Abstract
AdnectinsTM are a new family of therapeutic proteins based on the 10th fibronectin type III domain, and designed to bind with high affinity and specificity to therapeutically relevant targets. Adnectins share with antibody variable domains a beta-sheet sandwich fold with diversified loops, but differ from antibodies in primary sequence and have a simpler, single-domain structure without disulfide bonds. As a consequence, Adnectins bind targets with affinity and specificity as high as those of antibodies, but are easier to manipulate genetically and compatible with bacterial expression systems. Adnectins that bind macromolecular targets with nanomolar and picomolar affinity have been selected using in vitro evolution methods, including mRNA display, phage display and yeast display. CT-322, a PEGylated, anti-angiogenic Adnectin that binds vascular endothelial growth factor (VEGF) receptor 2 and blocks its interaction with VEGF A, C and D, is being evaluated in Phase II clinical trials for efficacy in several oncology indications.
Keywords: 10Fn3, Adnectin, Engineered therapeutic proteins, mRNA display, PROfusion
Antibodies and designed target-binding proteins
Over the past 20 years, antibodies that bind therapeutically relevant targets have become the fastest growing class of protein drugs, with ∼30 approved therapeutic antibodies on the market and ∼200 currently in clinical trials (Beck et al., 2010). Two of the reasons for this success are the high affinity and exquisite specificity of antibodies, which allow therapy to be targeted to specific cells or signaling pathways. In addition, since adaptive antibodies are part of the natural human response to infection, antibody drugs of mostly human molecular origin tend to have relatively low immunogenicity, a long half-life in the bloodstream and low toxicity due to limited off-target effects.
Discovery of therapeutic antibodies is a lengthy and complex process. The traditional hybridoma technology (Kohler and Milstein, 1975) consists of evoking a rodent immune response against the antigen of interest; screening myeloma-fused B-cells to identify those producing functional antibodies; and replacing the majority of the active rodent antibody sequence with analogous human sequence, preserving only the residues involved in target binding (humanization). Humanization is not required when the immunized rodents are transgenic and carry genetic material that encodes a human antibody repertoire (Lonberg, 2008). The most common alternative process, in vitro evolution, starts with construction of a library of antibody fragments, which can be obtained by capturing the natural diversity of antibody variable domains from human donors, by diversifying their sequences synthetically, or by combining the two approaches. The resulting library is then used to select the combination of variable heavy and light chains that bind the target antigen, using a display technology such as phage display (Bradbury and Marks, 2004; Thie et al., 2008), yeast display (Chao et al., 2006), mRNA display or ribosome display (Lipovsek and Pluckthun, 2004; Groves and Osbourn, 2005). Finally, the selected variable domains are reformatted into full-length antibodies. Engineered full-length monoclonal antibodies identified by either hybridoma technology or by in vitro evolution contain both variable domains that mediate target recognition and constant domains that mediate effector function such as recruitment of other components of the immune system. Almost invariably, engineered full-length antibodies are produced in mammalian cell culture.
The success of therapeutic monoclonal antibodies has sparked a growing interest in creating streamlined molecules that retain the tight and specific target binding, low toxicity and low immunogenicity of antibodies, but are faster to discover as well as easier and less expensive to manufacture. In addition, there is an interest in developing smaller target-binding proteins that may penetrate tissues faster, and that lack the Fc-mediated effector function, which is unnecessary in a simple antagonist of receptor–ligand interactions or in a delivery vehicle for a toxic payload. The final objective for the next generation of target-binding therapeutic proteins is modularity: the ability for proteins with different binding specificities to be genetically linked in order to generate bi- or multi-specific molecules, an engineering task that is challenging for traditional, full-length antibodies.
