Preface
Prions are unusual proteinaceous infectious agents that are typically associated with a class of fatal degenerative diseases of the mammalian brain. However, the discovery of fungal prions, which are not associated with disease, suggests that we must now consider the impact of these factors on basic cellular physiology in a different light. Fungal prions are epigenetic determinants that can alter a range of cellular processes, including metabolism and gene expression pathways, and these changes can lead to a range of prion-associated phenotypes. The mechanistic similarities between prion propagation in mammals and fungi suggest that prions are not a biological anomaly but instead are a new appreciated and perhaps ubiquitous regulatory mechanism.
Introduction
Cellular phenotypes are a ‘readout’ of the complex interplay of genetic and epigenetic determinants that ultimately define a unique proteome and thereby specify cellular identity. However, modulation of the proteome itself is emerging as a key concept in our understanding of the molecular basis of phenotypic traits. Many studies over the past two decades have highlighted how regulated changes in protein modifications, such as phosphorylation and glycosylation, contribute to cellular phenotypes by altering protein abundance, function and localization. Such changes can in turn impact on complex regulatory pathways that control cellular phenotypes. But post-translational modifications are not the whole story; changes in protein conformation might also explain many phenotypic switches albeit by a mechanism that is not yet fully understood.
The prototypical and perhaps most extensively characterized example of protein conformation-based, inherited phenotypic traits are those defined by proteinaceous infectious particles known as prions. These factors were originally identified as infectious entities associated with a group of transmissible neurodegenerative diseases in mammals1, 2 known as transmissible spongiform encephalopathies (TSEs) — such as Creudzfeldt–Jakob disease (CJD) and kuru in humans, scrapie in sheep and bovine spongiform encephalopathy (BSE) in cattle — the causative agent of which is resistant to treatments that damage nucleic acids.1, 3 The fact that prions could act as infectious agents despite the absence of a nucleic acid genome led to the formulation of the “protein-only” or prion hypothesis.1, 3 According to this idea, the TSE agent is a self-perpetuating conformer of a host protein PrP (prion protein).1 The infectious conformer of this protein (PrPSc) was predicted to recruit and convert the normal conformer PrPC into the PrPSc form through contacts between specific regions of the protein, thereby ‘replicating’ the agent during infection. A wealth of genetic and biochemical data now support this concept of conformational replication, leading to its near universal acceptance.
In addition to mammalian PrP, prions have also been found in two species of fungi, the yeast Saccharomyces cerevisiae and the multicellular fungus Podospora anserina (BOX 1).4 This intriguing collection of functionally unrelated proteins can, like PrP, individually adopt a range of physical forms and transition between these states under physiological conditions. In all systems, these physical transitions specify new phenotypes, which may result from alterations to the normal function of the protein (gain-of-function and/or loss-of-function), the cellular response to the new protein conformation and/or the rate of accumulation of the altered form. Remarkably, these alternative conformers, each with a distinct yet stable three dimensional shape, are self-replicating and can be transferred between cells or organisms, allowing the associated traits to be transmitted as infectious diseases, as occurs in mammals5, or inherited through cell division, as occurs in fungi.4 Although the mechanistic basis of prion propagation and transmission are emerging concepts in all systems, it is clear that these processes exist and ensure a level of genetic stability for prion-based epigenetic determinants that is in line with that of nucleic acid-based genetic determinants.
Box 1. Fungal prions, cytoplasmic inheritance and epigenetic regulation.
In a genetic cross between haploid [PRION+] and [prion−] strains of Saccharomyces cerevisiae, typically the resulting diploid is [PRION+], and all meiotic progeny also carry the prion determinant.4 If the associated phenotype was controlled by a loss-of-function nuclear gene mutation then typically it would be recessive in a diploid, and a 2 [PRION+]: 2 [prion−] segregation pattern would be evident amongst the meiotic progeny. This non-Mendelian mode of inheritance shown by prions is indicative of transfer through the cytoplasm, which can also occur in other epigenetic systems, for example, mitochondrial petite mutations.157 Inheritance of the [PRION+] determinant generally results in the establishment of a new stable genetic state that can be maintained and propagated over many generations. In most cases inheritance of the [PRION+] determinant will result in a change of phenotype when compared with the [prion−] cell (Table 1) that is not accompanied by a change in the nucleotide sequence of the prion protein-encoding gene, or for that matter, any other nuclear gene. Consequently, prions can rightly be viewed as epigenetic determinants that can affect cellular processes (see Table 1). Prion-based epigenetic systems may have evolved because they can rapidly modify a cellular phenotype in response to a changing environment without introducing a change in the sequence and function of the genome.
In this Review we examine the expanding range of cellular processes and complex phenotypes that are determined by these epigenetic elements in fungi and in mammals and discuss how the process of conformational self-replication provides a framework for understanding the molecular basis of prion-associated phenotypes. As the number of identified prion proteins continues to grow, we suggest that the prion mechanism has now moved from the realm of a disease-causing biological anomaly to one of a novel regulator of cell phenotype.
Prion-associated phenotypes in fungi
Fungal prions have been identified through classical genetic studies, sequence-based algorithms and genetic screens (Box 2)6-9. To date, eight proteins are confirmed fungal prions (Table 1), and at least another 20 proteins may potentially fall into this class. Remarkably, fungal prions affect the activity and/or regulation of several cellular processes, with many playing important roles in global gene regulation either at the transcriptional or post-transcriptional level (Table 1). Because of their roles in regulating the expression of genetic information, prion proteins could indirectly affect a wide range of co-regulated cellular processes.
Box 2. Can you identify a prion protein from its amino acid sequence?
Although prions that affect the host phenotype or cause disease have only been identified and verified in fungi and mammals, it is unlikely they are restricted to these two groups of organisms. One important question that we are now getting near to answering is whether we can identify a prion protein from its sequence. Soon after their discovery in fungi it emerged that there were some sequence features shared by the various yeast prion proteins particularly the presence of a region of the protein (the prion-forming domain; PrD) that contained an atypically high density of Gln and Asn residues and that was conformationally flexible.8, 9 A high density of Gln residues in a protein makes it prone to aggregate, but it is the high density of Asn residues that is crucial for prion propagation.9 Remarkably, it is the amino acid composition rather that its sequence that is crucial in determining prion potential.158 In addition to Gln-Asn content, some prion-forming proteins, such as Sup35 and to a lesser extent Rnq1, have short oligopeptide repeats with sequence similarity to a series of octapeptide repeats found in PrP, suggesting that such sequences may be important for prion propagation.143 One approach has identified 200 proteins in S. cerevisiae with this architecture, although many of these do not satisfy the genetic and/or biochemical criteria to be designated as prions.9 The fact that PrD of the Podospora anserina HET-s prion protein lacks both Asn and Gln residues, but can be efficiently propagated as the [Het-s] prion in either P.anserina32 or S.cerevisiae159 suggests that alternative mechanisms must exist for propagating fungal prions.
Table 1.
Prions in fungi
| Determinant | Protein | Cellular function | Phenotype | Ref |
|---|---|---|---|---|
| [URE3] | Ure2 | Transcriptional co-repressor | Nitrogen source utilisation | 7 |
| [PSI+] | Sup35 | Translation termination factor (eRF3) |
Nonsense suppression Resistance to toxic compounds |
12, 13 |
| [PIN+]/[RNQ+]* | Rnq1 | Unknown | De novo prion formation | 35 |
| [SWI+] | Swi1 | Subunit of the SWI/SNF chromatin remodelling complex |
Repression of transcription | 23 |
| [OCT+] | Cyc8 | Transcriptional co-repressor | Derepression of transcription | 24 |
| [MOT3+] | Mot3 | Transcriptional co-repressor | Derepression of transcription | 9 |
| [MCA+] | Mca1 | Metacaspase, regulates apoptosis |
None reported | 6 |
| [Het-s] ** | HET-s | Unknown | Vegetative incompatibility Cell death |
32 |
| [ISP+] | Sfp1 | Transcriptional activator | Translational accuracy | 30 |
The prion form of Rnq1 was originally designated as [RNQ+]. Subsequently, Rnq1 was identified as the prion protein that can induce de novo [PSI+] formation and was dubbed [PIN+] ([PSI+] inducibility). Although other Gln-Asn-rich proteins can act as [PIN+] prions and induce de novo [PSI+] formation, it is generally assumed that [PIN+] is the same as [RNQ+].
All prions bar [Het-s] are found in Saccharomyces cerevisiae. [Het-s] is found in Podospoa anserina.
