G. We have found that the hAGT gene has three additional SNPs (A/G at −1670, C/G at −1562, and T/G at −1561). Variants −1670A, −1562C, and −1561T almost always occur with −6A, and variants −1670G, −1562G, and −1561G almost always occur with −6G. Therefore, the hAGT gene may be subdivided into either −6A or −6G haplotypes. We show that these polymorphisms affect the binding of HNF-1α and glucocorticoid receptor to the promoter, and a reporter construct containing a 1.8-kb hAGT gene promoter with −6A haplotype has 4-fold increased glucocorticoid-induced promoter activity as compared with −6G haplotype. In order to understand the physiological significance of these haplotypes in an in vivo situation, we have generated double transgenic mice containing either the −6A or −6G haplotype of the hAGT gene and the human renin gene. Our ChIP assay shows that HNF-1α and glucocorticoid receptor have stronger affinity for the chromatin obtained from the liver of transgenic mice containing −6A haplotype. Our studies also show that transgenic mice containing −6A haplotype have increased plasma AGT level and increased blood pressure as compared with −6G haplotype. Our studies explain the molecular mechanism involved in association of the −6A allele of the hAGT gene with hypertension.
Keywords: Gene Expression, Gene Regulation, Promoters, Transcription Factors, Transcription Regulation, Angiotensinogen, Blood Pressure, Hypertension, Single Nucleotide Polymorphism, Transgenic
Introduction
The renin-angiotensin system plays an important role in the regulation of blood pressure. The octapeptide angiotensin-II is one of the most active vasopressor agents and is obtained from angiotensinogen (AGT)2 by the combined proteolytic action of renin and angiotensin-converting enzyme. AGT is primarily synthesized in the liver and to a lesser extent in the fat, kidney, brain, heart, adrenal, and vascular walls (1). The plasma concentration of AGT is close to the Michaelis constant of the enzymatic reaction between renin and AGT (2). For this reason, a rise in plasma AGT levels can lead to a parallel increase in the formation of angiotensin-II that may ultimately result in hypertension. Injection of recombinant AGT increases plasma AGT level and blood pressure in rats (3). Overexpression of the AGT gene increases blood pressure in transgenic animals (4), and AGT gene knock-out mice have reduced blood pressure (5). Kim et al. (6) have introduced up to four copies of the AGT gene in mice with each copy of the gene, resulting in a successive increase in blood pressure. These results directly demonstrated that small increases in plasma AGT level could increase blood pressure in a gene dose-dependent manner.
The AGT gene is associated with essential hypertension in Caucasians (7), Japanese (8), and Asian Indian subjects (9). Although several investigators have confirmed these initial association studies, some studies have refuted this claim (reviewed in Ref. 10). However, a number of large studies and meta-analysis have confirmed this association (10–12). This gene may also be involved in cardiac hypertrophy (13), coronary atherosclerosis (14), and microangiopathy-related cerebral damage (15, 16). Initially, it was shown that variant M235T is associated with essential hypertension and increased plasma AGT level in hypertensive patients. However, because amino acid 235 is located far away from the renin cleavage site, this polymorphism does not explain the molecular mechanism involved in increased plasma AGT level. hAGT gene also has an A/G polymorphism at the −6-position. Inoue et al. (17) have shown that molecular variants 235T and −6A are in linkage disequilibrium, and reporter constructs containing the human AGT gene promoter with −6A have increased promoter activity compared with −6G. This observation suggests that increased plasma AGT levels by 235T in hypertensive patients may actually be due to increased transcriptional activity of the human AGT gene by −6A allele. Two large genome-wide analyses have been performed recently by the Wellcome Trust consortium (52) and the Framingham Heart study consortium (18) to identify genes involved in human hypertension using the Affymetrix 500K SNP platform. However, this platform has neither M235T nor −6A/G polymorphism and therefore could not evaluate the role of this SNP in hypertension.
In order to prove the role of −6A allele in hypertension, Sigmund's group used a knock-in approach at the hypoxanthine-guanine phosphoribosyltransferase locus and generated transgenic mice containing either −6A or −6G in the 13.5-kb hAGT gene (containing 1.2 kb of the promoter and all of the exons and introns) (19). However, analysis of these transgenic mice showed that variation at nucleotide −6 has no effect either on the blood pressure or on the tissue-specific expression of the hAGT gene. This suggested that polymorphism at the −6-position may only be a marker and not a functional polymorphism. We have found that the hAGT gene has three additional SNPs (A/G at −1670, C/G at −1562, and T/G at −1561), and variants −1670A, −1562C, and −1561T almost always occur with −6A, and variants −1670G, −1562G, and −1561G almost always occur with −6G. Thus, the hAGT gene may be subdivided into −6A and −6G haplotypes. We have also shown that an oligonucleotide containing −1670A has stronger affinity for hepatocyte nuclear factor-1α (HNF-1α) as compared with −1670G, and an oligonucleotide containing −1562C and −1561T has stronger homology with glucocorticoid-responsive element (GRE) as compared with −1562G and −1561G. In order to understand the role of these polymorphic sites on transcriptional regulation of the hAGT gene, we have synthesized reporter constructs containing a 1.8-kb promoter with either −6A or −6G haplotype and used them for transient transfection in human liver and kidney cells. We have shown that the reporter construct with −6A haplotype has 1.4–1.6-fold increased basal promoter activity and 4-fold increased glucocorticoid-induced promoter activity as compared with the reporter construct containing −6G haplotype. These experiments have suggested that the −6A allele of the hAGT gene may be associated with increased plasma AGT level and with increased blood pressure due to increased binding of HNF-1α and glucocorticoid receptor (GR) to the hAGT gene promoter containing variants −1670A, −1562C, and −1561T that always occur with −6A. In order to understand the role of these SNPs in the expression of the hAGT gene in a tissue-specific manner and in the regulation of blood pressure in an in vivo situation, we have recombineered 180-kb-long BAC DNA (containing 116 kb of the 5′-flanking region, all five exons and four introns, and 54 kb of the 3′-flanking region of the hAGT gene) that contains −6G haplotype. We exchanged the 1.8-kb hAGT gene promoter in this BAC with the 1.8-kb fragment obtained from the genomic DNA from a hypertensive subject containing −6A haplotype. We used these BAC DNAs and produced transgenic mice containing either −6A or −6G haplotype of the hAGT gene. Because mouse renin is not able to cleave hAGT, we have made double transgenic mice containing human renin gene and either −6A or −6G haplotype of the hAGT gene to understand the physiological role of these haplotypes in an in vivo situation. We show here that double transgenic mice containing −6A allele of the hAGT gene have increased plasma AGT level and increased blood pressure as compared with transgenic animals containing −6G allele.
MATERIALS AND METHODS
Cell Culture
Human hepatoma cells (HepG2) and human kidney cells (HK-2) were grown as a monolayer in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in an atmosphere of 5% CO2 at 37 °C. Cells were grown to 80% confluence in 6-well plates for transient transfections.
Plasmid Construction
The reporter construct pHAGT2.6luc (or pHAGT2572luc), and its deletion constructs pHAGT1748luc, pHAGT1657luc, pHAGT1624luc, pHAGT1604luc, pHAGT1544luc, pHAGT1459luc, and pHAGT1223luc were synthesized by PCR amplification of the human AGT gene using TTAGCTAGCAGCACATCTTACATGGTGGC, TTAGCTAGCTAGCCACAGATAAGCT, TTAGCTAGCAGGCTTTAAAGAGCCGA, TTAGCTAGCGCACAAGGTTGT, TTAGCTAGCCAGTGCTAGATCCTT, TTAGCTAGCCCACTTGCTGATCCTCA, TTAGCTAGCAGGCAGGAGCACTTTGA, and TTAGCTAGCCCAGACAAGTGATT as the respective forward primers with the NheI restriction site and TCGAAGCTTCACAGCTCAGTTACATCT as the reverse primer with the HindIII restriction site. The amplified fragments contained the nucleotide sequence −2581 to +98, −1748 to +98, −1651 to +98, −1618 to +98, −1598 to +98, −1541 to +98, −1457 to +98, and −1223 to +98, respectively. The PCR-amplified product was subcloned in the pGL3 basic vector that lacks eukaryotic promoter and enhancer sequences (Promega, Madison, WI). The mutant plasmid pHAGT1618lucmutC/EBP and pHAGT1618lucmutGRE were obtained by site-specific mutagenesis using the Stratagene site-directed mutagenesis kit. The nucleotide sequence of oligonucleotides used for mutation of the CCAAT/enhancer-binding protein (C/EBP) site was CAAGGTTGTTATTTCATCAG, and the sequence for the mutation of GRE was TCCAGGTCCACTTGTGCTAC (mutated nucleotides are shown in boldface italic type).
Transient Transfections
Transfections in HepG2 and HK-2 cell cultures in 6-well plates were carried out using the Lipofectamine reagent (Polyfect, Qiagen, Valencia, CA) using the manufacturer's protocol. Briefly, 250 ng of reporter constructs and 50 ng of RSV β-gal was used in each experiment. RSV-GR was co-transfected to increase the expression of glucocorticoid receptor (20). After 4 h of transfection, the medium was changed to the serum-free medium, and after 24 h of transfection, cells were treated for an additional 24 h with dexamethasone (100 nm). The procedures for cell extract preparation and luciferase or β-gal activity were carried out as described previously (21).
Gel Mobility Shift Assay
The probes for the electrophoretic mobility shift assay (EMSA) were chemically synthesized, annealed, and radiolabeled at the 5′-ends by polynucleotide kinase using [γ-32P]ATP. The EMSA protocol was followed as described previously (22). Nuclear extracts for gel mobility shift assays were prepared by modification of a method described previously (23). Antibodies against C/EBP and HNF-1α were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Oligonucleotides
Double-stranded oligonucleotides containing −1670A, −1670G, consensus HNF-1α, −1606C/EBP(T)WT, −1606C/EBP(C)MUT, and consensus C/EBP binding sites were obtained by annealing AATAGTGAATGAA, AATAGTGAGTGAA, GTTAATGAATGAC, AAGTTGTTGCTTAATCAG, AAGTTGTCGCTTAATCAG, and TGCAGATTGCGCAATCTGCA with their respective complementary oligonucleotides (mutated nucleotides are shown in boldface italic type).