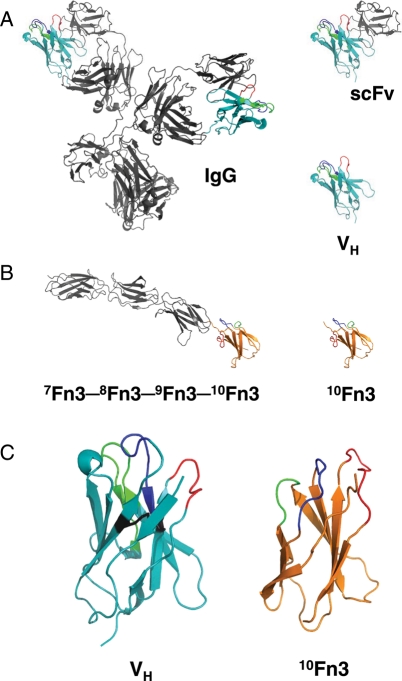
These considerations first led to the development of small engineered antibody fragments, including single-chain antibodies (Huston et al., 1991), which comprise two genetically fused antibody variable domains, one from the heavy chain and one from the light chain; and domain antibodies (Holt et al., 2003) and nanobodies (Vincke et al., 2009), which contain a single variable domain of human or humanized camelid origin, respectively (Fig. 1a).
Fig. 1.
Engineered antibodies and 10Fn3-based target-binding proteins in context. (A) Comparison of three-dimensional structures of a full-length monoclonal, IgG, antibody (PDB ID: 1ITGY; Harris et al., 1998), single-chain antibody (scFv) and domain antibody (VH; shown in teal). Complementarity-determining regions of the heavy-chain variable domains are shown in blue (CDR-H1), green (CDR-H2) and red (CDR-H3). (B) Comparison of three-dimensional structures of a fragment of human fibronectin (type III domains 7−10) (PDB ID: 1FNF; Leahy et al., 1996) and of a single 10th fibronectin type III domain (10Fn3; shown in orange). The three loops analogous to CDRs typically diversified in 10Fn3-based libraries are shown in blue (BC), green (DE) and red (FG). (C) Detailed comparison of three-dimensional structures of VH and of 10Fn3, showing diversified loops [colored as in (a) and (b)] and the disulfide bond in the VH domain (black).
In an even further departure from natural antibodies, the drive for simplicity, modularity, ease of production and favorable biophysical properties has led to the invention of designed target-binding proteins, also known as engineered scaffolds and antibody mimics (Binz and Pluckthun, 2005; Gebauer and Skerra, 2009). Designed binding proteins are relatively small, single-chain and single-domain proteins with engineered sequence diversity that allows high-affinity, specific binding to a wide range of target antigens. They are highly stable, soluble and well expressed in microbial systems. They can be selected from in vitro-generated libraries in their final therapeutic format, ensuring a fast discovery process, and they can be linked to create multi-specific therapeutics. Owing to their small size, most therapeutic designed target-binding proteins need to be modified to avoid fast renal clearance. Popular examples of this approach are chemical modification such as PEGylation and genetic linkage to a protein domain that binds a component of plasma, such as human serum albumin (Holt et al., 2008).
Adnectins, a family of designed proteins based on the framework of the 10th human fibronectin type III domain (10Fn3; Fig. 1b and c), are among the earliest advanced designed binding proteins, and have progressed the furthest in clinical studies.
10Fn3 as the starting point for Adnectin design
The 10th fibronectin type III domain was utilized as the starting point for the design of a family of target-binding proteins due to its structural similarity to antibody variable domains, suitability for modular assembly into multi-functional molecules, favorable biophysical properties and its abundance in human blood and extracellular matrix, which demonstrates that inherently this scaffold is not toxic or immunogenic.
Despite the lack of significant sequence homology, antibody variable domains and 10Fn3 (Main et al., 1992; Dickinson et al., 1994) have similar structures. As is illustrated in Fig. 1c, both antibody variable domains and 10Fn3 are sandwiches of two anti-parallel beta sheets, with solvent-accessible loops at each pole of the domain. One structural difference between the two is that, whereas the two beta sheets in antibody domains are linked through a disulfide bridge, 10Fn3 contains no disulfides or free cysteines (Fig. 1C). As a consequence, 10Fn3 retains its high thermostability [as manifested in a melting temperature above 80°C and free energy of unfolding between 6 and 9 kcal/mol (Plaxco et al., 1997; Cota et al., 2000; Batori et al., 2002)] under reducing conditions, and can be produced with a high yield in bacteria. Another difference is that antibody variable domains are somewhat larger than 10Fn3, with two more beta strands and an extra turn, which is not typically involved in antigen binding.