[PSI+] prion-based loss-of-function phenotypes
The epigenetic [PSI+] determinant is the prion form of Sup35, which is required to terminate mRNA translation and to release the polypeptide chain from the ribosome. [PSI+] enhances the efficiency of tRNA-mediated nonsense suppression10 and +1/−1 ribosomal frameshifting11 in S. cerevisiae, two phenotypes that could be ascribed to loss-of-function or modification-of-function mutations in Sup35. In [PSI+] cells, Sup35 is mainly found in large aggregates,12, 13 suggesting that conversion of Sup35 to its prion form compromises its activity in translation termination by sequestration. This reduction in release activity allows near cognate tRNAs to mistranslate a stop codon as sense. Sup35 proteins from many closely related Saccharomyces species retain the ability to behave as prions when expressed in the S. cerevisiae cytoplasm,14 indicating that [PSI+] could be a newly appreciated but conserved epigenetic mechanism for regulating both translation termination and reading-frame maintenance. This change in translational fidelity affects a broad range of cellular phenotypes either directly or indirectly.
The [PSI+]-dependent suppression of specific nonsense mutations generates readily assayed changes in phenotype. However, there are several complex phenotypes that are [PSI+] dependent but not simply attributable to suppression of a specific nonsense mutation, including altered sensitivity to a range of physical stresses such as heat15 and toxic chemical agents.16, 17 Such prion-mediated effects likely reflect a more global perturbation of the proteome that could arise through specific effects on a regulatory factor or alternatively through the extension of an ORF by stop codon read-through, which could modify the function of the encoded protein or inactivate it.
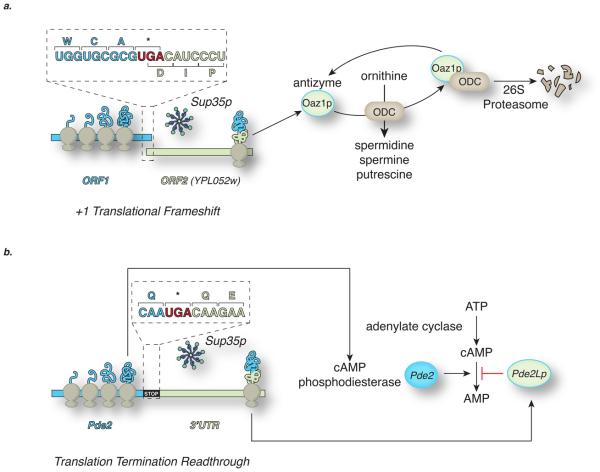
Despite the existence of opposing forces that act to ensure translational fidelity, such as the context of the stop codon18 and the nonstop mRNA decay pathway,19 recent studies have uncovered support for each of these mechanisms. For example, antizyme, encoded by OAZ1, is a negative regulator of polyamine biosynthesis that is translated from two overlapping out-of-frame ORFs by a +1 frameshift (Figure 1a).20 [PSI+] cells have increased antizyme levels and, correspondingly, reduced polyamine levels, a physiological state that may account for a large proportion of [PSI+]-dependent phenotypes.21 Also, the PDE2 ORF, encoding a high-affinity cAMP phosphodiesterase, is extended in [PSI+] cells leading to a destabilization of the encoded enzyme and resulting in an increase in cAMP levels (Figure 1b).22
Figure 1. Cellular consequences of termination codon readthrough and +1 ribosomal frameshifting mediated by the [PSI+] prion.
The [PSI+] prion compromises the function of the translation termination release factor Sup35, leading to the readthrough of stop codons or translational frameshifting. This epigenetic loss of Sup35 function creates new phenotypes based on the altered translation of specific mRNA transcripts.
a. OAZ1 in yeast consists of two overlapping open reading frames (ORF1 and YPL052w), with the latter in the +1 frame with respect to ORF1.20 During translation the ribosomes can either terminate or shift into the +1 frame at the ORF1 UGA codon, thus by-passing the stop codon and continuing translation into the YPL052w ORF. This +1 frameshift increases as polyamine levels rise. The resulting fused protein, antizyme, regulates the ubiquitin-dependent degradation of ornithine decarboxylase (ODC), which mediates the synthesis of the polyamines spermidine, spermine and putrescine. In [PSI+] cells there is a significant reduction in the levels of functional release factor, leading to an increase in the pause time at the UGA codon. This in turn leads to increased levels of +1 frameshifting and hence antizyme synthesis, resulting in a reduction in ODC and polyamine synthesis when compared with a [psi−] cell.21
b. As a component of the cAMP-dependent protein kinase signalling system in yeast, Pde2p controls the basal levels of cAMP by hydrolysing cAMP to AMP. Readthough of the native UGA codon gives rise to a form of Pde2p (Pde2Lp) that has no phosphodiesterase activity. Consequently [PSI+] cells have approximately two-fold higher levels of cAMP than [PSI+] cells.22
Other prion-based loss-of-function phenotypes in fungi
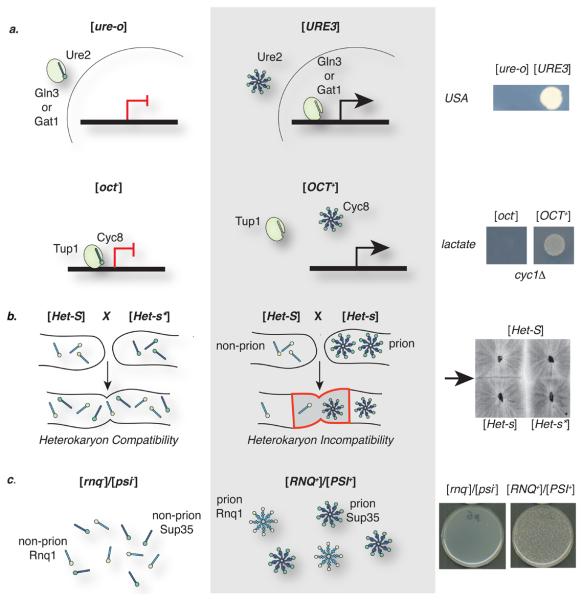
Five of the established yeast prion proteins (Ure2/[URE3], Swi1/[SWI+], Cyc8/[OCT+], Mot3/[MOT3+] and Sfp1/[ISP+]; Table 1; Figure 2), normally function to co-ordinately regulate transcription. For example, Swi123 and Cyc824 regulate the transcription of ~400 different genes, many of which contribute to a range of key cellular pathways. The newly described [MOT3+] prion9 also has a broad repertoire of target genes, including those important for cell wall remodelling under anaerobic conditions and for ergosterol biosynthesis, potentially affecting vacuole function,25 and Sfp1 coordinates the expression of ribosomal proteins and factors that control ribosome biogenesis.26 The transcriptional effects of these factors are mediated by their association with larger protein complexes (for example, the SWI–SNF chromatin remodelling complex, the Cyc8–Tup1 and Mot3–Rox1 co-repressor complexes, and Sfp1 regulation by TORC1). The switch to the prion form may affect these associations and, in so doing, alter the expression of a large portion of the yeast transcriptome. This idea has been experimentally confirmed for strains carrying the [OCT+] prion in which the Cyc8-Tup1 repressor complex becomes inactive and genes such as CYC7 (encoding iso-2-cytochrome C) and SUC2 (encoding invertase) become derepressed (Figure 2a).24
Figure 2. Fungal prions and their associated phenotypes.
Prion and non-prion conformers confer unique phenotypes by altering the normal activity of the prion protein. Several transcriptional regulators in yeast have been identified as prions. The [URE3] prion alters the capacity of Ure2 to associate with the transcriptional activators Gln3 and Gat1 in the cytoplasm (left), which occurs in the non-prion [ure-o] state, allowing these activators to translocate into the nucleus (centre), where they up-regulate genes that allow [URE3] cells to use the poor nitrogen source ureidosuccinate (USA) in the presence of ammonia (right). Figure kindly provided by C. Cullin (Institut de Biochimie Génétique Cellulaires, Bordeaux). The [OCT+] prion compromises the ability of the Cyc8 transcriptional repressor to associate with its co-repressor Tup1 (oval, left), leading to the upregulation of many genes, including CYC7 (centre), which allows the use of lactate in cyc1Δ strains (right). Figure kindly provided by S. Liebman (University of Illinois-Chicago).
b. The [Het-s] prion allows the Het-s protein to acquire a new activity to induce cell death (compare left and centre) following mating with a P. anserina strain that expresses the Het-S allele, leading to a block in heterokaryon formation, which leads to a barrier band of cell death (arrow, right). Figure kindly provided by S. Saupe (Institut de Biochimie Génétique Cellulaires, Bordeaux).
c. The [PIN+] (also known as [RNQ+]) prion allows Rnq1p to support the de novo formation of the [PSI+] prion especially when the levels of Sup35 protein are artificially elevated (center and right).
