Abstract
BAK is a key protein mediating mitochondrial outer membrane permeabilization; however, its behavior in the membrane is poorly understood. Here, we characterize the conformational changes in BAK and MCL-1 using detergents to mimic the membrane environment and study their interaction by in vitro pulldown experiments, size exclusion chromatography, titration calorimetry, and NMR spectroscopy. The nonionic detergent IGEPAL has little impact on the structure of MCL-1 but induces a conformational change in BAK, whereby its BH3 region is able to engage the hydrophobic groove of MCL-1. Although the zwitterionic detergent CHAPS induces only minor conformational changes in both proteins, it is still able to initiate heterodimerization. The complex of MCL-1 and BAK can be disrupted by a BID-BH3 peptide, which acts through binding to MCL-1, but a mutant peptide, BAK-BH3-L78A, with low affinity for MCL-1 failed to dissociate the complex. The mutation L78A in BAK prevented binding to MCL-1, thus demonstrating the essential role of the BH3 region of BAK in its regulation by MCL-1. Our results validate the current models for the activation of BAK and highlight the potential value of small molecule inhibitors that target MCL-1 directly.
Keywords: Membrane, Mitochondrial Apoptosis, NMR, Protein Conformation, Protein-Protein Interactions, BAK, MCL-1, Detergent
Introduction
The BCL-2 family of proteins plays a major role in regulating apoptosis by regulating the integrity of the mitochondrial outer membrane (1). Its deregulation can result in cancer, neurodegeneration, heart disease, and diabetes (2). This family is divided into three subgroups according to their sequence similarities and functions. The antiapoptotic proteins (BCL-2, BCL-w, BCL-XL, MCL-1, and A1) share up to four BCL-2 homology (BH)2 regions (BH1–4) and prevent cell death. The proapoptotic effectors (BAX and BAK) contain three BH regions (BH1–3) and directly mediate mitochondrial outer membrane permeabilization, which leads to apoptosis. The proapoptotic BH3-only proteins (BID, BIM, BAD, BMF, BIK, PUMA, NOXA, HRK/DPS, NIP3, bNIP3, and MULE) share only a common BH3 region and initiate apoptosis indirectly. In this study, we selected one protein from each group (MCL-1, BAK, and BID) and examined their interactions as part of the BCL regulatory network.
BAK targets the outer membrane of mitochondria and endoplasmic reticulum through its C-terminal transmembrane (TM) helix (3–5). Its soluble N-terminal domain, cBAK (6), adopts the α-helical bundle conformation that is conserved across all multiple BH BCL-2 family members. When apoptosis is induced by factors such as staurosporine, etoposide, cisplatin, anti-Fas antibody, or detergents (3, 7–9), BAK undergoes a conformational change, termed BAK activation, that is characterized by the insertion of additional α-helical elements into the membrane and the assembly of BAK into higher order oligomers (10, 11). The BH1 and BH3 regions of BAK have been reported to be important for its role in apoptosis (12, 13), but their specific functions are not fully understood. Further structural studies are needed to elucidate the conformational changes involved in BAK activation.
MCL-1 inhibits the proapoptotic activity of BH3-only proteins by accommodating their BH3 regions in its hydrophobic groove (14–17). It has also been reported as an endogenous BAK inhibitor, whose inhibition can be disrupted by the tumor suppressor p53 and NOXA (18–20). The regulatory mechanism remains unclear, whereas the deregulation is implicated in various hematopoietic and lymphoid cancers (21–23), which makes it an ideal therapeutic target for cancer treatment. The elucidation of the interaction between MCL-1 and BAK will provide more information for MCL-1-targeted drug design and screening.
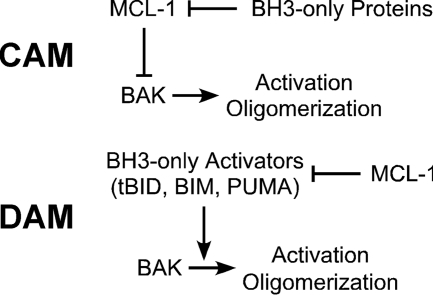
BID protein is the convergent point for intrinsic and extrinsic apoptosis. It responds to cellular stress and initiates BAX/BAK-mediated apoptosis (24–27). As for its activating mechanisms, there are two popular models in literature (Fig. 1). The competitively activating model (CAM) (15) proposes that BH3-only proteins release BAK from complexes with MCL-1 or BCL-XL by competitively binding to the pocket of the antiapoptotic proteins (19, 28, 29). The directly activating model (DAM) posits that BH3-only activators including tBID, BIM, and PUMA directly bind and activate BAK, and the role of the antiapoptotic proteins is to sequester the BH3-only activators to prevent apoptosis (4, 25–27, 30). The detection of direct protein-protein interactions may help to resolve the longstanding debate over CAM and DAM (29, 31–33).
FIGURE 1.
Models of BAK activation. In CAM, BH3-only proteins release BAK from its complexes with MCL-1 or BCL-XL by binding competitively to the pocket of the antiapoptotic proteins (19, 28, 29). In the DAM, BH3-only activators including tBID, BIM, and PUMA directly bind and activate BAK, and the role of the antiapoptotic proteins is to sequester the BH3-only activators to prevent apoptosis (4, 25–27, 30).