Loops BC, DE and FG of 10Fn3 are structurally analogous to the antibody complementarity-determining regions (CDR) H1, H2 and H3, respectively, and are thus the obvious candidates for diversification to generate artificial target-binding surfaces. The three loops at the opposite pole of 10Fn3, AB, CD and EF, are also candidates for diversification (Koide et al., 2002; Bloom and Calabro, 2009). Sequence alignment of 10Fn3 to other fibronectin type III domains reveals a divergence in loop sequence and length (Dickinson et al., 1994), suggesting that the 10Fn3 fold is likely to be compatible with artificial diversification of the six loops. The tolerance of five of the six 10Fn3 loops, AB, BC, CD, DE and FG, to re-engineering has been confirmed by the stability of 10Fn3 mutants following insertion of four glycines into these loops (Batori et al., 2002) or following random mutagenesis of BC and FG loops (Olson and Roberts, 2007). Even more relevant is the ability of several different groups to select and engineer 10Fn3-derived Adnectins with mutated BC, DE and FG loops, and, in one case, with mutated AB loop (Table I) (Koide et al., 1998, 2002, 2007; Xu et al., 2002; Richards et al., 2003; Karatan et al., 2004; Parker et al., 2005; Getmanova et al., 2006; Lipovsek et al., 2007; Gilbreth et al., 2008; Hackel et al., 2008; Olson et al., 2008; Liao et al., 2009; Hackel and Wittrup, 2010; Hackel et al., 2010; Wojcik et al., 2010). Whereas thermostability and solubility of 10Fn3 can decrease once its loops are replaced to allow target binding, the extremely high stability of the wild type means that even destabilized variants can be sufficiently stable for therapeutic applications (Parker et al., 2005; Hackel et al., 2008, 2010; Hackel and Wittrup, 2010). In addition, several studies have demonstrated the utility of approaches based on in vitro selection and on directed engineering that can increase the stability of wild-type 10Fn3 and its target-binding mutants (Koide et al., 2001; Dutta et al., 2005; Olson et al., 2008).
Table I.
Published selections of 10Fn3-based target-binding proteins (in chronological order of publication)
| Reference | Target | Display method | Size of naïve library | Diversi-fication method | ABa | BCa | DEa | FGa | Aff. mat.b | Scaff. mut.c | Lowest Kd (M)d | Tm (°C)e | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Koide et al. (1998) | Ubiquitin | Phage display | 108 | NNKf | — | AVTVR X5 | GRGDSPAS X5 | No | No | 10−6 (IC50) | NDg | ||
| Koide et al. (2002) | Estrogen receptor α-ligand | Yeast two-hybrid | 106 | NNK/NNSf | — | — | — | RGDSPAS X7 | No | No | ND | ND | Conformation-specific, binds receptor in yeast cells |
| Koide et al. (2002) | Estrogen receptor α ligand | Yeast two-hybrid | 105 | NNK/NNSf | TPTS TPX7TS | — | — | — | No | No | ND | ND | Fewer solutions than for FG-based library; not conformation-specific |
| Xu et al. (2002) | TNF-α | mRNA display | 1012 | NNSf | — | DAPAVTV X7 | GSKS X4 | GRGDSPASSK X10 | Yes | Multiple | 10−11 | ND | Spontaneous deletions in loops favored |
| Richards et al. (2003) | αvβ3 integrin | Phage display | 109 | NNKf | — | — | — | GRGDSPAS XRGDXXXX | No | No | 10−9 (IC50) | ND | Inhibit αvβ3-cell binding |
| Karatan et al. (2004) | Src SH3 | Phage display | 109 | NNKf | — | AVTVR X5 | — | GRGDSPAS X5 | No | Yes | 10−7 | ND | Detect western blots |
| Getmanova et al. (2006) | VEGF-R2 | mRNA display | 1013 | NNSf | — | DAPAVTV X7 | GSKS X4 | GRGDSPASSK X10 | Yes | Rare | 10−10 | 32–62h | VEGF antagonists; cell-based IC50= 10−9 M |