Chromatin Immunoprecipitation (ChIP) Assays
The ChIP assay was performed using the ChIP assay kit from Upstate Biotechnology, Inc. (Lake Placid, NY). The protocol was followed as described previously (22). Finally, the immunoprecipitated DNA (1 μl) and the input DNA (1 μl) were subjected to 35 cycles of PCR amplification using CAGGCACAGTGGAAACT as a forward and ACTGCAGTAACAAGTC as a reverse primer. This amplified 203-bp product from the upstream region of the human AGT gene promoter (−1546 to −1748) spanning HNF-1α, C/EBP, and GRE was analyzed on 2% Metaphore agarose gel.
Preparation and Modification of hAGT BAC
Human BAC containing the AGT gene (RP11-505D24) was obtained from Invitrogen. BAC was grown in low salt LB medium (5 g/liter NaCl, 10 g/liter tryptone, and 5 g/liter yeast extract) plus 12.5 μg/ml chloramphenicol at 37 °C, and BAC DNA was purified by using the Nucleobond BAC Maxiprep kit (BD Biosciences). The promoter sequence in the wild type AGT BAC contained −6G haplotype and was modified by recombineering using the galK two-step procedure in SW102 cells (24). SW102 cells were grown at 32 °C. First, the 1.8 kb of the promoter region of the hAGT gene was deleted by inserting the galK cassette. The following PCR primers were used for this purpose (AGT homology is underlined): AAAGACCAGGATCTTTGTTTTGTTCCCTGACATATGCTGAGCACCAGGAACCTGTTGACAATTAATCATCGGCA and AGCTGAGGGGGCCCCCGGCTTACCTTCTGCTGTAGTACCCAGAACAACGGTCAGCACTGTCCTGCTCCTT. Next, the galK cassette was replaced with a 1.8-kb promoter region from the DNA of a hypertensive subject containing the −6A haplotype of the hAGT gene. The genomic DNA from the hypertensive subject was amplified by using AAAGACCAGGATCTTTGTTTTGTTCCCTGACATATGCTGAGCACCAGGAA as a forward primer and AGCTGAGGGGGCCCCCGGCTTACCTTCTGCTGTAGTACCCAGAACAACGG as a reverse primer to produce a 1.8-kb region of the promoter. Wild type and modified BAC DNAs were analyzed by fingerprinting after treatment with SpeI. The modified region of the AGT BAC was finally confirmed by direct sequencing. The nucleotide sequences of the promoter, all of the exons, and the 3′-flanking region were sequenced to confirm that they differ only in the promoter region and contained −6A or −6G haplotypes, respectively.
Generation of Transgenic Mice
The hAGT BAC plasmids were used for microinjection to generate transgenic mice using standard procedure. The presence of the hAGT gene was confirmed by PCR amplification of the tail DNAs of transgenic animals using human AGT gene-specific primers. The forward and reverse primers for the amplification of the hAGT gene were CAGCAGTGAAACTCTGC and TTCAGTCATCACCGTGC, respectively, to produce a 342-bp amplification product. Chimeras were bred to C57BL/6 mice and then successfully backcrossed to C57BL/6 for at least seven generations. Copy number of the AGT gene in transgenic and C57 mice was determined by quantitative PCR using human and mouse AGT-specific primers, and animals with single copy genes were selected for future experiments. Initially, four founders from each injection were selected, but after gene copy analysis, one line was established from each group for further work. Nucleotide sequence analysis of the hAGT gene promoter from the tails confirmed that transgenic mice with −6A haplotype have variants −6A, −20A, −217A, −532T, −793A, −1074T, −1178G, −1561T, −1562C, and −1670A, whereas transgenic mice with −6G haplotype have variants −6G, −20A, −217G, −532C, −793G, −1074G, −1178A, −1561G, −1562G, and −1670G. All experiments involving animals were approved by the institutional review board of the New York Medical College.
Generation of Double Transgenic Mice
Female transgenic mice containing either −6A or −6G haplotype of the hAGT gene were crossed with male transgenic mice containing the human renin gene (PAC-hRen) developed by Sinn et al. (25). The genetic background of the PAC-hRen mice was originally B6SJL (C57BL/6J × SJL/JF2), but they have since been backcrossed for at least five generations with C57BL/6J prior to breeding with hAGT mice. The double transgenic mice were genotyped for the human AGT and renin genes by PCR amplification of the DNA isolated from their tails. The primers for the amplification of the hAGT gene were the same as used in the analysis of single transgenic mice. The forward and reverse primers for the amplification of the hRen gene were CTCTTCGATGCTTCGGATTC and TGGCAGAGTAGGGTGTTCCT, respectively, to produce a 250-bp product.
Quantitative RT-PCR of hAGT and Renin mRNA
Liver and kidney from 8-week-old male transgenic mice containing either −6A or −6G haplotype of the hAGT gene and C57 control were harvested following CO2 asphyxiation and stored in Allprotect tissue reagent (Qiagen). RNA was isolated using the RNeasy Plus minikit (Qiagen). One microgram of RNA was reverse-transcribed into cDNA using the Revert Aid first strand cDNA synthesis kit (Fermentas). Quantitative real-time RT-PCR was performed using Brilliant II SYBR Green QPCR master mix (Stratagene) and a Stratagene thermocycler (Mx3005P, Agilent Technologies). Primers for mouse and human AGT (catalog nos. PPM04219A and PPH01807A), mouse and human renin (catalog nos. PPM0748A and PPH07193A), and mouse GAPDH (catalog no. PPM02946A) genes were purchased from SuperArray Bioscience Corp. Following a 95 °C incubation for 10 min, 40 cycles of PCR (95 °C for 30 s, 60 °C for 30 s) were then performed using 1 μl of cDNA, 50 nm PCR primers, and 12.5 μl of SYBR Green PCR Master Mix in 25-μl reactions. Threshold cycles for three replicate reactions were determined using MxPro-Mx3005P software (version 4.10), and relative transcript abundance was calculated following normalization with human GAPDH.
Western Blotting
Blood samples were collected by cardiac puncture from mice immediately after CO2 asphyxiation. Approximately 250 μl of plasma was collected from each mouse by centrifuging 500 μl of blood sample at 12,000 rpm for 15 min at 4 °C. Liver protein extracts were prepared from the tissue stored in AllProtect reagent (Qiagen) using a protein isolation kit (Qiagen). Liver protein extracts (25 μg) and equal volumes of plasma samples from transgenic mice were fractionated by SDS-PAGE (10% polyacrylamide) in duplicate and transferred to Immobilon-P transfer membrane (Millipore Corp.) to detect hAGT protein. Membranes were blocked in 10% nonfat dry milk (Bio-Rad), and one membrane was immunoblotted with a 1:1500 dilution of commercially available monoclonal antibodies for hAGT (catalog no. H00000183-M01, Abnova Corp.), whereas the other was incubated with a 1:10,000 dilution of mouse albumin antibody (catalog no. A90134P, Bethyl Laboratories). Immune complexes were detected by HRP-conjugated anti-mouse IgG (catalog no. I1904-25C, U.S. Biologicals) using the SuperSignal West Pico chemiluminescence assay (Pierce) according to the manufacturer's protocol. Densitometric analysis of protein bands was performed by Quantity One quantitation software from Bio-Rad.
Immunohistochemistry
Mice were perfused with normal saline. Liver was excised in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde-PBS for 3 h at room temperature, kept overnight at 4 °C in 30% sucrose solution, and then embedded in OCT compound (Sakura Finetek, Torrence, CA). Croystat sections (8-μm thickness) were taken on slides and treated with blocking solution (5% goat serum in PBS with 0.2% Triton X-100) for 1 h at room temperature. Sections were then incubated overnight at 4 °C with mouse anti-human primary antibody for AGT (Abnova), followed by washes with PBS (four times for 10 min each) and incubation (2 h) with secondary Cy3-conjugated goat anti-mouse antibody (Abnova) at room temperature. Slides were finally washed with PBS (four times for 10 min each) and mounted. Immunofluorescence was visualized using a Zeiss Axioplan-2 fluorescent microscope. Images were captured and analyzed using AxioVision 2 multichannel image processing software (Zeiss, Göttingen, Germany).
In Vivo ChIP Assay
The ChIP assay was performed using the ChIP assay kit from Upstate Biotechnology, Inc. Mice were perfused with normal saline. Liver was excised and washed in PBS, minced into smaller pieces, fixed with 1% formaldehyde for 20 min at room temperature, and washed with chilled PBS followed by lysis. The DNA was fragmented by sonication, and 10 μl of the chromatin solution was saved as input. A 5-μg amount of anti-GR or HNF-1α or RNApol antibody or rabbit immunoglobulin G was added to tubes containing 900 μl of sonicated chromatin solution, and the mixture was incubated overnight at 4 °C. The antibody complexes were captured with protein A-agarose beads and subjected to serial washes (as described in the manufacturer's protocol). The chromatin fraction was extracted with SDS buffer and reverse cross-linked at 65 °C for 4–6 h. The DNA was then purified using a Qiagen miniprep column. The immunoprecipitated DNA (1 μl) and the input DNA (1 μl) were subjected to 35 cycles of PCR amplification using −314AGTfor (CTCAGGCTGTCACACACCTA) as a forward and −6AGTrev (TCTTCCCCTGGCCGGGTCACGAT) as a reverse primer when GR or RNA polymerase antibodies were used for immunoprecipitation. This amplified 317-bp fragment spanning the GR and RNA polymerase binding site (located between −314 and +3) of the human AGT gene promoter. We used −1757AGTfor (CAGGCACAGTGGAAACTCTCC) as a forward primer and −1554AGTrev (AGTAACAAGTCCACCTGGAC) as a reverse primer when HNF-1α antibody was used. This amplified the 205-bp fragment containing the HNF-1α binding region of the human AGT gene promoter. The PCR-amplified products were analyzed on 2% agarose gel.
Plasma Angiotensin II Level
Plasma angiotensin-II levels were determined by ELISA. Equal amounts of plasma from control (non-transgenic) and double transgenic mice containing either −6A or −6G haplotype of the hAGT gene were passed through phenylsilylsilica extraction columns (ALPCO Diagnostics, Salem, NH). The reversed phase extraction method was applied to extract the plasma as per the manufacturer's protocol. A peptide enzyme immunoassay for Ang II was performed in extracted plasma samples using an enzyme immunoassay kit (Peninsula Laboratories) as per the manufacturer's protocol. Plasma angiotensin-II levels were calculated as described by the manufacturer using a standard curve.