[URE3], the first fungal prion to be discovered, also exemplifies the phenotypic consequences of a transcriptional regulator – in this case the Ure2 protein - switching to a prion form. In its soluble non-prion form, Ure2 binds to two transcriptional activators (Gln3 and Gat1) in the cytoplasm, preventing their transit into the nucleus.27, 28 This sequestration represses the transcription of target genes and prevents the use of poor nitrogen sources while preferred sources, such as ammonia, are available (Figure 2a). In [URE3] cells, Ure2 is aggregated in the cytosol and cannot retain Gln3 and Gat1, resulting in activation of the transcriptional programme needed to use alternative nitrogen sources, thereby phenocopying loss-of-function ure2 mutations. Ure2 has also been implicated in cellular responses to heavy metals and oxidative stress that are independent of its transcriptional regulatory role29, so the switch to the [URE3] form could affect additional cellular phenotypes.
Prion-based gain-of-function phenotypes in fungi
The [PSI+] and [URE3] prions establish heritable loss-of-function phenotypes by inactivating their protein determinants, but there are also several examples of prion-mediated phenotypes in fungi linked with an apparent gain of function, including the well-studied [Het-s] prion of P. anserina and [ISP+] in S. cerevisiae30. [Het-s] was first identified as a non-Mendelian genetic element that controlled vegetative incompatibility.31 When a strain of the fungus carrying the [Het-s] prion meets a strain that expresses an allele of the het-s gene (het-S) that encodes a non-prion-forming form of the protein (called HET-S), the mixed heterokaryon formed on cell fusion dies (Figure 2b).32 This cell death blocks horizontal transmission of the prion and may represent a form of innate immunity.33 Although the mechanism of cell death remains unclear, this event is only triggered if the [Het-s] prion is present in one of the two strains; a cross between a het-s strain expressing the non-prion soluble form of the protein ([Het-s*]) or a Δhets strain is compatible.31, 32 Intriguingly, the incompatibility is dependent on the ability of Het-S to adopt a prion conformation in the presence of the [Het-s] prion, suggesting that these mixed complexes are either themselves the toxic form or indirectly induce toxicity upon co-aggregation.34
An apparent gain-of-function prion phenotype is also linked to Rnq1 (rich in N and Q) of S. cerevisiae. Although the cellular function of Rnq1 remains unknown, the aggregated prion form of this protein (originally designated [RNQ+])35 supports the de novo formation of the [PSI+] prion (Figure 2c).36, 37 On the basis of this phenotype, [RNQ+] is typically referred to as [PIN+] for [PSI+] inducibility and is the only prion reported to exist in non-laboratory strains of S. cerevisiae to date.38, 39 Exactly why the prion form of Rnq1 but not its soluble form can facilitate de novo prion formation remains an open question. However, in vitro studies have shown that preformed aggregates of Rnq1 can initiate the assembly of Sup35 prion polymers by a direct cross-seeding mechanism,40 although [PIN+] is not required for the continued propagation of [PSI+] or any other prion in vivo.41 Remarkably, [URE3] can provide [PIN+] activity in the absence of Rnq1,36 indicating that the same prion form can simultaneously contribute both loss-of-function and gain-of-function phenotypes.
Are fungal prions of benefit to the host?
While the prion mechanism provides a means to profoundly alter protein function and cellular phenotype in fungi, the question remains: does this process bring benefit or harm to the organism? Given the range of possible biological functions and phenotypes associated with fungal prions, the answer is likely to be equally complex. Two fungal prions, [PIN+]38 and [Het-s]42 have been isolated in wild strains suggesting that their presence may be beneficial, but two others, [URE3] and [PSI+], have not, raising the possibility that they are deleterious.38, 39 While the adverse effects of [URE3] and [PSI+] on yeast growth under some conditions supports this view;29, 43 the failure to detect these prions in natural isolates may not be a sufficient criterion to discount a potentially beneficial role for these phenotypic states. In the case of [PSI+], Sup35 homologues in Saccharomyces species have retained the ability to adopt a prion conformation in a laboratory setting,14 and under these conditions, the [PSI+] state is beneficial in the short term (i.e. in response to chemical or physical threats)15, 17, 21 and perhaps in the long-term, through adaptation and ultimately evolution of new genetic traits.16, 17, 44, 45 A more broad and systematic survey of prion determinants in natural isolates and in diverse niches will undoubtedly shed light on the costs and benefits of these protein conformational and phenotypic switches for fungi.
Prion-associated phenotypes in mammals
While fungal prion proteins adopt self-replicating conformers that are largely benign to the host under standard laboratory conditions, the appearance of similar conformers in mammals is most typically associated with emergence of disease. Despite this clearly disadvantageous phenotype, mounting evidence suggests that prion conversions in mammals, like their fungal counterparts, are a mechanism for regulating protein function.
Gain-of-function amyloids
In addition to prions, a broader group of endogenous mammalian proteins can undergo a prion-like self-replicating change in conformation, leading to the build-up of amyloid deposits in vivo. These amyloidoses can be distinguished from the prion diseases by their lack of infectivity. However, recent studies have uncovered a prion-like behaviour for some of these proteins within single organisms, where the amyloid form can be propagated from one cell to a neighboring cell by exogenous transfer of these aggregates.46 Among these proteins are the amyloids associated with Alzheimer's disease (amyloid-β),47 Huntington's disease (polyglutamine)48 and Parkinson's disease (α-synuclein).49 Because these prion-like amyloids lack a complete infectious cycle, they have been referred to as ‘prionoids’.50
How do these diseases relate to the normal function of the protein? The emerging consensus suggests that the toxicity associated with the amyloidoses represents a newly acquired activity linked to the alternative conformation.51 This hypothesis is consistent with our current understanding of the gain-of-function phenotypes of some fungal prions and the identification of proteins in bacteria and an invertebrate, for which the “normal” biological activity is associated with the amyloid form.52, 53 Recently, this concept of “functional” amyloid has been extended to vertebrates. For example, amyloid formation by a proteolytic fragment of the transmembrane glycoprotein Pmel17 is required for melanosome maturation and stimulates the synthesis of melanin,54, 55 and peptide and protein prohormones are stored in an amyloid form in pituitary secretory granules.56 In the case of the prohormones, amyloid formation is reversible,56 as is amyloid formation by an SH3 domain in vitro.57 Therefore, the amyloid form can, in principle, act as a mechanism to regulate protein function in mammals.
PrP as a disease-causing agent in vertebrates
In the case of the TSEs, a clear and irrefutable role for PrP in the appearance, phenotypic manifestation and spread of disease has been established, primarily through studies in transgenic mice. PrP is the major constituent of biochemically enriched preparations of the TSE agent,2 and mice devoid of PrP cannot replicate the infectious agent and are resistant to TSEs upon challenge.58 In addition, strong evidence supports a direct role for PrP in clinical disease. Depletion of PrP post-infection extends incubation times and reverses both neuropathology and behavioural defects induced by the infection.59, 60 These observations are consistent with a functional role for PrP in TSE aetiology. But, important questions remain: what is the molecular basis of TSE-associated neurotoxicity, and how does it relate to the normal function of PrP?
In fungi, the classification of prion-associated phenotypes as gain of function or loss of function is defined in relation to the null phenotype; however, a similar comparison for PrP and the TSEs has proven more complex. Early attempts to identify a function for PrP were hampered by the absence of overt phenotypic defects in PrP-null mice and of conserved structural or sequence motifs.58 Despite these initial setbacks, subsequent studies have shown that PrP-null mice differ from their wildtype counterparts in many activities, including circadian cycles, neuroprotection, synaptic function, lymphocyte activation, cell adhesion, stem cell renewal and proliferation, and olfaction,58, 61-63 and two recent studies have uncovered new roles for PrP. During zebrafish development, knockdown of either of the duplicated PrP genes induces loss of cell adhesion and altered localization of E-cadherin and Fyn Tyr kinase, phenotypes that are suppressed by expression of murine PrP.64 In adult mice, regulated proteolysis and expression of PrP on the neuronal cell surface is required for maintenance of myelination through a non-cell autonomous route.65 Together, these studies may suggest that PrP has a pleiotropic role in vivo, perhaps mediating its broad effects through an activity in cell signalling pathways.66
Can TSE pathogenesis be linked to a loss-of-function or a gain-of-function phenotype for PrP? Our current state of knowledge suggests that the answer to that question lies somewhere in-between the two possibilities. While the absence of neurodegeneration in PrP-null mice was originally considered to be incompatible with a loss-of-function model for prion diseases, subsequent studies have shown that expression of some PrP fragments induces spontaneous neurodegeneration in PrP-null mice.58, 67, 68 Based on these observations, PrP toxicity during TSE infection could be explained as a loss of some PrP functions but not others.66 Consistent with this idea, PrP must be expressed on the surface of neurons to mediate TSE pathogenesis following infection,69-71 suggesting that normal localization of the protein is required to elicit neurotoxicity. However, a gain-of-function model for the TSEs, as has been suggested for the amyloidoses, cannot be ruled out at this point. In this model, conformation conversion of PrP to the prion state would be predicted to be neurotoxic to the host. The putative neurotoxic species was originally proposed to be PrPSc, which accumulates during the terminal stages of disease and in purified preparation of the infectious agent;2 however, the presence of PrPSc correlates poorly with clinical disease.72, 73 Indeed, mice that are heterozygous for a PrP disruption progress to the terminal stage of disease more slowly than their wildtype counterparts despite the accumulation of similar levels of PrPSc.74 Thus, any gain-of-function model for the role of PrP in the TSEs must identify a novel and biologically active species of this protein.