To gain a deeper understanding of the BAK regulation, we developed a mini-system consisting of BAK, MCL-1, and BID-BH3 peptides with detergents to mimic the membrane environment of the cell. We used NMR spectroscopy, pulldown assays, and size exclusion chromatography to assess protein-protein binding and structural transitions. We find that detergent is required for the direct interaction between BAK and MCL-1 in vitro and that heterodimerization occurs through binding of the BH3 region of BAK to the BH3-binding site of MCL-1.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
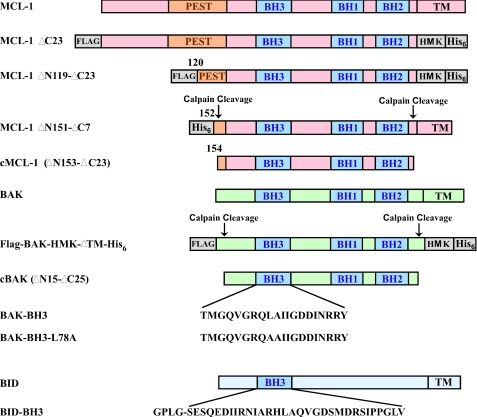
The protein fragments used are summarized in Fig. 2. FLAG-BAK-HMK-ΔTM-His6 (residues 2–186 from NCBI: AAA74466), calpain-proteolysed BAK (cBAK, residues 16–186 from NCBI: AAA74466), cBAK-L78A, core domain of MCL-1 (also calpain-proteolysed MCL-1, cMCL-1, residues 163–326 from NCBI: AAF64255) and BID-BH3 peptide (residues 76–106 from NCBI: NP_001187 with an N-terminal extension of GPLG) were purified as described previously (6, 15, 34). Escherichia coli strain BL21 (DE3) containing the corresponding plasmid was grown in LB media at 37 °C to an optical density of 0.8 at 600 nm and then induced by 1 mm isopropyl-β-d-thiogalactoside at 30 °C for 4 h. For NMR samples, cells were grown in M9 media supplemented with 15N ammonium chloride. The soluble proteins were purified by batch Ni2+-NTA or glutathione S-transferase affinity chromatography (Qiagen), followed by Sephadex G-75 size exclusion chromatography (GE Healthcare) and Q-Sepharose anion-exchange chromatography (GE Healthcare). A calpain digestion step was further applied to produce cBAK, cBAK-L78A, and cMCL-1, and calpain was removed by Q-Sepharose anion-exchange chromatography. BAK-BH3 (TMGQVGRQLAIIGDDINRRY) and BAK-BH3-L78A (TMGQVGRQAAIIGDDINRRY) peptides were synthesized chemically (Sheldon Biotechnology Centre, McGill University) and purified on a C18 reverse-phase column. All proteins and peptides were stored in 20 mm HEPES (pH 6.5) and 1 mm DTT buffer by flash freezing in liquid nitrogen and stored at −80 °C.
FIGURE 2.
Summary of MCL-1, BAK, and BID constructs used in this study. The domain organization, BCL homology regions (BH1–3), calpain protease cleavage sites, TM helices, and BH3 sequences of the different protein and peptide constructs are shown. MCL-1 has an N-terminal PEST domain that has been suggested to regulate its turnover. Exogeneously added His6 tags, FLAG epitope, and heart muscle kinase (HMK) phosphorylation sites are shown in gray. The BID-BH3 peptide was produced biosynthetically and includes four extra amino acids.
Site-directed Mutagenesis
The mutation L78A in the BH3 region of BAK was generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The mutagenesis was carried out by using FLAG-BAK-HMK-ΔTM-His6 in pET29b(+) plasmid as a template and using oligonucleotides (CAGGTGGGACGGCAGGCCGCCATCATCGGGGAC) and (GTCCCCGATGATGGCGGCCTGCCGTCCCACCTG) as PCR primers. The construct was confirmed by DNA sequencing.
Analytical Size Exclusion Chromatography
MCL-1 ΔN151-ΔC7, cBAK, and their mixture were prepared at 2 mg/ml (for the mixture, the concentration for each protein is 2 mg/ml) in 20 mm HEPES (pH 6.5), 1 mm DTT and with or without 0.1% IGEPAL (tert-octylphenoxy poly(oxyethylene)ethanol, average molecular weight 603; Sigma-Aldrich). The size exclusion running buffer contained 25 mm HEPES (pH 6.5), 150 mm NaCl, 1 mm DTT, and with or without 0.1% IGEPAL. A 30-μl sample was injected for Superdex 200 PC (3.2/30) analytical runs. The chromatographs were recorded and analyzed on Waters Millenium HPLC software. The identities of the proteins in the peak fractions were confirmed by SDS-PAGE.
Ni2+-NTA Pulldown Assay
Three buffers: buffer N, buffer W and buffer E, each containing 50 mm Tris (pH 6.8), 500 mm NaCl, 1% glycerol, 1 mm NaN3, with the addition 10 mm or 30 mm or 250 mm imidazole, respectively, were used in pulldown assays. Detergents were added as indicated. The Ni2+-NTA resin was first equilibrated with buffer N and excess buffer removed to obtain a 50:50 (resin:buffer) slurry. To 50 μl of this slurry, 50 μl of 1 mg/ml FLAG-BAK-HMK-ΔTM-His6, cMCL-1, or a 1:1 mixture of FLAG-BAK-HMK-ΔTM-His6:cMCL-1 were applied in the presence of different concentrations of IGEPAL or CHAPS as indicated. After an incubation time of 30 min at room temperature, the slurry was spin down at 7,000 rpm for 30 s, and the supernatant was removed. The resin in the pellet was washed three times with 500 μl of buffer W and eluted with 45 μl of buffer E. The elution was then mixed with 15 μl of 4× SDS buffer for SDS-PAGE analysis. For BID-BH3 peptide competition assays, all the buffers contained 0.1% IGEPAL. The FLAG-BAK-HMK-ΔTM-His6·cMCL-1 complex was first immobilized on Ni2+-NTA resin, and then the resin slurry was prepared similarly. A range of BID-BH3 peptide concentrations were applied to the resin slurry followed by incubation for 30 min at room temperature, centrifugation at 7,000 rpm for 30 s, and removal of the supernatant. The resin was washed three times with buffer W and eluted with buffer E. The elution and supernatant were then mixed with 4× SDS buffer and analyzed by SDS-PAGE.