Blood Pressure Measurement
All mice were fed with standard mice chow and had access to water ad libitum. Blood pressure (BP) was measured in the conscious state by telemetry. A radiotelemetric system from Data Science International (St. Paul, MN) was used for this procedure. Briefly, mice were anesthetized with ketamine and xylazine (90 and 10 mg/kg, respectively), and the left carotid artery was isolated. The tip of the telemetric catheter (model TA11PA-C10) was then inserted into the carotid artery and advanced into the aortic arch, with the telemetric device main body positioned into a subcutaneous pocket into the right flank. After 1 week of recovery from the surgical procedure, BP readings were recorded every 10 min using a Data-Science International instrument as described previously (26). Mean BP values were calculated for every hour from the values taken over 4 days. Animals were then treated with angiotensin receptor blocker, losartan (Sigma-Aldrich) (30 mg/kg/day), in drinking water. The blood pressure was then measured 24 h after losartan treatment.
Statistical Analysis
The unpaired t test was used to compare relative luciferase activities of reporter constructs in transient transfection experiments. All experiments were conducted in triplicate in three independent transfections. Data are expressed as means ± S.E. from each group containing six mice. Differences between group means were determined by a two-factor analysis of variance, followed by a Newman-Keuls post hoc analysis (NCSS 2007); p < 0.05 was considered significant (NCSS LLC, Kaysville, UT). Statistically significant results are marked by an asterisk (p < 0.05).
RESULTS
Variant −6A of the hAGT Gene Is Associated with Hypertension in Caucasian Subjects
In order to confirm a previous observation that −6A allele is associated with hypertension in Caucasian subjects, we analyzed genomic DNA from 293 Caucasian subjects with normal blood pressure and 354 Caucasian subjects with hypertension (mean age 59 ± 10 years, equal number of male and female subjects) for A/G polymorphism at −6 of the hAGT gene. All of these subjects were recruited from the outpatient department of Westchester Medical Center (Valhalla, NY). All case and control subjects gave informed consent before participating in the research. The research protocol was approved by the institutional review board at New York Medical College. All cases were diagnosed as having primary hypertension, and patients with secondary hypertension, diabetes mellitus, or ischemic heart disease were excluded. The criteria for hypertension were defined as a systolic BP of >140 mm Hg, a diastolic BP of >90 mm Hg, or under antihypertensive therapy. Blood pressure was measured twice with the subject seated with a 5-min interval. The normotensives (with systolic BP/diastolic BP of <140/90 mm Hg) without a history of hypertension and without diabetes mellitus were recruited from the same population and matched for sex and age. All participants completed a standard questionnaire on personal medical history and family history of hypertension. Genomic DNA was isolated from peripheral blood samples, and −6A/G polymorphism of the hAGT gene was analyzed by PCR amplification of DNA followed by BstNI restriction analysis as described (9). All patients and control subjects were in Hardy-Weinberg equilibrium. The frequency of the −6A allele in hypertensive patients was 0.51 as compared with 0.40 in the normotensive population, which is highly significant (p = <.001 and OR = 2.2, confidence interval 1.49–3.31). Statistical analysis based on genotype also suggested a highly significant role of the −6A allele in hypertension in this cohort (p < 0.001).
Upstream Region of the hAGT Gene Promoter Increases Glucocorticoid-induced Promoter Activity
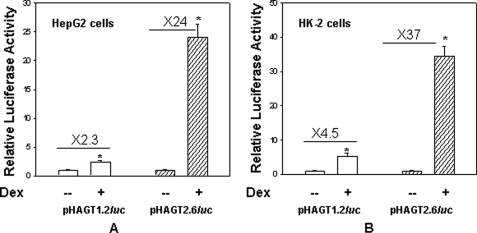
Because previous studies have shown that glucocorticoid treatment increases expression of the AGT gene in liver (20, 27), we were interested in identifying glucocorticoid-responsive elements in the promoter of the hAGT gene. We therefore synthesized reporter constructs that contained either 1253 bp (pHAGT1.2-luc) or 2572 bp (pHAGT2.6-luc) of the hAGT gene promoter attached to the luciferase gene. These reporter constructs were then transiently transfected in HepG2 cells along with RSV-GR (an expression vector containing rat GR coding sequence), and promoter activity was analyzed after dexamethasone treatment as described previously (27–29). Results of this experiment showed that whereas the reporter construct with the 1.2-kb promoter gave only 2.3-fold glucocorticoid-induced promoter activity, the reporter construct with the 2.6-kb promoter gave 24-fold increased promoter activity upon transient transfection in human liver cells (Fig. 1A). Similarly, the reporter construct with the 1.2-kb promoter gave only 4.5-fold glucocorticoid-induced promoter activity, whereas the reporter construct with the 2.6-kb promoter gave 37-fold glucocorticoid-induced promoter activity upon transient transfection in human kidney cells (Fig. 1B). This experiment suggested that nucleotide sequence located between 1.3 and 2.6 kb of the promoter is responsible for glucocorticoid-induced promoter activity of the hAGT gene.
FIGURE 1.
Dexamethasone treatment increases the promoter activity of phAGT2.6-luc as compared with phAGT1.2-luc upon transient transfection in HepG2 and HK-2 cells. HepG2 (A) or HK-2 cells (B) were transiently transfected with reporter construct phAGT1.2-luc (open bar) or phAGT2.6-luc (filled bar) and RSV-GR. After 24 h, cells were treated in the absence or presence of 100 nm Dex as described under “Materials and Methods.” Luciferase activity was determined after 24 h of Dex treatment. Error bars, S.E.
Deletion Constructs Show That Glucocorticoid-induced Promoter Activity Is Located between −1541 and −1748
In order to identify the promoter region responsible for glucocorticoid-induced promoter activity of the hAGT gene, we synthesized deletion constructs using pHAGT2.6-luc. These constructs contained 1748, 1651, 1618, 1598, 1541, and 1457 nucleotides from the transcriptional initiation sites attached to the luciferase gene (supplemental Fig. 1A). These deletion constructs were then used for transient transfection in HepG2 cells in the absence and presence of dexamethasone. Results of this experiment suggested that nucleotide sequence located between 1748 and 1541 is mainly responsible for the glucocorticoid-induced promoter activity (supplemental Fig. 1B).
Nucleotide Sequence Located between −1541 and −1748 Contains Binding Sites for Multiple Liver-enriched Transcription Factors
In order to identify nucleotide sequence that may be involved in glucocorticoid-induced promoter activity, we determined the nucleotide sequence of the hAGT gene located between −1748 and −1541. The nucleotide sequence of this region of the promoter revealed homology with binding sites of multiple liver-enriched transcription factors, such as HNF-3, HNF-1, and C/EBP, as well as GRE and SP1 (supplemental Fig. 2A). Binding of multiple transcription factors in this region of the promoter may be necessary for glucocorticoid-induced expression of the hAGT gene because previous studies have suggested that interaction of multiple transcription factors is crucial for glucocorticoid-induced promoter activity of a gene (30–32).
Nucleotide Sequence Located between −1541 and −1748 of the hAGT Gene Contains Three Additional SNP Sites
Previous studies have shown that the hAGT gene has eight SNPs in the 1.2-kb promoter (located at −6, −20, −217, −532, −776, −793, −1074, and −1178) that are involved in transcriptional regulation of this gene. We were therefore interested in examining whether nucleotide sequence located between −1748 and −1541 of the hAGT gene promoter contains additional SNPs that may modulate its transcriptional activity. We therefore determined nucleotide sequence of this region of the promoter from five normotensive and five hypertensive subjects by amplification and direct sequencing of this region of the promoter. Results of this experiment showed that this region of the promoter has three new SNPs (A/G at −1670, C/G at −1562, and T/G at −1561) (supplemental Fig. 2B). The position of these SNPs is also shown by an asterisk at the top of each nucleoside (supplemental Fig. 2A).
Variant −6A Almost Always Occurs with Variants −1561T, −1562C, and −1670A
Because variant −6A of the hAGT gene is associated with increased plasma AGT level and increased blood pressure, we were interested in finding whether −6A is in linkage disequilibrium with three new SNPs identified in the upstream region of the hAGT gene promoter. We therefore sequenced DNA from 100 subjects in these regions of the promoter. Results of this analysis suggested that variant −6A always occurred with variants −1561T, −1562C, and −1670A, and variant −6G always occurred with −1561G, −1562G, and −1670G (only four samples with −6A had −1561G and three samples had −6G and −1561T). Therefore, the hAGT gene may be subdivided into two haplotypes: −6A haplotype containing variants −6A, −1561T, −1562C, and −1670A; and −6G haplotype containing variants −6G, −1561G, −1562G, and −1670G.
Nucleotide Sequence of the hAGT Gene Containing −1670A Binds Strongly to Liver-enriched Transcription Factor HNF-1α as Compared with −1670G
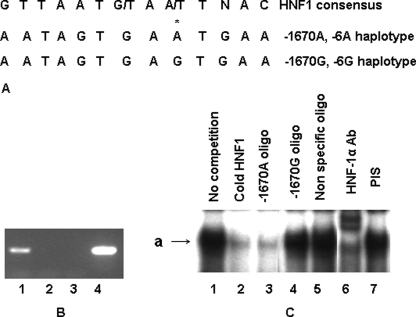
Because an A/G polymorphic site is located between −1748 and −1651 of the hAGT gene promoter and deletion of this region affects glucocorticoid-induced promoter activity, we were interested in finding whether nucleotide sequence around this polymorphic site binds to a transcription factor. Transfac analysis suggested that indeed nucleotide sequence containing −1670A has stronger homology with the consensus HNF-1 binding site as compared with −1670G (Fig. 2A). In order to confirm that HNF-1 binds to this region of the promoter, we performed a ChIP assay using HepG2 cells. Results of this assay show that HNF-1α binds to this region of the promoter (Fig. 2B). In order to analyze whether variants −1670A and −1670G bind differentially to HNF-1α, we performed an EMSA using radiolabeled oligonucleotides containing −1670A in the presence of HepG2 nuclear extract. Results of this experiment (Fig. 2C) showed that the complex formed by radiolabeled oligonucleotide containing consensus HNF-1 site (lane 1) (marked as a) was removed in the presence of cold oligonucleotide containing consensus HNF-1 binding site (lane 2) or self-oligonucleotide (lane 3) but not with an oligonucleotide containing −1670G (lane 4) or a nonspecific oligonucleotide (lane 5). This complex was HNF-1-specific, as shown by the fact that it was supershifted in the presence of HNF-1α antibody (lane 6) and not in the presence of preimmune serum (lane 7). Results of this experiment therefore suggested that oligonucleotide containing −1670A shows greater affinity for HNF-1α as compared with the oligonucleotide containing −1670G.
FIGURE 2.