| Lipovsek et al. (2007) | Lysozyme | Yeast display | 108 | 20-codon mix | — | DAPAVTV X7 | — | GRGDSPA X7 | Yes | Rare | 10−10 | ND | Likely BC-FG disulfide |
| Koide et al. (2007) | MBP, hSUMO4, ySUMO | Phage, yeast display | 1010 | Tyr/Ser (TMT) | — | PAVTVR (Y/S)4–8VS | GSKS (G/Y/S)(Y/S)3–7 | VTGRGDSPA (Y/S)9–13 | No | D3S, D7K | 10−8 | ND | Crystal structure of MBP-Fn fusion |
| Hackel et al. (2008) | Lysozyme | Yeast display | 107 | NNBf | — | DAPAVTVR X6–9 | GSKST X4–7 | GRGDSPASSK X5,6,8,10 | Yes | Multiple | 10−12 | 51–60 | Extensive affinity maturation |
| Olson et al. (2008) | Phospho-IκBα | mRNA display | 1013 | NNSf | — | DAPAVTV X7 | — | GRGDSPASSK X10 | No | ▵(1−7)i Yes | 10−8 | ND | Optimized for solubility; phospho-specific |
| Liao et al. (2009) | SARS N protein | mRNA display | 1012 | NNSf | — | DAPAVTV X7 | — | GRGDSPASSK X10 | No | ▵(1−7) Yes | 10−9 | ND | Inhibit viral replication |
| Gilbreth et al. (2008) | MBP | Phage, yeast display | 1010 | 9-codon mix biased towards Tyr, Ser, Gly | — | PAVTVR X4–8VS | GSKS X4–8 | VTGRGDSPA X9–13 | No | D3S, D7K | 10−8 | ND | Higher affinity than in MBP binders using binary Y/S diversity. Crystal structures. |
| Hackel and Wittrup (2010) | IgG | Yeast display | 108 | NNB (X); Tyr/Ser | — | DAPAVTVRY X6–9Y (Y/S)7–10 | GSKST X4–7 gsX0,1,3stj | GRGDSPASSK X5,6,8,10 (Y/S)6,7,8,10 | Yes | Multiple | 10−10 (NNB) 10−8 (S/Y) | 45–64 | NNB diversity and wt-or-shorter loops favored |
| Wojcik et al. (2010) | Abl SH2 | Phage display | 1010 | 19-codon mix biased toward Tyr, Ser, Gly | — | PAVTVR X4–8VX | GSKS (G/Y/S)(Y/S)3 | VTGRGDSPASSK X7–13 | Yes | D3S, D7K | 10−8 | ND | Inhibits Abl autophosphorylation and signaling. Crystal structure |
| Hackel et al. (2010) | EGFR, A33, HSA, FcγIIa, FcγIIIa, IgG | Yeast display | 108 | NNB, Tyr/Ser, ‘G4’k | — | DAPAVTVRY X7–10 | GSKST gsX0,1,3stj | GRGDSPASSK X5,6,8,10 | Yes | Multiple | 10−10 | 53–73 | ‘G4’ library outperformed NNB and Y/S |
aAB, BC, DE, FG: 10Fn3 loops diversified to make the original, naïve library. The peptide sequence shown in the top line marks the wild-type 10Fn3 positions replaced by a mixture of residues; the second line defines the nature of diversification. bAffinity maturation used—i.e. populations or individual clones selected from the naïve library were re-diversified and subjected to another selection to yield variants with even higher affinity for the target. cScaff. mut.: Scaffold mutations, i.e. mutations outside the intentionally diversified loops, were found in selected variants. dLowest Kd: Dissociation constant of the highest-affinity variant found in the selection. eTm, melting temperatures of selected variants. fNNK, NNS, NNB: Diversified codons commonly used to encode any natural amino-acid residue. In all cases, the first and the second position in the codon are synthesized using the ‘N’ phosphoramidite mix, which is an equimolar mixture of all four nucleotides (A, G, C and T). The third position is synthesized using mix ‘K’ (G or T), ‘S’ (G or C) or B (C, G or T). gND, not determined. hParker et al. (2005). i▵(1−7): The seven N-terminal residues were deleted during library construction. jg, s, t: 50% the wt G, S or T, 50% any other amino-acid residue; encoded at nucleotide level. k‘G4’: complex diversity scheme encoded by nucleotide mixes and designed (i) to approximate amino-acid distribution in antibody CDR-H3 and (ii) for higher probability of wild-type residues in positions deemed important for domain stability.