Conformational self-replication pathway
Although the exact mechanism by which misfolding of PrP alters the function of the protein to mediate TSE pathogenesis remains an open question, mounting evidence suggests that the process of conformational self-replication itself provides a robust framework for understanding the molecular basis of prion phenotypes. The prion hypothesis originally predicted that PrPSc catalyzed the conversion of the PrPC in the context of a heterodimer of the two forms.1, 3 However, subsequent studies have revealed that soluble PrPC binds to an oligomer of PrPSc, which stimulates remodelling upon incorporation into these complexes.5 Such a mechanism will progressively increase the size of prion complexes; however, without evoking a mechanism for generating new templates, this seeded polymerization process cannot account for the exponential increase in infectious titre observed over the course of disease in mammals or for the mitotic stability of prion propagation in yeast. What has emerged from mathematical models of prion propagation in both mammals and yeast is that, in addition to the seeded polymerization step, there must also be on-going polymer fragmentation to generate new templates (‘propagons’; Figure 3).75 This prediction is now well-supported by experimental studies with yeast prions, in which the fragmentation of prion polymers is catalysed by the molecular chaperone Hsp104 in conjunction with co-chaperones such as Hsp40 and Hsp70.76 In mammalian cells no orthologue of Hsp104 has been described, and it remains to be established how prion complexes are fragmented.77
Figure 3. How prions are propagated in the cell.
Conformationally flexible prion proteins are converted from their normal non-prion form to the self-replicating prion form by associating with existing oligomeric complexes of the same protein in the prion form. These extended polymers may then be dissociated into smaller units (polymer fragmentation) either enzymatically in yeast or mechanically in mammals to produce transmissible oligomers (propagons). This process of conformational self-replication is limited in efficiency by the degradation of normal conformers or of dissociated prion monomers and by the formation of large, non-transmissible aggregates ([Agg+]). Changes in the rates of these dynamic transitions create a range of protein-based phenotypes from a single prion protein.
Unlike a simple heterodimeric conversion mechanism, a multi-step pathway of conformational self-replication allows for the formation of intermediate states that could alter prion protein function and thereby explain some aspects of prion biology. For example, although the accumulation of PrPSc is inconsistent with a direct role for this conformer in TSE pathogenesis, TSE incubation times correlate with PrP expression level.74, 78, 79 How might PrP expression induce disease independently of PrPSc formation? One possibility is that a transient intermediate on the pathway to PrPSc formation mediates toxicity, a concept that may also explain pathogenesis in other non-transmissible neurodegenerative diseases5, 51, 80 and the stability of prion phenotypes in fungi.81 According to this idea, the rate of replication of the infectious PrP agent strongly influences the rate of formation of the transient toxic species, with efficient replication, such as that observed in the presence of increased PrP expression, allowing the accumulation of the intermediate form to levels sufficient to cause clinical disease.
According to this dynamic model of prion phenotypes, any condition that alters the efficiency of conformational self-replication has the potential to impact prion-associated phenotypes by modulating the proportion of the protein that is found in state that determines the phenotype. Such fine-tuning, in turn, allows an expansion of the range of phenotypes that may be conferred by a single protein without an underlying change in the genetic make-up of the host. Intriguingly, this framework provides a molecular explanation for many enigmatic aspects of prion biology.
Prion strains and evolution of their phenotypes
Prion proteins in both mammals and yeast can adopt a range of self-replicating conformers known as prion strains, which are thought to confer distinct phenotypes by assembling into aggregates with different physical properties.5, 82, 83 These differences are believed to specify unique rates of conversion of the soluble protein to the prion state and of fragmentation of prion complexes to generate new propagons, and thereby the efficiency of conformational self-replication, providing a molecular basis for the phenotypes.83
However, multiple prion strains may arise in individual mammals84, 85 through co-infection86, 87 or strain “mutation”/adaptation, where prion-associated phenotypes “evolve” in response to new conditions, such as the presence of compounds that interfere with prion propagation in vivo or to transfer to a new host.10, 88-92 The existence of mixtures of prion strains in vivo raises the possibility that prion phenotypes may not simply reflect the physical properties of a single conformer but rather the collective and dynamic behaviour of the various forms present. Indeed, prion strains have markedly different disease characteristics alone than when present in a mixture, including changes in incubation periods,84, 93 the efficacy of inoculation routes94 and transmission rates to other species.84, 85, 93 Likewise, prion disease pathology, duration and clinical symptoms in humans are altered by the coexistence of prion strains.95, 96 Thus, the interplay between prion strains directly affects prion phenotypes.
How do prion strains interact to produce observable phenotypes? As prion strains can only stably persist if their rates of replication counteract processes that lead to their decline, such as degradation in mammals or dilution through cell division in yeast,75, 83 the relative efficiencies of conformational self-replication for the interacting strains seems to be crucial in establishing the phenotype of prion mixtures. In mammals, co-inoculation with multiple prion strains that replicate at different rates almost invariably results in selection of the faster replicating strain,97, 98 but when two different prion strains are introduced at different times (known as superinfection), the outcome is influenced by the interval between inoculations, the inoculum dosage and the routes of inoculation (see below).99, 100 Conditions that allow a slower replicating prion strain to establish an infection before the introduction of a faster replicating prion strain through the same route either delay or completely block superinfection. These experiments suggest that prion strains compete for a limiting host component necessary for establishing a prion infection,101, 102 and the competition between existing strains may similarly be affected by the efficiency with which each conformer is replicated in a given tissue.94 Considering the process of conformational self-replication (Figure 3), this component is likely to be the non-prion state protein,102 a suggestion that is supported by studies in yeast showing that the prion strains that most efficiently incorporate non-prion state protein are phenotypically dominant (Figure 4A).83, 87, 103
Figure 4. Prion mixtures, strain competition and the species barrier.
a. Mixtures of prion strains in the same cell compete for non-prion state protein. Differences in the rate of fragmentation or conversion allow strains to establish phenotypic dominance by increasing their steady-state concentrations. In extreme cases, this competition leads to loss of one strain.
b. Sequence variants of prion proteins dominantly inhibit conformational replication of other alleles by incorporating into prion complexes and inhibiting further conformational conversion or fragmentation. Variants interfering with conversion exert their effects at substochiometic levels as they function at the site of conversion, whereas variants interfering with prion aggregate dynamics exert their effects at stochiometric levels since they must incorporate throughout the complex.
c. Incompatibility between donor and recipient prion proteins creates a barrier to interspecies transmission. This barrier may reflect an inability of the proteins to associate (not shown) or alternately to adopt compatible conformations. For strains capable of traversing a species barrier, the recipient prion protein may adopt an identical conformation (selection), leading to the conservation of strain identity on transfer, or, alternately, may adopt a partially compatible conformation that leads to a change in strain (adaptation).
Prion resistance and attenuation
The physical interaction between prion proteins is an essential event in conformational self-replication. Thus, it is perhaps not surprising that variations in the amino acid sequence of prion proteins affect their phenotypes in both mammals and yeast. Naturally occurring PrP polymorphisms in animals and man alter TSE characteristics and, in extreme cases, the susceptibility to prion disease.104 Emerging evidence suggests that sequence variants exert their effects by altering the efficiency of conformational self-replication, but they do so by targeting different steps in the process (Figure 4B).