A, nucleotide sequence around −1670 has homology with the HNF-1 binding site. Line 1 shows the consensus HNF-1 binding site, line 2 shows the nucleotide sequence containing −1670A, and line 3 shows the nucleotide sequence containing −1670G. B, ChIP assay shows that nucleotide sequence around −1670 binds to HNF-1α in HepG2 cells. Immunoprecipitated DNA from HepG2 cells was used to amplify a 203-bp sequence from the hAGT gene promoter (−1546 to −1748) containing the HNF-1α binding site. Lane 1, PCR product obtained in the presence of HNF-1α antibody; lane 2, PCR product obtained in the presence of preimmune serum; lane 3, PCR product obtained in the presence of HNF-1α antibody from another region of the promoter that does not contain the HNF-1 site; lane 4, PCR product obtained from the genomic DNA without immunoprecipitation (positive control) (lane 4). C, EMSA shows that nucleotide sequence containing −1670A has stronger homology with the HNF-1α binding site as compared with −1670G. EMSA was performed using radiolabeled oligonucleotide containing −1670A in the presence of HepG2 nuclear extract. Lane 1, radiolabeled oligonucleotide containing −1670A without competitor; lane 2, competition with a 50-fold excess of cold oligonucleotide containing consensus HNF-1 binding site; lane 3, competition with a 50-fold excess of cold oligonucleotide containing −1670A; lane 4, competition with a 50-fold excess of cold oligonucleotide containing −1670G; lane 5, competition with a 50-fold excess of cold oligonucleotide containing nonspecific oligonucleotide; lane 6, reaction in the presence of HNF-1α antibody; lane 7, reaction in the presence of preimmune serum (PIS).
Nucleotide Sequence Located between −1624 and −1604 Contains a C/EBP Binding Site That Plays an Important Role in Glucocorticoid-induced Promoter Activity
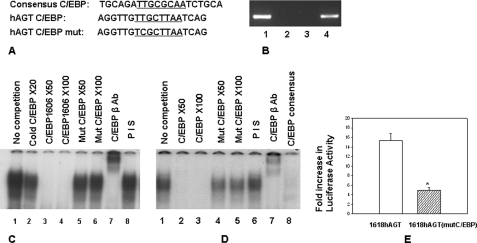
Our transient transfection assay using different deletion constructs showed that removal of nucleotide sequence between −1618 and −1598 reduces glucocorticoid-induced promoter activity (supplemental Fig. 1). Nucleotide sequence located in this region of the promoter has strong homology with the C/EBP site (Fig. 3A) (core binding sequence is underlined). Our ChIP assay showed that C/EBPβ binds to this region of the promoter (Fig. 3B). In order to further confirm that this sequence binds to C/EBP, we performed an EMSA (Fig. 3C). A complex obtained with radiolabeled oligonucleotide containing the consensus C/EBP site in the presence of HepG2 extract (lane 1) was removed in the presence of a 20-fold excess of cold self-oligonucleotide (lane 2) and a 50- or 100-fold excess of cold oligonucleotide containing nucleotide sequence located between −1606 and −1614 (labeled CEBP 1606) (lanes 3 and 4) but only slightly reduced in the presence of mutated CEBP1606 (lanes 5 and 6); this complex was supershifted in the presence of C/EBPβ antibody (lane 7) but not in the presence of preimmune serum (lane 8). In another experiment, radiolabeled oligonucleotide CEBP1606 formed a complex with HepG2 extract (Fig. 3D, lane 1), which was removed in the presence of a 50- or 100-fold excess of cold self-oligonucleotide (lanes 2 and 3) but only slightly reduced in the presence of mutated CEBP 1606 (lanes 4 and 5); this complex was not removed or supershifted in the presence of preimmune serum (lane 6) but was supershifted in the presence of C/EBPβ antibody (lane 7) and removed in the presence of cold oligonucleotide containing consensus C/EBP site (lane 8). Results of these experiments suggested that nucleotide sequence located between −1606 and −1614 binds to the C/EBP family of transcription factors. In order to confirm the role of this C/EBP binding site in glucocorticoid-induced promoter activity of the hAGT gene, we performed site-specific mutagenesis in the reporter construct pHAGT1618-luc to mutate the C/EBP site and performed a transient transfection assay. Results of this experiment show that whereas wild type construct had 15-fold dexamethasone-induced promoter activity, reporter construct containing mutation of the C/EBP site located between −1606 and −1614 had only 5-fold dexamethasone-induced promoter activity (Fig. 3E). These experiments confirmed that the C/EBP site located around −1610 is necessary for glucocorticoid-induced promoter activity of the hAGT gene.
FIGURE 3.
A, sequence homology between the consensus C/EBP binding site and −1600 region of the hAGT gene promoter. Line 1, consensus C/EBP binding site; line 2, nucleotide sequence located between −1596 and −1613 of the hAGT gene; line 3, oligonucleotide sequence containing mutated C/EBP binding site. B, ChIP assay shows that C/EBP binds to the −1600 region of the hAGT gene promoter in HepG2 cells. The ChIP assay was performed as described in the legend to Fig. 6B except that C/EBPβ antibody was used for immunoprecipitation. Shown are PCR product obtained in the presence of C/EBPβ antibody (lane 1), PCR product in the presence of preimmune serum (lane 2), PCR product obtained in the presence of C/EBPβ antibody from another region of the promoter that does not contain C/EBP site (lane 3), and PCR product obtained from the genomic DNA without immunoprecipitation (positive control) (lane 4). C, EMSA confirms that nucleotide sequence located between −1596 and −1613 of the hAGT gene promoter competes with the C/EBP binding site. EMSA was performed using radiolabeled oligonucleotide containing the consensus C/EBP binding site in the presence of HepG2 extract and competed with an oligonucleotide containing either wild type or mutated C/EBP site located in the −1596 to −1613 region of the hAGT gene promoter. Lane 1, complex obtained in the presence of radiolabeled oligonucleotide containing consensus C/EBP site; lane 2, competition with a 20-fold excess of cold C/EBP consensus oligonucleotide; lanes 3–6, competition in the presence of a 50- and 100-fold excess of an oligonucleotide containing either wild type or mutant C/EBP sequence from the hAGT gene; lane 7, reaction in the presence of C/EBPβ antibody; lane 8, reaction in the presence of preimmune serum (PIS). D, EMSA confirms that nucleotide sequence located between −1596 and −1613 of the hAGT gene promoter binds to C/EBPβ. EMSA was performed using radiolabeled oligonucleotide containing the C/EBP site located between −1596 and −1613 of the hAGT gene promoter in the presence of HepG2 extract. Lane 1, complex obtained in the presence of radiolabeled oligonucleotide alone; lanes 2–5, competition in the presence of a 50- and 100-fold excess of wild type and mutant cold C/EBP oligonucleotides; lane 6, reaction in the presence of preimmune serum; lane 7, reaction in the presence of C/EBPβ antibody; lane 8, reaction in the presence of a 50-fold excess of cold consensus C/EBP oligonucleotide. E, mutation of C/EBP site located between −1596 and −1613 of the hAGT gene promoter reduces glucocorticoid-induced promoter activity. Reporter constructs containing 1618 bp of the hAGT gene promoter with either wild type or mutated C/EBP site (located between −1596 and −1613) were transiently transfected in HepG2 cells, and glucocorticoid-induced promoter activity was analyzed as described above. The empty box shows glucocorticoid-induced promoter activity of the wild type construct, and the filled box shows glucocorticoid-induced promoter activity of the mutated construct. Error bars, S.E.
Nucleotide Sequence Containing SNPs at −1561 and −1562 Has Homology with GRE
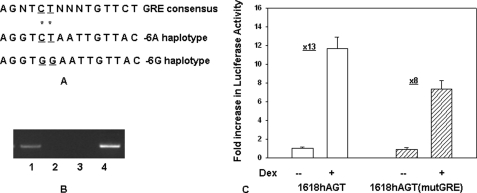
We were next interested in determining whether nucleotide sequence of the hAGT gene promoter containing SNPs at −1562 and −1561 binds to a transcription factor that may modulate glucocorticoid-induced promoter activity. Nucleotide sequence containing −1562C and −1561T (that is in linkage disequilibrium with −6A) (Fig. 4A, lane 2) has stronger homology with consensus GRE (Fig. 4A, lane 1) as compared with the nucleotide sequence containing −1562G and −1561G (that is in linkage disequilibrium with −6G) (Fig. 4A, lane 3). We next performed a ChIP assay using GR antibody and HepG2 cells. Results of this experiment confirmed that indeed GR binds to this region of the promoter (Fig. 4B). Because variants −1562C and −1561T had stronger homology with GRE, we wanted to examine their role in GR-induced promoter activity. We therefore mutated −1562C to −1562G and −1561T to −1561G by site-specific mutagenesis in the reporter construct containing pHAGT1618-luc and performed transient transfection in HepG2 cells. Results of this experiment showed that mutation of −1562C and −1561T reduced glucocorticoid-induced promoter activity from 13-fold to 8-fold (Fig. 4C).
FIGURE 4.
A, nucleotide sequence containing −1562C and −1561T of the hAGT gene has stronger homology with consensus GRE as compared with −1562G and −1561G. Line 1, nucleotide sequence of consensus GRE; line 2, nucleotide sequence of the hAGT gene containing −1562C and −1561T (−6A haplotype); line 3, nucleotide sequence of the hAGT gene containing −1562G and −1561G (−6G haplotype). *, polymorphic sites. B, ChIP assay shows that GR binds to the nucleotide sequence around −1560 of the hAGT gene promoter in HepG2 cells. The ChIP assay was performed as described in the legend to Fig. 5B except that GR antibody was used for immunoprecipitation. Shown are the PCR product obtained in the presence of GR antibody (lane 1), PCR product in the presence of preimmune serum (lane 2), PCR product obtained in the presence of GR antibody from another region of the promoter that does not contain the GR binding site (lane 3), and PCR product obtained from the genomic DNA without immunoprecipitation (positive control) (lane 4). C, mutation of the GRE site around −1560 of the hAGT gene promoter reduces glucocorticoid-induced promoter activity. Reporter constructs containing 1618 bp of the hAGT gene promoter with either wild type GRE (containing −1562C and −1561T) or mutated GRE (containing −1562G and −1561G) were transiently transfected in HepG2 cells, and glucocorticoid-induced promoter activity was analyzed as described above. Empty boxes show the promoter activity of the wild type construct in the absence and presence of Dex. Filled boxes show the promoter activity of the mutated construct in the absence and presence of Dex. Error bars, S.E.