Another advantage of 10Fn3-based target-binding proteins is that they are naturally well suited for multimerization to generate multi-functional binding molecules. In nature, 10Fn3 is a component of fibronectin, a long, single-chain polypeptide comprised type I, II and III domains separated by short linkers, with limited interactions between adjacent domains (Fig. 1b). A genetically linked string of Adnectins with different specificities closely mimics this natural arrangement and is thus likely to be compatible with independent folding, stability and function of the individual binding domains. In contrast, antibody variable domains are parts of a more compact, three-dimensional structure, with extensive packing and functional inter-dependence between the variable light and heavy domains (Fig. 1a).
The functional role of 10Fn3 in human fibronectin, integrin binding, is mediated by the RGD tripeptide found in the FG loop (Leahy et al., 1996); once that sequence is changed by site-directed mutagenesis or library diversification, 10Fn3 is stripped of its natural physiological function, and thus is expected to behave as an inert, non-toxic and non-immunogenic carrier of engineered, target-binding loops.
The process of Adnectin discovery
The steps that lead from human 10Fn3 to therapeutic Adnectins include:
design and construction of 10Fn3-based libraries (Xu et al., 2002);
selection of target-binding molecules from the library using in vitro display (Xu et al., 2002; Getmanova et al., 2006);
screening of selected sequences for predicted immunogenicity (De Groot et al., 2008);
screening of individual selected, bacterially produced proteins for favorable biophysical properties, including detailed binding kinetics, epitope-mapping, stability and solubility;
screening of the proteins for in vitro activity in cell-based assays;
optimization of selected Adnectins by focused re-diversification and re-selection (Xu et al., 2002; Getmanova et al., 2006);
modification for improved pharmacokinetics;
characterization of biological efficacy in animal models (Dineen et al., 2008; Mamluk et al., 2010);
clinical trials in human patients (Tolcher et al., 2010).
In addition, detailed structural characterization of Adnectin-target complexes using X-ray crystallography continues to inform the discovery process, especially library design and optimization strategies.
The two earliest 10Fn3-based library designs, published by Koide et al. (Koide et al., 1998) and Xu et al. (Xu et al., 2002), were guided primarily by structural and sequence alignments between 10Fn3, antibody variable domains and other Fn3 domains. The Koide library diversified five residues in the BC loop and five residues in a shortened FG loop, whereas the Xu library diversified seven residues in the BC loop, four residues in the DE loop and 10 residues in the full-length FG loop. In both cases, the diversified residues were encoded by a mixture of nucleotides that allowed any amino-acid residue to appear in any diversified position, with different amino acids represented at different frequencies due to the uneven redundancy of the genetic code. Koide et al used phage display to select proteins that bound ubiquitin with low-micromolar affinity, whereas Xu et al used mRNA display (PROfusion™) to select Adnectins that bound TNF-alpha with low-nanomolar affinity (after primary selection) and sub-nanomolar affinity (after affinity maturation). The library described by Xu et al became the first source of therapeutic Adnectins for Adnexus (now a Bristol-Myers Squibb R&D company); the selection from a larger library of the same design for binding to vascular endothelial growth factor receptor 2 (VEGF-R2) gave rise to CT-322, the Adnectin currently in clinical trials against glioblastoma multiforme, non-small cell lung cancer and metastatic colorectal cancer (Getmanova et al., 2006; Dineen et al., 2008; Mamluk et al., 2010).
Since the publication of the first 10Fn3-based libraries, library design has increased in complexity and sophistication, both in the choice of residues to diversify and in the ratio of amino-acid residues allowed in each diversified position. Several groups have published a variety of successful combinations of 10Fn3-base libraries and display methods (Table I). Target-binding molecules with low nanomolar to picomolar affinity have been selected from libraries of between 107 and 1013 different variants generated by the diversification of the three CDR-like loops of human 10Fn3, BC, DE and FG, using phage, yeast or mRNA display.
In several of the studies, diversity in the loop length as well as in loop sequence appeared to have contributed to high affinity of selected variants (Xu et al., 2002; Koide et al., 2007; Hackel et al., 2008, 2010; Hackel and Wittrup, 2010; Wojcik et al., 2010), and two studies identified selected pairs of cysteines predicted to be sufficiently close in space to form interloop disulfide bonds (Lipovsek et al., 2007; Hackel et al., 2010).