The most well-characterized sequence variant of human PrP is the Met/Val 129 polymorphism.105, 106 Although both PrP129 homozygotes (Met/Met or Val/Val) and heterozygotes are susceptible to prion disease, the genotype at this position affects the pattern of PrPSc accumulation in the brain and the clinical symptoms, incubation period and duration of disease.104 Disease progression is always more rapid in homozygotes than in heterozygotes, providing a potential molecular explanation for the over-representation of homozygotes in some TSEs.104 The Met/Val129 polymorphism has no effect on the structure or stability of native PrP,107-109 but it does affect conformational self-replication in vitro. Under certain conditions, mixture of Met129 and Val129 PrPs (as would occur in heterozygotes) slows the rate of amyloid growth110, 111 and favours the formation of an oligomeric intermediate that cannot be directly converted to the amyloid state.110, 112, 113 Together, these observations suggest that mixtures of Met129 and Val129 variants may alter disease progression by limiting the rate of conversion of PrP to the prion form.
Heterozygous interference appears to be a general phenomenon, as both animals and humans heterozygous for other PrP alleles show a similar overdominance.114-119 Perhaps the most intriguing of these polymorphisms are Glu/Lys 219 in humans and Gln/Arg 171 in sheep, which confer resistance to TSEs.104, 118, 120, 121 The protective effects of these variants correlate with their ability to dominantly inhibit conformational self-replication by the other PrP alleles.122, 123 Notably, this dominant inhibition occurs independently of trans-factors, suggesting an incompatibility in the interactions between these allelic variants.124, 125 Although the mechanisms by which PrP sequence variants inhibit conformational self-replication are currently unclear, the alleles can be distinguished by the ratios, relative to wildtype protein, at which they become effective inhibitors.123, 124 According to a mathematical model, these differences in effective inhibitory concentrations may reflect a targeting of distinct events in conformational self-replication. For example, inhibitors that theoretically act by binding to the ends of linear prion complexes and blocking PrP conversion are predicted to be effective at lower concentrations than inhibitors that would interfere with other aspects of conformational self-replication such as fragmentation, which would require binding along the length of the aggregate (Figure 4B).126
How changes in replication rates translate into altered phenotypic states requires an assessment of their effects within the context of a living organism, and the yeast prion models provide avenues to further explore these questions. Dominant inhibitory mutations have been isolated in the fungal prions and, as is the case for PrP polymorphisms, these mutations interfere with conformational self-replication to varying extents, creating a range of protein-based traits.127, 128 For the Sup35/[PSI+] prion, some substitutions, known as [PSI+]-no-more (PNM) mutations, induce prion loss, and others, known as antisuppressor (ASU) mutations, modestly decrease the efficiency of conformational self-replication, preserving the prion form but allowing the accumulation of soluble and functional Sup35, which reverses the prion phenotype.127 The most extensively studied PNM mutant is PNM2, which encodes a Sup35 Gly58Asp mutant.128, 129 The Sup35 Gly58Asp mutant incorporates into wild type Sup35 prion complexes and can even support prion propagation on its own.87, 130-132 However, PNM2 induces prion loss over many generations,129, 132 suggesting that this variant interferes with the replication of prion complexes and/or their transmission to daughter cells, ideas that can be directly tested through an assessment of prion protein dynamics in vivo.
Species barrier
In animal models of prion infectivity, a barrier to interspecies transmission has long been appreciated133, 134, but renewed interest in this phenomenon has arisen with the realization that variant CJD appeared in the human population following transfer of BSE from cattle.135, 136 Mechanistic studies now suggest that species barriers are the outcomes of interactions between prion strains and sequence variants of PrP, which reflect the rate of conformational self-replication in the recipient.80
Species barriers may manifest as a complete block of disease development84, 137 or, alternatively, as a prolonged incubation period following first passage that is shortened on subsequent passage within the same species.86, 133, 134 In both cases, replication of the infectious species occurs during the asymptomatic phase,138, 139 but the toxic species apparently does not reach the threshold concentration required for clinical disease within the organism's natural lifespan.140 This alteration to the efficiency of conformational self-replication has been linked to differences in the PrP sequences of the donor and recipient. For example, the species barriers between hamster or mink and mice are abolished by heterologus expression of the donor PrP in mice,84, 141 and, strikingly, this effect has been linked to identity at two PrP residues: 170 and 174.142 What is the molecular basis of this requirement for sequence compatibility? Prion protein sequence likely contributes to species barriers in multiple ways. First, it determines the efficiency of interaction between the donor and recipient proteins. Second, it defines the range of conformations that a prion protein may adopt,5, 143 and third, it determines the stability with which a given conformer is replicated.92 Indeed, the identity of residues at PrP positions 170 and 174 define the structure of a loop that can form an intermolecular interface common to amyloidogenic proteins,144 and variations at position 226 correlate with the stability of PrP strains.92 Thus, interspecies transmissibility may be operationally defined by the ability of the recipient protein to efficiently replicate the conformation imposed by the template.
Perhaps the strongest evidence to support the idea that the efficiency of conformational replication creates the barrier to interspecies prion transmission is the observation of asymmetry in prion conversions. For example, mouse PrP can seed the formation of amyloid by Syrian hamster PrP in vitro but not vice versa.145 A comparable asymmetry has been observed between Sup35 homologues from the yeasts Kluyveromyces lactis, Saccharomyces paradoxus, or Saccharomyces bayanus and S. cerevisiae14, 146, 147, between Ure2 homolgues from S. bayanus and S. cerevisiae,148 and between fragments of Rnq1.149 Although these reciprocal cross-seeding reactions involve the same two proteins, they differ in which protein is present in the prion form and, therefore, serves as the template for conformational conversion. Consistent with this model, there is a close correlation between seeding and structure: PrP variants, which can seed one another, spontaneously form amyloid fibres of similar secondary and quaternary structure in vitro, 150 and PrP and Sup35 molecules adopt different structures depending on the template provided.150, 151
The concept of a conformational replication barrier to interspecies transmission also explains an early observation that such transitions are often accompanied by a change in prion strain.152, 153 The emergence of a new strain could reflect the selection of a compatible strain from a mixture present in the initial inoculum (Figure 4C)86 or, alternately, a conversion of one conformer to another. Indeed, biological clones of prion strains undergo adaptation and selection following transfer to a new species in vivo and in vitro98, 154 (Figure 4C). Studies in vitro suggest that these transitions may proceed through mixed complexes with heterogenous structures, reflecting only a partial compatibility between the accessible conformations of the two proteins.155 Such mixed complexes could alter the physical properties of prion aggregates and thereby the efficiency of replication in a manner analogous to that described above for dominant inhibitory mutants. Indeed, interspecies transmissions that lead to the emergence of new strains tend to require more passages to stabilize the length of the incubation period in the new host than those that retain their strain identities.98, 153, 154
Conclusions
The prion hypothesis first emerged as a radical proposal to explain the transmission of a brain disease in animals. The possibility that by simply undergoing a change in its tertiary conformation, the PrP protein could switch from a benign to a self-replicating pathogenic form that induced a long-term and progressive neurodegeneration, was nothing short of heretical. Almost 40 years on, the hypothesis has turned to accepted dogma, but a wealth of recent studies now suggest that the prion concept is entering a new realm of even greater relevance, in which this process is no longer considered simply as an explanation for an unusual and invariably fatal disease. The realisation that there are a number of prions in fungi that can radically change the phenotype of the ‘infected host’ necessitates a rethinking of the role of prions. We would suggest that prions have evolved as epigenetic regulators of phenotype rather than as disease-causing agents. Thus, paralleling the underlying process of conformational self-replication, the prion mechanism provides a robust yet dynamic system to modulate protein function and thereby cellular phenotypes. The plasticity of this pathway, its epigenetic nature, and its potential to create a continuum of related phenotypes highlight its utility as a common regulatory process that we are only now beginning to appreciate. To date the impact of prions on species other than fungi has been restricted to the role of PrPSc in mammalian brain degeneration and death. However, the possibility that synaptic activity in sensory neurons in the brain may be regulated via a prion-like mechanism53, 156 provide the first clue that prion-mediated control of complex cellular processes might also exist in higher eukaryotes.
Acknowledgements
The research on yeast prions carried out in the Tuite laboratory is supported by funding from the Biotechnology and Biological Sciences Research Council and The Wellcome Trust and in the Serio laboratory by the National Institutes of Health (NIGMS) and National Science Foundation (ADVANCE). We thank members of the Tuite and Serio labs for critical reading of this manuscript prior to submission. We also thank Sven Saupe, Susan Liebman, and Christophe Cullin for providing images used in Figure 2. We apologize that due to space constraints we were unable to directly cite all primary studies that have contributed to our understanding of prion biology and its physiological effects.