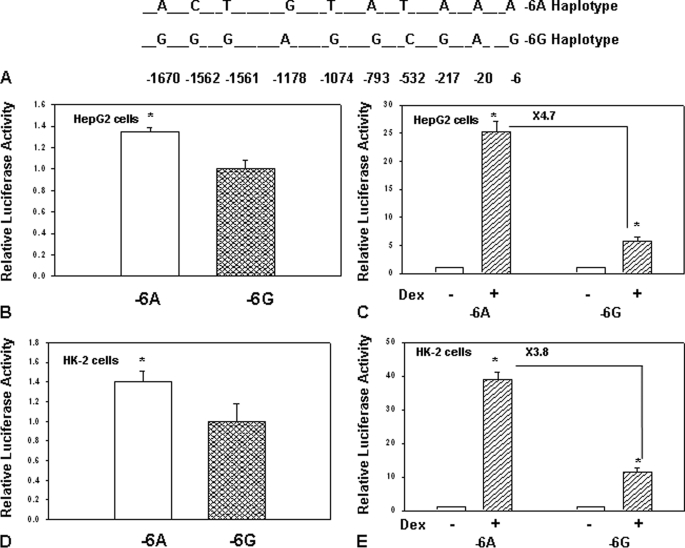
Reporter Construct Containing Haplotype −6A of hAGT Gene Has Increased Basal and Glucocorticoid-induced Promoter Activity as Compared with Haplotype −6G upon Transient Transfection in Human Liver and Kidney Cells
In order to understand the role of −6A and −6G alleles in the context of the 1.8-kb promoter of the hAGT gene upon transcriptional regulation, we synthesized reporter constructs using genomic DNA from a hypertensive subject (that contained −6A allele) and a normotensive subject (that contained −6G allele). Because previous studies have shown that the hAGT gene contains eight polymorphic sites in the 1.2-kb region of the promoter, we determined the nucleotide sequence of the entire region of the promoter present in these constructs. The positions of different SNPs in the 1.8-kb promoter of reporter construct containing −6A haplotype and −6G haplotype are shown in Fig. 5A. These reporter constructs were then transiently transfected in human liver cells (HepG2) and human kidney (HK-2) cells to determine their basal and glucocorticoid-induced promoter activity. Basal and glucocorticoid-induced promoter activity of reporter construct pHAGT1748-luc containing −6A and −6G haplotypes in HepG2 cells is shown in Fig. 5, B and C, and promoter activity in HK-2 cells is shown in Fig. 5, D and E. Results of these experiments showed that the reporter construct containing 1.8 kb of the hAGT gene promoter with −6A haplotype has 40–50% increased basal promoter activity and 4–6-fold increased glucocorticoid-induced promoter activity upon transient transfection in human liver and kidney cells as compared with −6G haplotype.
FIGURE 5.
A, position of different SNPs in 1.8-kb promoter of −6A and −6G haplotypes of the hAGT gene. SNPs located at positions −6, −20, −217, −532, −793, −1074, −1178, −1561, −1562, and −1670 in −6A and −6G haplotypes of the hAGT gene promoter are shown. B, reporter construct containing 1.8 kb of hAGT gene promoter with −6A haplotype has 1.4-fold increased promoter activity as compared with −6G haplotype upon transfection in HepG2 cells. The empty box shows the promoter activity of −6A haplotype, and the filled box shows the promoter activity of −6G haplotype. C, reporter construct containing 1.8 kb of hAGT gene promoter with −6A haplotype has 4.7-fold increased glucocorticoid-induced promoter activity as compared with −6G haplotype upon transient transfection in HepG2 cells. Empty boxes show the promoter activity in the absence of Dex, and filled boxes show the promoter activity in the presence of Dex. D, reporter construct containing 1.8 kb of hAGT gene promoter with −6A haplotype has 1.5-fold increased promoter activity as compared with −6G haplotype upon transfection in HK-2 cells. The empty box shows the promoter activity of −6A haplotype, and the filled box shows the promoter activity of −6G haplotype. E, reporter construct containing 1.8 kb of the hAGT gene promoter with −6A haplotype has 3.8-fold increased glucocorticoid-induced promoter activity as compared with −6G haplotype upon transfection in HK-2 cells. Empty boxes show the promoter activity in the absence of Dex, and filled boxes show the promoter activity in the presence of Dex. Error bars, S.E.
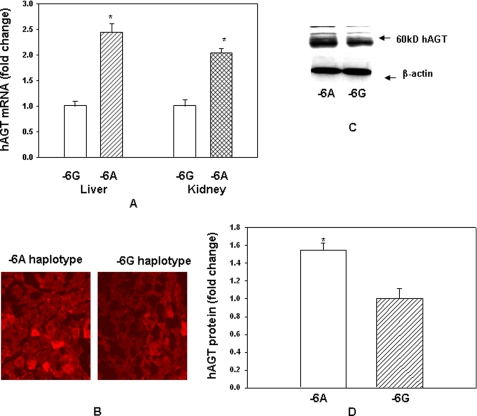
Angiotensinogen mRNA Is Increased in the Liver and Kidney of Transgenic Mice Containing −6A Haplotype of the hAGT Gene as Compared with the −6G Haplotype
In order to understand the role of −6A and −6G haplotypes in the expression of the hAGT gene in an in vivo situation, we generated double transgenic mice containing the human renin gene and either −6A or −6G haplotype of the hAGT gene as described under “Materials and Methods.” We started with a 180-kb BAC containing −6G haplotype of the hAGT gene (supplemental Fig. 3). This BAC was recombineered to produce another BAC with −6A haplotype of the hAGT gene. These BACs were then used to generate transgenic mice containing either −6A or −6G haplotype of the hAGT gene. These mice were then crossed with transgenic mice containing the human renin gene to obtain double transgenic mice. Because liver and kidney are the most important sites for the expression of AGT gene, we examined the effect of −6A and −6G haplotypes on hAGT mRNA level in these tissues of transgenic mice by quantitative RT-PCR. Quantitation of RNA level in −6A haplotype was performed using RNA level in −6G haplotype as 1. Results of these experiments show that the AGT mRNA level in the liver of transgenic mice containing −6A haplotype is increased about 2.5-fold, and in the kidney, it was increased 2.1-fold as compared with the transgenic mice containing −6G haplotype (Fig. 6A). We also confirmed the overexpression of hAGT protein in the liver of −6A haplotype compared with −6G haplotype by immunohistochemistry. Results of this experiment showed that the hAGT gene is expressed more abundantly in the liver from a transgenic animal containing −6A haplotype as compared with −6G haplotype (Fig. 6B). Finally, we examined the AGT protein level in the liver of transgenic mice by Western blot analysis. A typical Western blot is shown in Fig. 6C, and quantitation of AGT protein level normalized to the β-actin is shown in Fig. 6D. hAGT mRNA level was increased about 2.5-fold in the liver of transgenic mice containing −6A haplotype as compared with −6G haplotype, and AGT protein level was increased about 1.6-fold.
FIGURE 6.
hAGT expression is increased in the liver and kidney of double transgenic mice containing −6A haplotype of the hAGT gene as compared with −6G haplotype. A, RNA from the liver and kidney of transgenic mice containing either −6A or −6G haplotype of the hAGT gene was analyzed by quantitative RT-PCR. B, immunohistochemistry of hAGT in sections from the liver of transgenic mice containing −6A or −6G haplotype of the hAGT gene. C, Western blot analysis of hAGT in the liver of transgenic mice containing −6A or −6G haplotype of the hAGT gene. D, quantitation of hAGT protein in livers of transgenic mice containing −6A or −6G haplotype of the hAGT gene as determined by Western blot. Quantitation was performed after normalization with β-actin. Error bars, S.E.
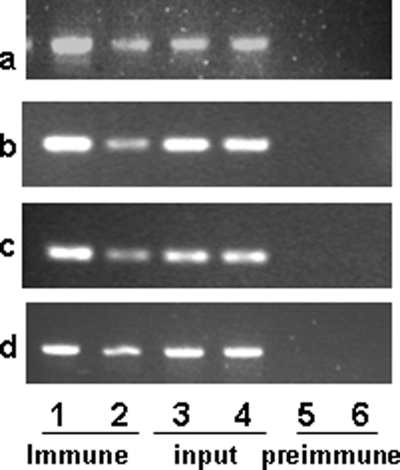
Chromatin Immunoprecipitation Assay Shows That HNF-1α, GR, and RNA Polymerase Have Higher Affinity for the Chromatin of Liver from Transgenic Mice Containing −6A Haplotype of the hAGT Gene as Compared with −6G Haplotype
Because (a) our studies described here have shown that nucleotide sequence of hAGT gene promoter containing −1670A has stronger homology with the HNF-1 consensus binding site as compared with −1670G, (b) our studies here have shown that nucleotide sequence containing −1562C and −1561T has stronger homology with consensus GRE as compared with −1562G and −1561G, and (c) previous studies have shown that nucleotide sequence containing −217A has stronger affinity for GR as compared with −217G (29), we wanted to confirm the effect of these polymorphisms on the binding of their cognate transcription factors using chromatin from the liver of double transgenic mice containing either the −6A or −6G haplotype of the hAGT gene. Results of our in vivo ChIP assay showed that HNF-1α binds more strongly to the nucleotide sequence around the −1670 region in the chromatin from the liver of transgenic mice containing −6A haplotype of the hAGT gene as compared with the −6G haplotype (Fig. 7a, compare lanes 1 and 2). Similarly, GR binds more strongly to the −1550 and −220 regions of the hAGT gene promoter in the chromatin of the liver from transgenic mice containing −6A haplotype as compared with −6G haplotype (Fig. 7, b and c). GR also binds to the −220 region of the promoter because we have previously shown that the hAGT gene has an A/G polymorphism at the −217-position, and GR shows stronger affinity for an oligonucleotide containing −217A as compared with −217G (29). Finally, we performed a ChIP assay using antibody against RNA polymerase to examine whether it binds more strongly to the hAGT gene promoter in the chromatin of liver containing −6A haplotype as compared with −6G haplotype. Results of this experiment showed that indeed RNA polymerase binds more strongly to the hAGT gene promoter in the chromatin of liver of transgenic mice containing −6A haplotype as compared with −6G haplotype (Fig. 7d). Taken together, results of this experiment confirmed that RNA polymerase, HNF-1α, and GR have stronger affinity for the hAGT gene promoter in the chromatin obtained from the liver of transgenic mice containing −6A haplotype as compared with −6G haplotype.
FIGURE 7.