Isothermal Titration Calorimetry (ITC) Measurements
ITC was carried out on a MicroCal VP-ITC titration calorimeter controlled by the VPViewer software (MicroCal, Inc., Northampton, MA). Experiments were performed in 50 mm Na·HEPES (pH 7.0) at 23 °C and consisted of 37 injections of 8 μl of 0.1 mm BAK-BH3 or BAK-BH3-L78A peptide into 1.45 ml of 0.01 mm human MCL-1 (four different constructs) to obtain a final peptide/protein ratio of 2:1. The calorimetric data were analyzed by ORIGIN software to determine thermodynamic binding constants.
NMR Titrations
NMR titrations were carried on 0.1 mm 15N-labeled cMCL-1 or 15N-labeled cBAK. Aliquots of unlabeled binding partners such as cMCL-1, cBAK, cBAK-L78A, BID-BH3, BAK-BH3-L78A, IGEPAL, and CHAPS stock solutions were titrated in until the saturation was achieved. The 1H-15N HSQC spectra were recorded on a Bruker Avance DRX600 MHz spectrometer at 30 °C, processed by NMRPipe (35), and visualized by NmrViewJ (version 8.0) (36). The backbone assignments of cMCL-1 and BAK in free form were determined previously (6, 15). Assignments in detergent were made by following chemical shift changes during titrations. The changes were calculated as ((Δ1H ppm)2 + (0.2 × Δ15N ppm)2)0.5) as a function of the residue number, and mapped onto the three-dimensional structures. The model of unliganded human cMCL-1 was calculated by Modeler 9 (version 2) (37) based on the structure of the Mus musculus MCL-1 protein 1WSX (38). The structure of cBAK was from Protein Data Bank code 2IMS (6).
RESULTS
BAK in Aqueous Solution Is Not Ready for Heterodimerization
Regulation of BAK by MCL-1 and BCL-XL has been proposed to occur through the engagement of its BH3 region into the hydrophobic groove of antiapoptotic proteins (18, 19). However, in the crystal structure of BAK (6), the BH3 region is partially buried in the protein interior and unavailable for binding without a major conformational change. To investigate the interaction of BAK with the inhibitor MCL-1, purified components were mixed in aqueous solution for 1 h at room temperature or overnight at 4 °C. Neither condition resulted in the formation of a MCL-1·BAK complex, as indicated by the failure of cMCL-1 to be retained on Ni2+-NTA resin by FLAG-BAK-HMK-ΔTM-His6 in a pulldown experiment, the absence of elution shifts in analytical size exclusion chromatography, and the absence of chemical shift changes in the NMR spectra of 15N-labeled cMCL-1 or 15N-labeled cBAK upon the addition of unlabeled partner (data not shown). Therefore, under these conditions, we conclude that MCL-1 and BAK do not interact with each other.
Conformational Changes of BAK and MCL-1 in Presence of IGEPAL
Pioneering studies on BCL-2 proteins revealed that specific detergents can induce conformational changes and/or their oligomerization (24, 34, 39, 40). Detergents can also induce BAK-mediated apoptosis (9). These led us to test the behavior of BAK and MCL-1 in the presence of detergents. We used both IGEPAL (a nonionic detergent) and CHAPS (a zwitterionic detergent) in our tests. Nonionic detergents have been reported to artificially promote the interactions between BCL-2 family of proteins (41), whereas CHAPS has been reported to reduce the dimerization between BAX and tBID (42).
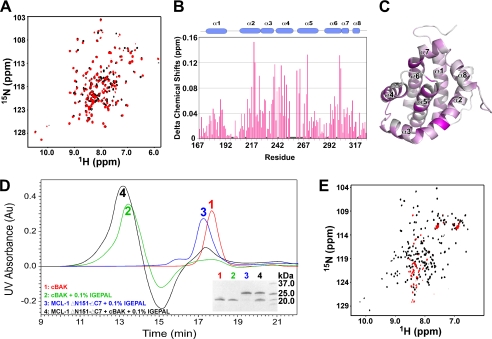
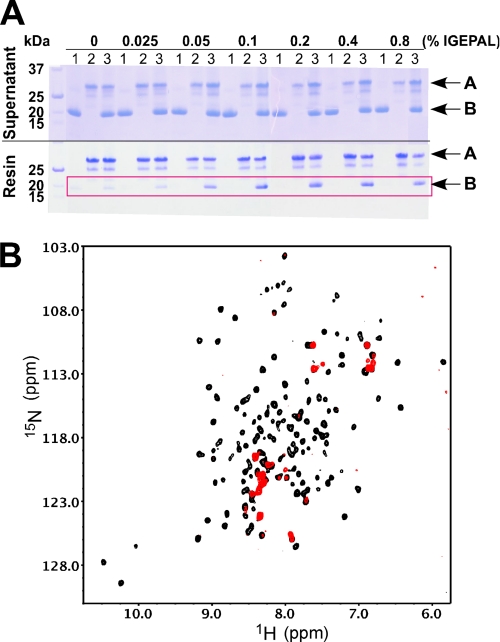
Upon the addition of a range of IGEPAL concentrations from four times below its critical micelle concentration (∼0.02% v/v) to 20 times above, only slight chemical shift changes were observed in 1H-15N HSQC spectra of 15N-labeled cMCL-1 (Fig. 3A and data not shown). The well dispersed pattern of peaks indicates that MCL-1 maintained its three-dimensional α-helical conformation. Analysis of the titration at 0.1% IGEPAL (Fig. 3, B and C) showed that the largest shifts, in the range of 0.1 to 0.16 ppm, came from the residues located in helix α2, α4, α5, and the end of α6, which comprise the hydrophobic groove for BH3 peptide binding. The cMCL-1 detergent interaction was transient and did not involve the formation of a protein·micelle complex or the oligomerization of cMCL-1, as shown by the absence of changes in the analytical size exclusion chromatography profile (Fig. 3D, blue trace, 17.2 min).
FIGURE 3.