GLOSSARY
- Conformer
One of a number of different stable tertiary structures that can be taken up by a single polypeptide chain
- De novo prion formation
The establishment of the prion form of a protein without benefit of acquired prion seeds or propagons or mutation i.e. spontaneous appearance of the prion form
- Epigenetic
A change in inherited phenotype resulting from a molecular event other than a change in the DNA sequence of the organism's genome e.g. DNA methylation chromatin remodelling
- Heterokaryon
Coexistence of two or more genetically different nuclei in a common cytoplasm
- Overdominance
A heterozygous phenotype that is outside of the range of phenotypes for either homozygous state
- Prion
A structurally altered form of a protein that can impose that form on a previously correctly folded form of the same protein and hence propagate the prion form in either heredity (yeast prions) or infectious (mammalian PrP) manner
- Prionoid
Proteins that show prion-like conformational self-replication activity but that lack infectivity
- Propagon
Conformation-replicating physical entities that are transmitted from mother to daughter cells and are required to maintain the [PRION+] phenotype. Seeds is commonly used as a synonym
- Protein conformation
The stable, native three-dimensional shape taken up by a polypeptide chain that determines the alternative functions of the protein
- +1/−1 ribosomal frameshift
A net shift in the translation of an open reading frame by one base either in the 3′ direction (+1) or 5′ direction (−1) directed by a specific signal in the mRNA sequence
- Strain “mutation” or adaptation
The conversion of one prion conformer to another, either spontaneously or in response to a new environment
- Superinfection
Sequential but temporally spaced inoculations of a host with more than one strain of an infectious agent
- tRNA-mediated nonsense suppression
The translation of a termination codon by a transfer RNA with a mutation in its anticodon. This can result in partial restoration of function to the encoded protein when the codon is located within the ORF i.e. a nonsense mutation
- Vegetative incompatibility
Death of a multinucleate heterokaryon arising from the fusion of two different strains of a fungus thereby preventing the transfer of cytoplasmic components from one strain to another
REFERENCES
- 1.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–44. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–4. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 4.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–8. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–6. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 6.Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci U S A. 2009;106:1892–6. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–9. doi: 10.1126/science.7909170. The first experimenatl proof that a non-Mendelian element (in this case [URE3]) in yeast can be explained by the ‘protein only’ prion hypothesis. [DOI] [PubMed] [Google Scholar]
- 8.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–5. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–58. doi: 10.1016/j.cell.2009.02.044. A comprehensive study that reveals the diversity of potential prions in yeast and provides direct experimental proof for [MOT3+], the prion form of the transcriptional co-repressor Mot3p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox B. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 11.Culbertson MR, Charnas L, Johnson MT, Fink GR. Frameshifts and frameshift suppressors in Saccharomyces cerevisiae. Genetics. 1977;86:745–64. doi: 10.1093/genetics/86.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–6. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 13.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–34. [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci U S A. 2007;104:2791–6. doi: 10.1073/pnas.0611158104. Using the Sup35/[PSI+] yeast prion system, the authors demonstrate that the barrier to interspecies prion transmission occurs at the point of conformational replication rather than simply the binding of two different conformers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–7. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 17.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–83. doi: 10.1038/35035005. An analysis of a variety of phenotypes that differentiate [PSI+] from [psi−] cells leads to the proposal that the [PSI+] prion provides the means to uncover hidden genetic variation and produce new heritable phenotypes. [DOI] [PubMed] [Google Scholar]
- 18.Williams I, Richardson J, Starkey A, Stansfield I. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:6605–16. doi: 10.1093/nar/gkh1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson MA, Meaux S, Parker R, van Hoof A. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc Natl Acad Sci U S A. 2005;102:10244–9. doi: 10.1073/pnas.0504557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palanimurugan R, Scheel H, Hofmann K, Dohmen RJ. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004;23:4857–67. doi: 10.1038/sj.emboj.7600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namy O, et al. Epigenetic control of polyamines by the prion [PSI+] Nat Cell Biol. 2008;10:1069–75. doi: 10.1038/ncb1766. The authors show that [PSI+] enhances the synthesis of antizyme, a regulator of polyamine synthesis, via a -1 frameshift event. This in turn leads to modulation of the levels of polyamines in the yeast cell that can lead to a range of phenotypes including some of those described by True & Lindquist (ref 17) [DOI] [PubMed] [Google Scholar]
- 22.Namy O, Duchateau-Nguyen G, Rousset JP. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol Microbiol. 2002;43:641–52. doi: 10.1046/j.1365-2958.2002.02770.x. [DOI] [PubMed] [Google Scholar]
- 23.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–5. doi: 10.1038/ng.112. The description of a yeast prion that is formed by a key chromatin remodeling factor and thus reveals a possible link between global transcriptional regulation and the ability of the Swi1 protein to undergo conformational conversion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–9. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hongay C, Jia N, Bard M, Winston F. Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. EMBO J. 2002;21:4114–24. doi: 10.1093/emboj/cdf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–63. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Courchesne WE, Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J Bacteriol. 1988;170:708–13. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunningham TS, Andhare R, Cooper TG. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J Biol Chem. 2000;275:14408–14. doi: 10.1074/jbc.275.19.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai R, Tate JJ, Cooper TG. Ure2, a prion precursor with homology to glutathione S-transferase, protects Saccharomyces cerevisiae cells from heavy metal ion and oxidant toxicity. J Biol Chem. 2003;278:12826–33. doi: 10.1074/jbc.M212186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogoza T, et al. Non-Mendelian determinant [ISP +] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci U S A. 2010;107:10573–7. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saupe SJ. A short history of small s: a prion of the fungus Podospora anserina. Prion. 2007;1:110–5. doi: 10.4161/pri.1.2.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A. 1997;94:9773–8. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paoletti M, Saupe SJ. Fungal incompatibility: evolutionary origin in pathogen defense? Bioessays. 2009;31:1201–10. doi: 10.1002/bies.200900085. [DOI] [PubMed] [Google Scholar]
- 34.Greenwald J, et al. The mechanism of prion inhibition by HET-S. Mol Cell. 2010;38:889–99. doi: 10.1016/j.molcel.2010.05.019. The authors show that the inhibition of the fibrillization of Podospora anserina HET-s by the closely related HET-S protein is not encoded by the structural differences per se. Rather, it reflects and effect on the stability and oligomerisation properties of the mixed aggregates formed between the two proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–72. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 36.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN] Cell. 2001;106:171–82. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 37.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–94. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 38.Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol Microbiol. 2003;49:1005–17. doi: 10.1046/j.1365-2958.2003.03608.x. [DOI] [PubMed] [Google Scholar]
- 39.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–80. doi: 10.1073/pnas.0504882102. This paper opens up the debate about the impact of yeast prions on the host and argues that because neither the [URE3] or [PSI+] prions are present in 70 different ‘natural’ strains they must have a negative effect on the host. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derkatch IL, et al. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A. 2004;101:12934–9. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derkatch IL, et al. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J. 2000;19:1942–52. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalstra HJ, Swart K, Debets AJ, Saupe SJ, Hoekstra RF. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci U S A. 2003;100:6616–21. doi: 10.1073/pnas.1030058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ter-Avanesyan MD, et al. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–92. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 44.Griswold CK, Masel J. Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet. 2009;5:e1000517. doi: 10.1371/journal.pgen.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph SB, Kirkpatrick M. Effects of the [PSI+] prion on rates of adaptation in yeast. J Evol Biol. 2008;21:773–80. doi: 10.1111/j.1420-9101.2008.01515.x. [DOI] [PubMed] [Google Scholar]