The published crystal structures of maltose-binding protein in complex with cognate 10Fn3 variants (Koide et al., 2007; Gilbreth et al., 2008) confirmed the pivotal role of tyrosines at the 10Fn3-antigen interface, at least where the diversified loop positions in the library were rich in tyrosine and serine, in agreement with the effect seen earlier in engineered antibodies (Fellouse et al., 2004, 2005). Still, when outcomes from similar selections were compared, libraries with a broader side-chain diversity appeared to have an advantage. In two separate selections from libraries of similar design, a library that allowed nine different amino-acid residues but favored tyrosine, serine and glycine (Gilbreth et al., 2008) yielded variants with higher affinity than did a binary tyrosine/serine library (Koide et al., 2007). Similarly, when two libraries with different diversity distributions were mixed and selected from simultaneously, the library that allowed the choice of any amino-acid residues in the diversified region yielded target-binding variants with higher affinity than did the library with diversity restricted to a tyrosine and serine (Hackel and Wittrup, 2010). A further study added a third, more complex library to the mix: diversity distribution in this third library both attempted to mimic the distribution of amino-acid residues in antibody CDR-H3 (with particularly high proportion of tyrosine (18%), serine, glycine, aspartic acid and arginine), and took into account the degree of preference for wild-type residues at each positions in the three 10Fn3, CDR-like loops (Hackel et al., 2010). This complex library, named ‘G4’, yielded about 90% of the target-binding variants against several different targets, with the remaining 10% of the variants originating from the fully diversified library using a random nucleotide mix, and none of the variants originating from the library with diversity restricted to a tyrosine and serine. These studies suggest that, whereas a high proportion of tyrosine in diversified loops is beneficial to binding, further benefit can be derived from the availability of a broad range of functional side chains.
Adnectins in the clinic
Of the properties critical for development of Adnectins as therapeutics, their ability to bind a wide range of targets is the best documented (Table I). Affinities as high as 1pM have been reported (Hackel et al., 2008), with numerous examples of Adnectins that bind targets in the sub- to low nanomolar range (Table I). This affinity range is comparable to that of therapeutic antibodies, and sufficient to target cells overexpressing the antigen and to inhibit many therapeutically relevant interactions. Several of the studies also report Adnectin binding or biological activity in cell-based assays (Richards et al., 2003; Getmanova et al., 2006; Wojcik et al., 2010) or high thermostability (Parker et al., 2005; Hackel et al., 2008, 2010; Hackel and Wittrup, 2010), both essential properties of a therapeutic-protein lead.
In our experience, high affinity for an appropriately formatted and biologically active target often translates into high activity in cell-based assays and into efficacy in animal studies. For example, the PEGylated Adnectin CT-322, which has a dissociation constant from VEGF-R2 of 11nM, inhibits VEGF-dependent proliferation of VEGF-R2 expressing cell lines and of human umbilical vein endothelial cells (Mamluk et al., 2010), and shows efficacy against xenographs in mice (Dineen et al., 2008; Mamluk et al., 2010). In Phase I clinical trials (Sweeney et al., 2008; Tolcher et al., 2010), CT-322 had an ∼100 h half-life in the bloodstream, was well tolerated, did not give rise to neutralizing antibodies and did not show signs of antibody-mediated clearance. In addition, administration of CT-322 altered physiological markers associated with VEGF-R2-dependent signaling, including elevating blood pressure and VEGF-A levels, indicating that the drug was biologically active in humans. Phase II trials are underway to evaluate the efficacy of CT-322, in combination with chemotherapy or with chemotherapy and radiation, against glioblastoma multiforme, non-small cell lung cancer and metastatic colorectal cancer (www.clinicaltrials.gov).
Future directions
CT-322 is only the first therapeutic Adnectin to emerge from a large series of Adnexus (Bristol-Myers Squibb) programs against therapeutic targets. Advanced library design and screening methods continue to generate high-potency Adnectins with excellent biophysical properties. Single-domain Adnectins like CT-322 are being followed by multi-domain Adnectins, with each domain contributing binding specificity for a different target. One such tandem Adnectin lead consists of an N-terminal Adnectin that binds epidermal growth factor receptor with 0.7 nM affinity and a C-terminal Adnectin that binds insulin-like growth factor 1 receptor with 0.1 nM affinity (Emanuel et al., 2010), leading to anti-proliferative effects through two separate pathways. Modularity of Adnectins is also being used to develop an all-protein therapeutic where improvement in pharmacokinetic properties is mediated by an Adnectin domain binding to human serum albumin instead of by chemical modification. In the longer term, the combination of modularity, small size and high stability of Adnectins also makes them excellent candidates for development of sustained-release products and other delivery alternatives to injection.