Behavior of BAK and MCL-1 in the presence of IGEPAL. A, 1H-15N HSQC spectra of 15N-labeled cMCL-1 without (black) and with (red) 0. 1% (v/v) IGEPAL. B, plot of IGEPAL-induced chemical shift changes by residue for cMCL-1. Prolines and residues that were unassigned in either the free or bound form are labeled with asterisks. The secondary structure (helices α1 to α8) of cMCL-1 is shown for reference. C, three-dimensional model of cMCL-1 colored according to the size of the chemical shift changes upon the addition of IGEPAL. The colors range from gray for no change to purple for the largest changes (0.155 ppm). D, analytical size exclusion profiles for cBAK (red), cBAK in 0.1% IGEPAL (green), MCL-1 ΔN151-ΔC7 in 0.1% IGEPAL (blue), and cBAK and MCL-1ΔN151-ΔC7 mixture in 0.1% IGEPAL (black). Peak fractions 1 to 4 were analyzed by SDS-PAGE (inset). E, 1H-15N HSQC spectra of 15N-labeled cBAK without (black) and with (red) 0.1% (v/v) IGEPAL.
Similar titrations with cBAK resulted in a very different behavior. At low IGEPAL concentrations, there was a gradual disappearance of peaks in the spectrum of 15N-labeled cBAK, and at 0.05% IGEPAL, the 1H-15N HSQC spectrum started to display a pattern typical of unfolded proteins (Fig. 3E, red, recorded in 0.1% IGEPAL). This suggests IGEPAL unfolds cBAK and the small number of signals observed (Fig. 3E, red) indicates that the majority of BAK inserts into the large detergent micelles. Additionally, the elution time of cBAK on analytical size exclusion chromatography shifts from 17.8 min (Fig. 3D, red trace) to 13.5 min (green trace). This high molecular weight peak could be either oligomers of cBAK or cBAK associated with IGEPAL micelles. Thus, we observed a strongly altered conformation of BAK in the presence of IGEPAL.
Behavior of BAK and MCL-1 in Presence of CHAPS
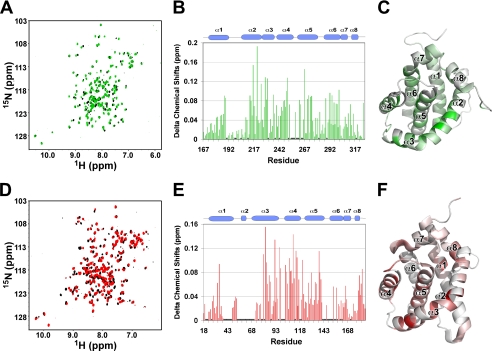
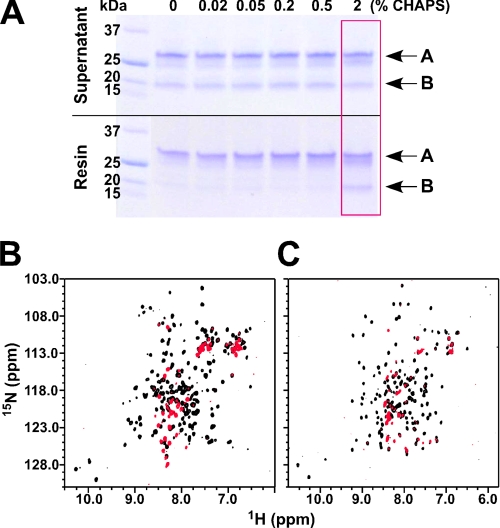
In comparison, the application of 2% (w/v) CHAPS generated little effects on both cMCL-1 and cBAK. Titrations with CHAPS induced only very minor chemical shift changes in the 1H-15N HSQC spectra of both proteins (Fig. 4, A and D). Even after overnight incubation (supplemental Fig. S1), the shifts were under 0.19 ppm for cMCL-1 (Fig. 4B) and 0.16 ppm for cBAK (Fig. 4E), which suggests that only minor conformational changes occurred. Perturbations of chemical shifts above 0.10 ppm occurred in the residues located in helices α2 to α5 and the end of α6 in cMCL-1 (Fig. 4C) and helices α3 to α5 in cBAK (Fig. 4F), indicating that the CHAPS interacts weakly with both proteins in their putative BH3-binding hydrophobic grooves.
FIGURE 4.
Behavior of cBAK and cMCL-1 in CHAPS. A, HSQC overlay of 15N-labeled cMCL-1 without (black) and with 2% CHAPS (red). B and E, plots of CHAPS-induced chemical shift changes by residue for cMCL-1 (B) and cBAK (E). Prolines and residues that were unassigned in either the free or bound form are labeled with asterisks. C and F, three-dimensional models of cMCL-1 (C) and cBAK (F) (6) colored according to the size of the chemical shift changes upon the addition of CHAPS. The colors range from gray for no change to green (0.19 ppm) or red (0.155 ppm). D, HSQC overlay of 15N-labeled cBAK without (black) and with 2% CHAPS for 20 min (red).
BAK Interacts with MCL-1 in Presence of Detergents
In light of the IGEPAL-mediated conformational changes in BAK, we tested whether either detergent allows the association of BAK with MCL-1. On analytical size exclusion chromatography (Fig. 3D), addition of MCL-1 ΔN151-ΔC7 to cBAK in 0.1% IGEPAL gave rise to a peak that eluted at 13.2 min (black trace), which suggests the formation of a MCL-1·BAK·micelle complex larger than either cBAK or MCL-1 ΔN151-ΔC7 alone in IGEPAL. Ni2+-NTA resin pulldown experiments showed that 0.1% IGEPAL was sufficient for FLAG-BAK-HMK-ΔTM-His6 to retain cMCL-1 on the Ni2+-NTA resin at an ∼1:1 ratio (Fig. 5A). In NMR titrations, the HSQC spectra of 15N-labeled cMCL-1 were strongly disturbed in the presence of both cBAK and IGEPAL (Fig. 5B). These results confirm a direct interaction between cBAK and cMCL-1.