- 46.Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11:301–7. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer-Luehmann M, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–4. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 48.Ren PH, et al. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol. 2009;11:219–25. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li JY, et al. Lewy bodies in grafted neurons in subjects with Parkinson's disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–3. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 50.Aguzzi A. Cell biology: Beyond the prion principle. Nature. 2009;459:924–5. doi: 10.1038/459924a. [DOI] [PubMed] [Google Scholar]
- 51.Glabe CG. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol Aging. 2006;27:570–5. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–47. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–35. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Fowler DM, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berson JF, et al. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol. 2003;161:521–33. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maji SK, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–32. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carulla N, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–8. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 58.Aguzzi A, Baumann F, Bremer J. The prion's elusive reason for being. Annu Rev Neurosci. 2008;31:439–77. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 59.Mallucci GR, et al. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–35. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 60.White MD, et al. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci U S A. 2008;105:10238–43. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steele AD, Lindquist S, Aguzzi A. The prion protein knockout mouse: a phenotype under challenge. Prion. 2007;1:83–93. doi: 10.4161/pri.1.2.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isaacs JD, Jackson GS, Altmann DM. The role of the cellular prion protein in the immune system. Clin Exp Immunol. 2006;146:1–8. doi: 10.1111/j.1365-2249.2006.03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson DA, Nixon RA. Sniffing out a function for prion proteins. Nat Neurosci. 2009;12:7–8. doi: 10.1038/nn0109-7. [DOI] [PubMed] [Google Scholar]
- 64.Malaga-Trillo E, et al. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009;7:e55. doi: 10.1371/journal.pbio.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bremer J, et al. Axonal prion protein is required for peripheral myelin maintenance. Nat Neurosci. 2010;13:310–8. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 66.Westergard CL, Christensen HM, Harris DA. The cellular prion protein (PrP ): its physiological function and role in disease. Biochim Biophys Acta. 2007;1772:629–44. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shmerling D, et al. Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell. 1998;93:203–14. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]
- 68.Baumann F, et al. Lethal recessive myelin toxicity of prion protein lacking its central domain. EMBO J. 2007;26:538–547. doi: 10.1038/sj.emboj.7601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mallucci G, et al. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–4. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 70.Brandner S, et al. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–43. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 71.Chesebro B, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–9. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 72.Nicoll AJ, Collinge J. Preventing prion pathogenicity by targeting the cellular prion protein. Infect Disord Drug Targets. 2009;9:48–57. doi: 10.2174/1871526510909010048. [DOI] [PubMed] [Google Scholar]
- 73.Radford HE, Mallucci GR. The Role of GPI-anchored PrPC in Mediating the Neurotoxic Effect of Scrapie Prions in Neurons. Curr Issues Mol Biol. 2009;12:119–128. [PubMed] [Google Scholar]
- 74.Bueler H, et al. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- 75.Masel J, Jansen VA, Nowak MA. Quantifying the kinetic parameters of prion replication. Biophys Chem. 1999;77:139–52. doi: 10.1016/s0301-4622(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 76.Haslberger T, Bukau B, Mogk A. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem Cell Biol. 2010;88:63–75. doi: 10.1139/o09-118. [DOI] [PubMed] [Google Scholar]
- 77.Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–32. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- 78.Prusiner S, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–86. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 79.Manson JC, Clarke AR, McBride PA, McConnell I, Hope J. PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration. 1994;3:331–40. [PubMed] [Google Scholar]
- 80.Laurent M. Bistability and the species barrier in prion diseases: stepping across the threshold or not. Biophys Chem. 1998;72:211–22. doi: 10.1016/s0301-4622(98)00135-5. [DOI] [PubMed] [Google Scholar]
- 81.Sindi SS, Serio TR. Prion dynamics and the quest for the genetic determinant in protein-only inheritance. Curr Opin Microbiol. 2009;12:623–30. doi: 10.1016/j.mib.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Legname G, et al. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci U S A. 2006;103:19105–10. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–9. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 84.Kimberlin RH, Walker CA. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol. 1978;39:487–96. doi: 10.1099/0022-1317-39-3-487. [DOI] [PubMed] [Google Scholar]
- 85.Marsh RF, Hanson RP. In: Slow Transmissible Diseases of the Nervous System. Prusiner SB, Hadlow WJ, editors. Academy Press; New York: 1979. pp. 451–460. [Google Scholar]
- 86.Dickinson AG. In: Slow virus diseases of animals and man. Kimberlin RH, editor. North-Holland Publishing Company; Amsterdam: 1976. pp. 209–241. [Google Scholar]
- 87.Derkatch IL, Bradley ME, Zhou P, Liebman SW. The PNM2 mutation in the prion protein domain of SUP35 has distinct effects on different variants of the [PSI+] prion in yeast. Curr Genet. 1999;35:59–67. doi: 10.1007/s002940050433. [DOI] [PubMed] [Google Scholar]
- 88.Ghaemmaghami S, et al. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009;5:e1000673. doi: 10.1371/journal.ppat.1000673. Quinacrine treatment induces the loss of some prion strains while promoting the amplification of others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manuelidis L, Fritch W, Xi YG. Evolution of a strain of CJD that induces BSE-like plaques. Science. 1997;277:94–8. doi: 10.1126/science.277.5322.94. [DOI] [PubMed] [Google Scholar]
- 90.Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010;327:869–72. doi: 10.1126/science.1183218. The frequency of prion strain mutation can be influenced by selective pressures, including host identity and exposure to chemical treatments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bruce ME. Scrapie strain variation and mutation. British Medical Bulletin. 1993;49:822–38. doi: 10.1093/oxfordjournals.bmb.a072649. [DOI] [PubMed] [Google Scholar]
- 92.Angers RC, et al. Prion Strain Mutation Determined by Prion Protein Conformational Compatibility and Primary Structure. Science. 2010 doi: 10.1126/science.1187107. The frequency of prion strain mutation for Chronic Wasting Disease has been linked to a change in PrP sequence that does not appear to alter the conformation of the prion form. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bessen RA, Marsh RF. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol. 1992;73:329–34. doi: 10.1099/0022-1317-73-2-329. [DOI] [PubMed] [Google Scholar]
- 94.Dickinson AG, Outram GW. In: Virus nonconventionnels et affections du systeme nerveux central. Court LA, Cathala F, editors. Masson; Paris: 1983. pp. 3–16. [Google Scholar]
- 95.Parchi P, et al. Incidence and spectrum of sporadic Creutzfeldt-Jakob disease variants with mixed phenotype and co-occurrence of PrPSc types: an updated classification. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0585-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cali I, et al. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effect on the phenotype and prion-type characteristics. Brain. 2009 doi: 10.1093/brain/awp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bruce M, Fraser H, McBride P, Scott J, Dickinson A. In: Prion Diseases of Humans and Animals. Prusiner S, Collinge J, Powell J, Anderton B, editors. Ellis Horwood; New York: 1992. pp. 497–508. [Google Scholar]
- 98.Bartz JC, Bessen RA, McKenzie D, Marsh RF, Aiken JM. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible mink encephalopathy. J Virol. 2000;74:5542–7. doi: 10.1128/jvi.74.12.5542-5547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dickinson AG, et al. Extraneural competition between different scrapie agents leading to loss of infectivity. Nature. 1975;253:556. doi: 10.1038/253556a0. [DOI] [PubMed] [Google Scholar]
- 100.Dickinson AG, Fraser H, Meikle VM, Outram GW. Competition between different scrapie agents in mice. Nat New Biol. 1972;237:244–5. doi: 10.1038/newbio237244a0. [DOI] [PubMed] [Google Scholar]
- 101.Dickinson AG, Outram GW. In: Slow Transmissible Diseases of The Nervous System. Prusiner SB, Hadlow WJ, editors. Academic Press; New York: 1979. pp. 13–31. [Google Scholar]
- 102.Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. Coinfecting prion strains compete for a limiting cellular resource. J Virol. 2010;84:5706–14. doi: 10.1128/JVI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99 Suppl 4:16392–9. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morales R, Abid K, Soto C. The prion strain phenomenon: molecular basis and unprecedented features. Biochim Biophys Acta. 2007;1772:681–91. doi: 10.1016/j.bbadis.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goldfarb L, et al. Patients wth Creutzfeldt-Jakob disease and kuru lack the mutation in the PRIP gene found in Gerstmann-Straussler syndrome, but they show a different double-allele mutation in the same gene. Am J Hum Genet. 1989;45:A189. [Google Scholar]
- 106.Owen F, Poulter M, Collinge J, Crow TJ. Codon 129 changes in the prion protein gene in Caucasians. Am J Hum Genet. 1990;46:1215–6. [PMC free article] [PubMed] [Google Scholar]