ChIP assay shows that GR, HNF-1α, and RNA polymerase bind strongly to the human AGT promoter from liver chromatin of transgenic mice containing −6A haplotype as compared with −6G haplotype. A ChIP assay was performed to examine the binding of HNF-1α, GR, and RNA polymerase to the liver chromatin in double transgenic mice containing −6A and −6G haplotypes of the hAGT gene. Immunoprecipitated DNA from the two haplotypes in the presence of antibodies against HNF-1α (a), GR (b and c), and RNA polymerase (d) was used to amplify nucleotide sequence containing their respective binding sites as described under “Materials and Methods.” In all panels (a–d), lanes 1 and 2, PCR-amplified product obtained from the ChIP of −6A and −6G haplotypes, respectively; lanes 3 and 4, PCR-amplified product using input genomic DNA from −6A and −6G haplotypes; lanes 5 and 6, PCR-amplified product in the presence of rabbit IgG for −6A and −6G haplotype, respectively. Chip assays were performed in triplicate.
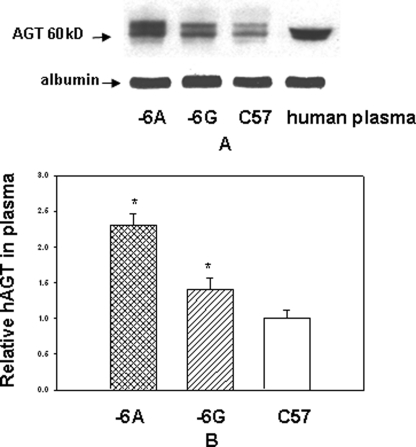
Plasma Angiotensinogen Level Is Increased in Transgenic Mice Containing −6A Haplotype of the hAGT Gene as Compared with the −6G Haplotype
We next determined if (a) hAGT protein in transgenic mice is correctly processed in the liver and secreted into plasma and (b) plasma AGT level is increased in transgenic mice containing −6A haplotype of the hAGT gene as compared with the −6G haplotype. Equal volumes of plasma from control C57 mice and male transgenic animals containing either −6A or −6G haplotype of the hAGT gene were analyzed by a Western blot assay along with human plasma as described (19) (Fig. 8A). Quantitation of AGT level was performed after normalization with mouse albumin (Fig. 8B). Results of this experiment showed that AGT level is increased by about 40% in the plasma of transgenic mice containing −6G haplotype as compared with the non-transgenic control animals. The plasma AGT level was increased by about 70% in transgenic mice containing −6A haplotype as compared with −6G haplotype (p = < 0.05). Electrophoretic mobility of hAGT protein in transgenic mice was similar to that of AGT in human plasma.
FIGURE 8.
Human AGT level is increased in plasma of double transgenic mice containing −6A haplotype of the hAGT gene as compared with −6G haplotype. A, equal amount of plasma (1 μg) from double transgenic mice containing either −6A or −6G haplotype of hAGT gene and hRen gene and from control C57 animals was analyzed by Western blot along with human plasma. The positions of molecular weight markers are shown on the left. B, histogram for the quantitation of protein level. Protein levels were normalized with albumin. AGT protein level in transgenic mice was calculated by assuming protein level in C57 mice as 1. Error bars, S.E.
Plasma Angiotensin-II Level Is Increased in Male Transgenic Mice Containing −6A Haplotype of the hAGT Gene as Compared with the −6G Haplotype
We next examined the effect of −6A and −6G haplotypes of the hAGT gene on plasma angiotensin-II level in double transgenic mice. Plasma was collected from control (non-transgenic), and male double transgenic mice containing either −6A or −6G haplotype of the hAGT gene, and plasma angiotensin-II levels were analyzed by ELISA. Results of this experiment (supplemental Fig. 4) showed that the plasma angiotensin-II level in transgenic mice containing −6G haplotype was increased by about 50% as compared with the non-transgenic littermate. On the other hand, plasma angiotensin-II level in transgenic mice containing −6A haplotype was increased by 100% as compared with the non-transgenic littermate (p = <0.05). Thus, plasma angiotensin-II level was increased by about 50% in double transgenic mice containing −6A haplotype of the hAGT gene as compared with −6G haplotype. Plasma angiotensin-II levels in single transgenic mice containing either the hAGT or hRen gene were the same as in control C57 mice (data not shown).
Mouse and Human Renin Level Is Decreased in Double Transgenic Mice Containing hRen and either −6A or −6G Haplotype of the hAGT Gene
Because it is known that an increased amount of AGT down-regulates renin expression, we were interested in examining the effect of −6A and −6G haplotypes of the hAGT gene on mouse and human renin gene expression in double transgenic mice. For this purpose, we analyzed the mRNA level of endogenous mouse renin and human renin gene in the kidney of non-transgenic and transgenic mice by quantitative RT-PCR. Results of this experiment showed that mouse renin mRNA level in transgenic mice containing −6G haplotype of the hAGT gene was about 40% less as compared with control C57 animals. On the other hand, renin mRNA level in transgenic mice containing −6A haplotype was 60% less as compared with control C57 animals (supplemental Fig. 5A). Human renin mRNA level in transgenic mice containing −6G haplotype of the hAGT gene was about 20% less and the mRNA level in −6A haplotype was 40% less as compared with control PAC-hRen animals (supplemental Fig. 5B). This suggested that although mouse as well as human renin gene expression is down-regulated by overexpression of the hAGT gene in transgenic mice, this effect is more pronounced in transgenic mice containing −6A haplotype as compared with the −6G haplotype.
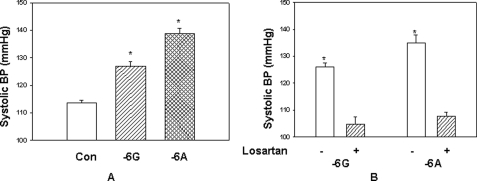
Blood Pressure Is Increased in Transgenic Mice Containing −6A Haplotype of the hAGT Gene as Compared with −6G Haplotype
Finally, we measured the effect of −6A and −6G haplotypes of the hAGT gene on blood pressure in double transgenic conscious male mice for 24 h using telemetry over a period of 2 weeks. Control C57 (non-transgenic) and double transgenic mice containing either −6A or −6G haplotype of the hAGT gene and hRen gene (3-month-old male mice) were surgically instrumented with telemetric probes. After 1 week of recovery from the surgical procedure, BP readings were recorded every 10 min using a Data-Science International instrument. Systolic blood pressure was measured over a period of 4 days, and mean values for daytime (6 a.m. to 6 p.m.) and nighttime (6 p.m. to 6 a.m.) as well as the average for the whole day (24 h) were calculated. A graph showing the mean BP values over a 24-h period of the −6A and −6G haplotypes of transgenic mice (number of animals = 6, days of recording = 4) is shown in Fig. 9A. Mean BP of control non-transgenic mice was 114 mm Hg (Con), mean BP of transgenic mice containing −6G haplotype was 127 mm Hg, and mean BP of transgenic mice containing −6A haplotype was 138 mm Hg (n = 6) (p < 0.05). Taken together, the results of this experiment showed that blood pressure of transgenic mice containing −6A haplotype was increased by 11 mm Hg as compared with transgenic mice containing −6G haplotype of the hAGT gene. Blood pressure of single transgenic mice containing either the hAGT or hRen gene was the same as that of control C57 mice. Because our experiments so far suggested that hAGT gene expression is increased in transgenic mice containing −6A haplotype that leads to increased plasma angiotensin-II level and increased blood pressure as compared with transgenic animals containing −6G haplotype, we next examined whether angiotensin receptor blockers reduce blood pressure in these animals. We used losartan as an AT1 receptor blocker and determined the blood pressure by the tail cuff method. Results from this experiment show that losartan reduces the systolic blood pressure in double transgenic mice containing both −6A and −6G haplotypes of the hAGT gene to or below the non-transgenic base-line level (Fig. 9B).
FIGURE 9.
A, blood pressure is increased in double transgenic mice containing −6A haplotype of the hAGT gene as compared with −6G haplotype. The mean of 24 h blood pressure of control (CON) and double transgenic mice containing −6G or −6A haplotype of the hAGT gene was measured by telemetry for 4 days. Each bar represents mean from six male animals taken over 4 days. B, blood pressure in transgenic mice is reduced by losartan treatment. Shown is the mean of 24 h blood pressure of double transgenic mice containing −6G or −6A haplotypes in the presence (+) and absence (−) of losartan. Error bars, S.E.
DISCUSSION
Although −6A allele of the hAGT gene is associated with increased plasma AGT level and increased blood pressure in humans, the molecular mechanism involved in this process is not clear. Previous in vitro and in vivo studies have shown that glucocorticoids play an important role in the expression of the AGT gene in the liver (33) and are the greatest inducers of its expression in adipocytes (34). Glucocorticoid treatment increases blood pressure in humans (35), and increased cortisol level is a marker for human hypertension (36). We have found that a reporter construct containing 1.8 kb of the hAGT gene promoter has about 10-fold increased glucocorticoid-induced promoter activity on transient transfection in human liver and kidney cells as compared with the reporter construct containing 1.2 kb of the promoter. Surprisingly, the hAGT gene has three new SNPs in this region of the promoter (A/G at −1670, C/G at −1562, and T/G at −1561). Variants −1670A, −1562C, and −1561T almost always occur with −6A; and variants −1670G, −1562G, and −1561G almost always occur with −6G. Therefore, the hAGT gene may be subdivided into −6A and −6G haplotypes. We have shown that a reporter construct containing 1.8 kb of the hAGT gene promoter with −6A haplotype has 50–60% increased basal promoter activity and 4–6-fold increased glucocorticoid-induced promoter activity upon transient transfection in human liver and kidney cells as compared with a reporter construct with −6G haplotype.
We have also shown that an oligonucleotide containing −1670A has stronger affinity for HNF-1α as compared with the same oligonucleotide containing −1670G, and deletion of the HNF-1α binding site reduces basal and glucocorticoid induced promoter activity upon transient transfection in liver cells. Although HNF-1 was originally discovered as a liver-specific transcription factor, recent studies have suggested that it is critical for diverse metabolic functions in pancreatic islets, liver, intestine, and kidneys (37). An intact and occupied HNF-1 site plays a crucial role in basal and IL-6-induced expression of the phosphoenolpyruvate carboxykinase gene in the kidney (38, 39). Therefore, variant −1670A (which always occurs with −6A) may increase the expression of the hAGT gene in the liver as well as the kidney of human subjects containing −6A haplotype by increased binding of HNF-1α to nucleotide sequence containing −1670A.