FIGURE 5.
Interaction between BAK and MCL-1 in the presence of IGEPAL. A, Coomassie-stained SDS-gels showing the supernatant (top) and proteins retained by the Ni2+-NTA resin (bottom) under the conditions indicated. Conditions 1–3, respectively, mean that cMCL-1, FLAG-BAK-HMK-His6, or their combination was loaded. Arrows labeled A show the position of FLAG-BAK-HMK-ΔTM-His6, and arrows labeled B show the position of cMCL-1. The red frame highlights the cMCL-1 retained by FLAG-BAK-HMK-ΔTM-His6 in the presence of IGEPAL. B, 1H-15N HSQC spectra of 15N-labeled cMCL-1 in 0.1% IGEPAL without (black) and with (red) unlabeled cBAK.
To our surprise, 2% CHAPS also sufficed to promote the association between MCL-1 and BAK. Fig. 6A shows that a concentration of CHAPS at 2% (above its critical micelle concentration of 0.5%) was sufficient for retention of cMCL-1 by FLAG-BAK-HMK-ΔTM-His6 on the Ni2+-NTA resin. The well dispersed 1H-15N HSQC spectra of both proteins in 2% CHAPS allowed us to detect the interaction using either 15N-labeled cBAK or cMCL-1 (Fig. 6, B and C). In both cases, the formation of a large high molecular weight complex of cBAK and cMCL-1 led to a loss of the majority of signals in the spectrum.
FIGURE 6.
Interaction between BAK and MCL-1 in CHAPS. A, Coomassie-stained SDS-gels showing the supernatant (top) and proteins retained by the Ni2+-NTA resin (bottom) in the presence of increasing concentrations of CHAPS as indicated. Arrows labeled A show the position of FLAG-BAK-HMK-ΔTM-His6, and arrows labeled B show the position of cMCL-1. The red frame indicates the condition in which the binding between BAK and MCL-1 was observed. B, overlay of 1H-15N HSQC spectra of 15N-labeled cBAK in 2% CHAPS without (black) and with unlabeled cMCL-1 (red). C, overlay of 1H-15N HSQC spectra of 15N-labeled cMCL-1 in 2% CHAPS without (black) and with unlabeled cBAK (red).
BAK-BH3 Peptide Can Engage Hydrophobic Groove of MCL-1
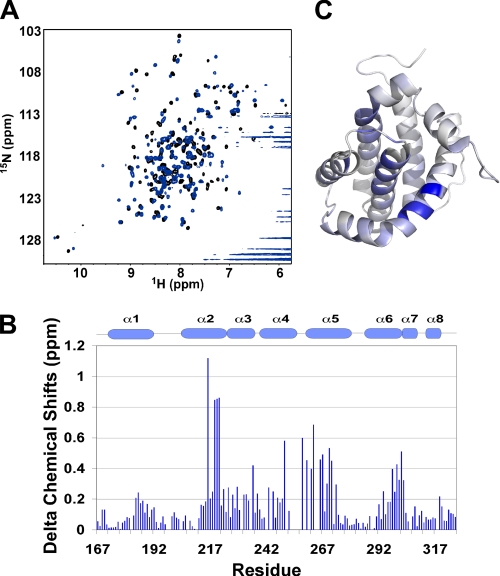
The observation of the MCL-1·BAK complex encouraged us to ask how BAK interacts with MCL-1. From experiments using cell lysates (18, 19), it was proposed that BAK associates with MCL-1 through its BH3 region. To verify this, we involved a peptide derived from the BH3 region of BAK, BAK-BH3, in our test. Titration of unlabeled BAK-BH3 into 15N-labeled cMCL-1 generated many chemical shift changes in the slow exchange regime, which is indicative of tight binding (Fig. 7A). In agreement with work from Hinds and co-workers (17), the BAK-BH3 peptide engages the hydrophobic groove of MCL-1 (Fig. 7, B and C). The dissociation constants for MCL-1 constructs with different N-terminal deletions (15) and BAK-BH3 peptides (wild-type and L78A mutant) were further tested by ITC in the absence of detergents (supplemental Fig. S2). As observed previously, removal of the N- and C-terminal extensions increased the affinity of MCL-1 for the BH3 peptides (15). The wild-type BAK-BH3 bound MCL-1 tightly with affinities ranging from 0.11 to 0.021 μm. In contrast, the mutation of L78A in BAK-BH3 decreased the affinity 160-fold (Table 1).
FIGURE 7.
MCL-1 interacts with BAK-BH3 peptide. A, 1H-15N HSQC spectra of cMCL-1 without (black) and with (blue) unlabeled BAK-BH3 peptide added. B, plot of BAK-BH3-induced chemical shift changes by residue for cMCL-1. Prolines and residues that were unassigned in either the free or bound form are labeled with asterisks. The secondary structure (helices α1 to α8) of cMCL-1 is shown for reference. C, three-dimensional model of cMCL-1 colored according to the size of the chemical shift changes upon the addition of BAK-BH3. The colors range from gray for no change to blue (1.12 ppm).
TABLE 1.