- 107.Liemann S, Glockshuber R. Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry. 1999;38:3258–67. doi: 10.1021/bi982714g. [DOI] [PubMed] [Google Scholar]
- 108.Hosszu LL, et al. The residue 129 polymorphism in human prion protein does not confer susceptibility to Creutzfeldt-Jakob disease by altering the structure or global stability of PrPC. J Biol Chem. 2004;279:28515–21. doi: 10.1074/jbc.M313762200. [DOI] [PubMed] [Google Scholar]
- 109.Apetri AC, Vanik DL, Surewicz WK. Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90-231. Biochemistry. 2005;44:15880–8. doi: 10.1021/bi051455+. [DOI] [PubMed] [Google Scholar]
- 110.Baskakov IV, Legname G, Baldwin MA, Prusiner SB, Cohen FE. Pathway complexity of prion protein assembly into amyloid. J Biol Chem. 2002;277:21140–8. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 111.Come J, Lansbury P. Predisposition of Prion Protein Homozygotes to Creudtzfeldt-Jakob Disease Can Be Explained by a Nucleation-Dependent Polymerization Mechanism. J Am Chem Soc. 1994;116:4109–10. [Google Scholar]
- 112.Tahiri-Alaoui A, Gill AC, Disterer P, James W. Methionine 129 variant of human prion protein oligomerizes more rapidly than the valine 129 variant: implications for disease susceptibility to Creutzfeldt-Jakob disease. J Biol Chem. 2004;279:31390–7. doi: 10.1074/jbc.M401754200. [DOI] [PubMed] [Google Scholar]
- 113.Tahiri-Alaoui A, Sim VL, Caughey B, James W. Molecular heterosis of prion protein beta-oligomers. A potential mechanism of human resistance to disease. J Biol Chem. 2006;281:34171–8. doi: 10.1074/jbc.M606606200. [DOI] [PubMed] [Google Scholar]
- 114.Jeong BH, et al. Association of sporadic Creutzfeldt-Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics. 2005;6:229–32. doi: 10.1007/s10048-005-0016-y. [DOI] [PubMed] [Google Scholar]
- 115.Westaway D, et al. Distinct prion proteins in short and long scrapie incubation period mice. Cell. 1987;51:651–62. doi: 10.1016/0092-8674(87)90134-6. [DOI] [PubMed] [Google Scholar]
- 116.Dickinson AG, Meikle VM, Fraser H. Identification of a gene which controls the incubation period of some strains of scrapie agent in mice. J Comp Pathol. 1968;78:293–9. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- 117.Foster JD, Dickinson AG. The unusual properties of CH1641, a sheep-passaged isolate of scrapie. Vet Rec. 1988;123:5–8. doi: 10.1136/vr.123.1.5. [DOI] [PubMed] [Google Scholar]
- 118.Goldmann W, Hunter N, Smith G, Foster J, Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J Gen Virol. 1994;75(Pt 5):989–95. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 119.Gordon W. ARS91-53: Report of Scrapie Seminar. US Deparment of Agriculture; 1964. pp. 53–68. [Google Scholar]
- 120.Shibuya S, Higuchi J, Shin RW, Tateishi J, Kitamoto T. Codon 219 Lys allele of PRNP is not found in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1998;43:826–8. doi: 10.1002/ana.410430618. [DOI] [PubMed] [Google Scholar]
- 121.Westaway D, et al. Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptible to natural scrapie. Genes Dev. 1994;8:959–69. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 122.Kaneko K, et al. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci U S A. 1997;94:10069–74. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Perrier V, et al. Dominant-negative inhibition of prion replication in transgenic mice. Proc Natl Acad Sci U S A. 2002;99:13079–84. doi: 10.1073/pnas.182425299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Geoghegan JC, Miller MB, Kwak AH, Harris BT, Supattapone S. Trans-dominant inhibition of prion propagation in vitro is not mediated by an accessory cofactor. PLoS Pathog. 2009;5:e1000535. doi: 10.1371/journal.ppat.1000535. Using PrP purified from mammalian cells and the PMCA approach, the authors demonstrate that dominant negative inhibition of prion propagation by PrP mutant can be recapitulated in vitro in the absence of trans factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee CI, Yang Q, Perrier V, Baskakov IV. The dominant-negative effect of the Q218K variant of the prion protein does not require protein X. Protein Sci. 2007;16:2166–73. doi: 10.1110/ps.072954607. Using recombinant protein purified from bacteria, the authors demonstrate that the Q218K variant of PrP interferes with amyloid formation by wildtype PrP in vitro, and their studies suggest that the inhibition occur not by an effect on binding but rather on conformational conversion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Masel J, Jansen VA. Designing drugs to stop the formation of prion aggregates and other amyloids. Biophys Chem. 2000;88:47–59. doi: 10.1016/s0301-4622(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 127.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–52. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 128.Young C, Cox B. Extrachromosomal Elements in A Super-Suppression System of Yeast. I. A Nuclear Gene Controlling The Inheritance of the Extrachromosomal Elements. Heredity. 1971;26:413–422. [Google Scholar]
- 129.Doel SM, McCready SJ, Nierras CR, Cox BS. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–70. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.King CY. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol. 2001;307:1247–60. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- 131.Kochneva-Pervukhova NV, et al. Mechanism of inhibition of [PSI+] prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J. 1998;17:5805–10. doi: 10.1093/emboj/17.19.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zlotnik I, Rennie JC. Experimental Transmission of Mouse Passaged Scrapie to Goats, Sheep, Rats and Hamsters. J Comp Pathol. 1965;75:147–57. doi: 10.1016/0021-9975(65)90005-8. [DOI] [PubMed] [Google Scholar]
- 134.Pattison I. NINDB Monograph, No2: Slow, Latent, and Temperate Virus Infections, National Institutes of Health; Washington, D.C.: 1965. pp. 249–57. [Google Scholar]
- 135.Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–90. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 136.Bruce ME, et al. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 137.Marsh RF, Burger D, Eckroade R, Zu Rhein GM, Hanson RP. A preliminary report on the experimental host range of the transmissible mink encephalopathy agent. J Infect Dis. 1969;120:713–9. doi: 10.1093/infdis/120.6.713. [DOI] [PubMed] [Google Scholar]
- 138.Hill AF, et al. Species-barrier-independent prion replication in apparently resistant species. Proc Natl Acad Sci U S A. 2000;97:10248–53. doi: 10.1073/pnas.97.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Race R, Chesebro B. Scrapie infectivity found in resistant species. Nature. 1998;392:770. doi: 10.1038/33834. [DOI] [PubMed] [Google Scholar]
- 140.Dickinson AG, Fraser H, Outram GW. Scrapie incubation time can exceed natural lifespan. Nature. 1975;256:732–3. doi: 10.1038/256732a0. [DOI] [PubMed] [Google Scholar]
- 141.Windl O, et al. Breaking an absolute species barrier: transgenic mice expressing the mink PrP gene are susceptible to transmissible mink encephalopathy. J Virol. 2005;79:14971–5. doi: 10.1128/JVI.79.23.14971-14975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sigurdson CJ, et al. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest. 2010;120:2590–9. doi: 10.1172/JCI42051. The authors link the barrier to interspecies prion transmission to the structure of a loop with the PrP protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–56. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 144.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–7. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 145.Vanik DL, Surewicz KA, Surewicz WK. Molecular basis of barriers for interspecies transmissibility of mammalian prions. Mol Cell. 2004;14:139–45. doi: 10.1016/s1097-2765(04)00155-8. [DOI] [PubMed] [Google Scholar]
- 146.Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell. 2001;7:1121–30. doi: 10.1016/s1097-2765(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 147.Chen B, et al. Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009;181:1159–67. doi: 10.1534/genetics.108.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kadnar ML, Articov G, Derkatch IL. Distinct type of transmission barrier revealed by study of multiple prion determinants of Rnq1. PLoS Genet. 2010;6:e1000824. doi: 10.1371/journal.pgen.1000824. Despite the absence of amino acid substitutions, the authors uncover a prion transmission barrier between fragments of the Rnq1 prion of S. cerevisiae, suggesting that protein conformation is the key determinant in prion compatibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jones EM, Surewicz WK. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 151.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pattison IH, Jones KM. Modification of a strain of mouse-adapted scrapie by passage through rats. Res Vet Sci. 1968;9:408–10. [PubMed] [Google Scholar]
- 153.Kimberlin RH, Walker CA, Fraser H. The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J Gen Virol. 1989;70 ( Pt 8):2017–25. doi: 10.1099/0022-1317-70-8-2017. [DOI] [PubMed] [Google Scholar]
- 154.Kimberlin RH, Cole S, Walker CA. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol. 1987;68 ( Pt 7):1875–81. doi: 10.1099/0022-1317-68-7-1875. [DOI] [PubMed] [Google Scholar]
- 155.Makarava N, Ostapchenko VG, Savtchenko R, Baskakov IV. Conformational switching within individual amyloid fibrils. J Biol Chem. 2009;284:14386–95. doi: 10.1074/jbc.M900533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Si K, Lindquist S, Kandel ER. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell. 2003;115:879–91. doi: 10.1016/s0092-8674(03)01020-1. The first clues using expression in yeast, of the existence of a prion-based mechanism in the sensory neurons of Aplysia (sea slug). In this case the translational regulator CPEB undergoes a conformational switch to a self perpetuating amyloid that potentially impacts on synpatic efficiency and thus long term facilitation. [DOI] [PubMed] [Google Scholar]
- 157.Bernardi G. Lessons from a small, dispensable genome: the mitochondrial genome of yeast. Gene. 2005;354:189–200. doi: 10.1016/j.gene.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 158.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–30. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]