Funding
The writing of this manuscript was funded by Adnexus, a Bristol-Myers Squibb R&D Company.
Acknowledgements
I am grateful to Eric Furfine, Ray Camphausen, Samta Kundu, John Edwards and Cliff Bechtold for their help with planning and revising the manuscript.
References
- Batori V., Koide A., Koide S. Protein Eng. 2002;15:1015–1020. doi: 10.1093/protein/15.12.1015. [DOI] [PubMed] [Google Scholar]
- Beck A., Wurch T., Bailly C., Corvaia N. Nat. Rev. Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- Binz H.K., Pluckthun A. Curr. Opin. Biotechnol. 2005;16:459–469. doi: 10.1016/j.copbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Bloom L., Calabro V. Drug Discov. Today. 2009;14:949–955. doi: 10.1016/j.drudis.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Bradbury A.R., Marks J.D. J. Immunol. Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M., Wittrup K.D. Nat. Protocols. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- Cota E., Hamill S.J., Fowler S.B., Clarke J. J. Mol. Biol. 2000;302:713–725. doi: 10.1006/jmbi.2000.4053. [DOI] [PubMed] [Google Scholar]
- De Groot A.S., McMurry J., Moise L. Curr. Opin. Pharmacol. 2008;8:620–626. doi: 10.1016/j.coph.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Dickinson C.D., Veerapandian B., Dai X.P., Hamlin R.C., Xuong N.H., Ruoslahti E., Ely K.R. J. Mol. Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Dineen S.P., Sullivan L.A., Beck A.W., Miller A.F., Carbon J.G., Mamluk R., Wong H., Brekken R.A. BMC Cancer. 2008;8:352. doi: 10.1186/1471-2407-8-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Batori V., Koide A., Koide S. Protein Sci. 2005;14:2838–2848. doi: 10.1110/ps.051603005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel S.L., Engle L.J., Cao C., et al. 101st Annual Meeting of American Association of Cancer Research; 17–21 April; Philadelphia. 2010. Abst 2586. [Google Scholar]
- Fellouse F.A., Wiesmann C., Sidhu S.S. Proc. Natl Acad. Sci. USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellouse F.A., Li B., Compaan D.M., Peden A.A., Hymowitz S.G., Sidhu S.S. J. Mol. Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Gebauer M., Skerra A. Curr. Opin. Chem. Biol. 2009;13:245–255. doi: 10.1016/j.cbpa.2009.04.627. [DOI] [PubMed] [Google Scholar]
- Getmanova E.V., Chen Y., Bloom L., Gokemeijer J., Shamah S., Warikoo V., Wang J., Ling V., Sun L. Chem. Biol. 2006;13:549–556. doi: 10.1016/j.chembiol.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Gilbreth R.N., Esaki K., Koide A., Sidhu S.S., Koide S. J. Mol. Biol. 2008;381:407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves M.A., Osbourn J.K. Expert Opin. Biol. Ther. 2005;5:125–135. doi: 10.1517/14712598.5.1.125. [DOI] [PubMed] [Google Scholar]
- Hackel B.J., Wittrup K.D. Protein Eng. Des. Sel. 2010;23:211–219. doi: 10.1093/protein/gzp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B.J., Kapila A., Wittrup K.D. J. Mol. Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B.J., Ackerman M.E., Howland S.W., Wittrup K.D. J. Mol. Biol. 2010;401:84–96. doi: 10.1016/j.jmb.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L.J., Skaletsky E., McPherson A. J. Mol. Biol. 1998;275:861–872. doi: 10.1006/jmbi.1997.1508. [DOI] [PubMed] [Google Scholar]
- Holt L.J., Herring C., Jespers L.S., Woolven B.P., Tomlinson I.M. Trends Biotechnol. 2003;21:484–490. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Holt L.J., Basran A., Jones K., Chorlton J., Jespers L.S., Brewis N.D., Tomlinson I.M. Protein Eng. Des. Sel. 2008;21:283–288. doi: 10.1093/protein/gzm067. [DOI] [PubMed] [Google Scholar]