Dissociation constants tested by ITC at 23 °C
| Peptides | MCL-1 Constructs |
|||
|---|---|---|---|---|
| MCL-1ΔC23 | MCL-1ΔN119-C23 | MCL-1ΔN151-C7 | cMCL-1ΔN153-C24 | |
| μm | μm | μm | μm | |
| BAK-BH3 | 0.11 | 0.048 | 0.023 | 0.021 |
| BAK-BH3-L78A | 19 | 8.0 | 3.3 | 3.3 |
cBAK Interacts with cMCL-1 through Its BH3 Region
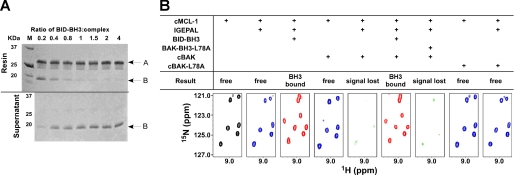
The binding of BAK-BH3 to MCL-1 directed us to test whether the BAK interacted in a similar manner to the BH3 peptide. The complex of FLAG-BAK-HMK-ΔTM-His6 and cMCL-1 was immobilized on Ni2+-NTA resin, and a high affinity BID-BH3 peptide (15) was used to compete for binding to cMCL-1. SDS-PAGE analysis of the pulldown experiment with immobilized BAK showed that increasing concentrations of the BID-BH3 peptide decreased the amount of cMCL-1 that was retained on the resin. When BID-BH3 was present at four times the concentration of the complex, cMCL-1 was fully released (Fig. 8A). This competition was also observed in NMR titrations in 0.1% IGEPAL. Upon the addition of the BID-BH3 peptide at a 4:1 ratio of 15N-cMCL-1·unlabeled cBAK complex, all of the signals for cMCL-1 were recovered and the spectrum was identical to that of the 15N-cMCL-1·unlabeled BID-BH3 complex (Fig. 8B and supplemental Fig. S3). Significantly, the mutant peptide BAK-BH3-L78A failed to retrieve these signals, suggesting that the engagement of the BH3 peptide into MCL-1 is necessary to disrupt the interaction with BAK. Parallel experiments with mutant cBAK-L78A showed that the mutant protein could not recruit cMCL-1 into IGEPAL micelles (Fig. 8B, far right panels). This confirms that the BAK BH3 region interacts with MCL-1. Similar conclusions were also drawn from the NMR titrations in 2% CHAPS. The complex could be disrupted by BID-BH3 peptide (supplemental Fig. S4A), and upon release, 15N-cBAK gave rise to a spectrum typical of an unfolded protein (supplemental Fig. S4B). This suggests that BAK loses its original α-helical fold when bound to MCL-1 via its BH3 region and is unable to refold when released.
FIGURE 8.
BID-BH3 peptides disrupts the complex of MCL-1·BAK. A, Coomassie-stained SDS-PAGE gels showing the results of the competition assay in 0. 1% IGEPAL. The upper panel gel shows proteins that were pulled down by the Ni2+-NTA resin, and the lower panel gel shows proteins that remained in the supernatant. The numbers above the gel show the ratio of BID-BH3 peptide to FLAG-BAK-HMK-ΔTM-His6·cMCL-1 complex when added for competition. The arrow labeled A shows the position of FLAG-BAK-HMK-ΔTM-His6, and arrows labeled B show the position of cMCL-1. B, slices of 1H-15N HSQC spectra (bottom) of 15N-labeled cMCL-1 in different conditions as shown. The spectra are color-coded according to the size of the spectral changes: black, free (control) cMCL-1 spectrum; blue, spectra with small chemical shift changes; pink, spectra with significant changes; green, spectra with absent signals).
DISCUSSION
BAX and BAK are the major proapoptotic effectors that undergo conformational changes and oligomerization to mediate mitochondrial outer membrane permeabilization (43, 44). To gain insight into this process, detergents have been used to mimic the native membrane environment of the protein. These studies have shown that BAX can be specifically activated by detergents to form higher order aggregates (24, 39, 45). Here, we describe the behavior of BAK in the presence of detergents. The nonionic detergent IGEPAL (0.1%) induced large changes in the spectrum of BAK (Fig. 3E), which likely reflects the formation of large complexes as observed for BAX in Triton X-100 (39, 41) and dodecyl-β-d-maltoside (46). Due to the large size of IGEPAL micelle, which is ∼90 kDa and equivalent to a BAK tetramer, we were unable to determine the oligomeric state of BAK in IGEPAL. In contrast, the conformation of BAK was not markedly affected by the zwitterionic detergent CHAPS (2%). Only minor perturbations were detected in the spectrum of BAK (Fig. 4, D–F) even after overnight incubation (supplemental Fig. S1). This indicates that BAK maintains its global fold with the detergent-induced changes restricted to the residues surrounding its putative BH3-binding hydrophobic groove. This result is consistent with reports that CHAPS has little effect on BAX oligomerization (45).
In this study, we also showed that the antiapoptotic protein MCL-1 undergoes only minor conformational changes in both detergents as revealed by the small changes in its NMR spectrum (Figs. 3, A–C, and 4, A–C) and its behavior in size-exclusion chromatography (Fig. 3D). Similar results were observed for BCL-XL and BCL-w at concentration of detergents below the critical micelle concentration; however, BCL-XL and BCL-w both display abrupt spectral changes at concentration of detergents above the critical micelle concentration (34, 40). This suggests that resistance to detergent-induced unfolding is not a general feature of antiapoptotic proteins.
At the level of protein-protein interactions, both IGEPAL and CHAPS were able to promote the association of BAK with MCL-1 (Figs. 5 and 6). Although this interaction has been observed within a native membrane environment (18–20), to our knowledge, this is the first time the interaction between the intact protein domains has been characterized in vitro. By peptide competition and site-directed mutagenesis, we showed that the interaction involves the BH3 region of BAK and the hydrophobic groove of MCL-1 (Fig. 8 and supplemental Figs. S3 and S4). This is quite consistent with conclusions from in vivo studies (18–20). The absence of binding without detergent suggests that the MCL-1-BAK interaction requires the exposure and accessibility of the BH3 region in BAK. This is relatively easy to imagine in the case of IGEPAL, where we observe large conformational changes in BAK. CHAPS does not strongly perturb the structure of BAK but likely increases the internal dynamics and decreases the energy barrier for the exposure of the BH3 region. Interestingly, the affinity of BAK for MCL-1 appeared to be less than the affinity of the isolated BAK-BH3 peptide; BAK was efficiently removed by BID-BH3, which has weaker affinity than BAK-BH3 (15). This suggests that the BH3 region of BAK is not fully accessible in the presence of detergents. This could be due to residual folding, steric hindrance due to the detergent micelles, or competing intramolecular interactions such as observed for MCL-1 where longer constructs display lower affinity than the core cMCL-1 domain (Table 1) (15).