The upstream region of the hAGT gene promoter has two polymorphic sites, T/G at −1561 and C/G at −1562, and nucleotide sequence containing −1562C and −1561T has stronger homology with consensus GRE as compared with the nucleotide sequence containing −1562G and −1561G. Our transient transfections show that reporter construct containing −1562C and −1561T has increased glucocorticoid-induced promoter activity as compared with the reporter construct containing −1562G and −1561G. We have previously shown that the hAGT gene has an A/G polymorphic site at −217, and reporter construct containing 1.2 kb of the promoter with −217A has increased glucocorticoid induced promoter activity as compared with −217G (29). Because transient transfection of reporter construct containing 1.8 kb of the promoter has 10-fold increased GR-induced promoter activity as compared with the reporter construct containing the 1.2-kb promoter, it seems that upstream GRE (located around −1560) is required for full GR-induced promoter activity. Previous studies have suggested that GREs may exist in the upstream region of the promoter and act as an enhancer. For example, a GRE is located (a) 6 kb upstream from the transcriptional initiation site in mouse HNF-4α1 promoter (40), (b) 11 kb upstream from the rat tyrosine aminotransferase promoter (41), and (c) 1.4 kb from the rat phosphoenolpyruvate carboxykinase gene promoter (30, 38). Our observation that deletion of the HNF-1 binding site around −1670 reduces GR-induced promoter activity suggests that an intact HNF-1 site is also required for glucocorticoid-induced expression of the hAGT gene. This observation is in accordance with previous studies suggesting the role of HNF-1 in GR-induced promoter activity of the HNF-4α1 gene (42). In addition, the HNF-1 binding site is required for glucocorticoid-induced transcription of the insulin-like growth factor binding protein-1 gene (43) and the phosphoenolpyruvate carboxykinase gene in the kidney (39).
Another finding of this study is that deletion of a 20-bp region of the promoter located between −1600 and −1620 drastically reduces glucocorticoid-induced promoter activity. This region of the promoter binds to the C/EBP family of transcription factors. Site-specific mutagenesis of this C/EBP binding site reduces GR-induced promoter activity and shows that this C/EBP site is important for glucocorticoid-induced expression of the hAGT gene. C/EBP family of transcription factors play an important role in liver- and adipose-specific gene expression, two tissues in which the AGT gene is abundantly expressed (44, 45). Glucocorticoids also induce the expression of C/EBPβ and C/EBPδ in hepatoma cells (46), primary culture of hepatocytes (47), and adipocytes (48). In addition, GR physically interacts with C/EBPβ and increases expression of a gene that contains the C/EBP binding site (32). C/EBP binding sites have been shown to be essential for GR-induced promoter activity of mouse HNF-4α1 (40) and arginase (32) genes.
In order to unambiguously prove the role of −6A allele in the expression of the hAGT gene in a tissue-specific manner and in the regulation of blood pressure in an in vivo situation, we have made transgenic mice containing either −6A or −6G haplotype of the hAGT gene present in 180-kb-long BAC DNA (containing 116 kb of the 5′-flanking region, all five exons and four introns, and 54 kb of the 3′-flanking region of the hAGT gene). The reason behind using BAC DNA was that because it contains 116 kb of the 5′-flanking region, it will have all of the cis-acting DNA elements required for tissue- and hormone-specific expression of the hAGT gene. Because there is a strict species specificity in the renin cleavage site and mouse renin is not able to cleave hAGT, we also made double transgenic mice containing the human renin gene and either the −6A or −6G haplotype of the hAGT gene. We show here that hAGT mRNA level is increased about 2.5-fold in the liver of male double transgenic mice containing −6A haplotype as compared with −6G haplotype. In order to confirm that an increase in mRNA level correspondingly increases its protein level, we also analyzed AGT protein level by Western analysis. Our results indicate that AGT protein level is also increased in the liver of transgenic mice containing −6A haplotype by about 1.6-fold as compared with the liver of transgenic mice containing −6G haplotype.
Because our EMSA experiment has shown that an oligonucleotide containing −1670A has stronger affinity with HNF-1α as compared with the same oligonucleotide containing −1670G, we performed a ChIP assay to confirm that HNF-1α binds strongly to the chromatin obtained from the liver of transgenic mice containing −6A haplotype as compared with −6G haplotype. Results of our in vivo ChIP assay have shown that indeed HNF-1α binds more strongly to the hAGT gene promoter in the chromatin obtained from the liver of transgenic mice containing −6A haplotype of the hAGT gene as compared with −6G haplotype. Our experiments suggest that transcription factors present in mouse cells correctly recognize the changes in cis-acting DNA elements present in the chromatin of two different haplotypes of a human gene and differentially regulate their transcription.
Because nucleotide sequence of the hAGT gene promoter containing −1562C and −1561T has stronger homology with consensus GRE as compared with −1562G and −1561G, we performed a ChIP assay to analyze the binding of GR in liver chromatin of transgenic mice containing either −6A or −6G haplotype of the hAGT gene. Our in vivo ChIP assay has shown that GR has stronger affinity for the chromatin obtained from the liver of transgenic animals containing −6A haplotype as compared with −6G haplotype. Glucocorticoid treatment increases blood pressure in humans (35), and increased cortisol level is a marker for human hypertension (36). Previous in vitro and in vivo studies have shown that glucocorticoids play an important role in the expression of the AGT gene in the liver (49) and are the greatest inducer of its expression in adipocytes (34). Overexpression of the AGT gene in adipocytes increases plasma AGT level and blood pressure in transgenic mice (50). AGT levels are increased in visceral adipose tissue as compared with the subcutaneous adipose tissue of human subjects and may be responsible for increased blood pressure in obese human subjects (51). Our experiments suggest that an increase in cortisol level (such as in stress and obesity) may increase the expression of the AGT gene in subjects containing −6A haplotype as compared with −6G haplotype. This may lead to increased tissue/plasma AGT level and increased blood pressure in subjects containing −6A haplotype of the AGT gene.
Because plasma AGT level is increased in hypertensive subjects containing the −6A allele of the hAGT gene as compared with the −6G allele (7), it was important to examine the plasma hAGT level in transgenic mice containing −6A and −6G haplotypes of the hAGT gene. We show that plasma AGT level is increased about 1.7-fold in transgenic mice containing the −6A haplotype of the hAGT gene as compared with the −6G haplotype. This increase in plasma AGT level may be responsible for increased blood pressure in transgenic mice containing −6A haplotype because previous studies have shown that plasma AGT level is directly associated with blood pressure. Kim et al. have shown that plasma AGT concentration is increased 1.45-fold between two-copy and four-copy AGT-containing transgenic mice, which results in an increase in blood pressure of about 8 mm Hg per copy number (6). Klett et al. (3) have also shown that administration of 1.2 mg/kg recombinant AGT to rats increases plasma AGT level by 1.4-fold within 30 min and increases blood pressure by 19 mm Hg. An increase in the plasma AGT level concurrently increased the plasma angiotensin-II level. Thus, plasma angiotensin-II level was increased by about 50% in transgenic mice containing −6A haplotype as compared with −6G haplotype. We have also shown that, as expected, mouse and human renin gene expression is down-regulated in the kidney of double transgenic mice as compared with the control animals. Even with this down-regulation of the renin gene expression, transgenic animals with −6A haplotype had increased blood pressure as compared with −6G haplotype.
We show here that systolic blood pressure is increased by 11 mm Hg in male double transgenic mice containing hRen gene and −6A haplotype of the hAGT gene as compared with −6G haplotype. Our results for blood pressure of transgenic mice containing −6A and −6G haplotypes of the hAGT gene differ from those of Cvetkovic et al. (19), who developed transgenic mice by inserting the hAGT gene containing either −6A or −6G in the context of 1.2 kb of the promoter at the hypoxanthine-guanine phosphoribosyltransferase locus. Probably three new polymorphisms identified in the upstream region of the promoter of hAGT gene that are present in our transgenic mice are responsible for this difference. The fact that this increase is due to an increased level of angiotensin-II is shown by our finding that angiotensin receptor blocker (losartan) reduced the blood pressure in transgenic mice.
In conclusion, we have shown that the hAGT gene promoter contains three new polymorphic sites at −1670, −1562, and −1561. Moreover, variants −1670A, −1562C, and −1561T almost always occur with −6A, and variants −1670G, −1562G, and −1561G almost always occur with −6G. Therefore, the hAGT gene may be subdivided into −6A and −6G haplotypes. We have shown that (a) HNF-1α and GR bind strongly to the promoter containing −6A haplotype of the hAGT gene, and (b) reporter construct containing 1.8 kb of the hAGT gene promoter with −6A haplotype has 50–60% increased basal and 4–6-fold increased glucocorticoid-induced promoter activity upon transient transfection in human liver and kidney cells as compared with −6G haplotype. We have generated double transgenic mice containing human renin and either −6A or −6G haplotype of the hAGT gene. Transgenic mice containing −6A haplotype have increased plasma AGT and angiotensin-II level and increased hAGT mRNA level in the liver as compared with transgenic mice containing −6G haplotype. Our in vivo ChIP assay shows that HNF-1α, GR, and RNA polymerase bind strongly to the chromatin obtained from the liver of transgenic mice containing −6A haplotype of the hAGT gene as compared with −6G haplotype. We also show that (a) transgenic mice containing −6A haplotype have 11 mm Hg increased systolic blood pressure as compared with the transgenic mice containing −6G haplotype, and (b) angiotensin receptor blocker reduces blood pressure in these transgenic mice. Our experiments for the first time explain the molecular mechanism involved in association of −6A allele of the hAGT gene with increased blood pressure in human subjects and provide a new experimental animal model that is very similar to human hypertension.
Supplementary Material
Acknowledgment
We are thankful to Dr. Curt Sigmund for providing hREN transgenic mice.
This work was supported, in whole or in part, by National Institutes of Health Grants HL081752 and HL66296 (to A. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- AGT
- angiotensinogen
- hAGT
- human AGT
- hRen
- human renin
- GRE
- glucocorticoid-responsive element
- GR
- glucocorticoid receptor
- C/EBP
- CCAAT/enhancer-binding protein
- BP
- blood pressure
- Dex
- dexamethasone
- BAC
- bacterial artificial chromosome.