- Huston J.S., Mudgett-Hunter M., Tai M.S., McCartney J., Warren F., Haber E., Oppermann H. Methods Enzymol. 1991;203:46–88. doi: 10.1016/0076-6879(91)03005-2. [DOI] [PubMed] [Google Scholar]
- Karatan E., Merguerian M., Han Z., Scholle M.D., Koide S., Kay B.K. Chem. Biol. 2004;11:835–844. doi: 10.1016/j.chembiol.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Kohler G., Milstein C. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Koide A., Bailey C.W., Huang X., Koide S. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- Koide A., Jordan M.R., Horner S.R., Batori V., Koide S. Biochemistry. 2001;40:10326–10333. doi: 10.1021/bi010916y. [DOI] [PubMed] [Google Scholar]
- Koide A., Abbatiello S., Rothgery L., Koide S. Proc. Natl Acad. Sci. USA. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A., Gilbreth R.N., Esaki K., Tereshko V., Koide S. Proc. Natl Acad. Sci. USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy D.J., Aukhil I., Erickson H.P. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Liao H.I., Olson C.A., Hwang S., Deng H., Wong E., Baric R.S., Roberts R.W., Sun R. J. Biol. Chem. 2009;284:17512–17520. doi: 10.1074/jbc.M901547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek D., Pluckthun A. J. Immunol. Methods. 2004;290:51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Lipovsek D., Lippow S.M., Hackel B.J., Gregson M.W., Cheng P., Kapila A., Wittrup K.D. J. Mol. Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- Lonberg N. Handb. Exp. Pharmacol. 2008;181:69–97. doi: 10.1007/978-3-540-73259-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main A.L., Harvey T.S., Baron M., Boyd J., Campbell I.D. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- Mamluk R., Carvajal I.M., Morse B.A., et al. MAbs. 2010;2 doi: 10.4161/mabs.2.2.11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson C.A., Roberts R.W. Protein Sci. 2007;16:476–484. doi: 10.1110/ps.062498407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson C.A., Liao H.I., Sun R., Roberts R.W. ACS Chem. Biol. 2008;3:480–485. doi: 10.1021/cb800069c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.H., Chen Y., Danehy F., Dufu K., Ekstrom J., Getmanova E., Gokemeijer J., Xu L., Lipovsek D. Protein Eng. Des. Sel. 2005;18:435–444. doi: 10.1093/protein/gzi050. [DOI] [PubMed] [Google Scholar]
- Plaxco K.W., Spitzfaden C., Campbell I.D., Dobson C.M. J. Mol. Biol. 1997;270:763–770. doi: 10.1006/jmbi.1997.1148. [DOI] [PubMed] [Google Scholar]
- Richards J., Miller M., Abend J., Koide A., Koide S., Dewhurst S. J. Mol. Biol. 2003;326:1475–1488. doi: 10.1016/s0022-2836(03)00082-2. [DOI] [PubMed] [Google Scholar]
- Sweeney C.J., Chriorean E.G., Mita M.M., Papadopoulos K.P., Silver B., Freed M., Gokemeijer J., Eaton E., Furfine E., Tolcher A.W. J. Clin. Oncol. 2008;26 abstr. 3523. [Google Scholar]
- Thie H., Meyer T., Schirrmann T., Hust M., Dubel S. Curr. Pharm. Biotechnol. 2008;9:439–446. doi: 10.2174/138920108786786349. [DOI] [PubMed] [Google Scholar]
- Tolcher A.W., Sweeney C.J., Papadopoulos K., et al. Clin. Cancer Res. 2010 in press. [Google Scholar]
- Vincke C., Loris R., Saerens D., Martinez-Rodriguez S., Muyldermans S., Conrath K. J. Biol. Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- Wojcik J., Hantschel O., Grebien F., Kaupe I., Bennett K.L., Barkinge J., Jones R.B., Koide A., Superti-Furga G., Koide S. Nat. Struct. Mol. Biol. 2010;17:519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Aha P., Gu K., et al. Chem. Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]