We can gain some limited insight into the structure of the heterodimeric complex in noting that the spectrum of BAK bound to cMCL-1 in CHAPS is similar to the spectrum of free BAK in IGEPAL. This suggests that the signals observed in the detergents most probably arise from disordered regions that do not interact with MCL-1 (supplemental Fig. S5).
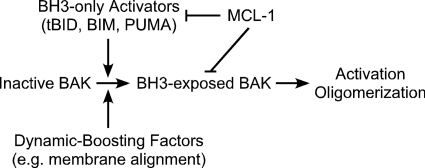
Our data add valuable information for evaluating the existing models of BAK activation. Taking a look at CAM and DAM in Fig. 1, we note that both models agree on the interaction between MCL-1 (antiapoptotic proteins) and BID (BH3-only proapoptotic proteins). The difference between these two models centers on whether MCL-1 or BID directly interact with BAK. Our data show that MCL-1 interacts not only with BH3-only proteins as represented by the structures of complexes of MCL-1 with BH3 peptides (14–17) but also with BAK. Thus, the function of MCL-1 is not limited to the inhibition of BH3-only proteins, and DAM does not encompass the whole regulatory pathway. We also observed that the interaction between BAK and MCL-1 is conditional on the exposure of the BH3 region of BAK; thus, BAK is not necessarily always kept in check by antiapoptotic proteins. How free BAK can be activated to promote apoptosis is not addressed by CAM. Direct activation through BH3-only protein binding (especially the BH3-only activators tBID, BIM, and PUMA) has been demonstrated as a component of DAM (47), whereas our observation of a complex of MCL-1·BAK in detergent suggests that some other means may activate BAK toward either mitochondrial outer membrane permeabilization or inhibition by antiapoptotic proteins. Thus, drawbacks exist for both models and point to a hybrid-activating model as outlined in Fig. 9. In this model, BAK can be activated by either the BH3-only activators or other factors (e.g. membrane alignment) that cause conformational changes; MCL-1 interacts both with BH3-only proteins and with BAK that has its BH3 region accessible. Although our study has been limited to studying the proteins in the presence of membrane mimics, we believe that the requirement for accessibility of the BAK BH3 applies to BAK and MCL-1 in their native environment.
FIGURE 9.
Hybrid-activating model. Inactive BAK is primed for conformational changes by either BH3-only activators or dynamic-boosting factors such as membrane alignment. Inhibition by MCL-1 is mediated by its interaction with both BH3-only proteins and BH3-exposed BAK.
The finding that a BID-BH3 peptide can displace BAK from MCL-1 highlights the potential therapeutic use of compounds that bind to MCL-1. The disruption of the MCL-1·BAK complex is a critical step to incite cells to undergo apoptosis. As an alternative to the well known tumor suppressor p53 that can disrupt complex formation by binding to BAK (18), our data show that the success of BID-BH3 and the failure of BAK-BH3-L78A to displace BAK from its complex with MCL-1 are dependent on their binding affinities to MCL-1 (Fig. 8, supplemental Fig. S4A, and Table 1) (15). This is consistent with the finding that obatoclax (GX15–070) is able to overcome MCL-1 mediated resistance to apoptosis (48, 49), whereas ABT-737 fails to do so (50–52). Our data suggest the need for high affinity small molecule inhibitors that target at MCL-1. As BAK released from the complex exists in an unfolded conformation (supplemental Fig. S4B) and may readily be able to undergo oligomerization to promote apoptosis, the therapeutic use of MCL-1 inhibitors should be particularly effective in transiting cells into apoptosis.
Supplementary Material
Acknowledgment
We acknowledge GeminX for providing the Flag-BAK-HMK-ΔTM-His6 and MCL-1 plasmids.
This work was supported by Canadian Institutes of Health Research Grant MOP-81277.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- BH
- BCL-2 homology region
- CAM
- competitively activating model
- cBAK
- calpain-cut BAK
- Ni2+ NTA
- nickel nitrilotriacetic acid
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- cMCL-1
- calpain-cut MCL-1
- DAM
- directly activating model
- ITC
- isothermal titration calorimetry
- IGEPAL
- tert-octylphenoxy poly(oxyethylene)ethanol
- HSQC
- heteronuclear single quantum coherence
- TM
- transmembrane.