REFERENCES
- 1.Corvol P., Soubrier F., Jeunemaitre X. (1997) Pathol. Biol. 45, 229–239 [PubMed] [Google Scholar]
- 2.Gould A. B., Green D. (1971) Cardiovasc. Res. 5, 86–89 [DOI] [PubMed] [Google Scholar]
- 3.Klett C. P., Granger J. P. (2001) Am. J. Physiol. Regul. Integr. Comp Physiol 281, R1437–R1441 [DOI] [PubMed] [Google Scholar]
- 4.Kimura S., Mullins J. J., Bunnemann B., Metzger R., Hilgenfeldt U., Zimmermann F., Jacob H., Fuxe K., Ganten D., Kaling M. (1992) EMBO J. 11, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanimoto K., Sugiyama F., Goto Y., Ishida J., Takimoto E., Yagami K., Fukamizu A., Murakami K. (1994) J. Biol. Chem. 269, 31334–31337 [PubMed] [Google Scholar]
- 6.Kim H. S., Krege J. H., Kluckman K. D., Hagaman J. R., Hodgin J. B., Best C. F., Jennette J. C., Coffman T. M., Maeda N., Smithies O. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2735–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeunemaitre X., Soubrier F., Kotelevtsev Y. V., Lifton R. P., Williams C. S., Charru A., Hunt S. C., Hopkins P. N., Williams R. R., Lalouel J. M. (1992) Cell 71, 169–180 [DOI] [PubMed] [Google Scholar]
- 8.Iwai N., Shimoike H., Ohmichi N., Kinoshita M. (1995) Hypertension 25, 688–693 [DOI] [PubMed] [Google Scholar]
- 9.Nejatizadeh A., Kumar R., Stobdan T., Goyal A. K., Gupta M., Javed S., Pasha M. Q. (2008) J. Hypertens. 26, 1094–1101 [DOI] [PubMed] [Google Scholar]
- 10.Dickson M. E., Sigmund C. D. (2006) Hypertension 48, 14–20 [DOI] [PubMed] [Google Scholar]
- 11.Castellano M., Glorioso N., Cusi D., Sarzani R., Fabris B., Opocher G., Zoccali C., Golin R., Veglio F., Volpe M., Mantero F., Fallo F., Rossi G. P., Barlassina C., Tizzoni L., Filigheddu F., Giacchè M., Rossi F. (2003) J. Hypertens. 21, 1853–1860 [DOI] [PubMed] [Google Scholar]
- 12.Sethi A. A., Nordestgaard B. G., Tybjaerg-Hansen A. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 1269–1275 [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen-Torvik L. J., North K. E., Gu C. C., Lewis C. E., Wilk J. B., Chakravarti A., Chang Y. P., Miller M. B., Li N., Devereux R. B., Arnett D. K. (2005) Hypertension 46, 1294–1299 [DOI] [PubMed] [Google Scholar]
- 14.Kretowski A., McFann K., Hokanson J. E., Maahs D., Kinney G., Snell-Bergeon J. K., Wadwa R. P., Eckel R. H., Ogden L., Garg S., Li J., Cheng S., Erlich H. A., Rewers M. (2007) Diabetes 56, 863–871 [DOI] [PubMed] [Google Scholar]
- 15.Schmidt H., Fazekas F., Kostner G. M., van Duijn C. M., Schmidt R. (2001) Stroke 32, 405–412 [DOI] [PubMed] [Google Scholar]
- 16.Schmidt H., Aulchenko Y. S., Schweighofer N., Schmidt R., Frank S., Kostner G. M., Ott E., van Duijn C. (2004) Stroke 35, 2592–2597 [DOI] [PubMed] [Google Scholar]
- 17.Inoue I., Nakajima T., Williams C. S., Quackenbush J., Puryear R., Powers M., Cheng T., Ludwig E. H., Sharma A. M., Hata A., Jeunemaitre X., Lalouel J. M. (1997) J. Clin. Invest. 99, 1786–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy D., Ehret G. B., Rice K., Verwoert G. C., Launer L. J., Dehghan A., Glazer N. L., Morrison A. C., Johnson A. D., Aspelund T., Aulchenko Y., Lumley T., Köttgen A., Vasan R. S., Rivadeneira F., Eiriksdottir G., Guo X., Arking D. E., Mitchell G. F., Mattace-Raso F. U., Smith A. V., Taylor K., Scharpf R. B., Hwang S. J., Sijbrands E. J., Bis J., Harris T. B., Ganesh S. K., O'Donnell C. J., Hofman A., Rotter J. I., Coresh J., Benjamin E. J., Uitterlinden A. G., Heiss G., Fox C. S., Witteman J. C., Boerwinkle E., Wang T. J., Gudnason V., Larson M. G., Chakravarti A., Psaty B. M., van Duijn C. M. (2009) Nat. Genet. 41, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cvetkovic B., Keen H. L., Zhang X., Davis D., Yang B., Sigmund C. D. (2002) Physiol. Genomics 11, 253–262 [DOI] [PubMed] [Google Scholar]
- 20.Brasier A. R., Ron D., Tate J. E., Habener J. F. (1990) Mol. Endocrinol. 4, 1921–1933 [DOI] [PubMed] [Google Scholar]
- 21.Narayanan C. S., Cui Y., Kumar A. (1998) Biochem. Biophys. Res. Commun. 251, 388–393 [DOI] [PubMed] [Google Scholar]
- 22.Jain S., Li Y., Patil S., Kumar A. (2007) Am. J. Physiol. Cell Physiol. 293, C401–C410 [DOI] [PubMed] [Google Scholar]
- 23.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warming S., Costantino N., Court DL, Jenkins N. A., Copeland N. G. (2005) Nucleic Acids Res. 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinn P. L., Davis D. R., Sigmund C. D. (1999) J. Biol. Chem. 274, 35785–35793 [DOI] [PubMed] [Google Scholar]
- 26.Butz G. M., Davisson R. L. (2001) Physiol. Genomics 5, 89–97 [DOI] [PubMed] [Google Scholar]
- 27.Campbell D. J., Habener J. F. (1986) J. Clin. Invest. 78, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brasier A. R., Tate J. E., Ron D., Habener J. F. (1989) Mol. Endocrinol. 3, 1022–1034 [DOI] [PubMed] [Google Scholar]
- 29.Jain S., Li Y., Patil S., Kumar A. (2005) J. Mol. Med. 83, 121–131 [DOI] [PubMed] [Google Scholar]
- 30.Cassuto H., Kochan K., Chakravarty K., Cohen H., Blum B., Olswang Y., Hakimi P., Xu C., Massillon D., Hanson R. W., Reshef L. (2005) J. Biol. Chem. 280, 33873–33884 [DOI] [PubMed] [Google Scholar]
- 31.Chakravarty K., Cassuto H., Reshef L., Hanson R. W. (2005) Crit. Rev. Biochem. Mol. Biol. 40, 129–154 [DOI] [PubMed] [Google Scholar]
- 32.Gotoh T., Chowdhury S., Takiguchi M., Mori M. (1997) J. Biol. Chem. 272, 3694–3698 [DOI] [PubMed] [Google Scholar]
- 33.Klett C., Ganten D., Hellmann W., Kaling M., Ryffel G. U., Weimar-Ehl T., Hackenthal E. (1992) Endocrinology 130, 3660–3668 [DOI] [PubMed] [Google Scholar]
- 34.Aubert J., Darimont C., Safonova I., Ailhaud G., Negrel R. (1997) Biochem. J. 328, 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker B. R., Phillips D. I., Noon J. P., Panarelli M., Andrew R., Edwards H. V., Holton D. W., Seckl J. R., Webb D. J., Watt G. C. (1998) Hypertension 31, 891–895 [DOI] [PubMed] [Google Scholar]
- 36.Litchfield W. R., Hunt S. C., Jeunemaitre X., Fisher N. D., Hopkins P. N., Williams R. R., Corvol P., Williams G. H. (1998) Hypertension 31, 569–574 [DOI] [PubMed] [Google Scholar]
- 37.Shih D. Q., Bussen M., Sehayek E., Ananthanarayanan M., Shneider B. L., Suchy F. J., Shefer S., Bollileni J. S., Gonzalez F. J., Breslow J. L., Stoffel M. (2001) Nat. Genet. 27, 375–382 [DOI] [PubMed] [Google Scholar]
- 38.Beale E. G., Clouthier D. E., Hammer R. E. (1992) FASEB J. 6, 3330–3337 [DOI] [PubMed] [Google Scholar]
- 39.Cassuto H., Olswang Y., Livoff A. F., Nechushtan H., Hanson R. W., Reshef L. (1997) FEBS Lett. 412, 597–602 [DOI] [PubMed] [Google Scholar]
- 40.Bailly A., Torres-Padilla M. E., Tinel A. P., Weiss M. C. (2001) Nucleic Acids Res. 29, 3495–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Espinás M. L., Roux J., Pictet R., Grange T. (1995) Mol. Cell. Biol. 15, 5346–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh D. S., Rechler M. M. (1997) Mol. Endocrinol. 11, 1822–1831 [DOI] [PubMed] [Google Scholar]
- 43.Suh D. S., Zhou Y., Ooi G. T., Rechler M. M. (1996) Mol. Endocrinol. 10, 1227–1237 [DOI] [PubMed] [Google Scholar]
- 44.Ramji D. P., Foka P. (2002) Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darlington G. J., Ross S. E., MacDougald O. A. (1998) J. Biol. Chem. 273, 30057–30060 [DOI] [PubMed] [Google Scholar]
- 46.Baumann H., Morella K. K., Campos S. P., Cao Z., Jahreis G. P. (1992) J. Biol. Chem. 267, 19744–19751 [PubMed] [Google Scholar]
- 47.Matsuno F., Chowdhury S., Gotoh T., Iwase K., Matsuzaki H., Takatsuki K., Mori M., Takiguchi M. (1996) J. Biochem. 119, 524–532 [DOI] [PubMed] [Google Scholar]
- 48.MacDougald O. A., Cornelius P., Lin F. T., Chen S. S., Lane M. D. (1994) J. Biol. Chem. 269, 19041–19047 [PubMed] [Google Scholar]
- 49.Corvol P., Jeunemaitre X. (1997) Endocr. Rev. 18, 662–677 [DOI] [PubMed] [Google Scholar]
- 50.Masuzaki H., Paterson J., Shinyama H., Morton N. M., Mullins J. J., Seckl J. R., Flier J. S. (2001) Science 294, 2166–2170 [DOI] [PubMed] [Google Scholar]
- 51.Atzmon G., Yang X. M., Muzumdar R., Ma X. H., Gabriely I., Barzilai N. (2002) Horm. Metab. Res. 34, 622–628 [DOI] [PubMed] [Google Scholar]
- 52.The Wellcome Trust Case Control Consortium (2007) Nature 447, 661–67817554300 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.