REFERENCES
- 1.Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green D. R., Kroemer G. (2004) Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 3.Griffiths G. J., Dubrez L., Morgan C. P., Jones N. A., Whitehouse J., Corfe B. M., Dive C., Hickman J. A. (1999) J. Cell Biol. 144, 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. (2000) Genes Dev. 14, 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 5.Breckenridge D. G., Germain M., Mathai J. P., Nguyen M., Shore G. C. (2003) Oncogene 22, 8608–8618 [DOI] [PubMed] [Google Scholar]
- 6.Moldoveanu T., Liu Q., Tocilj A., Watson M., Shore G., Gehring K. (2006) Mol. Cell 24, 677–688 [DOI] [PubMed] [Google Scholar]
- 7.Mandic A., Viktorsson K., Molin M., Akusjärvi G., Eguchi H., Hayashi S. I., Toi M., Hansson J., Linder S., Shoshan M. C. (2001) Mol. Cell. Biol. 21, 3684–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., Shimizu S., Sakamaki K., Yonehara S., Tsujimoto Y. (2004) J. Biol. Chem. 279, 33865–33874 [DOI] [PubMed] [Google Scholar]
- 9.Sawai H., Domae N. (2009) Biochem. Biophys. Res. Commun. 378, 529–533 [DOI] [PubMed] [Google Scholar]
- 10.Korsmeyer S. J., Wei M. C., Saito M., Weiler S., Oh K. J., Schlesinger P. H. (2000) Cell Death Differ. 7, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 11.Reed J. C. (2006) Cell Death Differ. 13, 1378–1386 [DOI] [PubMed] [Google Scholar]
- 12.Dewson G., Kratina T., Sim H. W., Puthalakath H., Adams J. M., Colman P. M., Kluck R. M. (2008) Mol. Cell 30, 369–380 [DOI] [PubMed] [Google Scholar]
- 13.Dai H., Meng X. W., Lee S. H., Schneider P. A., Kaufmann S. H. (2009) J. Biol. Chem. 284, 18311–18322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fire E., Gullá S. V., Grant R. A., Keating A. E. (2010) Protein Sci. 19, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q., Moldoveanu T., Sprules T., Matta-Camacho E., Mansur-Azzam N., Gehring K. (2010) J. Biol. Chem. 285, 19615–19624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czabotar P. E., Lee E. F., van Delft M. F., Day C. L., Smith B. J., Huang D. C., Fairlie W. D., Hinds M. G., Colman P. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day C. L., Smits C., Fan F. C., Lee E. F., Fairlie W. D., Hinds M. G. (2008) J. Mol. Biol. 380, 958–971 [DOI] [PubMed] [Google Scholar]
- 18.Leu J. I., Dumont P., Hafey M., Murphy M. E., George D. L. (2004) Nat. Cell Biol. 6, 443–450 [DOI] [PubMed] [Google Scholar]
- 19.Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. (2005) Genes Dev. 19, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillissen B., Wendt J., Richter A., Müer A., Overkamp T., Gebhardt N., Preissner R., Belka C., Dörken B., Daniel P. T. (2010) J. Cell Biol. 188, 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derenne S., Monia B., Dean N. M., Taylor J. K., Rapp M. J., Harousseau J. L., Bataille R., Amiot M. (2002) Blood 100, 194–199 [DOI] [PubMed] [Google Scholar]
- 22.Alvi A. J., Austen B., Weston V. J., Fegan C., MacCallum D., Gianella-Borradori A., Lane D. P., Hubank M., Powell J. E., Wei W., Taylor A. M., Moss P. A., Stankovic T. (2005) Blood 105, 4484–4491 [DOI] [PubMed] [Google Scholar]
- 23.Cavarretta I. T., Neuwirt H., Untergasser G., Moser P. L., Zaki M. H., Steiner H., Rumpold H., Fuchs D., Hobisch A., Nemeth J. A., Culig Z. (2007) Oncogene 26, 2822–2832 [DOI] [PubMed] [Google Scholar]
- 24.Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 25.Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 26.Cartron P. F., Gallenne T., Bougras G., Gautier F., Manero F., Vusio P., Meflah K., Vallette F. M., Juin P. (2004) Mol. Cell 16, 807–818 [DOI] [PubMed] [Google Scholar]
- 27.Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 28.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 29.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 30.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 31.Certo M., Del Gaizo Moore V., Nishino M., Wei G., Korsmeyer S., Armstrong S. A., Letai A. (2006) Cancer Cell 9, 351–365 [DOI] [PubMed] [Google Scholar]
- 32.Häcker G., Weber A. (2007) Arch. Biochem. Biophys. 462, 150–155 [DOI] [PubMed] [Google Scholar]
- 33.Chipuk J. E., Green D. R. (2008) Trends Cell Biol. 18, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denisov A. Y., Chen G., Sprules T., Moldoveanu T., Beauparlant P., Gehring K. (2006) Biochemistry 45, 2250–2256 [DOI] [PubMed] [Google Scholar]
- 35.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 36.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 37.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 38.Day C. L., Chen L., Richardson S. J., Harrison P. J., Huang D. C., Hinds M. G. (2005) J. Biol. Chem. 280, 4738–4744 [DOI] [PubMed] [Google Scholar]
- 39.Hsu Y. T., Youle R. J. (1998) J. Biol. Chem. 273, 10777–10783 [DOI] [PubMed] [Google Scholar]
- 40.Losonczi J. A., Olejniczak E. T., Betz S. F., Harlan J. E., Mack J., Fesik S. W. (2000) Biochemistry 39, 11024–11033 [DOI] [PubMed] [Google Scholar]
- 41.Hsu Y. T., Youle R. J. (1997) J. Biol. Chem. 272, 13829–13834 [DOI] [PubMed] [Google Scholar]
- 42.Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 43.Lindsten T., Ross A. J., King A., Zong W. X., Rathmell J. C., Shiels H. A., Ulrich E., Waymire K. G., Mahar P., Frauwirth K., Chen Y., Wei M., Eng V. M., Adelman D. M., Simon M. C., Ma A., Golden J. A., Evan G., Korsmeyer S. J., MacGregor G. R., Thompson C. B. (2000) Mol. Cell 6, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivashyna O., García-Sáez A. J., Ries J., Christenson E. T., Schwille P., Schlesinger P. H. (2009) J. Biol. Chem. 284, 23935–23946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bleicken S., Classen M., Padmavathi P. V., Ishikawa T., Zeth K., Steinhoff H. J., Bordignon E. (2010) J. Biol. Chem. 285, 6636–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhai D., Jin C., Satterthwait A. C., Reed J. C. (2006) Cell Death Differ. 13, 1419–1421 [DOI] [PubMed] [Google Scholar]
- 49.Nguyen M., Marcellus R. C., Roulston A., Watson M., Serfass L., Murthy Madiraju S. R., Goulet D., Viallet J., Bélec L., Billot X., Acoca S., Purisima E., Wiegmans A., Cluse L., Johnstone R. W., Beauparlant P., Shore G. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19512–19517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 51.van Delft M. F., Wei A. H., Mason K. D., Vandenberg C. J., Chen L., Czabotar P. E., Willis S. N., Scott C. L., Day C. L., Cory S., Adams J. M., Roberts A. W., Huang D. C. (2006) Cancer Cell 10, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S., Dai Y., Harada H., Dent P., Grant S. (2007) Cancer Res. 67, 782–791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.